Abstract
Chemical aminoacylation of orthogonal tRNA allows for the genetic encoding of a wide range of synthetic amino acids without the need to evolve specific aminoacyl-tRNA synthetases. This method, when paired with protein expression in the Xenopus laevis oocyte expression system, can extract atomic scale functional data from a protein structure to advance the study of membrane proteins. The utility of the method depends on the orthogonality of the tRNA species used to deliver the amino acid. Here, we report that the pyrrolysyl tRNA (pylT) from Methanosarcina barkeri fusaro is orthogonal and highly competent for genetic code expansion experiments in the Xenopus oocyte. The data show that pylT is amendable to chemical acylation in vitro; it is then used to rescue a cytoplasmic site within a voltage-gated sodium channel. Further, the high fidelity of the pylT is demonstrated via encoding of lysine within the selectivity filter of the sodium channel, where sodium ion recognition by the distal amine of this side-chain is essential. Thus, pylT is an appropriate tRNA species for delivery of amino acids via nonsense suppression in the Xenopus oocyte. It may prove useful in experimental contexts wherein reacylation of suppressor tRNAs have been observed.
Introduction
The method of in vivo nonsense suppression in Xenopus laevis oocytes via chemically aminoacylated tRNA has enabled the site-specific encoding of over 100 different amino acids into ion channels and other proteins1–3. This expression system is advantageous because the oocyte faithfully manufactures and traffics diverse ion channel and receptor proteins, where established techniques allow their analysis from the macroscopic to the level of single proteins. Noncanonical amino acids (ncAAs) have allowed atomic-level insights into structure, function, and pharmacology of ion channels. The system’s flexibility arises from the facile attachment of dinucleotide-amino acid substrates to truncated tRNA via enzymatic ligation4. That is, the same species of tRNA can be used for encoding the amino acid needed for the experimental inquiry, in contrast to co-injecting an aminoacyl-tRNA synthetase for ncAA aminoacylation5–7. For this reason, chemical acylation of tRNAs is widely used for genetic code expansion in Xenopus oocytes.
This approach continues to be useful for obtaining high-resolution functional details from a variety of ion channel and receptors. Notable examples of its use on post-synaptic ligand gated channels include the advancing of the energetic basis for ligand recognition8,9, main-chain chemistry in channel gating10, protein thermodynamics in channel activation9; as well as the application to voltage-gated ion channels11–17.
Multiple tRNA species have been used for delivery in the oocyte system, the most common being a mutated version of the glutamine tRNA from Tetrahymena thermophila, commonly termed THG7318. The utitlity of this tRNA is derived from the fact that is a natural amber (TAG) suppressor, therefore eliminating the need to alter the anticodon for nonsense suppression application. The specific motivation for the G73 mutation was to obscure recognition of the THG73 tRNA by endogenous glutamine synthetases in the oocyte expression system, thus increase its orthogonality. Although THG73 is orthogonal, multiple groups including ours have reported that it is susceptible under some experimental conditions to reacylation by endogenous glutaminyl-tRNA synthetases19–22. Efforts to further mutate THG73 to increase orthogonality have only been partially successful and are unable to completely eliminate in situ. The potential “error” introduced by tRNA reacylation is significant. Depending the functional tolerance at the site of incorporation within the target protein, misincorporation may lead to a mixed population of glutamine and the ncAA at the encoding site (introduced stop codon, usually TAG; amber codon)19,20. This variability can be controlled for by careful analysis of conditions performed in parallel with non-acylated tRNA (tRNA-CA), varied length of incubation following injection and limited abundance of tRNA, which provides an experimental window in which ncAA rescue precedes any such unintended readthrough event. However, if such an experimental window cannot be found, the encoding site must be abandoned22.
Pyrrolysine, the so-called “22nd amino acid,” is encoded by methanogenic archaea and bacteria using a tRNA that naturally recognizes the amber codon TAG23,24. This unique tRNA (pylT) displays exceptional orthogonality in bacteria and in mammalian cells and has been used to encode ncAAs via evolved aminoacyl-tRNA synthetases that recognize an amino acid of interest25. Overall, tRNA-synthetase pairs have enabled the encoding of more than 100 ncAAs in diverse environments including cell free translation17,26, mammalian cells27, bacteria28, and even whole animals29. However, the evolution of aminoacyl-tRNA sythetases to discern derivatized isosteric analogs from their natural amino acids counterparts (often necessary for atomic-level insights) has proven challenging. Here, we assayed the orthogonality of pylT via in vitro chemical aminoacylation and injection into Xenopus oocytes (Fig. 1), using high-resolution ion channel function as a sensitive and quantitative readout of rescue and readthrough.
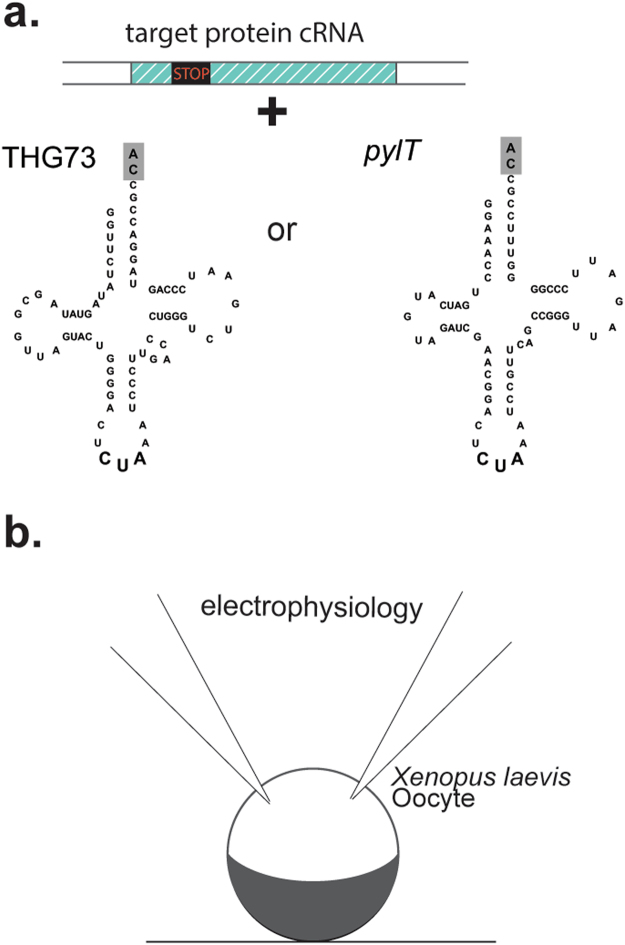
Figure 1.
Misacylation of orthogonal tRNA for genetic code expansion. (a) Target protein cRNA containing a site-directed stop codon (nonsense codon) is subsequently suppressed by an orthogonal tRNA - THG73 or pyrrolysine tRNA (pylT). The grey highlighted nucleotides “CA” represent ligated pdCpA (see methods). (b) Co-injection of tRNA and cRNA into the Xenopus oocyte enables electrophysiological two-electrode voltage clamp characterization of ion channels and receptors containing ncAA.
Results
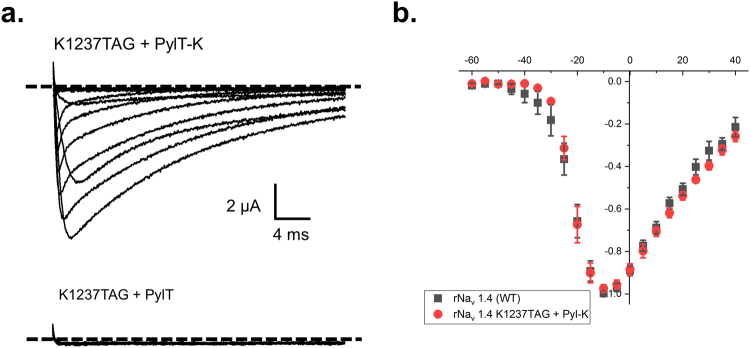
The sodium channel conducting state is strongly coupled to the transmembrane potential, thus one can precisely measure the flow of sodium conductance through the channel through standard electrophysiological approaches, Fig. 1. Voltage-gated sodium channel activation is characterized by transient inward, rapidly inactivating ionic currents. When cells are bathed in physiological recording solutions, e.g. 140 mM extracellular sodium, inward sodium currents appear as downward deflections in response to transient depolarization30,31; thus the level of recorded negative current is directly indicative of the number of full-length functional channels at the cell surface. To begin to scrutinize the orthogonality of the pylT, we chose amino acid position S571 in the human cardiac sodium channel hNav1.5 as a model site for encoding. Serine 571 is located in an unstructured intracellular loop between two domains of the channel and importantly has been mutated to other amino acids with diverse side-chain chemistries via conventional mutagenesis without compromising channel function32. We co-injected Xenopus laevis oocytes hNav 1.5 S571 cRNA (complementary RNA generated via in vitro transcription) with acylated and non-acylated tRNAs, and measured currents using the Two-Electrode Voltage Clamp technique after 24 hours33. When THG73 was used as the carrier tRNA for this position, significant hNav1.5 current was generated in the presence or absence of an appended tyrosine amino acid (i.e., whether we ligated the pdCpA, which lacks an amino acid, or the pdCpA-Tyrosine substrate) (Fig. 2, top panels). There was, in fact, no significant difference in the two conditions after 24hrs (Table 1), eliminating the prospect of adjusting injection conditions to abrogate readthrough while sparing rescue. Injection of S571TAG hNav1.5 cRNA alone did not elicit significant current, signifying that this site lacks appreciable ‘intrinsic’ bleedthrough at the level of the cRNA (Table 1)19–21. Therefore, in these conditions, it is possible that THG73-CA is acylated by an endogenous aminoacyl sythetase, and the resultant acylated tRNA supports the incorporation of an amino acid at position S571. By contrast, co-injection of pylT and hNav1.5-S571TAG yielded sodium currents that were strictly dependent on tRNA acylation, (Fig. 2, lower panels). Specifically, no sodium currents were detected for the condition for hNav1.5-S571TAG with non-acylated pylT (pylT-CA). In contrast, robust voltage-dependent sodium currents were seen when channel cRNA was co-injected with an tyrosine-acylated pylT (pylT-Tyr).
Figure 2.
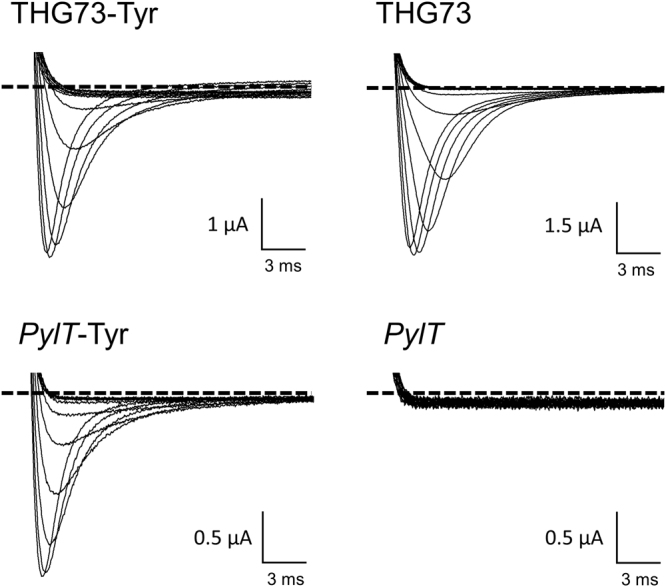
Rescue of an introduced stop codon into the human cardiac voltage gated sodium channel. Voltage-induced currents upon co-injection of hNav 1.5 S571TAG with either acylated (left) or unacylated tRNA (right) variants THG73 (top) or pylT (bottom). Xenopus oocytes expressing sodium channel variants were subjected to membrane depolarizations (steps from −80 mV to −20 mV). Traces show development of rapidly inactivating sodium currents as downward deflections. The level of zero current for each cell is indicated by a black dashed line.
Table 1.
Quantification of Nav currents of hNav1.5-S571TAG when co-injected with THG73 or pylT tRNA.
| Injection condition (24 hr) | Current at −20 mV (µA) | Std. Deviation | N-value | P-value (vs. like tRNA) |
|---|---|---|---|---|
| THG73 | −3.68 | 2.36 | 6 | — |
| THG73-Tyr | −2.26 | 1.53 | 5 | 0.13 |
| pylT | 0.011 | 0.0021 | 5 | — |
| pylT-Tyr | −1.31 | 0.44 | 5 | 0.0025 |
| S571TAG | −0.0073 | 0.0065 | 4 | — |
To confirm the in vitro enzymatic ligation of pdCpA and pdCpA- amino acid substrates to pylT, ligated and unligated tRNA samples assayed via denaturing TBE-Urea gels. Ligation of the pdCpA substrate is indicated by a gel shift corresponding to a two nucleotide increase in tRNA length. As indicated in Fig. 3, both pdCpA and pdCpA-amino acid substrates were efficiently ligated by the T4 RNA ligase. Therefore, the lack of observed reacylation-based readthrough in the oocyte expression system was not due to a lack of ligation of the pdCpA dinucleotide to truncated pylT.
Figure 3.
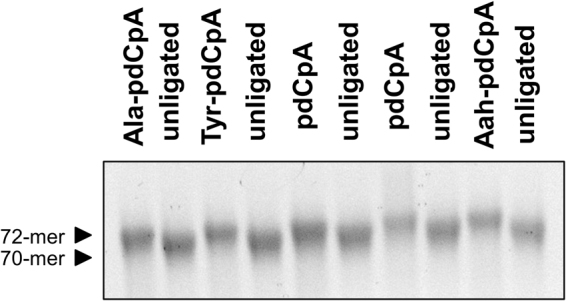
pylT can be efficiently ligated to dinucleotide-amino acid substrates in vitro. TBE UREA tRNA gels show successful ligation of substrates to pylT. Approximately 2 µg of tRNA was run per well. Each lane represents an independent ligation. Note the consistent gel shift representative of ligation of the dinucleotide-amino acid substrate to the truncated pylT. Abbreviations: Ala (alanine), Tyr (tyrosine), Aah (alpha-hydroxy alanine). Examples of additional substrate types are available in the Supplementary Figures.
The encoding fidelity of pylT was evaluated independently by rescuing a stop codon at position K1237 which resides in the sodium ion-selectivity filter of the rat skeletal muscle sodium channel, rNav 1.4. This unique structural feature promotes the selective passage of sodium through the channel over other monovalent cations, namely potassium. Mutagenesis and functional studies demonstrate that, while this site when mutated produces functional channels, the lysine amino acid at this position is absolutely necessary to support selectivity of sodium ions through the pore34,35. As a consequence of this functional prerequisite, any other amino acid encoded at this site, even the charged congener arginine, results in altered channel selectivity. Using standard electrophysiological approaches, ion channel selectivity can easily be quantified. Altered sodium ion selectivity of rNav1.4-K1237 is evidenced by a shift in the so-called reversal potential from +60 mV, the Nernst potential for sodium, to near 0 mV, the voltage where electrochemical gradients are balanced for a non-selective pore36. To demonstrate the encoding fidelity of the pylT, we coinjected rNav1.4-K1237TAG cRNA and pylT-lysine and observed voltage-dependent currents of size −4.9 ± 1.1 µA at −20 mV (N = 5) (Fig. 4A). Currents resulting from co-injection of rNav1.4 K1237TAG and full length (pdCpA-ligated) pylT were negligible (−0.12 ± 0.13 µA, N = 10, Fig. 4B). Importantly, the rescued channels displayed a reversal potential of +64.6 ± 3.9 mV (N = 5, Fig. 4). This value is in close agreement with that of WT rNav1.4 recorded in parallel (+66.6 ± 2.1 mV, N = 4, p = 0.72 between conditions), confirming the strict encoding of lysine at K1237TAG.
Figure 4.
pylT enables the faithful encoding of lysine into position K1237 of rNav 1.4. (a) Example traces of rNav 1.4-K1237TAG cRNA coinjected with either lysine-acylated (top) or unacylated full length (bottom) PylT. Oocytes were held at −100 mV and pulsed from −80 mV to +40 mV with 30 ms depolarizing steps. Voltage gated sodium channel activity is evidenced by increasingly large downward deflections in the traces in response to depolarization. The level of zero current for each cell is indicated by a dashed line. (b) Normalized current-voltage relationship plots comparing the WT rNav 1.4 channel to that of the rNav 1.4 K1237TAG rescued with lysine.
Discussion
Taken together, our results demonstrate that pylT is orthogonal in the Xenopus oocyte, and that it is useful for genetic code expansion experiments. We used voltage-gated sodium channels as exemplar proteins for this purpose, but we have also begun to enlist pylT as a tRNA for delivery of ncAAs into voltage-gated potassium channels, chloride channels, and enzymatic pumps, and these efforts have thus far been met with similar results. Thus, pylT may be used as an alternative to THG73 in cases where reacylation of THG73 poses a significant challenge. However, regardless of the species being used, we regard it advantageous to test for bleedthrough via co-injection of cRNA of interest with the pdCpA-ligated tRNA as there is no substitute for the empirical support provided by this negative control. It is also highly advantageous that pylT has been shown to be amenable to recoding of its anticodon from TAG to TGA and TAA37. Therefore, it may be used in future studies wherein site-specific dual suppression is desired16. Finally, it bears mentioning that there are additional tRNA species that have been shown to be orthogonal as part of coevolved tRNA- aminoacyl-tRNA synthetase pairs. Two such pairs have been sucessfully used in Xenopus oocytes6,7. These suppressor tRNAs therefore represent good candidates to test for amenability for chemical aminoacylation in future studies.
Methods
Molecular Biology
The S571TAG mutation was made into a pcDNA3.1 human Nav1.5 construct38 and the K1237TAG mutation was made into a pBSTA-based rat Nav1.4 construct22 using standard methods. For direct comparison of THG73 and pylT in hNav1.5, tRNA was generated and purified using the exact same procedure described by our group in detail in a recent report26. The transcription of template oligonucleotides generated tRNA with the following sequences: for THG73:
GGUUCUAUAGUAUAGCGGUUATUACUGGGGACUCUAAAUCCCUUGACCCUGGGUCUGAAUCCCAGUAGGACCGC
for pylT:
GGAAACCUGAUCAUGUAGAUCGAACGGACUCUAAAUCCGUUCAGCCGGGUUAGAUUCCCGGGGUUUCCGC
For experiments assaying the fidelity of encoding in the selectivity filter of rNav1.4, the pylT 70mer tRNA was synthesized by Integrated DNA Technologies (Coralville, IA). In all cases tRNA was reconstituted in 10 mM HEPES pH 7.2 and 3 mM MgCl2 and it was refolded in a thermocyler using a protocol with a denaturation step (94 °C for 3 min), followed by a linear ramp down to 4 °C over 20 minutes. Ligation reaction conditions, purification of acylated tRNA, and reconstitution of acylated tRNA were done as recently described26. Denaturing TBE-Urea gels were run as described in4 except that precast Mini-Protean gels were used (Biorad, Hercules CA).
Electrophysiology
Xenopus laevis oocytes were obtained through Ecocyte, Inc (Austin TX USA). For rescue of hNav1.5 and rNav1.4, we injected 25 nl of 1 ng/nl Nav cRNA and 25 nl of 25 μg of a tRNA pellet resuspended in 2.5 μl 3 mM cold NaOAc. Recordings were made approximately 24 hours later. For Nav1.5 experiments only, we also injected 12.5 ng of the rat β1 cRNA. For WT Nav1.4, we injected 2 ng of cRNA and recorded currents 24 hours later. All recordings were in oocyte Ringer’s essentially as described previously22. Currents were analysed via Clampfit 9.2, Molecular Devices, (Sunnyvale, CA). Reversal potentials were derived by fitting the linear component of the current-voltage relationship (+20 to +40 mV), and solving for the y-intercept. Statistical analysis was by unpaired student’s t-test, and exact p-values are noted in the text or tables.
Data availability
The raw data pertinent to this study is available from the authors upon reasonable request.
Electronic supplementary material
Acknowledgements
This work has been supported by NIH/NINDS High Impact Neuroscience Research Grant NS104617- “The facility for atomic Mutagenesis”. CAA is supported by NIH (GM106568, GM122420), is an American Heart Association Established investigator (5EIA22180002) and a member of the Membrane Protein structural Dynamics Consortium (NIH GM087519). DTI is supported by INFIEL17F0 from the Cystic Fibrosis Foundation. JDL is supported by Emily’s Entourage Foundation.
Author Contributions
D.T.I., J.D.L., and C.A.A. conceived the study, designed experiments, and wrote the paper. D.T.I., J.D.L., J.D.G., and G.D.G. conducted the experiments.
Competing Interests
The authors declare no competing interests.
Footnotes
Daniel T. Infield and John D. Lueck contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-23201-z.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Dougherty DA, Van Arnam EB. In vivo incorporation of non-canonical amino acids by using the chemical aminoacylation strategy: a broadly applicable mechanistic tool. Chembiochem: a European journal of chemical biology. 2014;15:1710–1720. doi: 10.1002/cbic.201402080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pless SA, Ahern CA. Unnatural amino acids as probes of ligand-receptor interactions and their conformational consequences. Annual review of pharmacology and toxicology. 2013;53:211–229. doi: 10.1146/annurev-pharmtox-011112-140343. [DOI] [PubMed] [Google Scholar]
- 3.Leisle L, Valiyaveetil F, Mehl RA, Ahern CA. Incorporation of Non-Canonical Amino Acids. Advances in experimental medicine and biology. 2015;869:119–151. doi: 10.1007/978-1-4939-2845-3_7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nowak MW, et al. In vivo incorporation of unnatural amino acids into ion channels in Xenopus oocyte expression system. Methods in enzymology. 1998;293:504–529. doi: 10.1016/S0076-6879(98)93031-2. [DOI] [PubMed] [Google Scholar]
- 5.Martin RP, Sibler AP, Dirheimer G, de Henau S, Grosjean H. Yeast mitochondrial tRNATrp injected with E. coli activating enzyme into Xenopus oocytes suppresses UGA termination. Nature. 1981;293:235–237. doi: 10.1038/293235a0. [DOI] [PubMed] [Google Scholar]
- 6.Kalstrup T, Blunck R. Dynamics of internal pore opening in K(V) channels probed by a fluorescent unnatural amino acid. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:8272–8277. doi: 10.1073/pnas.1220398110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhu S, et al. Genetically encoding a light switch in an ionotropic glutamate receptor reveals subunit-specific interfaces. Proc Natl Acad Sci USA. 2014;111:6081–6086. doi: 10.1073/pnas.1318808111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nowak MW, et al. Nicotinic receptor binding site probed with unnatural amino acid incorporation in intact cells. Science. 1995;268:439–442. doi: 10.1126/science.7716551. [DOI] [PubMed] [Google Scholar]
- 9.Lummis SC, et al. Cis-trans isomerization at a proline opens the pore of a neurotransmitter-gated ion channel. Nature. 2005;438:248–252. doi: 10.1038/nature04130. [DOI] [PubMed] [Google Scholar]
- 10.England PM, Zhang Y, Dougherty DA, Lester HA. Backbone mutations in transmembrane domains of a ligand-gated ion channel: implications for the mechanism of gating. Cell. 1999;96:89–98. doi: 10.1016/S0092-8674(00)80962-9. [DOI] [PubMed] [Google Scholar]
- 11.Santarelli VP, Eastwood AL, Dougherty DA, Horn R, Ahern CA. A cation-pi interaction discriminates among sodium channels that are either sensitive or resistant to tetrodotoxin block. The Journal of biological chemistry. 2007;282:8044–8051. doi: 10.1074/jbc.M611334200. [DOI] [PubMed] [Google Scholar]
- 12.Ahern CA, Eastwood AL, Dougherty DA, Horn R. An electrostatic interaction between TEA and an introduced pore aromatic drives spring-in-the-door inactivation in Shaker potassium channels. The Journal of general physiology. 2009;134:461–469. doi: 10.1085/jgp.200910260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahern, C. A., Eastwood, A. L., Dougherty, D. A. & Horn, R. Electrostatic contributions of aromatic residues in the local anesthetic receptor of voltage-gated sodium channels. Circ Res 102, 86–94, 10.1161/CIRCRESAHA.107.160663 (2008). [DOI] [PubMed]
- 14.Pless SA, Galpin JD, Niciforovic AP, Kurata HT, Ahern CA. Hydrogen bonds as molecular timers for slow inactivation in voltage-gated potassium channels. eLife. 2013;2:e01289. doi: 10.7554/eLife.01289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pless SA, Galpin JD, Niciforovic AP, Ahern CA. Contributions of counter-charge in a potassium channel voltage-sensor domain. Nature chemical biology. 2011;7:617–623. doi: 10.1038/nchembio.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lueck, J. D. et al. Atomic mutagenesis in ion channels with engineered stoichiometry. Elife510.7554/eLife.18976 (2016). [DOI] [PMC free article] [PubMed]
- 17.Kim RY, et al. Atomic basis for therapeutic activation of neuronal potassium channels. Nature communications. 2015;6:8116. doi: 10.1038/ncomms9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Saks ME, et al. An engineered Tetrahymena tRNAGln for in vivo incorporation of unnatural amino acids into proteins by nonsense suppression. The Journal of biological chemistry. 1996;271:23169–23175. doi: 10.1074/jbc.271.38.23169. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez EA, Lester HA, Dougherty DA. Improved amber and opal suppressor tRNAs for incorporation of unnatural amino acids in vivo. Part 1: minimizing misacylation. RNA. 2007;13:1703–1714. doi: 10.1261/rna.666807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rodriguez EA, Lester HA, Dougherty DA. Improved amber and opal suppressor tRNAs for incorporation of unnatural amino acids in vivo. Part 2: evaluating suppression efficiency. RNA. 2007;13:1715–1722. doi: 10.1261/rna.667607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodriguez EA, Lester HA, Dougherty DA. In vivo incorporation of multiple unnatural amino acids through nonsense and frameshift suppression. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:8650–8655. doi: 10.1073/pnas.0510817103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pless SA, et al. Asymmetric functional contributions of acidic and aromatic side chains in sodium channel voltage-sensor domains. The Journal of general physiology. 2014;143:645–656. doi: 10.1085/jgp.201311036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hao B, et al. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science. 2002;296:1462–1466. doi: 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- 24.Ibba M, Soll D. Genetic code: introducing pyrrolysine. Curr Biol. 2002;12:R464–466. doi: 10.1016/S0960-9822(02)00947-8. [DOI] [PubMed] [Google Scholar]
- 25.Wan W, Tharp JM, Liu WR. Pyrrolysyl-tRNA synthetase: an ordinary enzyme but an outstanding genetic code expansion tool. Biochim Biophys Acta. 2014;1844:1059–1070. doi: 10.1016/j.bbapap.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leisle, L. et al. Cellular encoding of Cy dyes for single-molecule imaging. Elife510.7554/eLife.19088 (2016). [DOI] [PMC free article] [PubMed]
- 27.Sakamoto K, et al. Site-specific incorporation of an unnatural amino acid into proteins in mammalian cells. Nucleic Acids Res. 2002;30:4692–4699. doi: 10.1093/nar/gkf589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, Brock A, Herberich B, Schultz PG. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 29.Chin JW. Expanding and reprogramming the genetic code of cells and animals. Annual review of biochemistry. 2014;83:379–408. doi: 10.1146/annurev-biochem-060713-035737. [DOI] [PubMed] [Google Scholar]
- 30.Hodgkin AL, Katz B. The effect of sodium ions on the electrical activity of giant axon of the squid. The Journal of physiology. 1949;108:37–77. doi: 10.1113/jphysiol.1949.sp004310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hodgkin AL, Huxley AF. A quantitative description of membrane current and its application to conduction and excitation in nerve. J Physiol. 1952;117:500–544. doi: 10.1113/jphysiol.1952.sp004764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Glynn P, et al. Voltage-Gated Sodium Channel Phosphorylation at Ser571 Regulates Late Current, Arrhythmia, and Cardiac Function In Vivo. Circulation. 2015;132:567–577. doi: 10.1161/CIRCULATIONAHA.114.015218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guan B, Chen X, Zhang H. Two-electrode voltage clamp. Methods Mol Biol. 2013;998:79–89. doi: 10.1007/978-1-62703-351-0_6. [DOI] [PubMed] [Google Scholar]
- 34.Backx PH, Yue DT, Lawrence JH, Marban E, Tamaselli GF. Molecular locatlization of an ion-binding site within the pore of mammalian sodium channels. Science J1 - sci. 1992;257:248–251. doi: 10.1126/science.1321496. [DOI] [PubMed] [Google Scholar]
- 35.Favre I, Moczydlowski E, Schild L. On the structural basis for ionic selectivity among Na+, K+, and Ca2+ in the voltage-gated sodium channel. Biophysical Journal. 1996;71:3110–3125. doi: 10.1016/S0006-3495(96)79505-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hille, B. Ion channels of excitable membranes. 3rd edn, (Sinauer, 2001).
- 37.Ambrogelly A, et al. Pyrrolysine is not hardwired for cotranslational insertion at UAG codons. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:3141–3146. doi: 10.1073/pnas.0611634104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pless SA, Galpin JD, Frankel A, Ahern CA. Molecular basis for class Ib anti-arrhythmic inhibition of cardiac sodium channels. Nature communications. 2011;2:351. doi: 10.1038/ncomms1351. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data pertinent to this study is available from the authors upon reasonable request.