Abstract
Estimating the post mortem interval (PMI) is still a crucial step in Forensic Pathology. Although several methods are available for assessing the PMI, a precise estimation is still quite unreliable and can be inaccurate. The present study aimed to investigate the immunohistochemical distribution and mRNA expression of hypoxia inducible factor (HIF‐1α) in post mortem gingival tissues to establish a correlation between the presence of HIF‐1α and the time since death, with the final goal of achieving a more accurate PMI estimation. Samples of gingival tissues were obtained from 10 cadavers at different PMIs (1–3 days, 4–5 days and 8–9 days), and were processed for immunohistochemistry and quantitative reverse transcription‐polymerase chain reaction. The results showed a time‐dependent correlation of HIF‐1α protein and its mRNA with different times since death, which suggests that HIF‐1α is a potential marker for PMI estimation. The results showed a high HIF‐1α protein signal that was mainly localized in the stratum basale of the oral mucosa in samples collected at a short PMI (1–3 days). It gradually decreased in samples collected at a medium PMI (4–5 days), but it was not detected in samples collected at a long PMI (8–9 days). These results are in agreement with the mRNA data. These data indicate an interesting potential utility of Forensic Anatomy‐based techniques, such as immunohistochemistry, as important complementary tools to be used in forensic investigations.
Keywords: forensic anatomy, forensic histopathology, HIF‐1α, hypoxia, post mortem interval
Introduction
The post mortem interval (PMI) is the amount of time that has elapsed since the time of death, and its determination has always been a fundamental issue in the forensic field. In standard forensic practice, PMI is usually estimated by assessing the evidence provided by the body itself, including the temperature, muscular and neuro‐muscular reactivity, post mortem lividity, autolysis, putrefaction and fluid biochemistry, as well as environmental markers. However, PMI estimation remains controversial, and it is often not possible to draw any definite conclusions concerning the time of death based on the appearance of a single post mortem change (Di Maio & Dana, 2007; Madea, 2016).
Many different approaches have been explored in recent years, and the literature is constantly expanded by newer strategies that are being considered as additional support so that a more accurate value can be provided by using a combination of different methods (Henssege et al. 2000a,b). Over the decades, histological and immunohistochemical analyses of different post mortem tissues have become increasingly important, as they might provide additional hints that may be useful in determining the time interval after death (Thaik‐Oo et al. 2002; Dettmeyer, 2011). For gingival tissue, PMI has been related to different histological changes (Pradeep et al. 2009; Yadav et al. 2015; Mahalakshmi et al. 2016), but the associated post mortem immunohistochemical changes seem to be overlooked.
The master regulator of cellular adaptation to a hypoxic environment is the transcription factor, hypoxia inducible factor‐1 (HIF‐1; Eskandani et al. 2017). Indeed, HIF‐1 is a heterodimeric transcription factor that is composed of two subunits: HIF‐1α (or its analogues HIF‐2α and HIF‐3α) and HIF‐1β subunits. HIF‐1β is constitutively expressed, whereas HIF‐1α is an oxygen‐sensitive subunit, and its expression is induced under hypoxic conditions (Masoud & Lin, 2015; Eskandani et al. 2017). HIF‐1α is constitutively transcribed and synthesized through a series of signalling events that involve several growth factors and other signalling molecules (Masoud & Lin, 2015). Under normal oxygen tension, HIF‐1α protein expression is negatively regulated by proteasomal degradation and ubiquitination in a pathway that involves von Hippel‐Lindau protein (pVHL), a tumour suppressor protein and one of the recognized components of an E3 ubiquitin protein ligase (Masoud & Lin, 2015). Hydroxylation by the prolyl‐4‐hydroxylases (PHDs) or HIF‐1 prolyl hydroxylases of one of two critical proline residues in their oxygen‐dependent degradation domain mediates their interaction with the von Hippel‐Lindau (VHL) E3 ubiquitin ligase complex that targets them for rapid proteasomal degradation (Bruick & McKnight, 2001; Hirsilä et al. 2005; Ng et al. 2011; Masoud & Lin, 2015). Because the hydroxylation action from PHDs requires the presence of oxygen, a hypoxic environment will inhibit the activities of the HIF‐P4Hs such that HIF‐1α escapes degradation, translocates to the nucleus and binds HIF‐β; this dimer then recognizes the HIF‐responsive elements in a number of hypoxia‐inducible target genes (Bruick & McKnight, 2001; Hirsilä et al. 2005; Masoud & Lin, 2015; Vasconcelos et al. 2016; Takedachi et al. 2017).
After death, the absence of blood flow leads to a rapid cellular oxygen deprivation, so a useful immunohistochemical marker for estimating the time of death should be one that is absent in physiological conditions and appears with hypoxia (Cecchi et al. 2014). On the basis of these observations, the aim of the study was to assess HIF‐1α and HIF‐1α mRNA expression in human gingival tissues at different PMIs.
Materials and methods
Cases
Ten samples of gingival tissues (0.5–1 cm2) were collected from maxillary gingiva adjacent to the first incisors during medico‐legal autopsies from cadavers of both sexes, with ages ranging from 18 to 60 years old. The PMI ranged between 1 and 10 days. Gingival tissue was sampled only in cases of trauma‐related death, when the gingiva at inspection did not show any transformative, traumatic or phlogistic alterations. In all cases investigated, the time of death was already known.
The control samples were gingival biopsies performed on clinically healthy individuals during the wisdom tooth extraction procedure and only when gingival inflammation was excluded.
On the basis of the PMI, the samples were grouped into different categories, as detailed in Table 1. The first group consisted of samples with a known PMI ranging from 1 to 3 days (short PMI – SPMI), the second of samples with a known PMI from 4 to 5 days (medium PMI – MPMI) and the third group of samples with a known PMI from 8 to 9 days (long PMI – LPMI).
Table 1.
On the basis of PMI, samples were divided into three main groups: SPMI, MPMI and LPMI
| Time since death (days) | No CASES | |
|---|---|---|
| SPMI | 1–3 | 2 |
| MPMI | 4–5 | 5 |
| LPMI | 8–9 | 3 |
| CTR | – | 3 |
CTR, control samples; LPMI, long post mortem interval; MPMI, medium post mortem interval; SPMI, short post mortem interval.
Sample collection and processing
For the immunohistochemistry analysis, gingival tissues were immediately fixed after collection in 4% formaldehyde (Sigma Aldrich, St Louis, MO, USA) in phosphate‐buffered saline (PBS) for 24 h at 4 °C; they were then dehydrated in a graded series of ethanol and embedded in paraffin wax (Fluka, Sigma‐Aldrich). Paraffin sections of 6 μm were obtained with an automated rotary microtome (Leica Microsystems Srl, Cambridge, UK) and collected on Superfrost glass slides (Carl Roth, Karlshure, Germany).
For mRNA expression analysis, gingival tissues were collected, and immediately frozen and stored at −80 °C until RNA isolation.
Immunohistochemistry
Paraffin sections were dewaxed and then washed three times for 5 min with PBS at pH 7.4. Endogenous peroxidase activity was blocked with 3% H2O2 (diluted in distilled water) at room temperature (RT) for 15 min. Non‐specific antibody binding was blocked with protein block milk (Invitrogen, Thermo Fischer Scientific, Monza, Italy) for 30 min at RT. The sections were incubated with the primary antibody against HIF‐1α (Invitrogen, Thermo Fisher Scientific, Monza, Italy), diluted 1 : 500 in blocking solution, at 4 °C overnight. To detect the antigen–antibody reaction, a secondary anti‐rabbit antibody was used, followed by diaminobenzidine tetrahydrochloride (DAB) as a substrate solution (Histofine Immunohistochemical staining kit, Nichirei Biosciences, Tokyo, Japan). Negative controls were performed by omitting the primary antibodies, and omitting the primary and secondary antibodies followed by DAB. Some sections were stained with haematoxylin and eosin following the manufacturer's instructions (Sigma Aldrich, St Louis, MO, USA). All samples were observed under a light microscope using an Eclipse E800 Nikon (Nikon, Tokyo, Japan). Representative images are shown.
The quantitative analysis of antibody‐stained areas, expressed as the relative amount of gingival tissues from cadavers compared with that from healthy donors, was assessed by area counting of five fields for each of three slides per sample at 60 × magnification in the Leica Qwin 3.0 software (Leica Microsystems Srl, Cambridge, UK), which allowed the antibody‐stained area to be selected and measured.
mRNA extraction and quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR)
The RNeasy Mini Kit (Invitrogen, Thermo Fisher Scientific, Monza, Italy) was used to extract RNA from gingival tissues, and 200 ng of total RNA was reverse transcribed into first‐strand cDNA using SuperScript™ III One‐Step RT‐PCR System (Invitrogen, Thermo Fisher Scientific, Monza, Italy). The HIF‐1α mRNA expression level was analysed using a real‐time‐PCR by a 7500 Real‐Time PCR machine (Applied Biosystems, Life Technologies, Monza, Italy). For mRNA quantification, a TaqMan assay (Life Technologies, Thermo Fischer Scientific, Monza Italy) specific for HIF‐1α (HIF‐1α; Hs00153153_m1) was used. The relative gene expression levels were normalized to that of glyceraldehyde 3‐phosphate dehydrogenase (GAPDH; Hs99999905_m1). Data are presented as fold changes relative to the levels in the control samples by using the formula, 2−ΔΔCT, as recommended by the manufacturer (User Bulletin number 2 P/N 4303859; Applied Biosystems).
Statistical analysis
Morphometric analyses of immunohistochemistry images and real time‐PCR values are presented as the mean ± SD, and each type of experiment was replicated three times. Student's t‐test with the Welch correction was applied to evaluate the differences between groups. Statistical analysis was performed using the GraphPad Prism 5.0 software (GraphPad Software, San Diego, CA, USA). P‐values < 0.05 were considered statistically significant.
Results
Immunohistochemical evaluation of HIF‐1α protein expression
Short PMI samples showed a high level of HIF‐1α expression that was mainly localized in the oral epithelium compared with the sub‐oral connective tissues (Fig. 1a,b). At higher magnification, a strong signal of the HIF‐1α protein was initially detected in the stratum basale of the oral mucosa, and it gradually decreased in the mid‐epithelium (stratum spinosum) and finally disappeared in the stratum granulosum (Fig. 1c,d). Several cells in the sub‐oral connective tissue were positive for HIF‐1α immunostaining (Fig. 1d).
Figure 1.
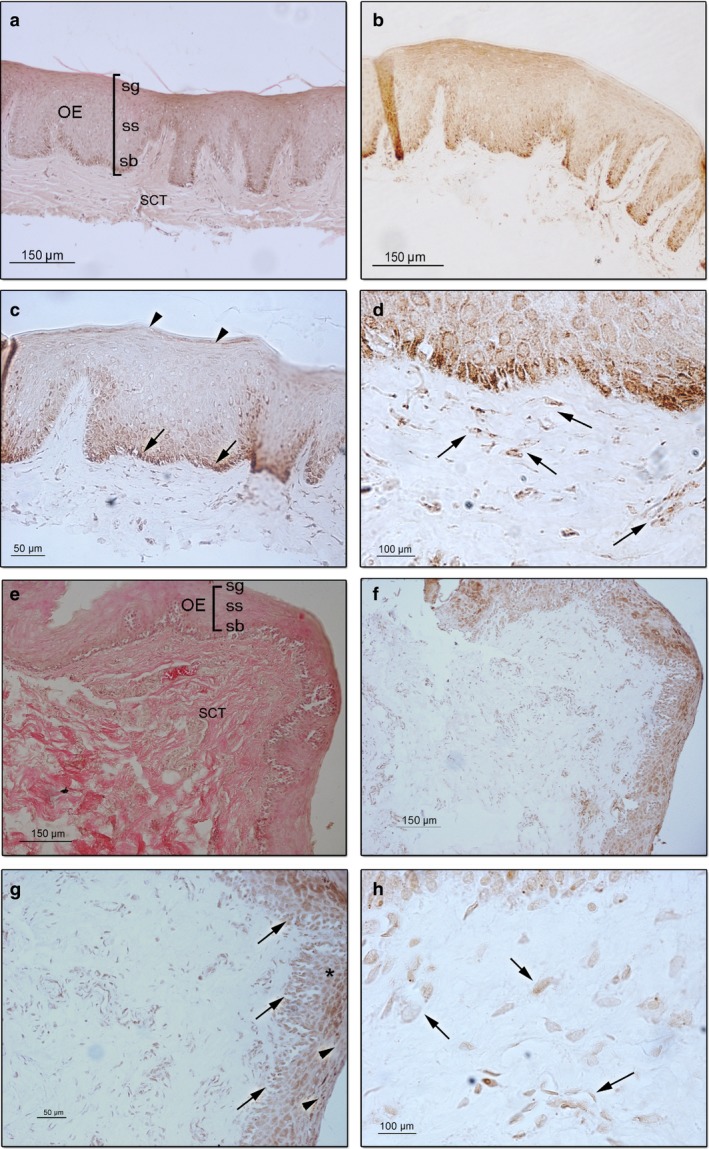
Representative sections of short post mortem interval (SPMI) gingival tissues showing hypoxia inducible factor (HIF‐1α) protein expression. (a) Haematoxylin–eosin staining of gingival mucosa showing the oral epithelium (OE), in which the stratum basale (sb), stratum spinoso (ss) and stratum granulosum (sg) and the sub‐oral connective tissues (SCT) are observed (magnification 10 ×; scale bar: 150 μm). (b) HIF‐1α protein is mainly localized in the OE (magnification 10 ×; scale bar: 150 μm). (c) A strong signal of the HIF‐1α protein is detected in the stratum basale (arrow), and it gradually decreased in the mid‐epithelium to finally disappeared in the stratum corneum (arrowhead; magnification 20 ×; scale bar: 50 μm). (d) Cells of the SCT showed a strong signal of HIF‐1α protein (arrow; magnification 100 ×; scale bar: 100 μm). Representative sections of oral medium post mortem interval (MPMI) gingival tissues showing HIF‐1α protein expression. (e) Haematoxylin–eosin staining of gingival mucosa showing the OE and the SCT (magnification 10 ×; scale bar: 150 μm). (f) HIF‐1α protein is localized in the OE (magnification 10 ×; scale bar: 150 μm). (g) HIF‐1α protein is detected in the stratum basale (arrow), and it gradually decreased in the mid‐epithelium (*) and stratum corneum (arrowhead; magnification 20 ×; scale bar: 50 μm). (h) SCT showing cells positive for HIF‐1α immunostaining (arrow; magnification 100 ×; scale bar: 100 μm).
Medium PMI oral mucosa showed positive staining for HIF‐1α expression that was mainly localized in the oral epithelium but had a lower signal intensity than did the SPMI samples (Fig. 1e,f). In the oral epithelium, the localization and distribution of the protein were very similar to those of the SPMI samples, with a high intensity of the protein in the basal layer and a gradual reduction in the mid‐layer and upper granular layer (stratum granulosum; Fig. 1f,g). The sub‐oral connective tissue showed positive stained cells with a lower intensity compared with that of the SPMI samples (Fig. 1h).
In all the LPMI samples analysed, no positive stained areas were detected (Fig. 2a,b). At higher magnification, a weak signal of HIF‐1α expression was detected in the basal layer of the oral epithelium, but no signal was detected in the mid‐layer or granular and upper granular layers of the oral epithelium (Fig. 2b,c). Cells of the sub‐oral connective tissues were negative for HIF‐1α immunostaining (Fig. 2d). Control samples showed no positive areas in either the oral epithelium or sub‐oral connective tissues (Fig. 2e–h).
Figure 2.
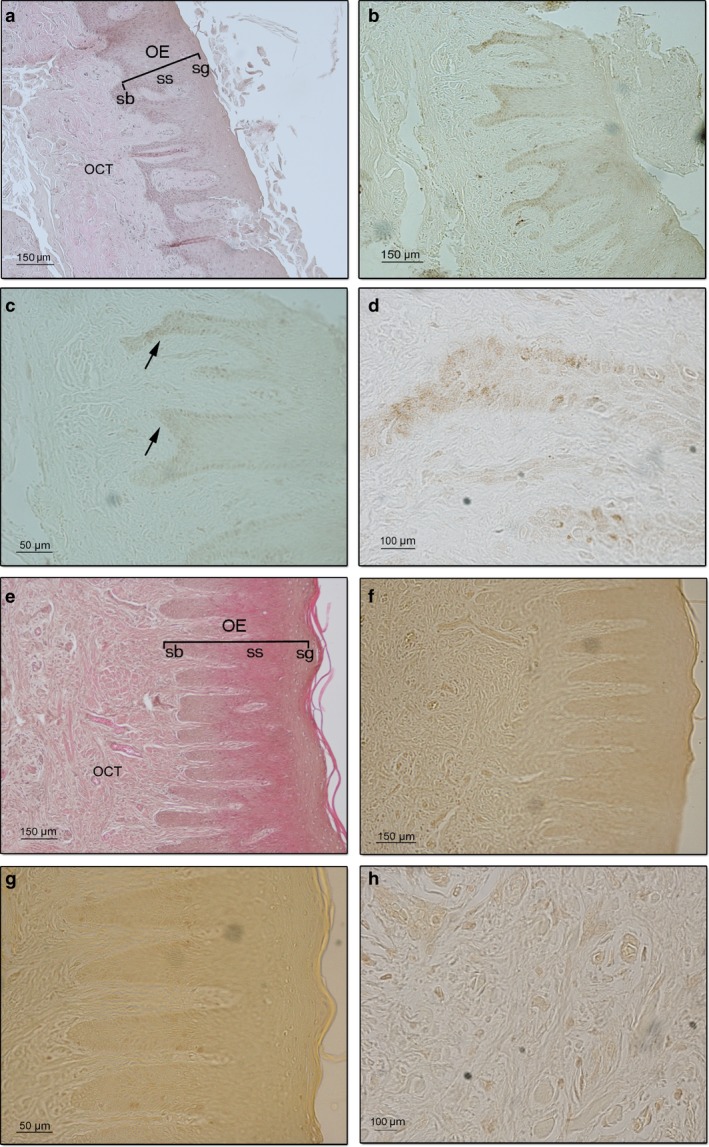
Representative sections of oral long post mortem interval (LPMI) gingival mucosa showing hypoxia inducible factor (HIF)‐1α protein expression. (a) Haematoxylin–eosin staining of gingival tissue showing the oral epithelium (OE) and the sub‐oral connective tissue (SCT; magnification 10 ×; scale bar: 150 μm). (b) No signal corresponding to HIF‐1α protein is observed (magnification 10 ×; scale bar: 150 μm). (c) A very weak signal of HIF‐1α protein is detected in the stratum basale (arrow; magnification 20 ×; scale bar: 50 μm). (d) No positive cells of the SCT are detected (magnification 100 ×; scale bar: 100 μm). Representative sections of gingival mucosa from healthy donors (control samples) showing HIF‐1α protein expression. (e) Haematoxylin–eosin staining of gingival mucosa showing the OE and the SCT (magnification 10 ×; scale bar: 150 μm). (f) No signal corresponding to HIF‐1α expression is observed in the oral mucosa (magnification 10 ×; scale bar: 150 μm). (g) At higher magnification, no positive cells are detected in the layers of the OE (magnification 20 ×; scale bar: 50 μm) or in the SCT (h) (magnification 100 ×; scale bar: 100 μm).
Morphometric analysis demonstrated a statistically significant increase in the HIF‐1α immunostained area in the SPMI group compared with those of the MPMI, LPMI and CTR groups (Fig. 3a; Table 2), and between the MPMI samples and both LPMI and CTR samples (Fig. 3a; Table 2). No significant differences were found between the LPMI and CTR groups (Fig. 3a; Table 2).
Figure 3.
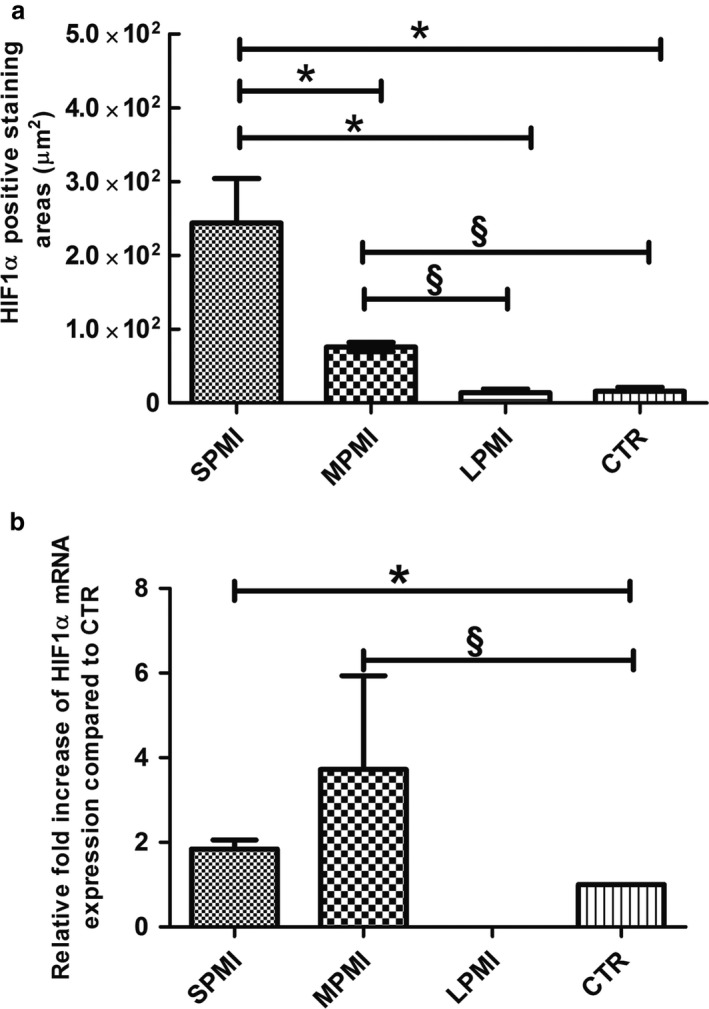
(a) Morphometric analysis of histological sections immunostained for hypoxia inducible factor (HIF‐1α) protein showing the positive stained area. The analysis was assayed by direct counting of five fields on three slides for each sample at 60 × magnification using the Leica Qwin software. In each group, the final data are the mean ± SD. *Represents a significant difference of short post mortem interval (SPMI) group against medium post mortem interval (MPMI), long post mortem interval (LPMI) and control (CTR) groups, P < 0.05; §represents a significant difference of MPMI group against LPMI and CTR groups, P < 0.05. (b) Quantitative reverse transcription‐polymerase chain reaction (qRT‐PCR) analysis of HIF‐1α mRNA expression in gingival tissues collected from bodies at different times of death and compared with control samples. Data are expressed as relative amount of fold increase of mRNA expression compared with control samples. Each individual assay was performed in triplicate, and the final data are expressed as the mean ± SD; *represents a significant difference of SPMI group against CTR group, P < 0.05; §represents a significant difference of MPMI group against CTR group, P < 0.05.
Table 2.
Mean values of HIF‐1α immunostained positive areas (µm2) and P‐value
| Mean (µm2) | SD | P‐value | |
|---|---|---|---|
| SPMI | 243 990 | 60 304 | 0.0216 (SPMI against MPMI) |
| 0.0042 (SPMI against LPMI) | |||
| 0.0044 (SPMI against CTR) | |||
| MPMI | 75 722.9 | 6366 | < 0.0001 (MPMI against LPMI) |
| < 0.0001 (MPMI against CTR) | |||
| LPMI | 13 954.5 | 4893 | 0.8242 (LPMI against CTR) |
| CTR | 15 699.4 | 5852 |
Student's t‐test with Welch correction was applied to evaluate the differences between all the groups. P‐values < 0.05 were considered statistically significant.
CTR, control samples; LPMI, long post mortem interval; MPMI, medium post mortem interval; SPMI, short post mortem interval.
qRT‐PCR analysis of HIF‐1α mRNA expression
The mRNA expression of HIF‐1α was evaluated by qRT‐PCR in the gingival tissues of cadavers, and the results were compared with those obtained for control samples (Fig. 3b). In SPMI gingival tissues, the mRNA of HIF‐1α showed a 1.8‐fold upregulation of expression compared with control samples (Fig. 3b), whereas the MPMI samples showed an upregulation of 3.7‐fold compared with control samples (Fig. 3b). No significant difference was found between SPMI and MPMI. No HIF‐1α mRNA expression was detected in the LPMI samples (Fig. 3b).
Discussion
Establishing PMI is of paramount importance in forensic practice, as it allows the moment of death to be retrospectively estimated. The capacity to estimate the time of death remains limited, and each method is potentially prone to errors; however, the literature is constantly expanded by new and innovative strategies that highlight all possible identifiable alterations that may occur after death to which a temporal value can be ascribed (Henssege & Madea, 2004). In this study, the levels of HIF‐1α protein and mRNA were investigated in gingival specimens to assess whether there might be a pattern of expression related to different PMIs. Gingivae were chosen to study HIF‐1α and HIF‐1α mRNA expression because the sampling method is quick, simple and not mutilating. Moreover, gingivae are usually protected from labial tissues, thus limiting the impact of environmental factors, which could be responsible for rapid tissue decay. Thus, gingival tissue, as a more stable region for investigation, represents the most suitable choice for protein and mRNA analyses.
Finally, HIF‐1α expression seems to be related to concurrent morphological cellular damage (Zhu et al. 2008) related to hypoxia. Indeed, HIF expression has been related to different causes of death in different organs, specific targets of hypoxia. In particular, increases in HIF expression have been observed: in myocardial tissue in cases of myocardial ischaemia (Blanco Pampìn et al. 2006) or cardiac death (Zhu et al. 2008); in the lungs in cases of mechanical asphyxia (Cecchi et al. 2014); and in the kidneys, especially in cases of acute circulatory failure (Zhao et al. 2006). However, to the best of our knowledge, gingival tissue is not a specific target of hypoxia in the peri mortem period; thus, there is no reason to hypothesize a different pattern of hypoxia related to HIF‐1α expression in gingival tissue that is associated with a specific cause of death.
According to a previous study (Takedachi et al. 2017), in our post mortem samples, HIF‐1α immunostaining was higher in the oral epithelium than it was in the sub‐oral connective tissues, thus reflecting the in vivo distribution observed in mice under hypoxic‐inflammatory conditions. Interestingly, as shown in the epithelial cells in a rat model, in which the level of HIF‐1α expression was relatively higher in the suprabasal layer, our results indicated a high staining reactivity for HIF‐1α that was detectable in the deeper layers of the oral mucosa and decreased gradually in the more superficial layers. This result could be ascribed to the fact that the basal cell layer has a higher degree of susceptibility to oxygen deprivation, whereas in the upper layers, the cellular activity is reduced with nuclear and cytoplasmic organelle loss.
Considering the pattern of expression of HIF‐1α, immunostaining was absent in the control group (time = 0), peaked at 1–3 days (SPMI samples), and gradually declined at 4–5 and 8–9 days after death (LPMI samples). This finding is consistent with the pattern of expression observed for the vascular endothelial growth factor, a similar immunohistochemical hypoxia‐related marker, whose levels in different organ tissues were measured to estimate PMI (Thaik‐Oo et al. 2002). The peak expression of HIF‐1α occurring within 3 days after death could be explained by the post mortem interruption of blood flow leading to a prolonged state of cellular oxygen deprivation, which is comparable to chronic ischaemic stress (Zhu et al. 2008), with concurrent HIF‐1α expression. The subsequent decline of HIF‐1α immunostaining is probably related to cellular necrosis and associated with transformative processes with HIF‐1α degradation concurrent to nuclear disruption (Kitanaka & Kuchino, 1999).
Other studies on myocardial (Zhu et al. 2008) and renal (Zhao et al. 2006) tissue demonstrated that HIF‐1α is a good marker of hypoxia because its expression is independent of gender, age and time since death. This latter point apparently contrasts our results; however, in contrast to studies on myocardial (Zhu et al. 2008) and renal (Zhao et al. 2006) tissues, our studied samples also considered PMIs longer than 2 days for gingival tissue, highlighting a peak of HIF‐1α expression occurring at 3 days after death and gradually decreasing until a PMI as long as 9 days.
Recent studies have demonstrated that RNA is more stable than previously thought; for this reason, it is widely used as a valuable tool in forensic pathology, including in identifying body fluids, estimating the age of biological stains and studying the mechanism of death (Sampaio‐Silva et al. 2013). Poòr et al. (2016) demonstrated that the RNA extracted from tissues of the oral cavity showed a low time‐dependent RNA degradation up to 21 days; for this reason it represents an ideal candidate for PMI estimation.
We successfully extracted RNA from post mortem samples up to 9 days, and qRT‐PCR analysis showed an amplification of HIF‐1α mRNA expression in SPMI and MPMI samples. However, no signal was detected in LPMI samples, which is in agreement with the weak signal of the protein evaluated by immunohistochemistry.
In summary, from our results, we can infer that gingival tissues demonstrate a relationship between the extent of HIF‐1α and HIF‐1α mRNA expression and the time elapsed from death that may serve as an additional basis for PMI estimation. In detail, our study demonstrated that SPMIs and MPMIs are characterized by an increase in HIF‐1α protein expression compared with LPMIs, where the protein expression is no longer detectable. The method suggested is a promising tool for distinguishing deaths that have occurred within 1 week from those that occurred more than 1 week ago.
Although further confirmation studies are required, these results seem to be encouraging and demonstrate that this approach, which is based on immunohistochemical and morphological methodologies, may have a high potential for being a useful tool, a valuable alternative or an addition to other methods in forensic PMI delimitation.
Furthermore, by using these basic Forensic Anatomy techniques, the identification of new markers and morphological parameters could become important complementary tools in traditional Forensic Pathology.
Conflict of interest
The authors have no conflict of interest to declare.
Author contributions
P.F. conceived the study and prepared the manuscript. M.C.M. took tissue samples, collected the data and drafted the manuscript. R.B.‐B. analysed the data. G.T. contributed to concept/design of the study, performed the staining and evaluation, and analysed the data. S.P. supervised and reviewed the manuscript. M.F. supervised the staining and evaluation, made the critical revision of the manuscript and gave final approval for publication.
References
- Blanco Pampìn J, Garcia Rivero SA, Otero Cepeda XL, et al. (2006) Immunohistochemical expression of HIF‐1alpha in response to early myocardial ischemia. J Forensic Sci 51, 120–124. [DOI] [PubMed] [Google Scholar]
- Bruick RK, McKnight SL (2001) A conserved family of prolyl‐4‐hydroxylases that modify HIF. Science 294, 1337–1340. [DOI] [PubMed] [Google Scholar]
- Cecchi R, Sestilli C, Prosperini G, et al. (2014) Markers of mechanical asphyxia: immunohistochemical study on autoptic lung tissues. Int J Legal Med 128, 117–125. [DOI] [PubMed] [Google Scholar]
- Dettmeyer R (2011) Staining techniques and microscopy In: Forensic Histopathology. Fundamentals and Perspectives. (ed. Springer ), pp 17–35. Berlin, Germany: Springer. [Google Scholar]
- Di Maio V, Dana S (2007) Time of death – decomposition In: Handbook of Forensic Pathology. (ed.Taylor and Francis ), pp 23–28. Boca Raton, FL: CRC Press. [Google Scholar]
- Eskandani M, Vandghanooni S, Barar J, et al. (2017) Cell physiology regulation by hypoxia inducible factor‐1: targeting oxygen‐related nanomachineries of hypoxic cells. Int J Biol Macromol 99, 46–62. [DOI] [PubMed] [Google Scholar]
- Henssege C, Madea B (2004) Estimation of the time since death in the early post‐mortem period. Forensic Sci Int 144, 167–175. [DOI] [PubMed] [Google Scholar]
- Henssege C, Althaus L, Bolt J, et al. (2000a) Experiences with a compound method for estimating the time since death. I. Rectal temperature nomogram for time since death. Int J Legal Med 113, 303–319. [DOI] [PubMed] [Google Scholar]
- Henssege C, Althaus L, Bolt J, et al. (2000b) Experiences with a compound method for estimating the time since death. II. Integration of non‐temperature‐based methods. Int J Legal Med 113, 320–331. [DOI] [PubMed] [Google Scholar]
- Hirsilä M, Koivunen P, Xu L, et al. (2005) Effect of desferrioxamine and metals on the hydroxylases in the oxygen sensing pathway. FASEB J 19, 1308–1310. [DOI] [PubMed] [Google Scholar]
- Kitanaka C, Kuchino Y (1999) Caspase‐independent programmed cell death with necrotic morphology. Cell Death Differ 6, 508–515. [DOI] [PubMed] [Google Scholar]
- Madea B (2016) Estimation of the time since death In: Forensic Pathology Advanced Forensic Series. (ed. Houck M.), pp 55–65. London, UK: Academic Press. [Google Scholar]
- Mahalakshmi V, Gururaj N, Sathya R, et al. (2016) Assessment of histological changes in antemortem gingival tissues fixed at various time intervals: a method of estimation of postmortem interval. J Forenisc Dent Sci 8, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masoud GN, Lin W (2015) HIF‐1α pathway: role, regulation and intervention for cancer therapy. Acta Pharm Sin B 5, 378–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KT, Li JP, Ng KM, et al. (2011) Expression of hypoxia‐inducible factor‐1α in human periodontal tissue. J Periodontol 82, 136–141. [DOI] [PubMed] [Google Scholar]
- Poòr VS, Lukàcs D, Nagy T, et al. (2016) The rate of RNA degradation in human dental pulp reveals post‐mortem interval. Int J Legal Med 130, 615–619. [DOI] [PubMed] [Google Scholar]
- Pradeep GL, Uma K, Sharada P, et al. (2009) Histological assessment of cellular changes in gingival epithelium in ante‐mortem and post‐mortem specimens. J Forensic Dent Sci 1, 61–65. [Google Scholar]
- Sampaio‐Silva F, Magalhaes T, Carvalho F, et al. (2013) Correction: profiling of RNA degradation for estimation of post mortem interval. PLoS ONE 8, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takedachi M, Iyama M, Sawada K, et al. (2017) Hypoxia‐inducible factor‐1α inhibits interleukin‐6 and ‐8 production in gingival epithelial cells during hypoxia. J Periodontal Res 52, 127–134. [DOI] [PubMed] [Google Scholar]
- Thaik‐Oo M, Tanaka E, Tsuchiya T, et al. (2002) Estimation of postmortem interval from hypoxic inducible levels of vascular endothelial growth factor. J Forensic Sci 47, 186–189. [PubMed] [Google Scholar]
- Vasconcelos RC, Costa Ade L, Freitas Rde A, et al. (2016) Immunoexpression of HIF‐1α and VEGF in periodontal disease and healthy gingival tissues. Braz Dent J 27, 117–122. [DOI] [PubMed] [Google Scholar]
- Yadav AB, Angadi PV, Kale AD, et al. (2015) Histological assessment of cellular changes in postmortem gingival specimens for estimation of time since death. J Forensic Odontostomatol 33, 19–26. [PMC free article] [PubMed] [Google Scholar]
- Zhao D, Zhu BL, Ishikawa T, et al. (2006) Quantitative RT‐PCR assays of hypoxia‐inducible factor‐1alpha, erythropoietin and vascular endothelial growth factor mRNA transcripts in the kidneys with regard to the cause of death in medicolegal autopsy. Leg Med (Tokyo) 8, 258–263. [DOI] [PubMed] [Google Scholar]
- Zhu BL, Tanaka S, Ishikawa T, et al. (2008) Forensic pathological investigation of myocardial hypoxia‐inducible factor‐1 alpha, erythropoietin and vascular endothelial growth factor in cardiac death. Leg Med (Tokyo) 10, 11–19. [DOI] [PubMed] [Google Scholar]


