Abstract
The aim of the present study was to reveal the potential molecular mechanisms of microsatellite instability (MSI) on the prognosis of gastric cancer (GC). The investigation was performed based on an RNAseq expression profiling dataset downloaded from The Cancer Genome Atlas, including 64 high-level MSI (MSI-H) GC samples, 44 low-level MSI (MSI-L) GC samples and 187 stable microsatellite (MSI-S) GC samples. Differentially expressed genes (DEGs) were identified between the MSI-H, MSI-L and MSI-S samples. Pathway enrichment analysis was performed for the identified DEGs and the pathway deviation scores of the significant enrichment pathways were calculated. A Multi-Layer Perceptron (MLP) classifier, based on the different pathways associated with the MSI statuses was constructed for predicting the outcome of patients with GC, which was validated in another independent dataset. A total of 190 DEGs were selected between the MSI-H, MSI-L and MSI-S samples. The MLP classifier was established based on the deviation scores of 10 significant pathways, among which antigen processing and presentation, and inflammatory bowel disease pathways were significantly enriched with HLA-DRB5, HLA-DMA, HLA-DQA1 and HLA-DRA; the measles, toxoplasmosis and herpes simplex infection pathways were significantly enriched with Janus kinase 2 (JAK2), caspase-8 (CASP8) and Fas. The classifier performed well on an independent validation set with 100 GC samples. Taken together, the results indicated that MSI status may affect GC prognosis, partly through the antigen processing and presentation, inflammatory bowel disease, measles, toxoplasmosis and herpes simplex infection pathways. HLA-DRB5, HLA-DMA, HLA-DQA1, HLA-DRA, JAK2, CASP8 and Fas may be predictive factors for prognosis in GC.
Keywords: gastric cancer, microsatellite instability, pathway, differentially expressed genes, co-expressed genes
Introduction
Microsatellite instability (MSI) is a form of genetic hyper-mutability on account of impairment of DNA mismatch repair (1,2), which comprises repeated nucleotides, predominantly GT/CT repeats. An increasing number of MSI target genes have been reported (3). Increasing studies have confirmed the importance of MSI in the pathogenesis of several types of cancer, including colon cancer (4,5), gastric cancer (GC) and ovarian cancer (6).
GC is one of the most common types of cancer and is the third leading cause of cancer-associated mortality around the world (7). MSI at a high level (MSI-H), a hallmark of hereditary nonpolyposis colorectal cancer, has been found in GC. MSI tends to increase from precancerous lesions to GC (8), and the MSI phenotype in early gastric cancer is an important precursor lesion of GC (9). It has been reported that MSI-H has different molecular characteristics, compared with MSI at a low level (MSI-L)/stable MSI (MSI-S); MSI-H tends to predict a better prognosis than MSI-L/MSI-S (10). Marrelli et al (11) provided evidence that, in patients with intestinal type non-cardia GC, the 5-year survival rate was significantly higher in the MSI-H group relative to the MSI-S group, and MSI status was a potential predictor of the long-term outcome of patients with intestinal type non-cardia GC. However, An et al observed a different outcome that MSI status did not appear to significantly affect the disease-free survival rate of patients with GC receiving 5-fluorouracil-based chemotherapy (12). Therefore, the association between GC prognosis and MSI status remains controversial. In addition, the underlying molecular mechanisms remain to be fully elucidated.
In the present study, a series of bioinformatics approaches were applied on GC samples with MSI to identify the possible genes and pathways involved in the prognosis of patients with GC and MSI. The differentially expressed genes (DEGs) were identified between MSI-H, MSI-L and MSI-S samples. Subsequently, the associations between the DEGs were analyzed using Pearson's correlation analysis for each MSI status, and three gene co-expression networks were constructed for the MSI-H, MSI-L and MSI-S samples, separately. Pathway enrichment analysis was performed for the identified DEGs, and pathway deviation scores were calculated to select the potential pathways associated with the prognosis of patients with GC. Furthermore, using the pathway deviation scores of these selected pathways, a Multi-Layer Perceptron (MLP) classifier was constructed for the survival prediction of patients with GC, which was then tested on an independent validation set of patients with GC. The present study aimed to provide further insights into the associations between MSI status and GC prognosis.
Materials and methods
RNAseq expression data preprocessing
The present study involved the secondary examination of an RNAseq expression profiling dataset downloaded from The Cancer Genome Atlas (TCGA; cancergenome.nih.gov/). It included 295 GC samples and the corresponding 20,532 genes. These GC samples comprised 64 MSI-H samples, 44 MSI-L samples and 187 MSI-S samples. The clinical information of the patients is shown in Table I. The raw data was standardized using z-score normalization (13).
Table I.
Demographic and clinical information of the training set and validation set.
| Dataset | Age (years) | Sex (M/F) | MSS/MSI-L/MSI-H | Survival status | OS (months) |
|---|---|---|---|---|---|
| Training | 65±5 | 182/113 | 187/44/64 | 230 alive/57 deceased | 12.7±13.4 |
| Validation | 67±6 | 61/39 | 90a/10 | NA | 10.1±12.6 |
MSS+MSIL; NA, survival times were unavailable; M, male; F, female; MSI, microsatellite instability; MSI-L, low-level MSI; MSI-H, high-level MSI; MSS, stable MSI; OS, overall survival.
DEG screening
The gene expression values of the MSI-H, MSI-L and MSI-S samples were compared by one-way analysis of variance function in R 3.2.0 (r-project.org/) (14) with P<0.05 as a strict threshold. The coefficient of variation (CV) of each gene was then calculated. The DEGs between the MSI-H, MSI-L and MSI-S samples were identified with 10% CV cut-off values.
Reverse Phase Protein Array (RPPA) data and analyses
The corresponding RPPA data was downloaded from the TCGA database (cancergenome.nih.gov/). The samples were classified into MSI-H, MSI-L and MSI-S groups as above. The protein expression levels were compared using Limma package 3.34.0 in R3.4.1 (bioconductor.org/packages/release/bioc/html/limma.html) (15). P<0.05 was set as the threshold.
Correlation analysis and construction of gene co-expression networks
Pearson's Correlation Analysis (16) was performed to analyze the associations between DEGs. The co-expressed genes were screened at Pearson's correlation coefficient (R) >0.5 or <0.5. With these co-expressed genes, three gene co-expression networks were constructed, with the genes represented as nodes, and Pearson's correlation coefficients between two genes presented as edges between two nodes. The degree of a gene was determined as the number of the edges possessed by a node. The topological properties of the networks were analyzed using Cytoscape software 3.6.0 (cytoscape.org/) (17).
Hierarchical cluster analysis
Unsupervised hierarchical cluster analysis of the DEGs in all samples was performed using the R heatmap 2 package in R version 3.2.0 (cran.r-project.org/web/packages/gplots/).
Pathway enrichment analysis
In order to elucidate the DEG-associated functional and metabolic pathways, the Database for Annotation, Visualization and Integrated Discovery (version 83.2; david.ncifcrf.gov/) (18) was utilized to perform Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis for the identified DEGs with P<0.05 as a strict cut-off value.
Pathway deviation scores
In order to screen the functional pathways showing different expression levels between the MSI-H, MSI-L and MSI-S samples, the pathway deviation scores of potential KEGG pathways were calculated based on the enriched genes in each potential pathway (19):
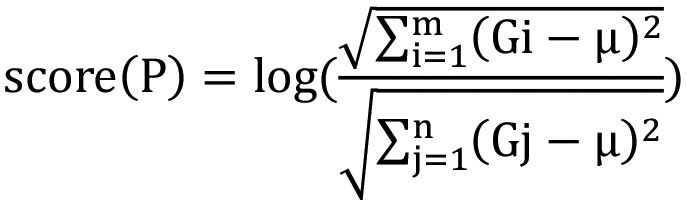 |
In the equation, m stands for the number of upregulated genes enriched in pathway P; n stands for the number of downregulated genes enriched in pathway P; µ stands for the averaged expression value of gene G in all samples. A higher score (P) indicates that a pathway is increasingly upregulated, whereas a lower score (P) signals that a pathway is increasingly downregulated.
MLP classifier
In order to determine the pathways and genes significantly associated with GC prognosis, MLP (20) was used to generate a classifier for predicting the outcomes of patients with GC. MLP is a feed-forward artificial neural network (ANN). The number of iterations was 1,000. The activation function used was sigmoid function. The ANNs were organized in a number of input nodes showing the deviation score of each pathway. The hidden layers contained five and three nodes; there was one output node showing the output of the network. The backpropagation algorithm (21) was used for training the MLP. The patients were divided into two groups; in the 'good outcome' group, patients had a survival time of ≥12 months and the final survival status was alive, whereas the 'poor outcome group' was defined as having survival time <12 months and the final survival status as deceased.
Testing of the classifier on an independent validation set
The dataset including 100 gastric cancer samples (22) downloaded from cBioPortal for Cancer Genomics (cbioportal.org/) was used as a validation set. The MLP classifier was tested on this validation set. A Receiver Operating Characteristic (ROC) curve was drawn using pROC package (version 1.9.1) in R 3.2.0 (23–25) to examine the performance of the classifier.
Results
DEGs
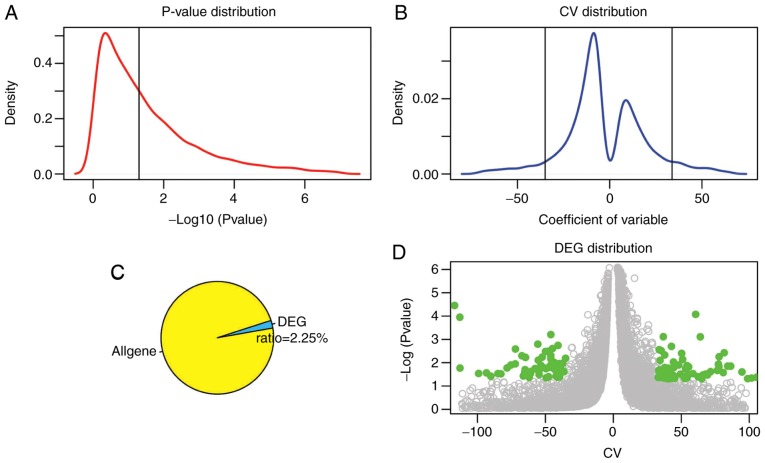
A total of 190 DEGs were screened between the MSI-H, MSI-L and MSI-S samples with CV >33.8 or <−35.1 and P<0.05 as the thresholds (Fig. 1). Fig. 1A and B revealed the distribution and threshold of the P-value and the CV of the genes, respectively. The majority of genes did not demonstrate significantly different expression (Fig. 1A) in the MSI-H patients there were an increased number of lower expression genes compared with the number of higher expressed genes (Fig. 1B). The DEGs account for 2.25% of all genes (Fig. 1C). Fig. 1D demonstrated the distribution of the DEGs in all genes using a volcano plot. The expression of these DEGs at the protein level was also determined in the RPPA dataset. There were 12 genes significantly differentially expressed at the protein level when using the cut-off value of P<0.05 (Table II). However, these expression changes require further validation in in vitro and in vivo experiments.
Figure 1.
DEG screening. (A) P-value distribution. The black vertical line represents the threshold of P=0.05 [−log (0.05)=1.3]. (B) CV distribution. Two black vertical lines represents 10% CV and 90% CV, respectively (−35.1, 33.8). (C) Ratio of DEGs in all genes. (D) DEG distribution; green points represent DEGs and grey points represent other genes. DEGs, differentially expressed genes; CV, coefficient of variation.
Table II.
Significantly differentially expressed genes at the protein level.
| Gene | P-value |
|---|---|
| TLDC1 | 0.0001 |
| CRABP2 | 0.0019 |
| C1ORF116 | 0.0075 |
| C6ORF132 | 0.0121 |
| CAPNS2 | 0.0213 |
| HSD3B2 | 0.0272 |
| GPR157 | 0.0290 |
| HEPN1 | 0.0306 |
| KLK12 | 0.0315 |
| HOXB2 | 0.0329 |
| FOXI2 | 0.0371 |
| C12ORF54 | 0.0479 |
Analysis of gene co-expression networks
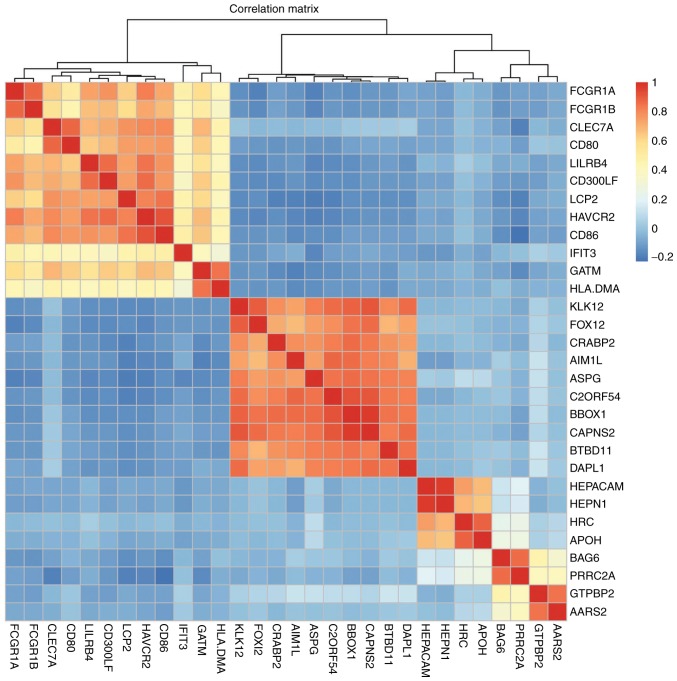
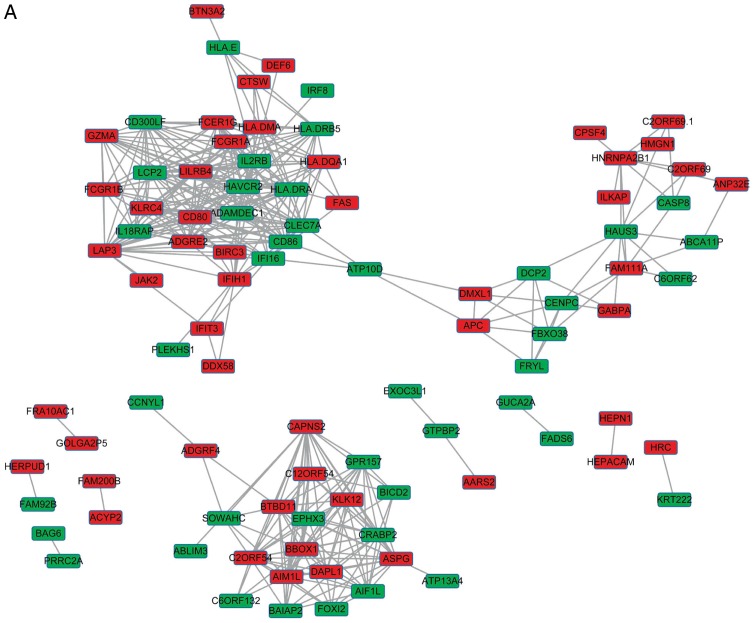
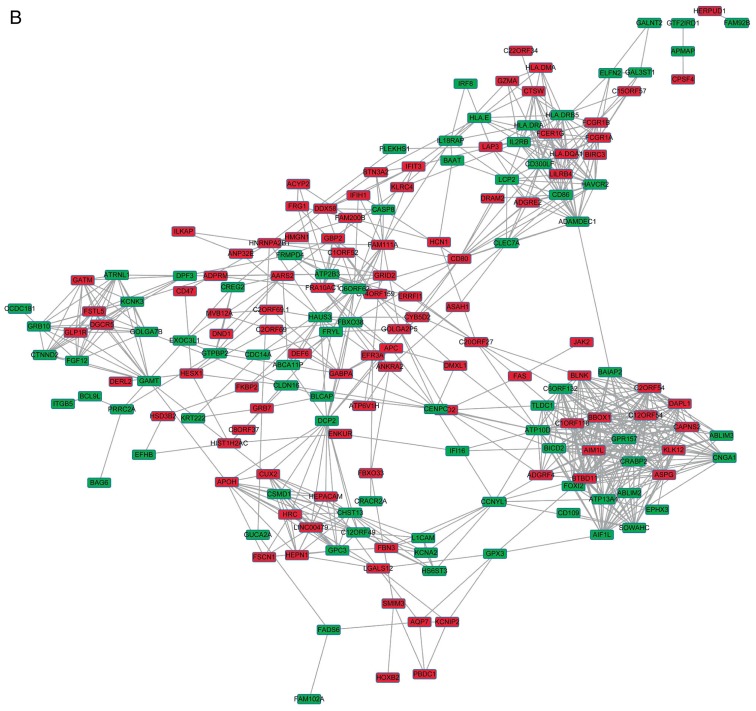
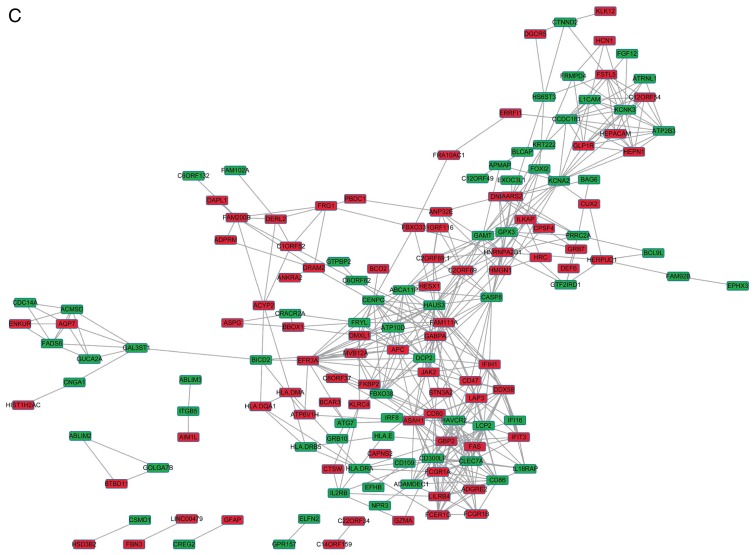
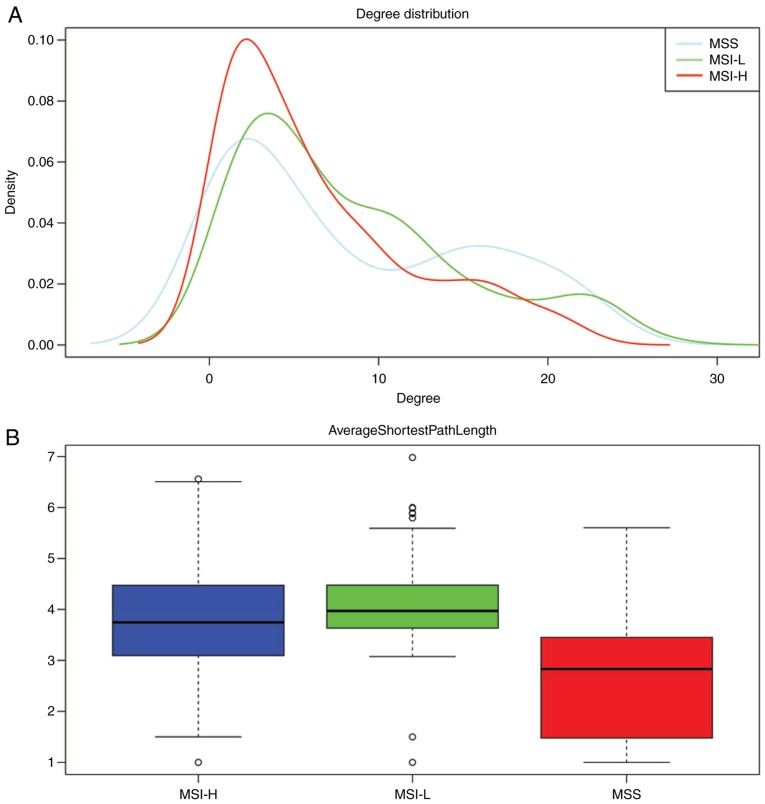
The DEGs with R >0.5 or <−0.5 were identified. The top 30 co-expressed genes based on the R values are shown in a heatmap (Fig. 2). As shown in Fig. 3, three co-expression networks were constructed for the co-expressed genes in the MSI-H, MSI-L and MSI-S samples, separately. The degree and average shortest path length of the three networks were analyzed (Fig. 4A and B) and genes tended to have higher degrees and longer average shortest path lengths in the MSI-H samples, compared with those in the MSI-L and MSI-S samples. This indicated that the correlations between DEGs were weaker in the MSI-H samples, compared with those in the MSI-L and MSI-S samples.
Figure 2.
Heatmap of top 30 co-expressed genes. Genes are shown on the horizontal and vertical axes. The red lattice represents a positive correlation, the blue lattice represents a negative correlation. The color bar indicates the R value.
Figure 3.
Gene co-expression networks of gastric cancer samples of different MSI status. (A) MSI-S samples; (B) MSI-L samples. (C) MSI-H samples. Red rectangles represent upregulated genes, green rectangles represent downregulated genes. MSI, microsatellite instability; MSI-S, stable MSI; MSI-L, low-level MSI; MSI-H, high-level MSI.
Figure 4.
Topological properties of 3 gene co-expressed networks. (A) Degree of genes in the three networks; (B) Average shortest path length of the three networks. MSI, microsatellite instability; MSS, stable MSI; MSI-L, low-level MSI; MSI-H, high-level MSI.
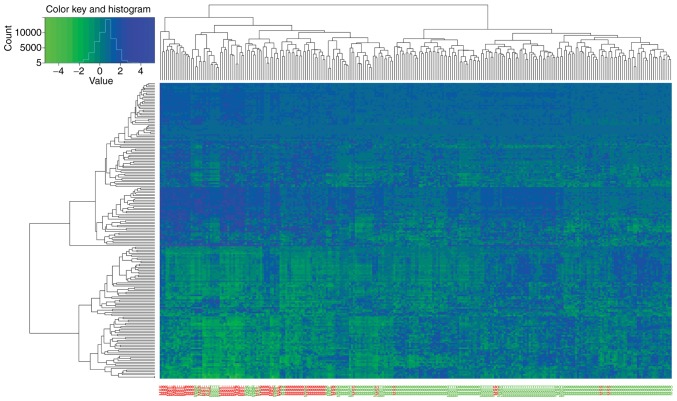
Unsupervised hierarchical cluster analysis of DEGs
The unsupervised hierarchical cluster analysis of the DEGs showed that the expression levels of DEGs were significantly different between GC patients in the good outcome group and patients in the poor outcome group (Fig. 5).
Figure 5.
Heatmap of unsupervised hierarchical cluster analysis of DEGs. Samples are on the vertical axis; genes are on the horizontal axis. Among samples on the vertical axis, the samples with a good outcome are in green, and the samples with a poor outcome are in red. In the heatmap, upregulated genes are shown in red and downregulated genes are shown in green.
Survival analysis of patients in the training set
As shown in Table III, 25 of the 36 MSI-L samples and 47 of the 168 MSI-S samples had a poor prognosis, with no significant difference in prognosis. By contrast, only 2/59 MSI-H samples had a poor prognosis (Table III). There was a significant difference in prognosis between the MSI-H and MSI-L/MSI-S samples.
Table III.
Outcome of patients in the training set.
| MSI | Prognosis
|
|
|---|---|---|
| Good (n) | Poor (n) | |
| MSI-H | 57 | 2 |
| MSI-L | 11 | 25 |
| MSS | 121 | 47 |
Good indicates patient survival at 12 months following diagnosis. Poor indicates patient succumbed to mortality 12 months following diagnosis. MSI, microsatellite instability; MSI-L, low-level MSI; MSI-H, high-level MSI; MSS, stable MSI.
Taking into account the similarity between the patients with MSI-L and MSI-S in prognosis and characteristics of the gene co-expression networks, these two groups of patients were combined as a single group in subsequent analysis.
Pathway functional annotation
KEGG pathway enrichment analysis was performed for the DEGs. As shown in Table IV, these DEGs were significantly enriched in pathways, including herpes simplex infection, intestinal immune network for IgA production pathway, leishmaniasis, antigen processing and presentation, and measles.
Table IV.
Significant pathways enriched with differentially expressed genes.
| Term | Count | P-value | Genes |
|---|---|---|---|
| Graft-vs.-host disease | 8 | 2.52E-08 | CD86, CD80, HLA-DRB5, FAS, HLA-E, HLA-DMA, HLA-DQA1, HLA-DRA |
| Allograft rejection | 8 | 5.89E-08 | CD86, CD80, HLA-DRB5, FAS, HLA-E, HLA-DMA, HLA-DQA1, HLA-DRA |
| Type I diabetes mellitus | 8 | 1.48E-07 | CD86, CD80, HLA-DRB5, FAS, HLA-E, HLA-DMA, HLA-DQA1, HLA-DRA |
| Autoimmune thyroid disease | 8 | 6.80E-07 | CD86, CD80, HLA-DRB5, FAS, HLA-E, HLA-DMA, HLA-DQA1, HLA-DRA |
| Viral myocarditis | 8 | 1.29E-06 | CD86, CD80, CASP8, HLA-DRB5, HLA-E, HLA-DMA, HLA-DQA1, HLA-DRA |
| Herpes simplex infection | 12 | 1.59E-06 | DDX58, HMGN1, IFIH1, GTF2IRD1, CASP8, HLA-DRB5, JAK2, FAS, HLA-E, HLA-DMA, HLA-DQA1, HLA-DRA |
| Tuberculosis | 10 | 5.85E-05 | FCGR1A, CASP8, HLA-DRB5, FCER1G, ATP6V1H, JAK2, CLEC7A, HLA-DMA, HLA-DQA1, HLA-DRA |
| Cell adhesion molecules | 9 | 7.58E-05 | CLDN16, CD86, CD80, HLA-DRB5, L1CAM, HLA-E, HLA-DMA, HLA-DQA1, HLA-DRA |
| Intestinal immune network for IgA production | 6 | 9.50E-05 | CD86, CD80, HLA-DRB5, HLA-DMA, HLA-DQA1, HLA-DRA |
| Phagosome | 9 | 1.28E-04 | FCGR1A, HLA-DRB5, ITGB5, ATP6V1H, CLEC7A, HLA-E, HLA-DMA, HLA-DQA1, HLA-DRA |
| Asthma | 5 | 2.05E-04 | HLA-DRB5, FCER1G, HLA-DMA, HLA-DQA1, HLA-DRA |
| Rheumatoid arthritis | 7 | 2.26E-04 | CD86, CD80, HLA-DRB5, ATP6V1H, HLA-DMA, HLA-DQA1, HLA-DRA |
| Influenza A | 9 | 3.11E-04 | DDX58, IFIH1, HLA-DRB5, JAK2, CPSF4, FAS, HLA-DMA, HLA-DQA1, HLA-DRA |
| Systemic lupus erythematosus | 8 | 3.50E-04 | HIST1H2AC, CD86, CD80, FCGR1A, HLA-DRB5, HLA-DMA, HLA-DQA1, HLA-DRA |
| Leishmaniasis | 6 | 6.70E-04 | FCGR1A, HLA-DRB5, JAK2, HLA-DMA, HLA-DQA1, HLA-DRA |
| Antigen processing and presentation | 6 | 9.15E-04 | KLRC4, HLA-DRB5, HLA-E, HLA-DMA, HLA-DQA1, HLA-DRA |
| Toxoplasmosis | 7 | 1.09E-03 | CASP8, HLA-DRB5, JAK2, BIRC3, HLA-DMA, HLA-DQA1, HLA-DRA |
| Staphylococcus aureus infection | 5 | 1.98E-03 | FCGR1A, HLA-DRB5, HLA-DMA, HLA-DQA1, HLA-DRA |
| Inflammatory bowel disease | 5 | 3.69E-03 | IL18RAP, HLA-DRB5, HLA-DMA, HLA-DQA1, HLA-DRA |
| HTLV-I infection | 7 | 4.18E-02 | IL2RB, HLA-DRB5, HLA-E, HLA-DMA, HLA-DQA1, HLA-DRA, APC |
| Measles | 5 | 4.34E-02 | DDX58, IL2RB, IFIH1, JAK2, FAS |
Term, pathway identity; count, number of genes enriched in a pathway.
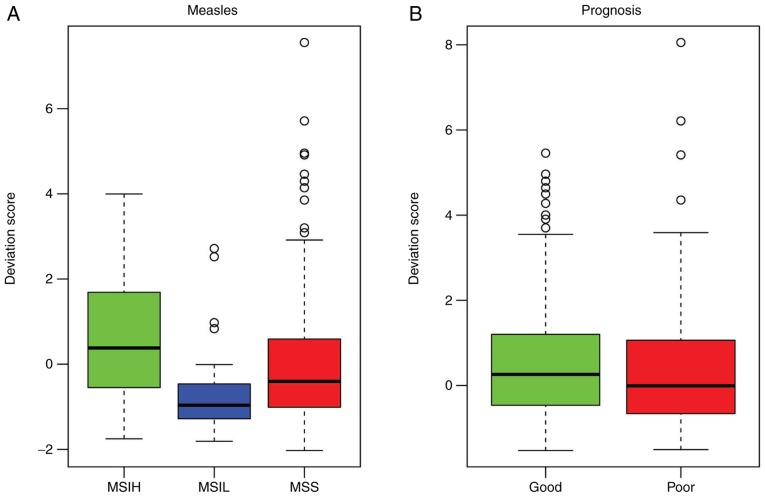
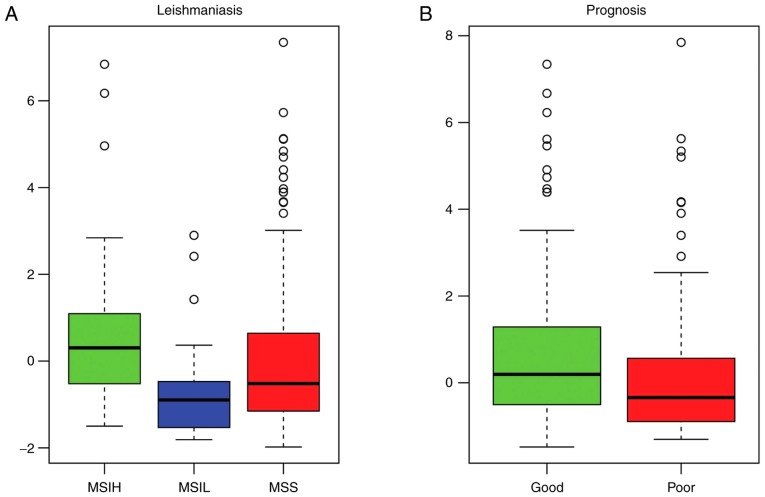
The pathway deviation scores of the significant enrichment pathways were calculated in order to investigate functional differences in the pathways between the different samples. The measles pathway (Fig. 6A and B) and leishmaniasis pathway (Fig. 7A and B) were upregulated in the MSI-L samples and downregulated in the MSI-H samples, compared with those in the MSI-S samples. In addition, the deviation score of the leishmaniasis pathway was higher in the good prognosis samples, compared with that in the poor prognosis samples (Fig. 7B).
Figure 6.
Pathway deviation score of measles pathway. (A) Pathway deviation score of measles pathway in MSI-H, MSI-L and MSS samples; (B) pathway deviation score of measles pathway in good outcome samples and poor outcome samples. MSI, microsatellite instability; MSI-L, low-level MSI; MSI-H, high-level MSI; MSS, stable MSI.
Figure 7.
Pathway deviation score of leishmaniasis pathway. (A) Pathway deviation score of leishmaniasis pathway in MSI-H, MSI-L and MSS samples; (B) pathway deviation score of leishmaniasis pathway in good outcome samples and poor outcome samples. MSI, microsatellite instability; MSI-L, low-level MSI; MSI-H, high-level MSI; MSIS, stable MSI.
In order to detecting the potential pathways significantly associated with the prognosis of patients with GC, 10 pathways with significantly different pathway deviation scores between the MSI-H and MSI-S/MSI-L samples (P<0.05), and between the good outcome and poor outcome samples (P<0.05) were selected from the 20 significant enrichment pathways (Table V), including measles, antigen processing and presentation, rheumatoid arthritis, phagosome, systemic lupus erythematosus, herpes simplex infection, inflammatory bowel disease, tuberculosis, type I diabetes mellitus, and toxoplasmosis.
Table V.
Analysis of pathway deviation scores.
| Pathway | P-value prognosis | P-value MSS |
|---|---|---|
| Measles | 2.03E-29 | 3.32E-03 |
| Antigen processing and presentation | 7.42E-19 | 4.80E-02 |
| Rheumatoid arthritis | 1.70E-13 | 1.25E-03 |
| Phagosome | 5.84E-12 | 2.48E-02 |
| Systemic lupus erythematosus | 1.31E-11 | 6.02E-03 |
| Herpes simplex infection | 1.18E-06 | 1.67E-02 |
| Inflammatory bowel disease | 1.14E-05 | 4.63E-02 |
| Tuberculosis | 3.57E-04 | 3.98E-04 |
| Type I diabetes mellitus | 1.28E-03 | 7.13E-03 |
| Toxoplasmosis | 1.64E-03 | 4.82E-03 |
| Cell adhesion molecules | 7.99E-03 | 9.63E-02 |
| Viral myocarditis | 1.11E-02 | 9.75E-03 |
| Asthma | 1.21E-02 | 2.01E-02 |
| HTLV I infection | 4.21E-02 | 3.54E-02 |
| Autoimmune thyroid disease | 8.06E-02 | 7.13E-03 |
| Allograft rejection | 8.86E-02 | 7.13E-03 |
| Staphylococcus aureus infection | 1.50E-01 | 4.63E-02 |
| Graft versus host disease | 2.26E-01 | 7.13E-03 |
| Leishmaniasis | 2.52E-01 | 1.03E-02 |
| Influenza A | 2.99E-01 | 1.79E-02 |
| Intestinal immune network for IgA production | 3.91E-01 | 7.66E-03 |
MSS, microsatellite stability.
MLP classifier construction
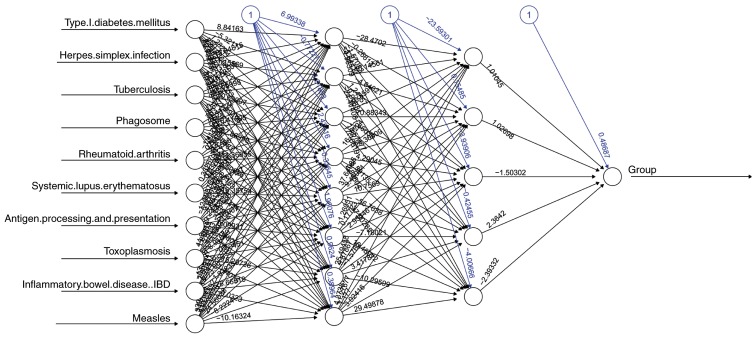
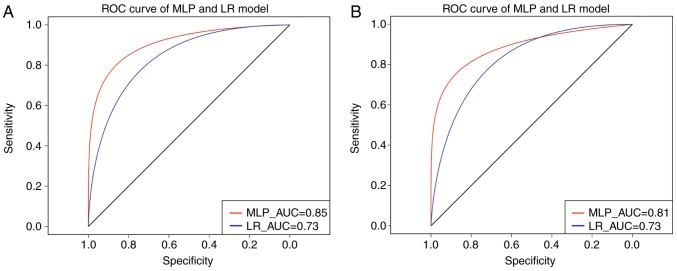
An MLP classifier was constructed with pathway deviation score input for predicting the prognosis of patients with GC (Fig. 8). In addition, the ROC curve method was used to assess the performance of the MLP classifier, compared with logistic regression (LR). As shown in Fig. 9A, the area under the curve (AUC) of the MLP classifier and LOR was 0.85 and 0.73, respectively, indicating that the performance of the classifier was superior to that of LOR in predicting the outcome of patients with GC.
Figure 8.
Multi-Layer Perceptron neural network. A black edge linking any two neural nodes (circles) represents the correlation between the two nodes, with the bias term indicated by the black number. Blue edges and numbers represent the weight value of the correlation between any two neural nodes.
Figure 9.
ROC curve of the MLP classifier. (A) Curve for training set; (B) curve for validation set. The vertical axis represents sensitivity and the horizontal axis represents specificity. ROC, Receiver Operating Characteristic; MLP, Multi-Layer Perceptron; AUC, area under the curve; LR, logistic regression.
The MLP classifier was further validated on an independent validation set of 100 GC samples. The clinical information of the 100 samples is shown in Table I. As shown in Fig. 9B, the MLP classifier had an elevated AUC value, compared with that in the LR model (0.81 vs. 0.73). This confirmed the superior performance of the MLP model, compared with the LOR model.
Survival analysis of the independent validation set
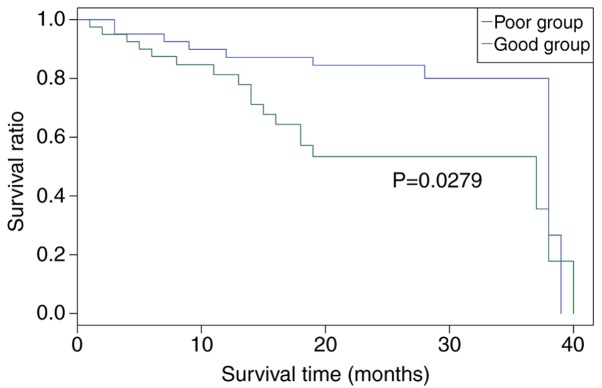
Using the survival durations of the samples, the 100 GC samples of the independent validation set were classified into good prognosis (survival ≥12 months) and poor prognosis (survival <12 months) groups using the MLP classifier. As shown in Fig. 10, the survival rate of patients was higher in the good outcome group, compared with that in the poor outcome group (P=0.0279). This finding confirmed the performance of the MLP classifier in predicting GC prognosis.
Figure 10.
Survival analysis of patients using the log-rank test in the validation set. The vertical axis represents ratio of patient survival; the horizontal axis represents the survival time.
Discussion
GC is one of the most frequent malignancies. It has been reported that MSI-H GC samples exhibit clinical and molecular features distinct from MSI-S GC samples (26,27). The present study found that, in the training set, 74 of 295 patients with GC had a poor prognosis. Of these 74 patients, only two patients were of MSI-H status, whereas the other 72 patients were of MSI-S/MSI-L status, which was in accordance with a previous finding that MSI-H GC tended to have improved overall survival rates, compared with MSI-S GC (11).
An MLP classifier, based on the pathway deviation score of 10 significant pathways, was constructed in the present study, which predicted the outcome of patients with GC with proficiency. The performance of the MLP classifier was verified on an independent validation set of 100 GC samples. The pathway deviation score of each pathway was significantly different between the MSI-H and MSI-S/MSI-L samples, and between the good prognosis and poor prognosis samples. These findings confirmed that MSI status was associated with the survival rate of patients with GC. In addition, the effect of MSI status on GC prognosis may be partly mediated by 10 significant pathways, comprising the measles, antigen processing and presentation, rheumatoid arthritis, phagosome, systemic lupus erythematosus, herpes simplex infection, inflammatory bowel disease, tuberculosis, type I diabetes mellitus, and toxoplasmosis pathways.
An increasing number of studies have established the integral role of inflammation and immune in the development of GC (28–30). Nissen et al found that inflammatory bowel disease was a risk factor for the development of GC (31). Consistent with these findings, the present study found that the inflammation- and immune-related antigen processing and presentation, and inflammatory bowel disease pathways were associated with the MSI status and the prognosis of patients with GC. Furthermore, these two pathways were significantly enriched with major histocompatibility complex (MHC), class II, DRβ5 (HLA-DRB5), MHC class II, DMα (HLA-DMA), MHC class II, DQα1 (HLA-DQA1), and MHC class II, DRα (HLA-DRA), which encode important MHC class II molecules on antigen-presenting cells. Ribeiro et al found that the expression of MHC class I-related chains A and B in tumors may be involved in the progression of GC and be of predictive value for prognosis in GC with large tumors (32). This suggested that HLA-DRB5, HLA-DMA, HLA-DQA1 and HLA-DRA may be of predictive value for survival rates of patients with GC.
Janus kinase 2 (JAK2), a member of the JAK family, is a non-receptor tyrosine kinase. Judd et al provided in vitro and in vivo evidence that GC cell growth was compromised by suppressing the JAK2/signal transducer and activator of transcription 3 pathway (33). It has been reported that the overexpression of JAK2 promotes GC cell migration and invasion (34). The caspase-8 (CASP8) gene encodes caspase-8 protein, which is critical in the execution-phase of cell apoptosis. Tumor necrosis factor receptor superfamily member 6 (Fas), known as apoptosis antigen-1, interacts with its natural ligand (FasL), leading to apoptosis in responsive cells (35). It has been demonstrated that CASP8 gene mutation may be involved in the pathogenesis of GC (36). Yang et al (37) reported that Fas signaling facilitated GC metastasis. Wang et al (38) found that the expression of Fas was downregulated in GC, and that CASP8 was associated with outcome in patients with GC. These findings suggest that JAK2, CASP8 and Fas are involved in the pathogenesis of GC. In the present study, JAK2 was significantly enriched in the measles, toxoplasmosis, and herpes simplex infection pathways. CASP8 and FAS were significantly enriched in the measles and toxoplasmosis pathways. This indicated that JAK2, CASP8 and Fas may be associated with GC prognosis mediated by MSI status.
The MLP classifier is based on the minimization of the number of misclassified vectors of a training set (empirical risk), whereas the support vector machine (SVM) minimizes a functional sum of the empirical risk and controls the ability of the machine to learn any training set without error (39). The comparison between MLP and SVM has been reported previously, and there is debate regarding the effect and choice of these two classifiers. However, one thing is certain for the two classifiers, that is, the performance of the SVM classifier is better when the sample size is smaller, whereas the MLP classifier performance is better when the sample size is larger. In the present study, the sample size was relatively large, and this is the reason why the MLP method was used in the present study. The results were satisfactory and achieved the aim of the study; the MLP classifier performed well in predicting the prognosis of patients with GC.
The present study has several limitations. It is a secondary study of an RNA-seq expression profiling dataset, and lacks experimental evidence to validate these findings. Additionally, the sample size of the independent validation set was limited by the number of available datasets.
In conclusion, the results of the present study suggested that MSI status may affect GC prognosis, partly through the antigen processing and presentation, inflammatory bowel disease, measles, toxoplasmosis, and herpes simplex infection pathways. In addition, HLA-DRB5, HLA-DMA, HLA-DQA1, HLA-DRA, JAK2, CASP8 and Fas may be recommended as potential predictive factors of prognosis in GC. These results contribute to an improved understanding of the association between MSI status and GC prognosis, and the underlying molecular mechanisms. Further experimental investigations are warranted to verify these findings.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Author's contributions
XH and DL designed the study, performed data analyses and wrote the manuscript. JW collected the data and organized the literature. JW and GW conceived and designed the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Yamamoto H, Adachi Y, Taniguchi H, Kunimoto H, Nosho K, Suzuki H, Shinomura Y. Interrelationship between microsatellite instability and microRNA in gastrointestinal cancer. World J Gastroenterol. 2012;18:2745–2755. doi: 10.3748/wjg.v18.i22.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yamamoto H, Imai K. Microsatellite instability: An update. Arch Toxicol. 2015;89:899–921. doi: 10.1007/s00204-015-1474-0. [DOI] [PubMed] [Google Scholar]
- 3.Imai K, Yamamoto H. Carcinogenesis and microsatellite instability: The interrelationship between genetics and epigenetics. Carcinogenesis. 2008;29:673–680. doi: 10.1093/carcin/bgm228. [DOI] [PubMed] [Google Scholar]
- 4.Kang J, Lee HW, Kim IK, Kim NK, Sohn SK, Lee KY. Clinical implications of microsatellite instability in T1 colorectal cancer. Yonsei Med J. 2015;56:175–181. doi: 10.3349/ymj.2015.56.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138:2073–2087. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Caliman LP, Tavares RL, Piedade JB, DE Assis AC. Evaluation of microsatellite instability in women with epithelial ovarian cancer. Oncol Lett. 2012;4:556–560. doi: 10.3892/ol.2012.776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 8.Li B, Liu HY, Guo SH, Sun P, Gong FM, Jia BQ. Microsatellite instability of gastric cancer and precancerous lesions. Int J Clin Exp Med. 2015;8:21138–21144. [PMC free article] [PubMed] [Google Scholar]
- 9.Sugimoto R, Sugai T, Habano W, Endoh M, Eizuka M, Yamamoto E, Uesugi N, Ishida K, Kawasaki T, Matsumoto T, Suzuki H. Clinicopathological and molecular alterations in early gastric cancers with the microsatellite instability-high phenotype. Int J Cancer. 2016;138:1689–1697. doi: 10.1002/ijc.29916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choi YY, Bae JM, An JY, Kwon IG, Cho I, Shin HB, Eiji T, Aburahmah M, Kim HI, Cheong JH, et al. Is microsatellite instability a prognostic marker in gastric cancer? A systematic review with meta-analysis. J Surg Oncol. 2014;110:129–135. doi: 10.1002/jso.23618. [DOI] [PubMed] [Google Scholar]
- 11.Marrelli D, Polom K, Pascale V, Vindigni C, Piagnerelli R, De Franco L, Ferrara F, Roviello G, Garosi L, Petrioli R, Roviello F. Strong prognostic value of microsatellite instability in intestinal type non-cardia gastric cancer. Ann Surg Oncol. 2016;23:943–950. doi: 10.1245/s10434-015-4931-3. [DOI] [PubMed] [Google Scholar]
- 12.An JY, Kim H, Cheong JH, Hyung WJ, Kim H, Noh SH. Microsatellite instability in sporadic gastric cancer: Its prognostic role and guidance for 5-FU based chemotherapy after R0 resection. Int J Cancer. 2012;131:505–511. doi: 10.1002/ijc.26399. [DOI] [PubMed] [Google Scholar]
- 13.San Segundo E, Tsanas A, Gómez-Vilda P. Euclidean distances as measures of speaker dissimilarity including identical twin pairs: A forensic investigation using source and filter voice characteristics. Forensic Sci Int. 2017;270:25–38. doi: 10.1016/j.forsciint.2016.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ahmed S, Brennan L, Eppig J, Price CC, Lamar M, Delano-Wood L, Bangen KJ, Edmonds EC, Clark L, Nation DA, et al. Visuoconstructional impairment in subtypes of mild cognitive impairment. Appl Neuropsychol Adult. 2016;23:43–52. doi: 10.1080/23279095.2014.1003067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu AN, Wang LL, Li HP, Gong J, Liu XH. Correlation between posttraumatic growth and posttraumatic stress disorder symptoms based on Pearson correlation coefficient: A meta-analysis. J Nerv Ment Dis. 2017;250:380–389. doi: 10.1097/NMD.0000000000000605. [DOI] [PubMed] [Google Scholar]
- 17.Van Parys T, Melckenbeeck I, Houbraken M, Audenaert P, Colle D, Pickavet M, Demeester P, Van de Peer Y. A cytoscape app for motif enumeration with ISMAGS. Bioinformatics. 2017;33:461–463. doi: 10.1093/bioinformatics/btw626. [DOI] [PubMed] [Google Scholar]
- 18.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 19.Wu T, Wang X, Li J, Song X, Wang Y, Wang Y, Zhang L, Li Z, Tian J. Identification of personalized chemoresistance genes in subtypes of basal-like breast cancer based on functional differences using pathway analysis. Plos One. 2015;10:e0131183. doi: 10.1371/journal.pone.0131183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cangelosi D, Pelassa S, Morini M, Conte M, Bosco MC, Eva A, Sementa AR, Varesio L. Artificial neural network classifier predicts neuroblastoma patients' outcome. Bmc Bioinformatics. 2016;17:347. doi: 10.1186/s12859-016-1194-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Galdino L, Semrau D, Lavery D, Saavedra G, Czegledi CB, Agrell E, Killey RI, Bayvel P. On the limits of digital back-propagation in the presence of transceiver noise. Opt Express. 2017;25:4564–4578. doi: 10.1364/OE.25.004564. [DOI] [PubMed] [Google Scholar]
- 22.Wang K, Yuen ST, Xu J, Lee SP, Yan HH, Shi ST, Siu HC, Deng S, Chu KM, Law S, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–582. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- 23.Carpenter J, Bithell J. Bootstrap confidence intervals: When, which, what? A practical guide for medical statisticians. Stat Med. 2000;19:1141–1164. doi: 10.1002/(SICI)1097-0258(20000515)19:9<1141::AID-SIM479>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 24.Carlone M, Cruje C, Rangel A, McCabe R, Nielsen M, Macpherson M. ROC analysis in patient specific quality assurance. Med Phys. 2013;40:042103. doi: 10.1118/1.4795757. [DOI] [PubMed] [Google Scholar]
- 25.Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, Müller M. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falchetti M, Saieva C, Lupi R, Masala G, Rizzolo P, Zanna I, Ceccarelli K, Sera F, Mariani-Costantini R, Nesi G, et al. Gastric cancer with high-level microsatellite instability: Target gene mutations, clinicopathologic features, and long-term survival. Hum Pathol. 2008;39:925–932. doi: 10.1016/j.humpath.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 27.Beghelli S, de Manzoni G, Barbi S, Tomezzoli A, Roviello F, Di Gregorio C, Vindigni C, Bortesi L, Parisi A, Saragoni L, et al. Microsatellite instability in gastric cancer is associated with better prognosis in only stage II cancers. Surgery. 2006;139:347–356. doi: 10.1016/j.surg.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 28.Deng Q, He B, Liu X, Yue J, Ying H, Pan Y, Sun H, Chen J, Wang F, Gao T, et al. Prognostic value of pre-operative inflammatory response biomarkers in gastric cancer patients and the construction of a predictive model. J Transl Med. 2015;13:16. doi: 10.1186/s12967-015-0409-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bornschein J, Kandulski A, Selgrad M, Malfertheiner P. From gastric inflammation to gastric cancer. Dig Dis. 2010;28:609–614. doi: 10.1159/000320061. [DOI] [PubMed] [Google Scholar]
- 30.Kim SY, Park C, Kim HJ, Park J, Hwang J, Kim JI, Choi MG, Kim S, Kim KM, Kang MS. Deregulation of immune response genes in patients with Epstein-Barr virus-associated gastric cancer and outcomes. Gastroenterology. 2015;148:137–147. doi: 10.1053/j.gastro.2014.09.020. [DOI] [PubMed] [Google Scholar]
- 31.Nissen LH, Assendorp EL, van der Post RS, Derikx LA, de Jong DJ, Kievit W, Pierik M, van den Heuvel T, Verhoeven R, Overbeek LI, et al. Impaired gastric cancer survival in patients with inflammatory bowel disease. J Gastrointestin Liver Dis. 2016;25:431–440. doi: 10.15403/jgld.2014.1121.254.nis. [DOI] [PubMed] [Google Scholar]
- 32.Ribeiro CH, Kramm K, Gálvez-Jirón F, Pola V, Bustamante M, Contreras HR, Sabag A, Garrido-Tapia M, Hernández CJ, Zúñiga R, et al. Clinical significance of tumor expression of major histocompatibility complex class I-related chains A and B (MICA/B) in gastric cancer patients. Oncol Rep. 2016;35:1309–1317. doi: 10.3892/or.2015.4510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Judd LM, Menheniott TR, Ling H, Jackson CB, Howlett M, Kalantzis A, Priebe W, Giraud AS. Inhibition of the JAK2/STAT3 pathway reduces gastric cancer growth in vitro and in vivo. Plos One. 2014;9:e95993. doi: 10.1371/journal.pone.0095993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xu Y, Zhou T, Si J, Wei Z. Inhibition of migration and invasion of gastric cancer cells by snail-regulated MiR-375 through JAK2 targeting. J Clin Oncol. 2014;32:56. doi: 10.1200/jco.2014.32.3_suppl.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neumann L, Pforr C, Beaudouin J, Pappa A, Fricker N, Krammer PH, Lavrik IN, Eils R. Dynamics within the CD95 death-inducing signaling complex decide life and death of cells. Mol Syst Biol. 2010;6:352. doi: 10.1038/msb.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Soung YH, Lee JW, Kim SY, Jang J, Park YG, Park WS, Nam SW, Lee JY, Yoo NJ, Lee SH. CASPASE-8 gene is inactivated by somatic mutations in gastric carcinomas. Cancer Res. 2005;65:815–821. [PubMed] [Google Scholar]
- 37.Yang Y, Zhao Q, Cai Z, Cheng G, Chen M, Wang J, Zhong H. Fas signaling promotes gastric cancer metastasis through STAT3-dependent upregulation of fascin. Plos One. 2015;10:e0125132. doi: 10.1371/journal.pone.0125132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X, Fu Z, Chen Y, Liu L. Fas expression is downregulated in gastric cancer. Mol Med Rep. 2016;15:627–634. doi: 10.3892/mmr.2016.6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bazzani A, Bevilacqua A, Bollini D, Brancaccio R, Campanini R, Lanconelli N, Riccardi A, Romani D. An SVM classifier to separate false signals from microcalcifications in digital mammograms. Phys Med Biol. 2001;46:1651–1663. doi: 10.1088/0031-9155/46/6/305. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.