Abstract
Human epidermal growth factor receptor 2 (HER2) plays an important role in the pathogenesis of various cancers. HER2 alterations have been suggested to be a therapeutic target in non‐small‐cell lung cancer (NSCLC), just as in breast and gastric cancers. We previously reported that the pan‐HER inhibitor afatinib could be a useful therapeutic agent as HER2‐targeted therapy for patients with NSCLC harboring HER2 alterations. However, acquired resistance to afatinib was observed in the clinical setting, similar to the case for other HER inhibitors. Thus, elucidation of the mechanisms underlying the development of acquired drug resistance and exploring means to overcome acquired drug resistance are important issues in the treatment of NSCLC. In this study, we experimentally established afatinib‐resistant cell lines from NSCLC cell lines harboring HER2 alterations, and investigated the mechanisms underlying the acquisition of drug resistance. The established cell lines showed several unique afatinib‐resistance mechanisms, including MET amplification, loss of HER2 amplification and gene expression, epithelial‐to‐mesenchymal transition (EMT) and acquisition of cancer stem cell (CSC)‐like features. The afatinib‐resistant cell lines showing MET amplification were sensitive to the combination of afatinib plus crizotinib (a MET inhibitor), both in vitro and in vivo. The resistant cell lines which showed EMT or had acquired CSC‐like features remained sensitive to docetaxel, like the parental cells. These findings may provide clues to countering the resistance to afatinib in NSCLC patients with HER2 alterations.
Keywords: acquired resistance, afatinib, human epidermal growth factor receptor 2, lung cancer, non‐small cell lung cancer
1. INTRODUCTION
Recent developments in the genomic characterization of tumors have contributed to the development of novel therapeutic approaches, and improved patient survival has been obtained with some molecular‐targeted therapies developed on the basis of the genetic profiles of tumors. For example, EGFR tyrosine kinase inhibitors (TKI), such as gefitinib and erlotinib, have been shown to exhibit marked antitumor activity in patients with non‐small cell lung cancer (NSCLC) carrying activating EGFR mutations.1 Other oncogenic alterations, such as ALK, KRAS, NRAS, BRAF, MET and FGFR, have also been identified in some subsets of NSCLC patients as possible treatment targets.2, 3, 4
Human epidermal growth factor receptor 2 (HER2) is a member of the HER family, which is composed of 4 receptor tyrosine kinases (RTK). It is frequently overexpressed in various human cancers, and many preclinical studies have demonstrated that overexpression of HER2 or mutations of the HER2 kinase domain play an important role in oncogenic transformation and tumorigenesis.5, 6, 7, 8 In the case of breast and gastric cancers, targeted therapies in patients with HER2‐positive tumors have been clinically proven to be effective.9, 10 Among NSCLC patients, the reported frequency of HER2 overexpression and HER2 amplification are 11%‐32% and 2%‐23%, respectively.11, 12, 13, 14 HER2 mutations are identified in approximately 2%‐4% of NSCLC and are usually mutually exclusive of other driver mutations.15, 16 However, it remains to be established whether HER2‐targeted therapy is of clinical benefit in patients with HER2‐altered NSCLC. Previous clinical trials have failed to show any clinical benefit of trastuzumab monotherapy or any additional effect of trastuzumab over conventional first‐line chemotherapy in HER2‐positive NSCLC patients.17, 18, 19 However, subsets of patients with high HER2 expression levels, HER2 gene amplification or HER2 mutations have been demonstrated to show a good response to HER2‐targeted treatment.17, 20, 21 In addition, a recent retrospective study showed that HER2‐targeted therapy was beneficial in combination with chemotherapy for patients with HER2‐mutant NSCLC.22 These findings suggest the potential usefulness of HER2‐targeted therapy in HER2‐altered NSCLC patients.
Afatinib, also known as BIBW2992, is an irreversible pan‐HER (EGFR, HER2, HER3 and HER4) inhibitor, and has recently been approved for the treatment of NSCLC patients carrying EGFR mutations in the tumor. We previously reported that afatinib could inhibit cell growth in both HER2‐amplified and HER2‐mutant NSCLC cells, just as in EGFR‐mutant NSCLC cells, through G1 arrest and apoptosis.23 In clinical settings, afatinib monotherapy was effective in 3 out of 3 assessable patients with an HER2‐mutated adenocarcinoma in an exploratory study of 5 patients.21 In addition, the recent retrospective studies showed the potential benefit of afatinib for patients with HER2‐mutant lung cancers.22, 24 While further prospective studies including randomized trials are required, afatinib could be approved as one of the therapeutic options for HER2‐altered NSCLC patients.
However, development of acquired resistance to afatinib, just like the case for other HER inhibitors, has been reported in most EGFR‐mutant NSCLC patients treated with afatinib,25, 26 and the situation is considered to be similar for HER2‐altered NSCLC patients. Therefore, overcoming acquired resistance to afatinib is a critical clinical issue.
In this study, we experimentally established various afatinib‐resistant cell lines from HER2‐altered NSCLC cell lines as the parental cell lines, and examined the molecular profiles of these cells to uncover the mechanisms of acquisition of the drug resistance, and investigated the treatment strategies for these cells with acquired resistance to afatinib.
2. MATERIALS AND METHODS
2.1. Reagents and cell cultures
Three NSCLC cell lines (Calu3, NCI‐H2170 [H2170] and NCI‐H1781 [H1781]), purchased from ATCC (Manassas, VA, USA), were used as the patent cell lines for this study. These cell lines were authenticated by ATCC using short‐tandem‐repeat polymorphism analysis, and used within 6 months of receipt. All the lung cancer cells were cultured in RPMI‐1640 media supplemented with 10% FBS, in a humidified incubator with 5% CO2 at 37°C. Afatinib‐resistant sublines were established by different methods: the parental cells were cultured with stepwise escalation of the concentration of afatinib in the culture medium (Calu3 and H2170: from 5 nmol/L to 2 μmol/L, H1781: from 5 nmol/L to 0.4 μmol/L) over a period of 6 months (stepwise escalation method), or use of a high concentration of afatinib from the start of culture (Calu3 and H2170: 2 μmol/L, H1781: 0.4 μmol/L) until 6 months (high concentration method). Finally, afatinib‐resistant sublines designated as H2170‐ARS, Calu3‐ARS and H1781‐ARS were established by the stepwise escalation method, and those designated as H2170‐ARH, Calu3‐ARH and H1781‐ARH were established by the high concentration method (Table 1). Acquired resistance was defined as an IC50 value of over a 100‐fold higher than that for the parental cell line. Afatinib and docetaxel were purchased from Synkinase (San Diego, CA, USA) and Wako Pure Chemical Industries (Osaka, Japan), respectively. Crizotinib and elacridar were purchased from Sigma‐Aldrich (St. Louis, MO, USA).
Table 1.
Afatinib‐resistant cell lines and the resistance mechanisms
| Cell lines | Afatinib exposure | HER2 amp/mut | MET amp | EMT | CSC markers |
|---|---|---|---|---|---|
| Calu3 | N/A | amp | − | − | − |
| Calu3‐ARS | Stepwise | amp | + | − | − |
| Calu3‐ARH | High | amp | − | + | − |
| H2170 | N/A | amp | − | − | − |
| H2170‐ARS | Stepwise | Loss of amp | − | − | − |
| H2170‐ARH | High | Loss of amp | − | − | − |
| H1781 | N/A | mut (G776VC) | − | − | − |
| H1781‐ARS | Stepwise | mut (G776VC) | − | − | + |
| H1781‐ARH | High | mut (G776VC) | − | − | + |
amp, amplification; CSC, cancer stem cell; EMT, epithelial‐to‐mesenchymal transition; mut, mutation; N/A, not applicable.
Details of the phospho‐receptor tyrosine kinase array and the cell proliferation assay are included in Document S1.
2.2. DNA and RNA extraction
Genomic DNA were extracted from the cell lines using the DNeasy Blood and Tissue Kit (Qiagen, Venlo, the Netherlands), and total RNA were extracted using the RNeasy Mini Kit (Qiagen). Complementary DNA (cDNA) was synthesized from total RNA using a High‐Capacity cDNA Reverse Transcription Kit (Thermo Fisher Scientific, Waltham, MA, USA), in accordance with the manufacturer's instructions.
2.3. Copy number analysis by quantitative real‐time PCR
Copy number gains (CNG) of MET, HER2 and EGFR were determined by quantitative real‐time (qRT)‐PCR using Taqman copy number assays (Thermo Fisher Scientific). TaqMan RNase P Control (Thermo Fisher Scientific) was used as the reference gene. The relative copy number of each sample was determined by comparing the ratio of the expression level of the target gene to that of the reference gene in each sample with the ratio in standard genomic DNA (Merck, Darmstadt, Germany). The catalog numbers of the TaqMan assays are shown in Table S1A. Based on the results of our previous study, we defined amplification as a value of greater than 4.27, 28
2.4. Fluorescence in situ hybridization assay
A dual‐color FISH assay was performed using the PathVysion HER‐2 DNA Probe Kit (Abbott Molecular, Des Plaines, IL, USA), in accordance with the manufacturer's instructions. Twenty metaphase spreads and 200 interphase nuclei were analyzed in each slide.
2.5. Direct sequencing
We determined the mutational status of the tyrosine kinase domain of HER2 by direct sequencing; the PCR conditions employed are shown in Table S1B.
2.6. Western blot analysis
The total cell lysate was extracted with lysis buffer, a mixture of RIPA buffer, phosphatase inhibitor cocktails 2 and 3 (Sigma‐Aldrich) and Complete Mini (Roche, Basel, Switzerland). Western blot analysis was carried out using the conventional method using the following primary antibodies: anti‐EGFR, phospho‐ (p‐) EGFR (Tyr1068), HER2, p‐HER2 (Tyr1221⁄1222), MET, p‐MET (Tyr1234⁄1235), AKT, p‐AKT (Ser473), p44⁄p42 MAPK, p‐p44⁄p42 MAPK, E‐cadherin, N‐cadherin, vimentin, ALDH1 (Cell Signaling Technology, Danvers, MA, USA) and b‐actin (used as the loading control) (Merck Millipore, Billerica, MA, USA). The secondary antibody was HRP‐conjugated anti‐mouse or anti‐rabbit IgG (Santa Cruz Biotechnology, Dallas, TX, USA). To detect specific signals, the membranes were examined using the ECL Prime Western Blotting Detection System (GE Healthcare, Amersham, UK) and LAS‐3000 (Fujifilm, Tokyo, Japan).
2.7. mRNA expression analysis by quantitative reverse‐transcription PCR
The gene expressions of the putative cancer stem cell (CSC) markers ALDH1A1, ABCB1, CD44, Oct‐4 and CD133 were analyzed by qRT‐PCR using the cDNA, TaqMan Gene Expression Assays, and the ABI StepOnePlus Real‐Time PCR Instrument (Thermo Fisher Scientific). The mRNA expression was calculated using the delta‐delta‐CT method. The glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) gene was used as the endogenous control. The catalog numbers of the TaqMan assays are shown in Table S1C.
2.8. Cell growth inhibition assay
Cells were cultured with or without afatinib for 72 hours and the sensitivity to the drug was determined by a modified MTS assay using CellTiter 96 Aqueous One Solution Reagent (Promega, Fitchburg, WI, USA), as previously described.28 The antiproliferative effect of each drug was expressed in terms of the IC50 value, which refers to the concentration of the drug required to inhibit cell proliferation by 50%.
2.9. Xenograft model
Six‐week‐old BALB/c‐nu/nu female mice were purchased from Charles River Laboratories (Yokohama, Japan). All mice were provided with sterilized food and water, and housed in a barrier facility under a 12:12‐h light: dark cycle. The Calu3‐ARS cells (5 × 106) were suspended in 100 μL of RPMI‐1640 medium with the Matrigel Basement Membrane Matrix (Corning, Corning, NY, USA) and injected subcutaneously into the backs of the mice. The volumes of the tumors on the backs of the mice were calculated using the empirical formula V = ½ × [(shortest tumor diameter)2 × (longest tumor diameter)2]. One week after the injection, the mice were randomly allocated to 4 groups that received vehicle, 20 mg/kg/d of afatinib, 10 mg/kg/d of crizotinib, or 20 mg/kg/d of afatinib plus 10 mg/kg/d of crizotinib. The drug solutions were prepared in 0.5 w/v (%) methyl cellulose. The vehicle/drugs were given orally as a suspension by gavage once daily, on 6 days per week. The tumor volumes were measured 3 times a week with calipers. After 3 weeks of treatment, the mice were killed, and the tumors were harvested and photographed.
All the animal experiments were carried out in accordance with protocols approved by the Animal Care and Use Committee, Okayama University (permission number: OKU‐2017353).
2.10. Statistical analysis
All data were analyzed using the JMP v 9.0.0 software (SAS Institute, Cary, NC, USA). Differences among groups were compared by t test. P < .05 was considered as denoting statistical significance. All tests were 2‐sided.
3. RESULTS
3.1. Genotypic mechanisms underlying the development of acquired resistance to afatinib
We established 6 afatinib‐resistant cell lines from the 3 parental NSCLC cell lines harboring HER2 alterations, including 2 HER2‐amplified cell lines (Calu3 and H2170) and 1 HER2‐mutant cell line (H1781). To investigate the genotypic changes in the cells with acquired resistance to afatinib, we examined the copy number of HER2, EGFR and MET, gain of which is well known as a mechanism underlying the resistance to HER inhibitors.29 Significant copy number gains of MET were detected in the Calu3‐ARS cells (Figure 1A). In addition, the HER2 copy number was almost lost in the H2170‐ARS and H2170‐ARH cells. Copy number analysis also revealed progressive decrease of the HER2 copy number of H2170‐ARS cells with increasing number of passages, eventually leading to the disappearance of the amplified HER2 alleles (Figure 1B). To investigate the mechanism underlying the progressive decrease of the copy number of HER2, we performed the FISH assay and found that all of H2170 parental cells harbored amplified HER2 alleles (Figure 1C). Furthermore, the degree of amplification was less in all H2170‐derived resistant subline cells than in H2170 parental cells. These findings suggested that the decrease in HER2 alleles occurred at the individual cell level. Next, we investigated the mutational status of the tyrosine kinase domain of the HER2 gene; this investigation, however, revealed no additional mutations in any of the afatinib‐resistant cell lines (Table 1). We also examined the expression levels of HER2, EGFR and MET protein and the phosphorylation levels of these proteins by western blotting (Figure 1D). Consistent with the results of the copy number analysis, the expressions of p‐MET and MET were upregulated in the Calu3‐ARS cells as compared to the corresponding parental cells. In the H2170‐ARS and H2170‐ARH cells, the expressions of p‐HER2 and HER2 were almost completely lost. In addition, downstream signals, such as p‐AKT and p‐MAPK activation, in the H2170‐ARS and H2170‐ARH cells were much weaker as compared to those in the corresponding parental cells. Furthermore, the phospho‐RTK array revealed that the activation of almost all the RTK was downregulated in the afatinib‐resistant cells as compared to the corresponding parental cells (Figure S1A), whereas the proliferative capability of the cells was maintained at a level similar to that of the corresponding parental cells (Figure S1B).
Figure 1.
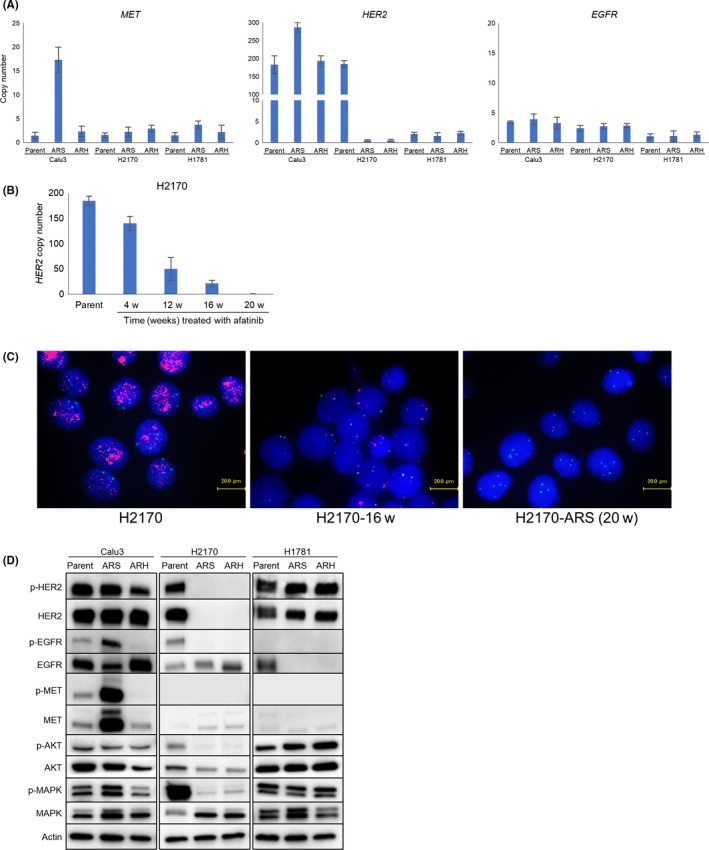
Genotypic analysis of the lung cancer cell lines harboring HER2 alterations and their afatinib‐resistant sublines. A, The copy numbers of MET,HER2 and EGFR were determined by a quantitative reverse‐transcription PCR assay. The MET copy number was significantly amplified in the Calu3‐ARS cells. The HER2 copy number was almost lost in the H2170‐ARS and H2170‐ARH cells. B, Copy number analysis using the H2170‐derived resistant subline showed progressive decrease of the HER2 copy number with an increasing number of passages, with eventual disappearance of amplified HER2 from the cells. w, weeks. C, FISH assay (red, HER2; green, CEP 17; blue, nucleus) showed that the progressive decrease of the HER2 copy number occurred at the individual cell level. H2170‐16w, H2170 cells treated with afatinib for 16 weeks. D, Western blot analysis showed that the expressions of p‐MET and MET were upregulated in the Calu3‐ARS cells, while the p‐HER2 and HER2 expressions were almost lost in the H2170‐ARS and H2170‐ARH cells
3.2. Phenotypic changes in the cells with acquired resistance to afatinib
To investigate the phenotypic changes following the development of acquired resistance to afatinib, we comparatively examined the expression levels of an epithelial marker (E‐cadherin) and mesenchymal marker (N‐cadherin and vimentin) in the resistant and corresponding parental cells. Western blot analysis revealed downregulation of E‐cadherin and upregulation of N‐cadherin and vimentin in the Calu3‐ARH cells (Figure 2A); furthermore, the resistant cells had acquired a spindle‐like shape in contrast to the morphology of the corresponding parental Calu3 cells (Figure 2B). These findings suggest the occurrence of epithelial‐to‐mesenchymal transition (EMT) in the Calu3‐ARH, which was considered as being the mechanism underlying the resistance to afatinib.
Figure 2.
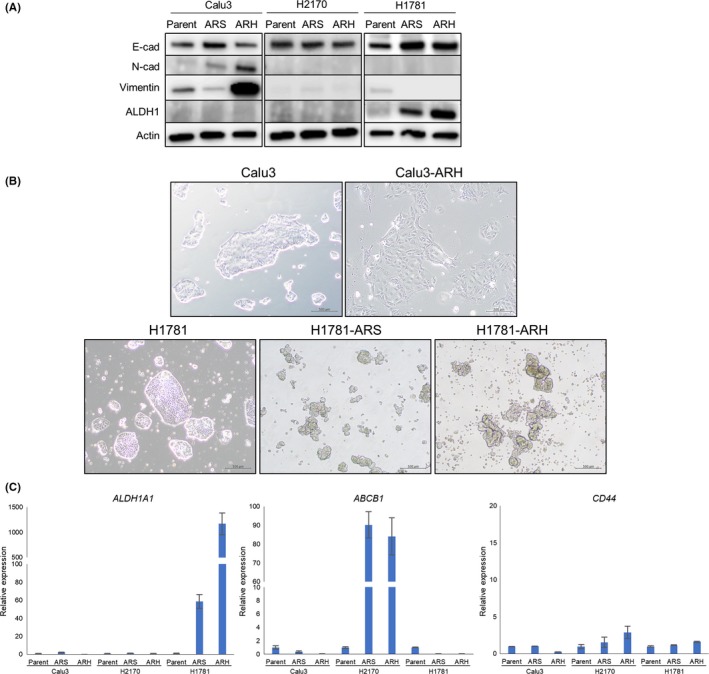
Phenotypic analysis of lung cancer cell lines harboring HER2 alterations and their afatinib‐resistant sublines. A, Western blot analysis showed that the Calu3‐ARH cells exhibited downregulation of E‐cadherin and upregulation of N‐cadherin and vimentin, while the H1781‐ARS and H1781‐ARH cells exhibited upregulation of ALDH1. B, The images of the cells show that the Calu3‐ARH cells exhibited a spindle‐like shape. The H1781‐ARS and H1781‐ARH cells were non‐adhesive and capable of forming sphere‐like clusters. C, mRNA expressions of the putative cancer stem cell markers ALDHA1,ABCB1 and CD44 were examined by quantitative reverse‐ transcription PCR. ALDH1A1 was overexpressed in the H1781‐ARS and H1781‐ARH cells, while ABCB1 was overexpressed in the H2170‐ARS and H2170‐ARH cells
Next, we investigated the expression levels of ALDH1A1, ABCB1, CD44, Oct‐4 and CD133, which are putative CSC markers in NSCLC (Figure 2C and Figure S2A). ALDH1A1 was found to be overexpressed in the H1781‐ARS and H1781‐ARH cells as compared to the parental H1781 cells. Western blot analysis also revealed overexpression of ALDH1A1 in the H1781‐ARS and H1781‐ARH cells (Figure 2A). These cell lines with acquired resistance of afatinib were non‐adhesive and capable of forming sphere‐like clusters (Figure 2B). We conducted a sphere formation assay to examine the cellular functional features of CSC‐like properties,28 and found that H1781‐ARS and H1781‐ARH cells acquired higher ability to form spheres in suspension culture compared with parental H1781 cells (Figure S2B). To examine the CSC‐like properties in vivo, we conducted tumor transplantation experiments.28, 30, 31 We subcutaneously implanted H1781, H1781‐ARS and H1781‐ARH cells to examine the tumor‐forming capability in NOD/SCID mice. We found that H1781‐ARS and H1781‐ARH cells exhibited higher tumorigenicity than parental H1781 cells. The H1781‐ARS and H1781‐ARH cells established larger tumors with shorter latencies than the parental H1781 cells (Figure S2C). These findings suggest that the H1781‐ARS and H1781‐ARH cells had acquired CSC‐like properties, which could be considered as the mechanism underlying the resistance to afatinib. ABCB1 was overexpressed in the H2170‐ARS and H2170‐ARH cells, but not in either the H1781‐ARS or H1781‐ARH cells (Figure 2C). CD44, Oct‐4 and CD133 overexpression was not detected in any of the resistant cell lines.
3.3. Drug sensitivities of the afatinib‐resistant cells showing MET amplification
The sensitivities of the cells to various drugs are shown in Table 2. The cell growth inhibition assay showed that the Calu3‐ARS cells showing MET amplification were insensitive to monotherapy with crizotinib, which is a MET inhibitor, but sensitive to combined treatment with afatinib plus crizotinib (Figure 3A). Western blot analysis revealed that afatinib inhibited the expression of p‐HER2 and crizotinib inhibited the expression of p‐MET in both the parental Calu3 and Calu3‐ARS cells. Both HER2 and MET were activated in the Calu3‐ARS cells; therefore, crizotinib monotherapy was not effective for inhibiting their downstream signals (Figure 3B).
Table 2.
IC50 values (μmol/L) against various agents in HER2‐altered NSCLC cell lines
| Cell lines | Afatinib | MET inhibitor | Docetaxel | |
|---|---|---|---|---|
| Crizotinib | Afatinib plus crizotinib (0.2 μmol/L) | |||
| Calu3 | 0.0029 | 4 | 0.033 | 0.0018 |
| Calu3‐ARS | >10a | 1.2 | 0.0088 | 0.00058 |
| Calu3‐ARH | 1.8a | 3.2 | 2.2a | 0.00013 |
| H2170 | 0.019 | N/A | N/A | 0.000048 |
| H2170‐ARS | 3.8a | N/A | N/A | 0.015a |
| H2170‐ARH | 2.6a | N/A | N/A | 0.015a |
| H1781 | 0.0075 | N/A | N/A | 0.0011 |
| H1781‐ARS | 0.82a | N/A | N/A | 0.0032 |
| H1781‐ARH | 1.1a | N/A | N/A | 0.0029 |
The IC50 value in each resistant cell line to that in the parental cell line is higher than 5‐fold. N/A, not applicable; NSCLC, non‐small‐cell lung cancer.
Figure 3.
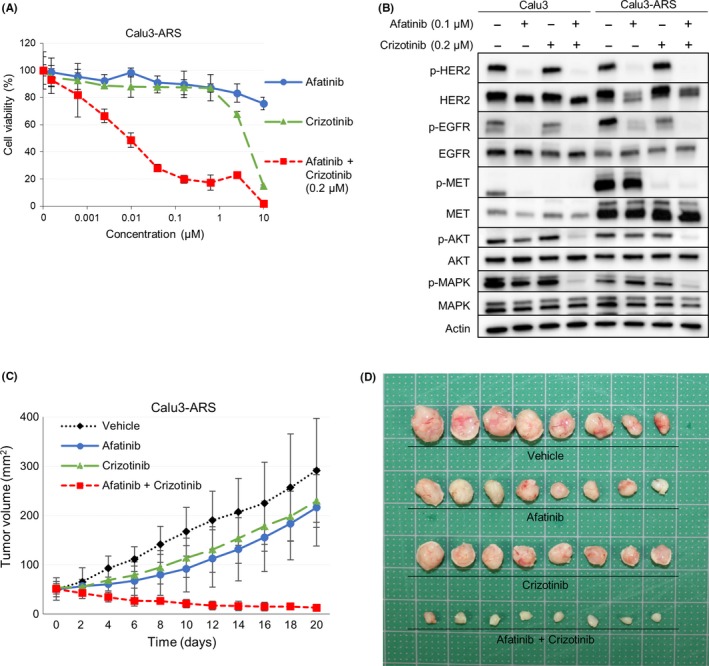
The combination of afatinib plus crizotinib (a MET inhibitor) was effective against the afatinib‐resistant cell line showing MET amplification. A, Cell growth inhibition assay revealed that the combination of afatinib plus crizotinib was highly effective against the Calu3‐ARS cells showing MET amplification. B, Western blot analysis showed that afatinib inhibited the expression of p‐HER2 and crizotinib inhibited the expression of p‐MET in both the parental Calu3 and Calu3‐ARS cells. C, Mice with Calu3‐ARS tumors were given vehicle, afatinib, crizotinib, or afatinib plus crizotinib. The tumor volumes were determined on the indicated days after the start of treatment. Data represent the mean ± SE (n = 8). *P < .05 vs vehicle control by the t test. D, Appearance of the Calu3‐ARS tumors after treatment at the time of sacrifice of the mice
On the basis of our in vitro data, we investigated the antitumor effect of crizotinib in a xenograft mouse model of afatinib‐resistant cells showing MET amplification. As shown in Figure 3C,D, combined treatment with afatinib plus crizotinib significantly inhibited the tumor growth of the Calu3‐ARS xenografts as compared to vehicle control or either afatinib or crizotinib alone (P < .01).
3.4. Sensitivity of the afatinib‐resistant cells to chemotherapeutic agents
The H2170‐ARS and H2170‐ARH cells, which had lost HER2 amplification, also acquired resistance to docetaxel (Table 2). To investigate whether their resistance mechanism was associated with overexpression of ATP‐binding Cassette sub‐family B Member 1 (ABCB1), which is well known to induce resistance to chemotherapy, we conducted the cell growth inhibition assay with or without ABCB1 inhibitor elacridar (Figure S3A,B). We found that elacridar recovered the sensitivity to docetaxel, but not to afatinib in H2170‐ARS and H2170‐ARH cells. These findings were similar to those in our previous report using docetaxel‐resistant NSCLC cells with EGFR‐mutations.32 In contrast, the other afatinib‐resistant cell lines, including Calu3‐ARH, H1781‐ARS and H1781‐ARH, which showed EMT or had acquired CSC‐like features, showed maintained sensitivity to docetaxel, because ABCB1 was not upregulated in these acquired resistance cell lines.
4. DISCUSSION
In this study, we established multiple cell lines that exhibited acquired resistance to the irreversible pan‐HER inhibitor afatinib, from 3 parental HER2‐altered NSCLC cell lines, and examined the mechanisms underlying the development of acquired resistance to afatinib. The resistance mechanisms varied, and included MET amplification and overexpression, loss of HER2 amplification and expression, EMT and acquisition of CSC‐like features. From our previous study using EGFR‐mutant NSCLC cells, we reported that the method of exposure of cell cultures to drugs influences the mechanisms of acquired resistance to HER inhibitors (gefitinib and afatinib).28, 33 Consistent with that report, in this study also, conducted using HER2‐altered NSCLC cells, the resistance mechanisms were affected by the manner of drug exposure. The resistance mechanism in the Calu3‐ARS cells established by the stepwise escalation method appeared to be MET amplification, whereas that in the Calu3‐ARH cells established by the high drug concentration method was considered to be possibly attributable to the acquisition of EMT features.
MET amplification is one of the major mechanisms underlying the acquisition of resistance to HER inhibitors in EGFR‐mutant NSCLC.28, 29, 33, 34 MET amplification has also been reported to contribute to trastuzumab resistance in HER2‐positive breast cancer cells.35 In this study, we identified MET amplification as one of the mechanisms underlying the acquisition of resistance of HER2‐altered NSCLC to the pan‐HER inhibitor afatinib. Importantly, we clarified that combined treatment with afatinib plus crizotinib, a MET inhibitor, was highly effective against HER2‐altered NSCLC showing acquired resistance to afatinib, both in vitro and in vivo. These results suggest the importance of reassessment of tumor cells after the acquisition of resistance to afatinib; if such reassessment in clinical cases reveals MET amplification, then combined treatment with afatinib plus crizotinib should be considered.
In afatinib‐resistant cell lines established from H2170 cells showing HER2 amplification, the copy number of HER2 decreased gradually during the course of exposure to afatinib. We previously reported loss of the EGFR‐mutant allele and progressive decrease of the EGFR copy number in an EGFR‐mutant NSCLC cell line during gefitinib exposure.28 However, to the best of our knowledge, loss of HER2 amplification during afatinib treatment has never been reported before in in vitro examinations. However, loss of HER2 amplification has sometimes been reported after trastuzumab treatment in clinical HER2‐positive breast or gastric cancer patients.36, 37, 38, 39 Our finding seems to be consistent with these latter clinical reports. Loss of HER2 amplification in the residual disease following neoadjuvant trastuzumab‐based therapy has been reported to be associated with a poorer relapse‐free survival when compared with maintained HER2 expression.40 The biological basis of these genetic alterations has not been clarified yet, and might include tumor heterogeneity, genetic drift toward a more aggressive tumor phenotype, or selective pressure of adjuvant therapy.38 Our in vitro experiments revealed that the decrease in HER2 alleles occurred at the individual cell level, suggesting that the tumor heterogeneity could not be the reason for acquired afatinib resistance.
The characteristics of the afatinib‐resistant cell lines with loss of HER2 amplification were very interesting. In these resistant cells, the activation of not only HER2 but also other RTK, and their downstream signals were much lower than those in the corresponding parental cell lines, although their proliferative ability was maintained. However, we have yet to clarify the resistance mechanisms in detail, and continued research into the resistance mechanisms and investigation of the appropriate treatment strategy for afatinib‐resistant tumors with loss of HER2 amplification is necessary.
As for the other afatinib‐resistant cell lines, Calu3‐ARH showed the features of EMT as the possible mechanism underlying afatinib resistance, and the afatinib‐resistant H1781‐ARS and H1781‐ARH cells had acquired CSC‐like properties. Although EMT and acquisition of CSC‐like features are well known as mechanisms underlying the acquisition of resistance to various drugs, this is the first report of their occurrence during the course of exposure to afatinib in HER2‐altered NCSLC cells. Interestingly, the Calu3‐ARS, H1781‐ARS and H1781‐ARH showing EMT or CSC‐like features remained sensitive to docetaxel, whereas we previously reported that EGFR‐mutant NSCLC cells with acquired resistance to EGFR‐TKI caused by EMT or acquisition of CSC‐like features exhibited resistance to docetaxel.28, 33 None of the Calu3‐ARS, H1781‐ARS or H1781‐ARH cells showed upregulation of ABCB1, which was considered as the reason for the afatinib‐resistant cells’ maintained sensitivity to docetaxel. Overexpression of ABCB1 has been reported to be strongly associated with EMT or the acquisition of CSC‐like features;41 therefore, the characteristics of the Calu3‐ARH, H1781‐ARS and H1781‐ARH may differ somewhat from those of EGFR‐TKI‐resistant cells showing EMT or CSC‐like features reported previously.28, 33 Moreover, the precise molecular mechanisms underlying how EMT or CSC‐like features lead to afatinib resistance remain unclear, and thorough investigation using resistance models established by us could yield clues to uncover these mechanisms.
In conclusion, we established 6 cell lines with acquired resistance to afatinib from 3 parental HER2‐altered NSCLC cell lines. The mechanisms of afatinib resistance in the established cell lines varied, and included MET amplification, loss of HER2 amplification, and EMT or acquisition of CSC‐like features, and the cells showed specific sensitivity patterns to various drugs, such as crizotinib and docetaxel. These findings underscore the importance of reassessment of cancer tumors after treatment and personalized medicine choices made based on such reassessment for cancer treatment.
CONFLICT OF INTEREST
There are no potential conflicts of interest to declare.
Supporting information
ACKNOWLEDGMENTS
We thank Dr Takehiro Matsubara (Biobank, Okayama University Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama, Japan) and Ms Fumiko Isobe (Department of Thoracic, Breast and Endocrinological Surgery, Okayama University Graduate School of Medicine, Dentistry and Pharmaceutical Sciences, Okayama, Japan) for their technical support.
Torigoe H, Shien K, Takeda T, et al. Therapeutic strategies for afatinib‐resistant lung cancer harboring HER2 alterations. Cancer Sci. 2018;109:1493–1502. https://doi.org/10.1111/cas.13571
Funding information
Management Expenses Grant.
REFERENCES
- 1. Mok TS, Wu YL, Thongprasert S, et al. Gefitinib or carboplatin‐paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361:947‐957. [DOI] [PubMed] [Google Scholar]
- 2. Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311:1998‐2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pao W, Girard N. New driver mutations in non‐small‐cell lung cancer. Lancet Oncol. 2011;12:175‐180. [DOI] [PubMed] [Google Scholar]
- 4. Kwak EL, Bang YJ, Camidge DR, et al. Anaplastic lymphoma kinase inhibition in non‐small‐cell lung cancer. N Engl J Med. 2010;363:1693‐1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chazin VR, Kaleko M, Miller AD, Slamon DJ. Transformation mediated by the human HER‐2 gene independent of the epidermal growth factor receptor. Oncogene. 1992;7:1859‐1866. [PubMed] [Google Scholar]
- 6. Neve RM, Lane HA, Hynes NE. The role of overexpressed HER2 in transformation. Ann Oncol. 2001;12(Suppl 1):S9‐S13. [DOI] [PubMed] [Google Scholar]
- 7. Wang SE, Narasanna A, Perez‐Torres M, et al. HER2 kinase domain mutation results in constitutive phosphorylation and activation of HER2 and EGFR and resistance to EGFR tyrosine kinase inhibitors. Cancer Cell. 2006;10:25‐38. [DOI] [PubMed] [Google Scholar]
- 8. Perera SA, Li D, Shimamura T, et al. HER2YVMA drives rapid development of adenosquamous lung tumors in mice that are sensitive to BIBW2992 and rapamycin combination therapy. Proc Natl Acad Sci USA. 2009;106:474‐479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Slamon DJ, Leyland‐Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783‐792. [DOI] [PubMed] [Google Scholar]
- 10. Bang YJ, Van Cutsem E, Feyereislova A, et al. Trastuzumab in combination with chemotherapy versus chemotherapy alone for treatment of HER2‐positive advanced gastric or gastro‐oesophageal junction cancer (ToGA): a phase 3, open‐label, randomised controlled trial. Lancet. 2010;376:687‐697. [DOI] [PubMed] [Google Scholar]
- 11. Tan D, Deeb G, Wang J, et al. HER‐2/neu protein expression and gene alteration in stage I‐IIIA non‐small‐cell lung cancer: a study of 140 cases using a combination of high throughput tissue microarray, immunohistochemistry, and fluorescent in situ hybridization. Diagn Mol Pathol. 2003;12:201‐211. [DOI] [PubMed] [Google Scholar]
- 12. Pellegrini C, Falleni M, Marchetti A, et al. HER‐2/Neu alterations in non‐small cell lung cancer: a comprehensive evaluation by real time reverse transcription‐PCR, fluorescence in situ hybridization, and immunohistochemistry. Clin Cancer Res. 2003;9:3645‐3652. [PubMed] [Google Scholar]
- 13. Cappuzzo F, Varella‐Garcia M, Shigematsu H, et al. Increased HER2 gene copy number is associated with response to gefitinib therapy in epidermal growth factor receptor‐positive non‐small‐cell lung cancer patients. J Clin Oncol. 2005;23:5007‐5018. [DOI] [PubMed] [Google Scholar]
- 14. Swanton C, Futreal A, Eisen T. Her2‐targeted therapies in non‐small cell lung cancer. Clin Cancer Res. 2006;12:4377s‐4383s. [DOI] [PubMed] [Google Scholar]
- 15. Stephens P, Hunter C, Bignell G, et al. Lung cancer: intragenic ERBB2 kinase mutations in tumours. Nature. 2004;431:525‐526. [DOI] [PubMed] [Google Scholar]
- 16. Shigematsu H, Takahashi T, Nomura M, et al. Somatic mutations of the HER2 kinase domain in lung adenocarcinomas. Cancer Res. 2005;65:1642‐1646. [DOI] [PubMed] [Google Scholar]
- 17. Gatzemeier U, Groth G, Butts C, et al. Randomized phase II trial of gemcitabine‐cisplatin with or without trastuzumab in HER2‐positive non‐small‐cell lung cancer. Ann Oncol. 2004;15:19‐27. [DOI] [PubMed] [Google Scholar]
- 18. Lara PN Jr, Laptalo L, Longmate J, et al. Trastuzumab plus docetaxel in HER2/neu‐positive non‐small‐cell lung cancer: a California Cancer Consortium screening and phase II trial. Clin Lung Cancer. 2004;5:231‐236. [DOI] [PubMed] [Google Scholar]
- 19. Clamon G, Herndon J, Kern J, et al. Lack of trastuzumab activity in nonsmall cell lung carcinoma with overexpression of erb‐B2: 39810: a phase II trial of Cancer and Leukemia Group B. Cancer. 2005;103:1670‐1675. [DOI] [PubMed] [Google Scholar]
- 20. Mazieres J, Peters S, Lepage B, et al. Lung cancer that harbors an HER2 mutation: epidemiologic characteristics and therapeutic perspectives. J Clin Oncol. 2013;31:1997‐2003. [DOI] [PubMed] [Google Scholar]
- 21. De Greve J, Teugels E, Geers C, et al. Clinical activity of afatinib (BIBW 2992) in patients with lung adenocarcinoma with mutations in the kinase domain of HER2/neu. Lung Cancer. 2012;76:123‐127. [DOI] [PubMed] [Google Scholar]
- 22. Mazieres J, Barlesi F, Filleron T, et al. Lung cancer patients with HER2 mutations treated with chemotherapy and HER2‐targeted drugs: results from the European EUHER2 cohort. Ann Oncol. 2016;27:281‐286. [DOI] [PubMed] [Google Scholar]
- 23. Suzawa K, Toyooka S, Sakaguchi M, et al. Antitumor effect of afatinib, as a human epidermal growth factor receptor 2‐targeted therapy, in lung cancers harboring HER2 oncogene alterations. Cancer Sci. 2016;107:45‐52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lai W‐CV, Lebas L, Milia J, et al. Afatinib in patients with metastatic HER2‐mutant lung cancers: an international multicenter study. J Clin Oncol. 2017; 35 (Suppl): abstr 9071 Available from URL: https://meetinglibrary.asco.org/record/145760/abstract (Accessed 3 June 2017) [Google Scholar]
- 25. Sequist LV, Yang JC, Yamamoto N, et al. Phase III study of afatinib or cisplatin plus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31:3327‐3334. [DOI] [PubMed] [Google Scholar]
- 26. Miller VA, Hirsh V, Cadranel J, et al. Afatinib versus placebo for patients with advanced, metastatic non‐small‐cell lung cancer after failure of erlotinib, gefitinib, or both, and 1 or two lines of chemotherapy (LUX‐Lung 1): a phase 2b/3 randomised trial. Lancet Oncol. 2012;13:528‐538. [DOI] [PubMed] [Google Scholar]
- 27. Soh J, Okumura N, Lockwood WW, et al. Oncogene mutations, copy number gains and mutant allele specific imbalance (MASI) frequently occur together in tumor cells. PLoS ONE. 2009;4:e7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shien K, Toyooka S, Yamamoto H, et al. Acquired resistance to EGFR inhibitors is associated with a manifestation of stem cell‐like properties in cancer cells. Cancer Res. 2013;73:3051‐3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Engelman JA, Zejnullahu K, Mitsudomi T, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039‐1043. [DOI] [PubMed] [Google Scholar]
- 30. Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755‐768. [DOI] [PubMed] [Google Scholar]
- 31. Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324:1670‐1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen H, Shien K, Suzawa K, et al. Elacridar, a third‐generation ABCB1 inhibitor, overcomes resistance to docetaxel in non‐small cell lung cancer. Oncol Lett. 2017;14:4349‐4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hashida S, Yamamoto H, Shien K, et al. Acquisition of cancer stem cell‐like properties in non‐small cell lung cancer with acquired resistance to afatinib. Cancer Sci. 2015;106:1377‐1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bean J, Brennan C, Shih JY, et al. MET amplification occurs with or without T790M mutations in EGFR mutant lung tumors with acquired resistance to gefitinib or erlotinib. Proc Natl Acad Sci USA. 2007;104:20932‐20937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shattuck DL, Miller JK, Carraway KL III, Sweeney C. Met receptor contributes to trastuzumab resistance of Her2‐overexpressing breast cancer cells. Cancer Res. 2008;68:1471‐1477. [DOI] [PubMed] [Google Scholar]
- 36. Xian Z, Zynger DL. HER2 loss after neoadjuvant treatment: is the adjuvant trastuzumab treatment feasible?‐reply. Hum Pathol. 2017;65:247‐248. [DOI] [PubMed] [Google Scholar]
- 37. Pietrantonio F, Caporale M, Morano F, et al. HER2 loss in HER2‐positive gastric or gastroesophageal cancer after trastuzumab therapy: implication for further clinical research. Int J Cancer. 2016;139:2859‐2864. [DOI] [PubMed] [Google Scholar]
- 38. Guarneri V, Dieci MV, Barbieri E, et al. Loss of HER2 positivity and prognosis after neoadjuvant therapy in HER2‐positive breast cancer patients. Ann Oncol. 2013;24:2990‐2994. [DOI] [PubMed] [Google Scholar]
- 39. Altundag K. HER2 loss after neoadjuvant treatment: is the adjuvant trastuzumab treatment feasible? Hum Pathol. 2017;65:247. [DOI] [PubMed] [Google Scholar]
- 40. Mittendorf EA, Wu Y, Scaltriti M, et al. Loss of HER2 amplification following trastuzumab‐based neoadjuvant systemic therapy and survival outcomes. Clin Cancer Res. 2009;15:7381‐7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sugano T, Seike M, Noro R, et al. Inhibition of ABCB1 overcomes cancer stem cell‐like properties and acquired resistance to MET inhibitors in non‐small cell lung cancer. Mol Cancer Ther. 2015;14:2433‐2440. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


