Abstract
The human fungal pathogen Cryptococcus neoformans undergoes many phenotypic changes to promote its survival in specific ecological niches and inside the host. To explore the role of chromatin remodeling on the expression of virulence-related traits, we identified and deleted seven genes encoding predicted class I/II histone deacetylases (HDACs) in the C. neoformans genome. These studies demonstrated that individual HDACs control non-identical but overlapping cellular processes associated with virulence, including thermotolerance, capsule formation, melanin synthesis, protease activity and cell wall integrity. We also determined the HDAC genes necessary for C. neoformans survival during in vitro macrophage infection and in animal models of cryptococcosis. Our results identified the HDA1 HDAC gene as a central mediator controlling several cellular processes, including mating and virulence. Finally, a global gene expression profile comparing the hda1Δ mutant versus wild-type revealed altered transcription of specific genes associated with the most prominent virulence attributes in this fungal pathogen. This study directly correlates the effects of Class I/II HDAC-mediated chromatin remodeling on the marked phenotypic plasticity and virulence potential of this microorganism. Furthermore, our results provide insights into regulatory mechanisms involved in virulence gene expression that are likely shared with other microbial pathogens.
Introduction
Pathogenic microorganisms must maintain the ability to adapt to environmental changes as well as to the specific cell stresses encountered during interaction with host cells. Chromatin remodeling is one mechanism by which eukaryotic microbes might direct these adaptations in a rapid manner. In eukaryotic cells, genomic DNA is folded with histone and non-histone proteins into chromatin, a highly dynamic organizational structure. The basic subunit of chromatin is the nucleosome, consisting of DNA surrounding two H3/H4 histone protein heterodimers and two H2A/H2B histone heterodimers1,2. These histone proteins are the targets of different post-translational modifications, leading to changes in the chromatin structure. The degree of chromatin remodeling resulting from these histone modifications regulates gene expression, offering a more rapid mechanism of adaptation than spontaneous genetic mutations3–5.
One of the best studied post-translational histone modification is the variable acetylation that occurs at lysine residues. Histone acetylation and deacetylation are regulated by the activities of opposing enzymes: the histone acetyltransferases (HATs) and histone deacetylases (HDACs)6. Chromatin remodeling is involved in several cell processes such as stress response, adaptation, immune response and carcinogenesis7–12. However, chromatin remodeling mechanisms are poorly understood in pathogenic microorganisms, particularly in the context of virulence.
HDACs catalyze the removal of acetyl groups, leading to chromatin condensation13,14. Therefore, these enzymes play crucial roles in regulating gene expression as they modulate the accessibility of chromatin to transcriptional regulators and other regulating factors14. HDACs are evolutionarily conserved and are found in plants, fungi, and animals, as well as archaea and eubacteria15,16. They have been associated with epigenetic phenotypic changes in many fungal species such as Ustilago maydis17, Aspergillus nidulans18, Candida albicans19–21, Schizosaccharomyces pombe22,23, Saccharomyces cerevisiae24, and Cryptococcus neoformans25,26. It has also been reported that HDACs play important roles in virulence-related processes and morphological changes in some fungi. For example, the plant fungal pathogen U. maydis displays altered development and pathogenesis in the setting of altered HDAC activity17. Additionally, the human fungal pathogen C. albicans requires intact HDAC function for the yeast-hyphal transition that is central to its pathogenesis20.
Phylogenetically, fungal HDACs are divided into three main classes. The “classical” HDAC family proteins fall into two classes: class I (Hos2- and Rpd3-like proteins) and class II (Hda1-like proteins)15,27,28. The “non-classical” or Class III HDACs are Sir2-like proteins16. The central enzymatic domains of Class I and II HDACs are similar to one another, containing a central Zn2+ atom29. The class III enzymes are more divergent and are nicotinamide adenine dinucleotide (NAD)-dependent30. The number of HDACs of each class encoded in the genome can vary widely between species. For example, the model fungi S. cerevisiae and S. pombe contain differing numbers of HDAC genes. The budding yeast S. cerevisiae has three class I HDACs (Rpd3, Hos2 and Hos1) and two class II HDACs (Hda1 and Hos3). In contrast, in the fission yeast S. pombe there are two class I HDACs (Clr6 and Hos2) and one class II HDAC (Hda1)6,27.
In previous experiments, we tested the effects of chemical inhibitors of Class I and II HDACs (Sodium butyrate and Trichostatin A, respectively) on the expression of virulence-associated phenotypes in the human fungal pathogen C. neoformans. This encapsulated yeast is frequently found in the environment in association with decaying vegetation31. However, when encountered by people with compromised immune systems, especially persons with AIDS32, this microorganism can cause a lethal infection of the central nervous system33. In addition to its clinical importance, this fungal pathogen displays a remarkable degree of phenotypic plasticity in response to host and environmental cues (reviewed in34). We demonstrated that these HDAC inhibitors affected the expression of several microbial phenotypes, including growth rate at 37 °C, expression of the anti-phagocytic polysaccharide capsule, and production of the antioxidant melanin pigment35.
Here, we conducted a complementary genetic investigation of HDACs in C. neoformans. Using available genome data, we performed a comprehensive characterization of Class I and II HDAC homologues in this species. By creating targeted mutations of each of these HDAC genes, we tested our hypothesis that individual HDAC proteins would control specific aspects of C. neoformans adaptation to various environmental cues.
Results
Identification and characterization of C. neoformans Class I and II HDAC genes
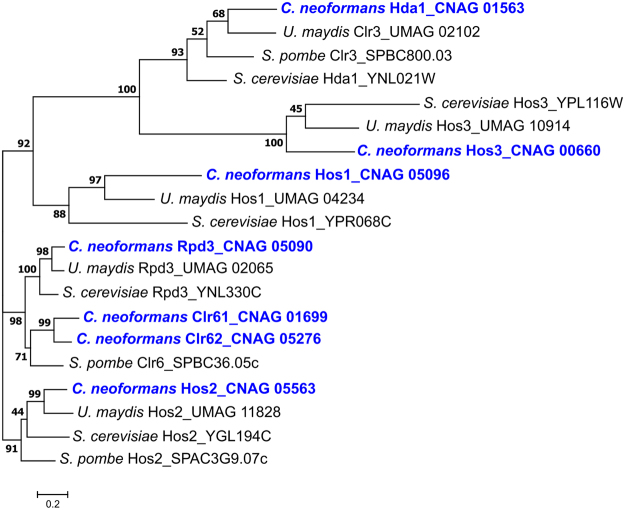
We used known HDAC gene sequences from three divergent fungal species (S. cerevisiae, S. pombe, and U. maydis) to conduct a genome-wide BLAST search and phylogenetic analysis of putative Class I and II HDAC genes in C. neoformans. Seven such genes were identified in the genome of the clinical C. neoformans strain H99 (Fig. 1). Two of the seven C. neoformans HDAC genes, encoding predicted Class I enzymes, had already been identified: HOS2 (CNAG_05563) and RPD3 (CNAG_05690)25. The other five predicted HDAC genes were named according to their closest relative in S. cerevisiae or S. pombe: Class I: HOS1 (CNAG_05096), CLR61 (CLR6 homologue-1; CNAG_01699), and CLR62 (CLR6 homologue-2; CNAG_05276); Class II: HDA1/CLR3 (CNAG_01563), and HOS3 (CNAG_00660). The C. neoformans HDA1/CLR3 gene was discussed in a recent manuscript as CLR3, encoding a possible histone modifier responsible for coordinating with Polycomb proteins to assist in the repression of subtelomeric gene expression26. However, two publications assigned the “CLR3” gene name to a distinct C. neoformans gene encoding a B-zip transcription factor required for capsule formation (CNAG_00871)36,37. Therefore, to avoid confusion in nomenclature, we refer to the CNAG_01563 gene by its closest homologue in S. cerevisiae, HDA1. All seven putative C. neoformans proteins identified in this manner possess domains characteristic of class I or II HDACs.
Figure 1.
Different fungal species contain varying numbers of genes encoding Class I and Class II histone deacetylases (HDACs). Maximum likelihood statistical method was used to demonstrate phylogenetic relationships for the predicted protein sequences for Class I and Class II HDACs in Cryptococcus neoformans, Saccharomyces cerevisiae, Schizosaccharomyces pombe and Ustilago maydis. The phylogenetic tree was created in MEGA7 with a WAG + G + I model and gamma shape parameters with bootstrap test of phylogeny applied with 500 replicates. Scale bar: 0.2 amino acid substitutions per site.
To ensure that these putative HDAC genes were actually expressed, we compared transcript levels for each gene after incubation of the wild-type strain in rich YPD medium versus incubation in minimal medium. The later condition was chosen since it induces a nutritional stress associated with capsule and melanin biosynthesis, two phenotypes that were impaired by pharmacological HDAC inhibition35. All seven HDAC genes were expressed and showed increased transcript accumulation when transitioned to minimal medium (Fig. S1).
To evaluate whether HDACs were relevant for C. neoformans virulence, we generated deletion mutants for each class I and II HDAC gene in strain H99 MATa and MATα genetic backgrounds. The HOS2 and RPD3 genes were previously described to be important for C. neoformans virulence in a murine model of cryptococcal infection25. We re-created these two mutants independently and used them as controls for comparison to the other mutant strains in our experimental conditions.
HDAC genes are required for stress tolerance and expression of virulence-associated phenotypes
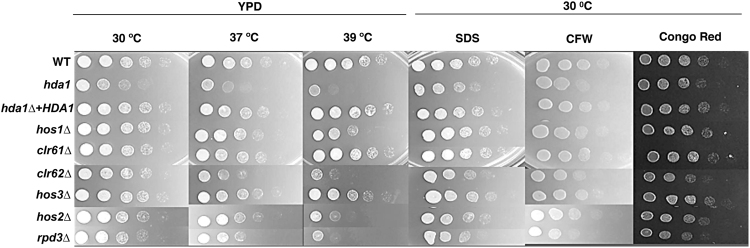
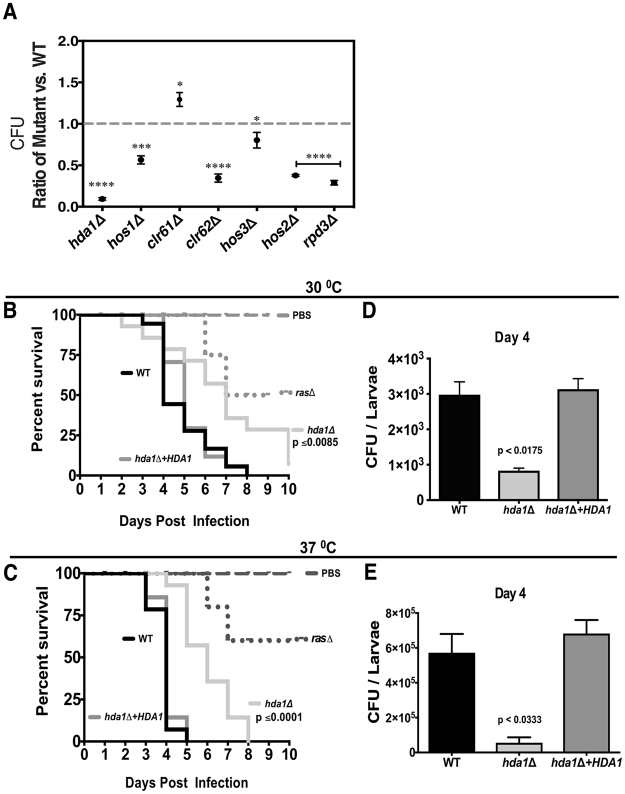
HDACs regulate different processes involved in adaptation and stress response17,23. We therefore tested the HDAC mutants for various phenotypes related to virulence and microbial differentiation (summarized in Table 1). Thermotolerance at 37 °C was compromised for the hda1Δ, hos1Δ, clr62Δ, hos2Δ and rpd3Δ HDAC mutants, being even more pronounced at 39 °C (Fig. 2). Additionally, growth inhibition in the presence of cell surface stresses (SDS, Calcofluor White and Congo Red) was observed for the hda1Δ, clr62Δ and rpd3Δ mutants, suggesting a defect in cell integrity in these strains (Fig. 2).
Table 1.
Phenotypic comparisons of HDAC mutants.
| Strain | 39 °C growth | Cell wall stressors | Capsule | Melanin | Protease activity | Survival in macrophages |
|---|---|---|---|---|---|---|
| Class I HDAC | ||||||
| rpd3∆ | − | − | + | + | + | − |
| hos2∆ | − | + | + | − | − | − |
| hos1∆ | − | + | ++ | + | + | − |
| clr61∆ | + | + | + | + | + | ++ |
| clr62∆ | − | − | − | + | + | − |
| Class II HDAC | ||||||
| hda1∆ | − | − | − | − | − | − |
| hos3∆ | + | + | − | + | + | − |
Summary of in vitro data presented in Results (“−” indicating defective phenotype, “+” indicating a phenotype similar to wild-type, and “++” indicating enhanced compared to wild-type).
Figure 2.
Temperature and cell stress-related phenotypes of the HDAC mutants. 5-fold serial dilutions for the indicated strains were spotted onto various media at the indicated temperatures. Growth was assessed after 48 hours of incubation. SDS (sodium dodecyl sulfate); CFW (calcofluor white).
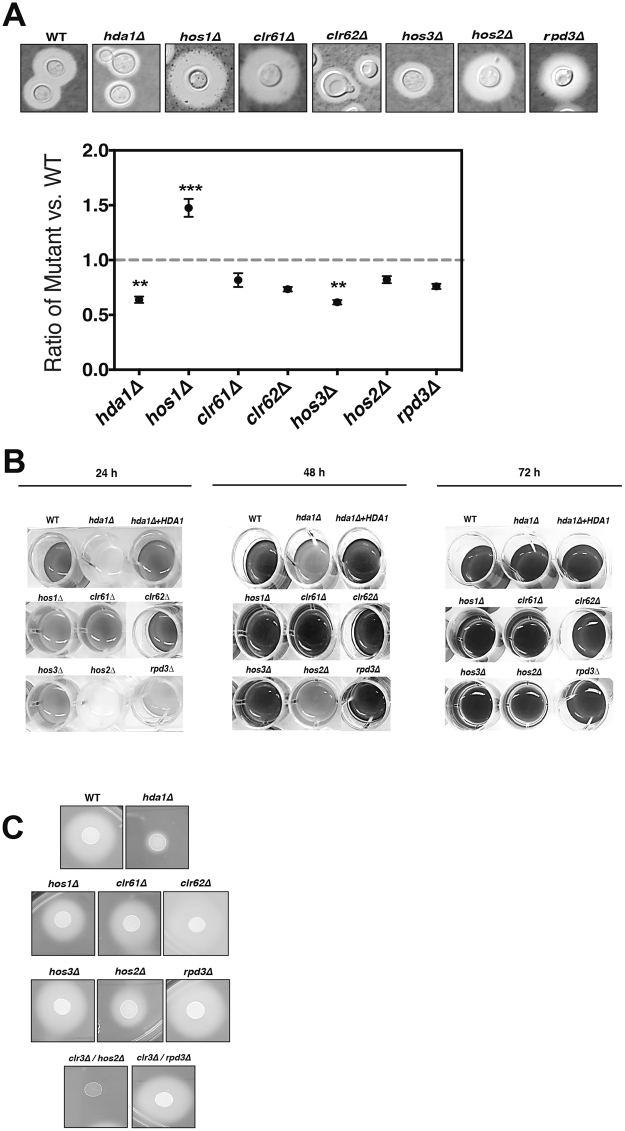
Since a marked reduction of C. neoformans capsule size was observed after chemical HDAC inhibition35, we hypothesized that specific HDAC mutants would correspondingly exhibit a hypocapsular phenotype. After incubation in capsule-inducing conditions, the hda1Δ, clr62Δ and hos3Δ mutants showed a significant reduction in capsule expansion (Fig. 3A), an overlapping but non-redundant set of mutants compared to those with stress-induced growth defects (Fig. 2). Capsule changes were assessed both by direct microscopy with India ink counter-stain as well as by quantitative determination of packed cell volume as a surrogate measure of total cell volume (Fig. S2A). In contrast, the hos1Δ mutant maintained a larger surface capsule than wild-type, suggesting that HDACs can play different or opposing functions in pathogen-related phenotypes.
Figure 3.
HDACs control C. neoformans virulence-associated phenotypes. (A) Capsule. Each strain was incubated in CO2-independent tissue culture medium for 72 hours to induce capsule formation. Capsule size was assessed using India ink counterstaining, and representative cells are displayed Capsule volume was quantified by assessing the packed cell volume of normalized cell suspensions, expressed as a ratio to wild-type. Data points represent averages of triplicate samples (+/− standard error). (B) Melanin. Indicated strains were incubated in minimal media with 1 mM L-DOPA at 30 °C, 150 rpm. Melanin production was assessed visually at 24, 48, and 72 hours. (C) Extracellular protease production. Indicated strains were spotted onto BSA agar and incubated at 30 °C for 3 days. The presence of a peripheral clear halo indicates protease activity.
Despite its reduction in surface capsule expression, the hda1Δ mutant displayed an apparent increase in total cell volume, perhaps a sign of delayed cell cycle progression (Fig. S2B). In this haploid yeast species, flow cytometry of propidium iodide-stained hda1Δ mutant cells demonstrated an enrichment in the population of cells with a 2n DNA content after incubation at 37 °C, suggestion a shift in cells in the G2/M stage of the cell cycle (Fig. S2C). Interestingly, we observed a similar delay in cell cycle progression when we treated C. neoformans cells with the Class I/II HDAC inhibitor sodium butyrate (NaBut)35.
Melanin production is an important virulence factor for C. neoformans, protecting the fungal cells from free radical-induced damage38. When incubated in melanin-inducing medium, we observed a marked delay in melanin production for the hda1Δ and hos2Δ mutants (Fig. 3B). Prolonged incubation resulted in eventual melanization of all HDAC mutant strains. The melanin defect in the hda1Δ mutant was fully suppressed in the hda1Δ + HDA1 reconstituted strain (Fig. 3B). These two melanin-deficient strains also produced decreased extracellular proteases, as determined by a reduced halo of clearing on BSA medium (Figs 3C, S3). Secreted proteases likely promote C. neoformans survival during interaction with host cells.
HDA1 plays a role in C. neoformans sexual reproduction
In other fungal species such as U. maydis and S. pombe, deletion of HDAC genes results in defective mating, emphasizing the importance of chromatin remodeling in sexual reproduction and development6,17,39,40. Additionally, the functional role of many HDACs has been linked to the yeast-to-hyphae transition in C. albicans, another form of fungal morphological transition41,42. We previously observed reduced C. neoformans mating hyphae production after chemical HDAC inhibition35, indicating that these enzymes are also likely involved in the mating and hyphal developmental processes in C. neoformans.
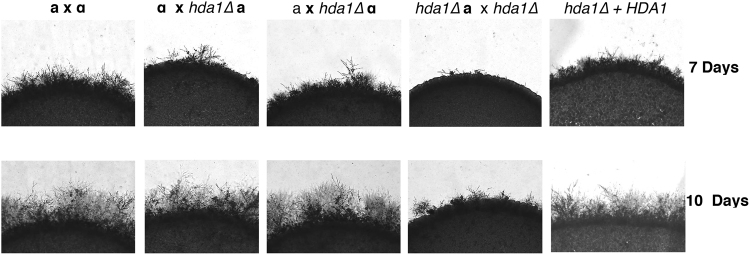
The individual HDAC mutants were crossed with a wild-type mating partner, and mating hyphae formation was monitored daily. For most HDAC mutants, no alterations in mating were observed (Fig. S4). However, the hda1Δ mutant showed a noticeable reduction in hyphae formation compared to wild-type (Fig. 4). This reduction in mating hyphae production is subtle when observed in unilateral crosses (hda1Δ × wild-type), but it is more pronounced in a bilateral cross (hda1Δ a × hda1Δ α) (Fig. 4). The impaired mating phenotype is completely restored to wild-type levels in the hda1Δ + HDA1 reconstituted strain. The HDA1 homolog also has been implicated in chromatin dynamics at the mating-type locus and in promoting epigenetic stability of heterochromatin in S. pombe23.
Figure 4.
HDA1 is required for mating. Overnight cultures of wild-type, hda1Δ mutant and hda1∆ + HDA1 reconstituted strains of opposite mating type (MATa or MATα) were mixed as indicated in equal amounts onto MS agar and incubated at room temperature, protected from the light. The edges of the mating mixtures were assessed for mating hyphae at 7 and 10 days (100×).
HDACs are required for C. neoformans pathogenicity
To assess the virulence of the HDAC mutants, we first examined the intracellular survival of all HDAC mutant strains in co-culture experiments with J774.1 murine macrophages. After 18 h of co-incubation, macrophages were lysed and the number of colony-forming units (CFUs) of surviving yeast cells was determined by quantitative culture. Phagocytosis rates did not significantly differ between these strains and wild-type, suggesting that altered intracellular survival most likely accounted for different CFUs after co-culture (data not shown). Although most of the mutants showed some degree of reduced survival in macrophages, the hda1Δ mutant was the most attenuated, consistent with its defects in multiple virulence-associated phenotypes. Interestingly, the clr61Δ mutant demonstrated better survival compared to wild-type in this in vitro model of host-pathogen interaction (Fig. 5A).
Figure 5.
The Hda1 HDAC protein is required for virulence in surrogate models of infection. (A) HDACs and survival in macrophages. Cells of each strain were co-incubated with J774A.1 macrophages (MOI: multiplicity of infection of 1:1). Macrophages were pre-activated with PMA (phorbol myristate acetate) and yeasts were opsonized with the 18B7 anti-GXM antibody. Viable colony-forming units (CFU) after 18 hours of co-cultures were assessed by quantitative culture, and the graphs represent the average ratio of each mutant strain normalized to the wild-type control (dashed line). Error bars represent standard errors of the mean. Statistical test: One-way ANOVA with Dunnett’s multiple-comparison test used to compare the means of results from three independent experiments. ****p < 0.0001; ***p < 0.001; *p < 0.05. (B,C) Galleria mellonella. The indicated strains were inoculated into larvae of the greater wax moth G. mellonella, and survival was monitored at 30 °C (B) and 37 °C (C). Mock infections with PBS injections were used as uninfected controls. Strains: Wild-type, hda1∆ mutant, hda1∆ + HDA1 reconstituted strain and the ras1∆ hypovirulent mutant43. Statistical test: Kaplan-Meier method. (D,E) In a separate experiment, 5 larvae from each group were assessed for fungal burden by quantitative culture on day 4 post infection. Statistical test: One-way ANOVA with Dunnett’s multiple-comparisons test.
We also used a well-characterized invertebrate model of host-fungal interaction to assess the virulence of the HDAC mutants in the larvae of the greater wax moth Galleria mellonella. This model assesses survival of the infected insect host as well as measuring persistence of the infecting fungal cells. Additionally, since this infection can be performed at both ambient temperature at 30 °C and 37 °C, we were able to distinguish the effects of altered thermotolerance from other contributors to virulence. The clr62∆ mutant was fully virulent in the Galleria infection model, even though it displayed reduced survival in macrophage co-culture (Fig. S5A). These observations suggest that the clr62∆ thermotolerance defect, and not its other more modest in vitro mutant phenotypes, explains its altered interaction with macrophages. Consistent with their persistence in macrophages, the clr61∆ and hos3∆ mutants displayed no detectable alteration in virulence in the Galleria infection model, with larvae succumbing to the infection at a similar rate as those infected with the isogenic wild-type strain (Fig. S5B,C). Moreover, quantitative cultures indicated higher levels of persistence of the clr61∆ mutant compared to wild-type, similar to its enhanced ability to survive within activated macrophages (Fig. S5F). Also consistent with the macrophage data, the hos2∆ and rpd3∆ mutants displayed reduced virulence in Galleria, with statistically significant reductions in host death rate due to infection (Fig. S5D,E). The hda1∆ mutant also displayed a marked reduction in survival in this infection model, similar to the highly attenuated ras1∆ mutant strain43. Importantly, the reduction in hda1∆ virulence was observed at both 30oC and 37oC, indicating that the thermotolerance defect alone cannot account for this strain’s hypovirulence (Fig. 5B,C). There was also a significantly reduced fungal burden for this strain at 4 days post-infection at both temperatures, consistent with its dramatic reduction in overall virulence (Fig. 5D,E).
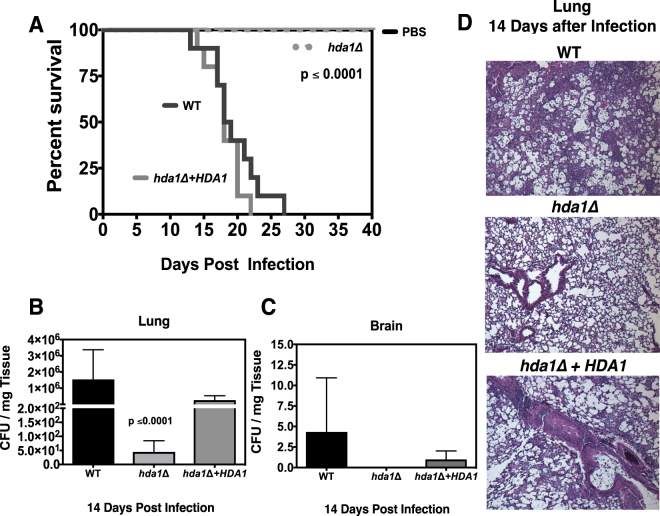
Given that the hda1Δ mutant displayed the most striking virulence attenuation among the HDAC mutants, we also tested this strain in the murine inhalation model of cryptococcosis. We intranasally inoculated female C57BL/6 mice with the hda1Δ mutant strain, as well as isogenic reconstituted and wild-type strains. During the course of infection, we serially assessed surrogate end-points of progressive infection known to correlate with impaired survival (weight loss, neurological symptoms, and inability to maintain self-care)44. In a subset of infected mice, the lungs were prospectively assessed by quantitative fungal culture and histopathology at days 7 and 14 post inoculation.
Mice infected with the wild-type and the reconstituted strain exhibited a median survival time of 18 days, and a maximal survival time of 26 days (Fig. 6A). In contrast, all mice infected with the hda1Δ strain survived to the end of the experiment at 40 days with no signs of clinical illness (Fig. 6A). The number of CFUs recovered from the lungs and brains of infected mice was significantly reduced for the hda1Δ mutant compared to wild-type (Fig. 6B,C and S6). Additionally, at both time points, histopathological assessment with H&E staining showed fewer fungal cells and only minimal lung inflammation for the animals infected with the hda1Δ mutant (Fig. 6D and S6). Overall, these observations corroborate the macrophage and G. mellonella assays, reinforcing the importance of Hda1 for virulence of C. neoformans.
Figure 6.
Effect of the hda1Δ mutant strain on virulence as assessed by the murine inhalational model of infection. (A) Ten C57BL/6 female mice were intranasally inoculated with the wild-type, hda1Δ, or hda1∆ + HDA1 strain to establish an infection. Animal survival was monitored for 40 days. Statistical test: Kaplan-Meier method. p < 0.0001 (B) Lungs and (C) brains of infected animals were harvested 14 days post infection, and quantitative cultures per mg tissue were performed. Statistical test: One-way ANOVA with Dunnett’s posttest. (D) Histopathological analysis of H&E-stained lung tissue from animals infected with the indicated strains after 14 days of infection.
Hda1 transcriptionally regulates genes required for adaptation and virulence
To identify the genes and processes regulated by Hda1, we used RNA-Seq to perform transcriptome analysis of the hda1Δ mutant compared to wild-type. For these experiments, we specifically chose incubation conditions (30 °C in minimal medium for 2 hours) to approximate those used for capsule and melanin induction as well as to match those used to measure HDAC gene transcript levels by RT-PCR.
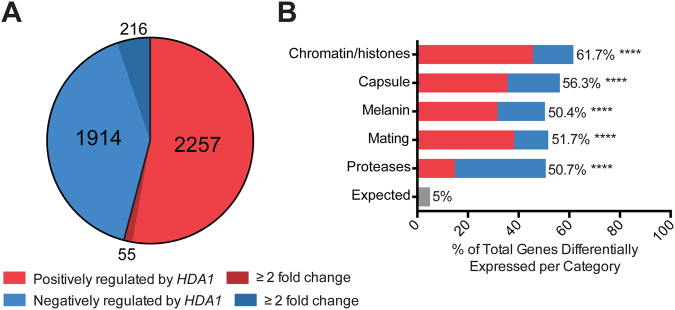
We used two independent methods of data analysis to ensure reproducibility of results. Using the TopHat2/Cufflinks analysis pipeline45,46, we found a large number of genes (4715) with a statistically significant difference in transcript abundance between the wild-type and hda1Δ mutant strains (Fig. S7). Using the Star/DESeq2 analysis method47–49, we documented a similar number of genes (4982) with a statistically significant difference in transcript levels between these strains (Fig. S7B). These data sets were highly correlated, with 4171 genes found to differ in transcript abundance by both methods (Fig. S7C). Of these 4171 genes, 1914 were negatively regulated by HDA1 (displayed increased mRNA levels in the hda1∆ mutant) and 2257 were positively regulated by HDA1 (displayed decreased mRNA levels in the hda1∆ strain) (Fig. 7A). Although statistically significant, most of these changes in transcript levels were small in absolute magnitude, with only 271 genes demonstrating a transcriptional difference between the two strains of greater than 2-fold (216 genes negatively regulated by HDA1, and 55 genes positively regulated by HDA1).
Figure 7.
The transcriptional signature of the hda1∆ mutant versus wild-type. (A) Hda1-dependent upregulated and downregulated genes in the hda1∆ mutant versus wild-type as determined by two independent RNA-Seq analysis pipelines. Graph represents values from STAR/DeSeq2 analysis. (B) Modified gene ontology enrichment analysis of Hda1-dependent genes. The percent of genes differentially expressed in the hda1∆ strain versus wild-type in each category is plotted on the x-axis. Colors represent the proportion of differentially expressed genes in each category positively or negatively regulated by HDA1. Statistical test: Chi-square of observed versus expected frequency of 5%; ****p < 0.0001.
We used the FungiDB database to perform a species-specific, modified gene ontology enrichment analysis (see Methods) to explore HDA1-dependent transcription of genes associated with cellular processes known to be affected by Hda1 protein function. These data are graphically represented in Fig. 7B, demonstrating notable enrichment of each functional category among the Hda1-regulated genes. There was a highly significant association between Hda1-regulation and genes involved in capsule, melanin, and mating, consistent with the observed hda1Δ mutant phenotypes. Importantly, the gene category most enriched among Hda1-dependent genes are those involved in chromatin/histone function (Fig. 7B).
Consistent with the hda1Δ mutant defect in melanin production, the LAC1 and LAC2 transcript levels, corresponding to two laccase genes encoding the main phenoloxidases required for melanin production, were both reduced in the hda1Δ mutant. Similarly, multiple genes involved in various aspects of encapsulation were also expressed at lower levels in the hda1Δ strain compared to wild-type (Table 2 and Fig. 7B). Interestingly, most of these capsule-associated genes were only marginally altered in expression. This observation suggests that the altered phenotypes of the hda1Δ mutant were due to a cumulative effect on the expression of multiple genes rather than a single gene. Additionally, the similarity in transcriptional changes in multiple capsule-associated genes suggests some degree of HDAC-mediated transcriptional co-regulation for genes involved in this very important cellular process.
Table 2.
Expression data for selected Hda1-regulated genes identified by modified gene ontology enrichment analysis.
| Gene ID | Gene name | Product Description | log2(fold change) |
|---|---|---|---|
| Chromatin/Histones | |||
| CNAG_00051 | SNT1 | Putative Set3c deacetylase complex subunit | −0.276468546 |
| CNAG_00063 | Histone H3 | −0.466103457 | |
| CNAG_00085 | ASF1 | Histone chaperone ASF1 | 0.224363607 |
| CNAG_00375 | GCN5 | Saga complex histone acetyltransferase | −0.338704771 |
| CNAG_00561 | Histone acetyltransferase type B catalytic subunit | −0.237158105 | |
| CNAG_00660 | HOS3 | Histone deacetylase HOS3 | −0.576126726 |
| CNAG_00718 | CAC2 | Chromatin assembly factor 1 subunit B | −0.137046165 |
| CNAG_00740 | SNF5 | Swi/Snf chromatin-remodeling complex subunit | −0.374848103 |
| CNAG_01013 | Chromatin binding protein | −0.66158404 | |
| CNAG_01018 | Histone-lysine N-methyltransferase Su(var)3–9 | −0.225072362 | |
| CNAG_01148 | FPR3 | Peptidyl-prolyl cis-trans isomerase | −0.664323021 |
| CNAG_01520 | Histone-arginine methyltransferase CARM1 | −0.507582078 | |
| CNAG_01563 | HDA1 | Histone deacetylase | −3.811428692 |
| CNAG_01648 | Histone H4 | −0.45507281 | |
| CNAG_01699 | CLR61 | Histone deacetylase | −0.160504318 |
| CNAG_01863 | SNF2 | Chromatin remodeling complex ATPase | −0.202397993 |
| CNAG_01972 | TAF10 | C2H2 zinc finger protein Zas1A | 0.151175426 |
| CNAG_02195 | Origin recognition complex subunit 1 | −0.746619743 | |
| CNAG_02215 | HAP3 | Transcriptional activator | −0.184025245 |
| CNAG_02536 | TAF6 | Transcription initiation factor TFIID subunit 6 | −0.1596263 |
| CNAG_02749 | Histone-lysine N-methyltransferase SUV420H | −0.450115461 | |
| CNAG_03188 | SET202 | Histone-lysine N-methyltransferase, H3 lysine-36 specific | −0.252801765 |
| CNAG_03203 | DNA polymerase epsilon p12 subunit | 0.371759788 | |
| CNAG_04168 | Histone H1/5 | −0.582447791 | |
| CNAG_05096 | HOS1 | Histone deacetylase | −0.123187881 |
| CNAG_05221 | Histone H2A.Z | −0.183930793 | |
| CNAG_05276 | CLR62 | Histone deacetylase | −0.176270846 |
| CNAG_05290 | SPT3 | Transcription initiation protein SPT3 | 0.159839958 |
| CNAG_05404 | Histone-lysine N-methyltransferase SUV39H | 0.135941792 | |
| CNAG_05428 | TAF5 | Transcription initiation factor TFIID subunit 5 | 0.16375121 |
| CNAG_06283 | LIV4 | Putative myb-like mRNA polymerase I termination factor | −0.479281412 |
| CNAG_06392 | SGF29 | Putative saga histone acetyltransferase complex component | −0.489834833 |
| CNAG_06544 | Non-histone chromosomal protein 6 | −0.61516928 | |
| CNAG_06597 | SPT8 | Transcriptional activator SPT8 | −0.224863088 |
| CNAG_06745 | Histone H3 | −0.576382711 | |
| CNAG_06746 | Histone H2B | −0.606684422 | |
| CNAG_06747 | Histone H2A | −0.53480499 | |
| CNAG_07027 | SPT2 | Protein SPT2 | 0.101538608 |
| CNAG_07565 | TAF9 | Transcription initiation factor TFIID subunit 9B | 0.194617264 |
| CNAG_07572 | ELP3 | Pol II transcription elongation factor | −0.32202168 |
| CNAG_07680 | HAP5 | Transcriptional activator HAP5 | 0.131411312 |
| CNAG_07807 | Histone H4 | −0.37405091 | |
| Capsule | |||
| CNAG_00600 | CAP60 | Capsule-associated protein | 0.472665779 |
| CNAG_00697 | UGE1 | UDP-glucose epimerase | 0.406541427 |
| CNAG_00701 | CAS31 | Protein involved in gxm O-acetylation | 0.195896704 |
| CNAG_00721 | CAP59 | Alpha-1,3-mannosyltransferase | −0.283339391 |
| CNAG_02581 | CAS33 | Hypothetical protein | −0.230488663 |
| CNAG_02805 | CAN1 | Carbonic anhydrase | 0.381708027 |
| CNAG_02885 | CAP64 | Capsule-associated protein | −0.411533956 |
| CNAG_03322 | UXS1 | UDP-glucuronic acid decarboxylase | −0.192984755 |
| CNAG_03426 | GMT2 | GDP-mannose transporter 2 | −1.301133952 |
| CNAG_03644 | CAS3 | Hypothetical protein | −0.680904969 |
| CNAG_03695 | CAS41 | Probable sugar phosphate/phosphate translocator; capsule biosynthetic protein | −0.289383393 |
| CNAG_03735 | CAP4 | Hypothetical protein | −0.583727866 |
| CNAG_03929 | CAS42 | Solute carrier family 35, member C2 | −0.140514137 |
| CNAG_04312 | MAN1 | Mannose-6-phosphate isomerase | −0.199177397 |
| CNAG_05023 | CAS91 | Putative maltose o-acetyltransferase | −0.396029552 |
| CNAG_05081 | PDE1 | Phosphodiesterase | −0.17141564 |
| CNAG_05139 | UGT1 | Solute carrier family 35 (UDP-sugar transporter), member A1/2/3 | 0.135099801 |
| CNAG_05144 | CAN2 | Carbonic anhydrase | −0.881416777 |
| CNAG_05222 | NRG1 | Transcriptional regulator Nrg1 | −0.265150815 |
| CNAG_05431 | RIM101 | pH-response transcription factor PacC/Rim101 | 0.181521283 |
| CNAG_05817 | GMT1 | GDP-mannose transporter 1 | −0.2343137 |
| CNAG_06016 | CAP6 | Hypothetical protein | 0.139069648 |
| CNAG_06524 | FRE3 | Ferric reductase | 4.348573694 |
| Melanin | |||
| CNAG_02434 | ATX1 | Putative copper ion transporter | −0.164690738 |
| CNAG_03464 | LAC2 | Laccase | −0.253171475 |
| CNAG_03465 | LAC1 | Laccase | −0.509427708 |
| CNAG_05081 | PDE1 | Phosphodiesterase, phosphodiesterase, variant | −0.17141564 |
| CNAG_05465 | GIB2 | Guanine nucleotide-binding protein subunit beta-like protein | −0.316312202 |
| CNAG_06415 | CCC2 | Cu2 -exporting ATPase | 0.152106422 |
| CNAG_06524 | FRE3 | Ferric reductase | 4.348573694 |
| CNAG_07701 | CTR2 | Putative copper ion transporter | 0.528660759 |
| Mating | |||
| CNAG_00293 | RAS1 | Ras-like protein | −0.198864829 |
| CNAG_01262 | GPB1 | Guanine nucleotide-binding protein subunit beta | −0.14439011 |
| CNAG_01452 | MAT3 | Mat3 pheromone repeat protein | −0.31601691 |
| CNAG_01730 | STE7 | MAP kinase kinase | −0.264935377 |
| CNAG_02756 | CDC43 | Geranylgeranyltransferase-I beta subunit | −0.21364078 |
| CNAG_02883 | RAC1 | Rho family protein | −0.428370761 |
| CNAG_03938 | CPR2 | Pheromone a factor receptor | 0.549162139 |
| CNAG_04119 | ROM2 | Rho guanyl-nucleotide exchange factor | 0.13250298 |
| CNAG_04761 | RAS2 | Ras family protein | −0.779751921 |
| CNAG_05465 | GIB2 | Guanine nucleotide-binding protein subunit beta-like protein | −0.316312202 |
| CNAG_05866 | PRM1 | Putative plasma membrane fusion protein | 1.12944946 |
| CNAG_05925 | CDC3 | Septin ring protein | −0.208014522 |
| CNAG_05970 | PAK1 | Ste/Ste20/PakA protein kinase | 0.070285615 |
| CNAG_06806 | ETF1alpha | Electron transfer flavoprotein alpha subunit | −0.523135543 |
| CNAG_06808 | STE3alpha | Pheromone a factor receptor | −0.657125254 |
| CNAG_06811 | RPL22alpha | Large subunit ribosomal protein L22e | −0.406283818 |
| CNAG_06812 | SPO14alpha | Phospholipase D1 | −0.323123919 |
| CNAG_06813 | CAP1alpha | Hypothetical protein | −0.701589794 |
| CNAG_06980 | STE11alpha | MAPKK kinase, Ste/Ste11 protein kinase | 0.317591405 |
| CNAG_07407 | MFalpha3 | Mating-type pheromone alpha | −0.974668369 |
| CNAG_07409 | RPO41alpha | DNA-directed mRNA polymerase, mitochondrial | −0.66659612 |
| CNAG_07410 | CID1alpha | Hypothetical protein | −0.197735218 |
| CNAG_07507 | STE50 | Protein kinase regulator | −0.274868604 |
| Protease | |||
| CNAG_00581 | Saccharopepsin | 0.406541427 | |
| CNAG_01343 | ATP-dependent Clp protease ATP-binding subunit ClpX | 0.397082268 | |
| CNAG_01688 | ATP-dependent metalloprotease | −0.454088763 | |
| CNAG_02239 | 26 S protease regulatory subunit 4 | 0.447325323 | |
| CNAG_02282 | Carboxypeptidase A4 | 0.450060236 | |
| CNAG_03904 | 26 S protease regulatory subunit 6B | 0.150163286 | |
| CNAG_04380 | Peptidase | −0.391189814 | |
| CNAG_04635 | Endopeptidase | 0.225975854 | |
| CNAG_04666 | 26 S protease regulatory subunit 8 | 0.186381652 | |
| CNAG_04906 | 26 S protease regulatory subunit 10B | 0.282517098 | |
| CNAG_05742 | STP1 | Putative site-2 protease | −0.354935589 |
| CNAG_05872 | Endopeptidase | 0.437070994 | |
| CNAG_06153 | 26 S protease regulatory subunit 6A-B | 0.401220915 | |
| CNAG_06410 | ATP-dependent Clp endopeptidase, proteolytic subunit ClpP | −0.20806081 | |
| CNAG_07520 | Endopeptidase | 0.661884424 | |
| HDA1 associated | |||
| CNAG_05333 | Hypothetical protein | 3.16455427 | |
| CNAG_06874 | HpcH/HpaI aldolase/citrate lyase | 0.721327† | |
| CNAG_07651 | DEAD-box ATP-dependent RNA helicase 26 | 0.924457338 | |
Log2(fold change) values for hda1∆ versus wild-type from DESeq2 analysis. Table amended from full list of differentially expressed genes in each category (Table S1). Negative changes represent genes positively regulated by Hda1; positive changes represent genes negatively regulated by Hda1.
†log2(fold change) value obtained from TopHat2/Cuffdiff analysis.
Similar trends of reduction in transcript abundance were evident for families of genes involved in mating and extracellular protease production (Table 2 and Fig. 7B). Additionally, several genes involved in transcription and chromatin remodeling, including other HDAC genes and the SAGA complex histone acetyltransferase (GCN5), were differently expressed in the hda1Δ mutant (Table 2).
Furthermore, we analyzed transcript levels of genes described to be regulated by Hda1. Hda1 was previously identified as a chromatin-related protein component of the C. neoformans Polycomb system, responsible for repression of gene expression in subtelomeric domains by recognizing H3K27 histone methylation26. In that study, HDA1 was associated with repression of the expression of the subtelomeric genes CNAG_05333, CNAG_06874 and CNAG_0765126. In our RNA-Seq analyses we found that the same group of genes displayed increased levels of mRNA in the hda1Δ mutant (Table 2), reinforcing the concept that fungal Hda1 orthologs play a role in the assembly of subtelomeric heterochromatin in fungi as diverse as the basidiomycete C. neoformans and the ascomycete S. pombe22,50.
Discussion
Chromatin remodeling by directed histone protein acetylation provides a rapid means to regulate transcription in response to changing environmental signals. This type of efficient and precise control of gene expression potentially allows microbial pathogens to maintain remarkable phenotypic plasticity in order to adapt to the many stresses encountered in the infected host. In fungi such as A. nidulans, C. albicans, S. cerevisiae and S. pombe, histone deacetylation and associated chromatin changes have been shown to mediate diverse cellular processes, including those associated with host adaptation and virulence18,20,21,24,51.
In the present work, we report a comprehensive characterization of the roles of Class I/II HDACs in the expression of virulence attributes of the opportunistic pathogenic fungus C. neoformans. Seven sequence homologues of S. cerevisiae and S. pombe class I and II HDAC genes were found in the C. neoformans genome. This number is higher than that described for many other fungal species. It also appears that there are two paralogous genes encoding homologs of the Clr6 HDAC (designated CLR61 and CLR62), corroborating the phylogenetic study of Nishida, et al.52. Our phenotypic studies of the respective paralog mutant strains suggest that these two genes have undergone some degree of neofunctionalization. For example, the clr61∆ mutant showed no impaired virulence attributes in several surrogate models of fungal virulence. In fact, this strain was more efficient in surviving inside macrophages in vitro and in G. mellonella larvae. On the other hand, the clr62∆ mutant was sensitive to high temperature and displayed reduced capsule compared to wild-type.
HDAC genes control specific development- and virulence-associated phenotypes
Individual mutant strains were obtained for each HDAC gene, and mutant strain virulence phenotypes were characterized in comparison to wild-type. We observed a large amount of phenotypic heterogeneity among these mutants, suggesting that individual HDAC proteins help to control the expression of a defined and specific set of genes. For example, the hda1Δ, clr62Δ and rpd3Δ mutant strains showed a reduction in the ability to grow on solid medium in the presence of surface-stressing agents such as SDS and CFW. In the presence of Congo red, a cell wall stressor, the growth reduction was subtle. Interestingly, recent investigators53 developed a chemical-genomic profiling approach to identify S. cerevisiae genes that may be related to growth under cell wall stress conditions and observed that specific HAT and HDAC genes were relevant to this process. Liu and collaborators observed growth defects at 37 °C for C. neoformans hos2Δ and rpd3Δ mutants25. In our assays, we observed that the rpd3Δ mutant presented impaired growth under cell wall stress conditions in addition to temperature, suggesting that the RPD3 gene plays a role in cell surface damage prevention.
We also demonstrated that hda1Δ, clr62Δ and hos3Δ strains displayed a significant reduction in the polysaccharide capsule expansion, while the hos1Δ mutant presented a larger capsule in comparison to the wild-type. These opposing effects on capsule formation by different C. neoformans HDAC genes suggest that histone acetylation might be a rapid and tunable way to precisely control encapsulation in response to environmental changes. This type of complex phenotypic regulation is plausible given the number of genes involved in this important physiological process34,54–58. Interestingly, the deletion of genes involved in histone acetylation (HATs) in C. neoformans also resulted in alterations in capsule size56,59. Even though HATs and HDACs exhibit opposite enzymatic function, these data together highlight the importance of the chromatin acetylation and deacetylation dynamics in the regulation of capsule expansion.
The hda1Δ mutant also displayed a subtle increase in the cell body volume. This observation is likely related to the increase in the G2/M cell population observed at 24 h of growth in minimal medium at 37 °C. The S. pombe Hda1 homolog is also involved cell cycle control60. Furthermore, the C. neoformans hda1Δ mutant shift in G2/M cell population phenocopied the results of treatment of the wild-type strain with the HDAC inhibitor sodium butyrate35.
We noticed a transient delay in melanin production for the hda1Δ and hos2Δ mutant strains. A melanization defect for the hos2Δ mutant had been previously reported25. It is possible that the delay in the melanin synthesis is related to the cell arrest in G2/M we observed for the hda1Δ mutant, since melanization in C. neoformans is influenced by cell density through a quorum sensing-related mechanism61,62. Altered expression of genes involved in the phenol oxidation step of melanin formation was also noted for the hda1Δ mutant strain.
The production of secreted proteases is an important virulence trait for C. neoformans, as they are involved in the growth and survival in the presence of antifungal drugs63 and in the invasion of the central nervous system64. We have shown that the hda1Δ and hos2Δ strains display compromised secreted protease activities and that the HDA1 and HOS2 genes seem to be redundant in the regulation of this phenotype. Also, the simultaneous deletion of both genes resulted in impaired thermotolerance (data not shown). In U. maydis these two genes act redundantly in regulating pathogenesis. Interestingly, in C. albicans HDA1 and HOS2 exert opposite roles in the control of morphogenesis21. We report for the first time that HDAC genes are necessary for full mating hyphae formation in C. neoformans, thus corroborating the data we obtained with chemical inhibitors of these enzymes35. The C. neoformans mating process involves a complex series of signaling and developmental events, including pheromone sensing, hyphal morphological transition, meiosis, and sporulation. Of note, pheromone gene expression was also altered in the hda1∆ mutant strain, suggesting a molecular mechanism for the observed deficient mating phenotypes.
HDAC mutants were studied in several infection models, including insects, isolated macrophages, and mice. Although the hda1Δ mutant showed the most prominently altered virulence in all of the models, we also noticed an in vivo survival defect in G. mellonella for the hos2Δ and rpd3Δ mutants. This result confirms prior findings of altered virulence for these mutant strains25. Distinct from the hda1Δ mutant, we did not observe any significant differences in the number of yeast cells recovered from larvae infected with the hos2Δ and rpd3Δ mutants versus the wild-type strain. This finding suggests that these fungal strains mediate virulence by multiple means, including the expression of factors that mediate host damage, as well as direct proliferation in vivo.
Class I/II HDACs function in the context of other epigenetic regulators
We hypothesized that Hda1 is involved in the adaptation and regulation of gene expression during infection. Sugiyama demonstrated that the S. pombe Hda1 homologue Clr3 takes part in a multiprotein complex involved in transcriptional silencing within telomeric regions50. Dumesic reported that C. neoformans Hda1/Clr3 co-precipitated with the Polycomb repressor complex and that, in the hda1∆/clr3∆ mutant strain, three genes related to subtelomeric regions were upregulated by qRT-PCR analysis26. We have corroborated the observed Hda1-repression of these same genes in our RNA-Seq experiment with a newly created hda1∆ mutant strain.
In addition to classical HDACs, other proteins participate in histone deacetylation and chromatin remodeling in fungi. Recently, Bouklas and collaborators65 evaluated the function of C. neoformans SIR2, a member of a large family of NAD+-dependent, non-classical HDACs collectively known as sirtuins30. The authors demonstrated that SIR2 loss resulted in a slightly impaired microbial fitness, which was rescued by Sir2 agonists. Additionally, Arras, et al.66 identified and deleted five C. neoformans sirtuin genes, demonstrating that two of the five deletion strains revealed mutant phenotypes in vitro. They found that three sirtuin genes (SIR2, HST3 and HST4) play a role in virulence in vivo, supporting the importance of chromatin remodeling in C. neoformans pathogenesis. Similar to these studies in non-classical HDACs, our targeted mutagenesis of C. neoformans Class I/II HDACs demonstrates that not all HDAC mutants regulate virulence-associated phenotypes in vitro. However, our data also suggest redundant or opposing roles for distinct HDACs for phenotypes such as encapsulation or melanin production. Therefore, having multiple HDACs may allow C. neoformans to precisely control the expression of specific virulence attributes. This type of fine control of gene expression is accomplished by HDACs that regulate FLO11 expression in S. cerevisiae24,67.
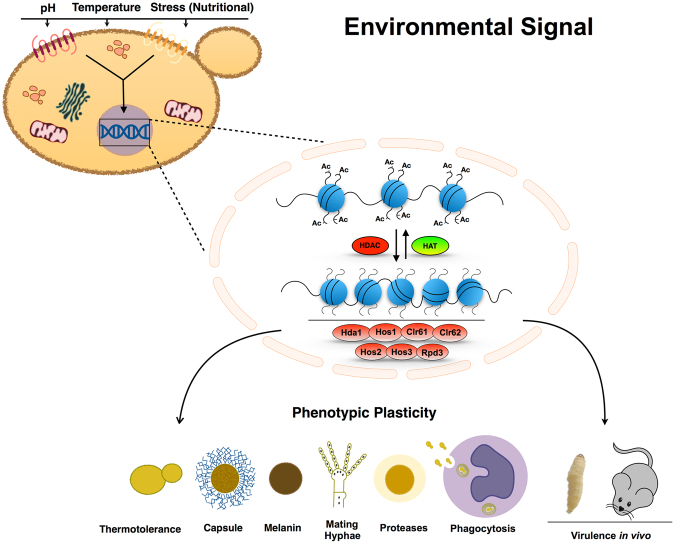
In summary, we have shown that the Class I and II histone deacetylases play distinct and overlapping roles in C. neoformans virulence processes (Fig. 8). These processes include thermotolerance, capsule formation, melanin synthesis, protease activity and cell wall integrity. We also demonstrated that HDACs are necessary for C. neoformans survival in multiple models of cryptococcal infection (Fig. 8). Among the HDAC genes, HDA1 controls multiple processes associated with fungal pathogenesis and development. The altered virulence of this mutant strain is likely due to its markedly reduced expression of capsule, melanin, and extracellular proteases, processes that are specifically required for microbial survival in the host. Finally, comparative transcriptional profiles of the hda1Δ and wild-type strains correlated mutant phenotypic changes with altered transcription of potentially relevant genes. The large number of genes with modest Hda1-dependent transcriptional changes suggest that the observed hda1Δ mutant phenotypes result from a complex and composite effect on the expression of multiple genes rather than a limited number of specific target genes.
Figure 8.
Model for HDAC regulation of the main virulence phenotypes in C. neoformans. Cells sense environmental cues (pH, high temperature, stress nutritional etc.) through membrane receptors and a signal is transmitted into the cell. The signal results in an adaptive response that requires chromatin remodeling mediated by the balance between HAT and HDAC activity. HDAC enzymes play a role in the regulation of the expression of all virulence phenotypes shown in the scheme. The loss of HDAC enzymes interferes with the correct adaptive response to environmental signals and impairs virulence factor expression.
Pharmacological manipulation of HDAC activity is being studied in several human diseases including malignancies and autoimmune diseases. For example, agents that control histone protein acetylation have been postulated as adjunctive therapy in HIV infection, potentially driving the virus out of latency and clearing infected patients of long-lived cellular reservoirs of infection68. In a similar manner, our data may contribute to the development of new therapeutic approaches for the treatment of fungal infections such as cryptococcosis.
Materials and Methods
Phylogenetic analysis of HDAC class I and II proteins
The predicted protein sequences of previously identified histone deacetylases for S. cerevisiae (Sc), S. pombe (Sp), and U. maydis (Um) were obtained from two fungal genome databases: the Broad Institute Fungal Genome Initiative (https://www.broadinstitute.org/fungal-genome-initiative, accessed 01/2015) and FungiDB (http://fungidb.org/fungidb/). These sequences were compared with HDAC gene homologues in C. neoformans (Cn), indicated by boxes. Multiple sequence alignments and phylogenetic analyses were performed using MUSCLE 3.7 (Multiple Sequence Comparison by Log-expectation): http://phylogeny.lirmm.fr/phylo_cgi/one_task.cgi? Task_type = muscle. A phylogenetic tree was prepared using MEGA769.
Strains, media, and growth conditions
Gene loci and strains used in this study are listed in Supplementary Tables S2 and S3. All strains were generated in the C. neoformans var. grubii strain H99 and were cultured on YPD (yeast extract [2%], peptone [1%], dextrose [2%]), unless stated otherwise. Fungal strains were stored in 15% glycerol at −80 °C until use.
Molecular biology
Gene deletion constructs were generated using PCR primer extension/overlap and biolistic transformation according to the double-joint PCR approach45 to create a deletion construct with a split neomycin (NEO) or nourseothricin (NAT) resistance marker to replace each genomic coding sequence of interest by homologous recombination, as previously described58. The flanking homologous regions of the deletion constructs were amplified using the primers listed in Table S4. All mutant strains were created from at least two independent transformations, and the transformants were selected in medium supplemented with the appropriate antibiotic (100 µg/ml NAT, 100 µg/ml NEO). Mutations were confirmed by PCR using primers directed against the 5′- and 3′-flanking regions to ensure precise replacement of the native locus with the mutant allele.
To confirm the previously documented association between Hos2 and Rpd3 with C. neoformans virulence and in vitro virulence-associated phenotypes, we created independent hos2Δ (FS10) and rpd3Δ (FS13) mutants based on those from the 2015 Madhani Cryptococcus mutant collection (Fungal Genetics Stock Center)25. First, the whole deletion cassette with the NAT resistance marker was amplified by PCR and introduced by biolistic transformation into the wild-type strain H99. Mutations among the transformants were confirmed by PCR. The clr61Δ mutant was obtained from the C. neoformans mutant library25, and the ORF deletion was confirmed by PCR using primers AA4491 + AA4492. Given the lack of discernible mutant phenotypes for the clr61 strain, either in vitro or in vivo, we elected not to create an independent mutant strain for this gene.
To create the hda1Δ mutant in the mating type a (MATa) strain background, the MATα hda1Δ mutant was crossed with the KN99 MATa wide-type strain on MS mating media. Spores were isolated by microdissection, and recombinant spores were identified by PCR and neomycin (NEO) resistance. The hda1Δ/hos2Δ and hda1Δ/rpd3Δ double mutants were created by crossing the hda1Δ MATa with the MATα hos2Δ or rpd3Δ strain on MS medium. We used similar mating methods (genetic crosses with strain KN99a, microdissection of spores, PCR confirmation of mutations) to create multiple clr62Δ, hos1Δ, and hos3Δ strains, to confirm the association between the specific HDAC gene mutation and the observed alterations of phenotypes in vitro.
The hda1Δ + HDA1 reconstituted strain was made by biolistically transforming the wildtype HDA1 locus into the hda1 mutant: the HDA1 locus was amplified using primers P11 (AAGG-AGATCT-GACACTTACGCTCTTT) and P12 (GTAG-TCTAGA-AGTTGTGTTCATCAGTCA) (BglII site underlined) for cloning into the BamHI site of the pCH233 plasmid containing the nourseothricin-resistance (NAT) marker.
Capsule Induction and Quantitation of Capsule Size
Capsules were visualized by negative staining with India ink. Capsule radius was calculated35 using ImageJ (Fiji) software70. At least 100 cells were measured for each assay, and the data were presented by relative quantification (mutant/wild-type capsule size). The packed cell volume (as a surrogate measure of capsule size) was calculated as previously described71,72.
Melanin assays
Melanin production was assessed by inoculation of the cells in a chemically defined minimal medium: 15 mM dextrose, 10 mM MgSO4, 29.4 mM KH2PO4, 13 mM glycine and 3 μM thiamine, pH 5.5, supplemented with 1 mM L-DOPA (Sigma-Aldrich) and incubated at 30 °C for 24 hours with shaking at 150 rpm. Melanization also was assessed on Niger seed agar72.
Protease activity
Each strain (105 cells) was inoculated on BSA agar medium (2% agar, 1% YNB medium, 1% BSA, 2% glucose) and incubated at 30 °C. The presence of a halo of clearance surrounding the colony was measured. The halo size was defined as the difference between (halo + colony diameter) minus (colony diameter). The diameter ratio (mutant halo size/wild-type halo size) was used for normalization of the data.
Macrophage assay
We measured the ability of the fungal cells to survive macrophage ingestion according to previous protocols73,74. J774 A.1 cells (5 × 104/well) were added to 96-well plates and activated by addition of 10 nM phorbol myristate acetate (PMA) and incubated for 1 h at 37 °C with 5% CO2. C. neoformans cells were opsonized with antibody 18B7 and added to the macrophages at a multiplicity of infection (MOI) of 1:1 (yeasts: macrophages), and the plates were incubated for 1 h at 37 °C in 5% CO2. The co-cultures were then washed three times with PBS to remove yeasts that were not internalized, and the plate was incubated at 37 °C in 5% CO2 for 18 h. Macrophages were lysed, and equal aliquots from each condition were quantitatively cultured on YPD agar and incubated at 30 °C for 2 days. Phagocytosis efficiency was measured for each strain, as previously described75, to ensure that altered engulfment did not account for differences in fungal recovery rates from this assay.
Virulence assessment in the G. mellonella model
G. mellonella larvae were infected (5 × 104 CFU) on the last left leg as previously described43. After infection, the caterpillars were placed at 30 or 37 °C, and monitored every 24 hours for a total of 10 days. After 4 days of infection, the 5 larvae pre-selected for CFU analysis were sacrificed, and hemolymph was quantitatively cultured on YPD + chloramphenicol (1 mg/ml).
Virulence assessment in the murine inhalation model of cryptococcosis
The virulence of the C. neoformans strains was assessed using the murine inhalation model of cryptococcosis44. Briefly, groups of 10 female C57BL/6 mice were anesthetized with isoflurane and inoculated intranasally with 1 × 105 fungal cells (in 25 μL sterile saline). The mice were monitored and sacrificed based on predetermined clinical endpoints that predict imminent mortality, using CO2 and bilateral thoracotomy. All studies were performed in compliance with American Veterinary Medical Association and Duke University institutional guidelines for animal experimentation.
Histopathologic analysis and brain and lung fungal burden
Five additional female C57BL/6 mice per strain, per time point, were infected as described above and sacrificed at seven and fourteen days post-infection. One lung from each mouse was inflated and harvested in 10% neutral buffered formalin at the indicated days post-infection. All lungs were then embedded in paraffin, cut into 5 μm thick slices, and stained with H&E by the Duke University Histopathology Core Facility. For CFU analysis, the remaining lung and brains were removed surgically, weighed and then macerated in PBS. Colonies were counted and normalized by the weight of each tissue; the mean of the replicates for each individual were used to calculate the fungal burden in each organ.
RNA preparation, sequencing and analysis
Three biological replicates of wild-type and hda1Δ mutant cells were incubated in YPD medium to mid-log phase and transferred to minimal medium for 2 h at 30 °C. RNA was extracted by using the Qiagen RNeasy Plant Minikit (Qiagen, Valencia, CA). Library preparation and RNA sequencing were performed by the Duke Sequencing and Genomic Technologies Shared Resource. Sequencing was performed on an Illumina HiSeq 2000/2500 instrument with 50 bp single end reads. To achieve sufficient sequence coverage for a reference transcriptome, the WT sample was sequenced with 72-bp paired-end reads. All raw and processed data have been deposited in NCBI’s Gene Expression Omnibus76 and are accessible through GEO Series accession number GSE109582 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE109582).
Data was analyzed by two independent methods to ensure rigor and reproducibility of the results. All reads were mapped to the C. neoformans reference genome using Tophat2 software45, and transcript quantitation was calculated via the Cufflinks/Cuffdiff pipeline46 using default parameters and a false discovery rate (FDR) of 10%. The C. neoformans H99 reference genome was obtained from the Sequencing Project at the Broad Institute of MIT and Harvard (accessed 03/31/2015). We excluded from the final analysis a limited number of genes (309) whose expression was unable to be interpreted either due to very low expression, ambiguous attribution of read assignments, or insufficient coverage. The annotation of gene function and location in this study was performed using FungiDB77–79.
To confirm and update our original analysis, a second independent alignment and differential expression analysis was performed following an RNA-Seq Bioconductor workflow48. This workflow utilizes STAR alignment software49 and the DESeq2 differential gene expression analysis package for R47 (FDR 10%). The C. neoformans strain H99 genome used for this analysis was obtained from NCBI (accessed July 2017). Similar to above, we excluded from our data analysis a limited number of genes (209) whose expression was unable to be interpreted either due to very low expression, ambiguous attribution of read assignments, or insufficient coverage.
Those genes that were significantly differentially regulated by both methods (4171), were analyzed by a modified gene ontology enrichment analysis using the FungiDB database. We generated a list of C. neoformans H99 genes associated with each cellular function of interest by using the “Search” function on FungiDB. The terms used in each search were “Chromatin”, “Histones”, “Capsule”, “Melanin”, “Mating”, and “Protease”. The lists generated were based on protein product descriptions (InterPro domains), user comments, PubMed citations, and phenotypic data included in the FungiDB database. Each functional gene list was compared to the significantly differentially regulated genes. The proportion/percent of significantly differentially regulated genes in each group was calculated, and enrichment was determined by a Chi-square test with a 5% expected frequency.
Statistical analysis
Data are expressed as means ± SE of at least triplicate samples. Statistical analysis and significance were performed by using the GraphPad Prism version 6.01 for Mac (GraphPad Software) or R version 3.4.0 for Mac, and considered significant if p-values were <0.05. Normality and variance assumptions were verified using the Shapiro-Wilk test. One-way ANOVA and Dunnett’s posttest was applied to compare variation related to the control wild-type. Two-way ANOVA and Tukey’s posttest were used to compare different groups with more than one variable. The 95% confidence interval was determined for all the experiments.
Ethics statement
All animal experimentation was performed according to established protocols approved by the Duke University Institutional Animal Care and Use Committee (IACUC). The minimum number of animals was used for each experiment to ensure statistical significance based pre-test predictions. All infections were performed after inhalational anesthesia with isoflurane. Daily animal care was overseen by Duke Vivarium veterinary staff. According to IACUC-approved protocols, animals were sacrificed by CO2 asphyxiation, followed by a secondary means of ensuring animal death (bilateral thoracotomy), according to institutional guidelines.
Data Availability
All data generated or analyzed during this study are included in this published article (or its Supplementary Information files). All microbiological strains, plasmids and sequencing data files are available to the scientific community upon request.
Electronic supplementary material
Acknowledgements
The authors would like to recognize the 2015 Madhani Cryptococcus mutant collection available at the Fungal Genetics Stock Center. These knockout strains were made freely available ahead of publication to the scientific community. This collection was created by the Madhani laboratory at UCSF and was supported by NIH grant R01AI100272. We also recognize the Duke Cancer Institute Flow Cytometry Shared Resource for flow cytometry support, as well as the Duke Sequencing and Genomic Technologies Shared Resource Core Facility for RNA-Sequencing. These studies were performed in partial fulfillment of graduate studies for Brandão FAS supported by CAPES/Brazil (99999.010237/2014-03), FAPDF/Brazil (PDJ -1191/2016), which are greatly appreciated. MJ Poças-Fonseca received a fellowship from CAPES/Brazil (99999.001884/2015-08). This work was supported by NIH R01 grant AI074677 and P01 grant AI104533 to JAA. JAA was a Burroughs Wellcome Fund New investigator in Molecular Pathogenic Mycology.
Author Contributions
F.B., M.J.P.F., and J.A.A. conceived and designed the experiments. F.B., S.E., K.O., K.P., C.B.N., L.F., A.L.B. conducted the experiments. F.B., S.E. and J.A.A. analyzed the results. F.B., S.E., M.J.P.F. and J.A.A. wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-21965-y.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 2.Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol. 2012;13:436–447. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 4.Krogan NJ, et al. Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Molecular and cellular biology. 2003;23:4207–4218. doi: 10.1128/MCB.23.12.4207-4218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 6.Ekwall K. Genome-wide analysis of HDAC function. Trends Genet. 2005;21:608–615. doi: 10.1016/j.tig.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 7.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dirscherl SS, Krebs JE. Functional diversity of ISWI complexes. Biochem Cell Biol. 2004;82:482–489. doi: 10.1139/o04-044. [DOI] [PubMed] [Google Scholar]
- 9.Venkatesh S, Workman JL. Histone exchange, chromatin structure and the regulation of transcription. Nat Rev Mol Cell Biol. 2015;16:178–189. doi: 10.1038/nrm3941. [DOI] [PubMed] [Google Scholar]
- 10.Tartey S, Takeuchi O. Chromatin Remodeling and Transcriptional Control in Innate Immunity: Emergence of Akirin2 as a Novel Player. Biomolecules. 2015;5:1618–1633. doi: 10.3390/biom5031618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helming KC, Wang X, Roberts CW. Vulnerabilities of mutant SWI/SNF complexes in cancer. Cancer Cell. 2014;26:309–317. doi: 10.1016/j.ccr.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hohmann AF, Vakoc CR. A rationale to target the SWI/SNF complex for cancer therapy. Trends Genet. 2014;30:356–363. doi: 10.1016/j.tig.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annual review of biochemistry. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 14.Thiagalingam S, et al. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann N Y Acad Sci. 2003;983:84–100. doi: 10.1111/j.1749-6632.2003.tb05964.x. [DOI] [PubMed] [Google Scholar]
- 15.Leipe DD, Landsman D. Histone deacetylases, acetoin utilization proteins and acetylpolyamine amidohydrolases are members of an ancient protein superfamily. Nucleic acids research. 1997;25:3693–3697. doi: 10.1093/nar/25.18.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17–31. doi: 10.1016/j.jmb.2004.02.006. [DOI] [PubMed] [Google Scholar]
- 17.Elias-Villalobos A, Fernandez-Alvarez A, Moreno-Sanchez I, Helmlinger D, Ibeas JI. The Hos2 Histone Deacetylase Controls Ustilago maydis Virulence through Direct Regulation of Mating-Type Genes. PLoS pathogens. 2015;11:e1005134. doi: 10.1371/journal.ppat.1005134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shwab EK, et al. Histone deacetylase activity regulates chemical diversity in Aspergillus. Eukaryot Cell. 2007;6:1656–1664. doi: 10.1128/EC.00186-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lohse MB, Johnson AD. Temporal anatomy of an epigenetic switch in cell programming: the white-opaque transition of C. albicans. Mol Microbiol. 2010;78:331–343. doi: 10.1111/j.1365-2958.2010.07331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hnisz D, Schwarzmuller T, Kuchler K. Transcriptional loops meet chromatin: a dual-layer network controls white-opaque switching in Candida albicans. Mol Microbiol. 2009;74:1–15. doi: 10.1111/j.1365-2958.2009.06772.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zacchi LF, Schulz WL, Davis DA. HOS2 and HDA1 encode histone deacetylases with opposing roles in Candida albicans morphogenesis. PLoS ONE. 2010;5:e12171. doi: 10.1371/journal.pone.0012171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aygun O, Mehta S, Grewal SI. HDAC-mediated suppression of histone turnover promotes epigenetic stability of heterochromatin. Nat Struct Mol Biol. 2013;20:547–554. doi: 10.1038/nsmb.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lorenz DR, Meyer LF, Grady PJ, Meyer MM, Cam HP. Heterochromatin assembly and transcriptome repression by Set1 in coordination with a class II histone deacetylase. Elife. 2014;3:e04506. doi: 10.7554/eLife.04506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robbins N, Leach MD, Cowen LE. Lysine deacetylases Hda1 and Rpd3 regulate Hsp90 function thereby governing fungal drug resistance. Cell Rep. 2012;2:878–888. doi: 10.1016/j.celrep.2012.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu OW, et al. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell. 2008;135:174–188. doi: 10.1016/j.cell.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dumesic PA, et al. Product binding enforces the genomic specificity of a yeast polycomb repressive complex. Cell. 2015;160:204–218. doi: 10.1016/j.cell.2014.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bjerling P, et al. Functional divergence between histone deacetylases in fission yeast by distinct cellular localization and in vivo specificity. Molecular and cellular biology. 2002;22:2170–2181. doi: 10.1128/MCB.22.7.2170-2181.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/bj20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finnin MS, et al. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- 30.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 31.Harrison TS. Cryptococcus neoformans and cryptococcosis. J Infect. 2000;41:12–17. doi: 10.1053/jinf.2000.0695. [DOI] [PubMed] [Google Scholar]
- 32.Rajasingham R, et al. Global burden of disease of HIV-associated cryptococcal meningitis: an updated analysis. Lancet Infect Dis. 2017;17:873–881. doi: 10.1016/S1473-3099(17)30243-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mitchell TG, Perfect JR. Cryptococcosis in the era of AIDS–100 years after the discovery of Cryptococcus neoformans. Clinical microbiology reviews. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O’Meara TR, Alspaugh JA. The Cryptococcus neoformans capsule: a sword and a shield. Clinical microbiology reviews. 2012;25:387–408. doi: 10.1128/CMR.00001-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brandao FA, et al. Histone deacetylases inhibitors effects on Cryptococcus neoformans major virulence phenotypes. Virulence. 2015;6:618–630. doi: 10.1080/21505594.2015.1038014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jung KW, et al. Systematic functional profiling of transcription factor networks in Cryptococcus neoformans. Nat Commun. 2015;6:6757. doi: 10.1038/ncomms7757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maier, E. J. et al. Model-driven mapping of transcriptional networks reveals the circuitry and dynamics of virulence regulation. Genome research, 10.1101/gr.184101.114 (2015). [DOI] [PMC free article] [PubMed]
- 38.Wang Y, Casadevall A. Susceptibility of melanized and nonmelanized Cryptococcus neoformans to nitrogen- and oxygen-derived oxidants. Infection and immunity. 1994;62:3004–3007. doi: 10.1128/iai.62.7.3004-3007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ekwall K, Nielsen O, Ruusala T. Repression of a mating type cassette in the fission yeast by four DNA elements. Yeast (Chichester, England) 1991;7:745–755. doi: 10.1002/yea.320070709. [DOI] [PubMed] [Google Scholar]
- 40.Hansen KR, et al. H3K9me-independent gene silencing in fission yeast heterochromatin by Clr5 and histone deacetylases. PLoS Genet. 2011;7:e1001268. doi: 10.1371/journal.pgen.1001268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu Y, Su C, Wang A, Liu H. Hyphal development in Candida albicans requires two temporally linked changes in promoter chromatin for initiation and maintenance. PLoS biology. 2011;9:e1001105. doi: 10.1371/journal.pbio.1001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lopes da Rosa J, Kaufman PD. Chromatin-mediated Candida albicans virulence. Biochim Biophys Acta. 2013;1819:349–355. doi: 10.1016/j.bbagrm.2011.08.007. [DOI] [PubMed] [Google Scholar]
- 43.Mylonakis E, et al. Galleria mellonella as a model system to study Cryptococcus neoformans pathogenesis. Infection and immunity. 2005;73:3842–3850. doi: 10.1128/IAI.73.7.3842-3850.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cox GM, Mukherjee J, Cole GT, Casadevall A, Perfect JR. Urease as a virulence factor in experimental cryptococcosis. Infection and immunity. 2000;68:443–448. doi: 10.1128/IAI.68.2.443-448.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq 2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Love MI, Anders S, Kim V, Huber W. RNA-Seq workflow: gene-level exploratory analysis and differential expression. F1000Res. 2015;4:1070. doi: 10.12688/f1000research.7035.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sugiyama T, et al. SHREC, an effector complex for heterochromatic transcriptional silencing. Cell. 2007;128:491–504. doi: 10.1016/j.cell.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 51.Bauer, I. et al. A Class 1 Histone Deacetylase with Potential as an Antifungal Target. MBio7, 10.1128/mBio.00831-16 (2016). [DOI] [PMC free article] [PubMed]
- 52.Nishida H, Matsumoto T, Kondo S, Hamamoto M, Yoshikawa H. The early diverging ascomycetous budding yeast Saitoella complicata has three histone deacetylases belonging to the Clr6, Hos2, and Rpd3 lineages. J Gen Appl Microbiol. 2014;60:7–12. doi: 10.2323/jgam.60.7. [DOI] [PubMed] [Google Scholar]
- 53.Garcia R, et al. Genomic profiling of fungal cell wall-interfering compounds: identification of a common gene signature. BMC Genomics. 2015;16:683. doi: 10.1186/s12864-015-1879-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bose I, Reese AJ, Ory JJ, Janbon G, Doering TL. A yeast under cover: the capsule of Cryptococcus neoformans. Eukaryot Cell. 2003;2:655–663. doi: 10.1128/EC.2.4.655-663.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Loftus BJ, et al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 2005;307:1321–1324. doi: 10.1126/science.1103773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Haynes BC, et al. Toward an integrated model of capsule regulation in Cryptococcus neoformans. PLoS pathogens. 2011;7:e1002411. doi: 10.1371/journal.ppat.1002411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alspaugh JA. Virulence mechanisms and Cryptococcus neoformans pathogenesis. Fungal Genet Biol. 2015;78:55–58. doi: 10.1016/j.fgb.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ost KS, O’Meara TR, Huda N, Esher SK, Alspaugh JA. The Cryptococcus neoformans alkaline response pathway: identification of a novel rim pathway activator. PLoS Genet. 2015;11:e1005159. doi: 10.1371/journal.pgen.1005159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.O’Meara TR, et al. Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule. PLoS pathogens. 2010;6:e1000776. doi: 10.1371/journal.ppat.1000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.George AA, Walworth NC. Escape from Mitotic Arrest: An Unexpected Connection Between Microtubule Dynamics and Epigenetic Regulation of Centromeric Chromatin in Schizosaccharomyces pombe. Genetics. 2015;201:1467–1478. doi: 10.1534/genetics.115.181792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eisenman HC, Chow SK, Tse KK, McClelland EE, Casadevall A. The effect of L-DOPA on Cryptococcus neoformans growth and gene expression. Virulence. 2011;2:329–336. doi: 10.4161/viru.2.4.16136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Albuquerque P, et al. Quorum sensing-mediated, cell density-dependent regulation of growth and virulence in Cryptococcus neoformans. MBio. 2013;5:e00986–00913. doi: 10.1128/mBio.00986-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bien CM, Chang YC, Nes WD, Kwon-Chung KJ, Espenshade PJ. Cryptococcus neoformans Site-2 protease is required for virulence and survival in the presence of azole drugs. Mol Microbiol. 2009;74:672–690. doi: 10.1111/j.1365-2958.2009.06895.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vu K, et al. Invasion of the central nervous system by Cryptococcus neoformans requires a secreted fungal metalloprotease. MBio. 2014;5:e01101–01114. doi: 10.1128/mBio.01101-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bouklas T, Jain N, Fries BC. Modulation of Replicative Lifespan in Cryptococcus neoformans: Implications for Virulence. Front Microbiol. 2017;8:98. doi: 10.3389/fmicb.2017.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Arras SDM, et al. Sirtuins in the phylum Basidiomycota: A role in virulence in Cryptococcus neoformans. Scientific reports. 2017;7:46567. doi: 10.1038/srep46567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barrales RR, Korber P, Jimenez J, Ibeas JI. Chromatin modulation at the FLO11 promoter of Saccharomyces cerevisiae by HDAC and Swi/Snf complexes. Genetics. 2012;191:791–803. doi: 10.1534/genetics.112.140301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Matalon S, Rasmussen TA, Dinarello CA. Histone deacetylase inhibitors for purging HIV-1 from the latent reservoir. Mol Med. 2011;17:466–472. doi: 10.2119/molmed.2011.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Granger DL, Perfect JR, Durack DT. Virulence of Cryptococcus neoformans: regulation of capsule synthesis by carbon dioxide. J. Clin. Invest. 1985;76:508–516. doi: 10.1172/JCI112000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee KT, et al. Systematic functional analysis of kinases in the fungal pathogen Cryptococcus neoformans. Nat Commun. 2016;7:12766. doi: 10.1038/ncomms12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.O’Meara, T. R., Holmer, S. M., Selvig, K., Dietrich, F. & Alspaugh, J. A. Cryptococcus neoformans Rim101 is associated with cell wall remodeling and evasion of the host immune responses. MBio410.1128/mBio.00522-12 (2013). [DOI] [PMC free article] [PubMed]
- 74.Esher, S. K. et al. Relative Contributions of Prenylation and Postprenylation Processing in Cryptococcus neoformans Pathogenesis. mSphere1, 10.1128/mSphere.00084-15 (2016). [DOI] [PMC free article] [PubMed]
- 75.Luberto C, et al. Roles for inositol-phosphoryl ceramide synthase 1 (IPC1) in pathogenesis of C. neoformans. Genes Dev. 2001;15:201–212. doi: 10.1101/gad.856001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Barrett T, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic acids research. 2013;41:D991–995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stajich JE, et al. FungiDB: an integrated functional genomics database for fungi. Nucleic acids research. 2012;40:D675–681. doi: 10.1093/nar/gkr918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 79.Wilkinson L. Exact and approximate area-proportional circular Venn and Euler diagrams. IEEE Trans Vis Comput Graph. 2012;18:321–331. doi: 10.1109/TVCG.2011.56. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (or its Supplementary Information files). All microbiological strains, plasmids and sequencing data files are available to the scientific community upon request.