Summary
Deep characterization of the frequencies, phenotypes and functionalities of liver and peripheral blood natural killer (NK), natural killer T (NKT) and T cells from healthy individuals is an essential step to further interpret changes in liver diseases. These data indicate that CCR7, a chemokine essential for cell migration through lymphoid organs, is almost absent in liver NK and T cells. CD56bright NK cells, which represent half of liver NK cells, showed lower expression of the inhibitory molecule NKG2A and an increased frequency of the activation marker NKp44. By contrast, a decrease of CD16 expression with a potential decreased capacity to perform antibody‐dependent cellular cytotoxicity was the main difference between liver and peripheral blood CD56dim NK cells. Liver T cells with an effector memory or terminally differentiated phenotype showed an increased frequency of MAIT cells,T‐cell receptor‐γδ (TCR‐γδ) T cells and TCR‐αβ CD8+ cells, with few naive T cells. Most liver NK and T cells expressed the homing markers CD161 and CD244. Liver T cells revealed a unique expression pattern of killer cell immunoglobulin‐like receptors (KIR) receptors, with increased degranulation ability and higher secretion of interferon‐γ. Hence, the liver possesses a large amount of memory and terminally differentiated CD8+ cells with a unique expression pattern of KIR activating receptors that have a potent functional capacity as well as a reduced amount of CCR7, which are unable to migrate to regional lymph nodes. These results are consistent with previous studies showing that liver T (and also NK) cells likely remain and die in the liver.
Keywords: liver, natural killer, natural killer T and T cells
Abbreviations
- ADCC
antibody‐dependent cellular cytotoxicity
- APC
allophycocyanin
- FITC
fluorescein isothiocyanate
- IFN‐γ
interferon‐γ
- KIR
killer cell immunoglobulin‐like receptors
- LMCs
human liver mononuclear cells
- NK
natural killer cells
- PB
peripheral blood
- PBMC
peripheral blood mononuclear cells
- PE
phycoerythrin
- PerCP
peridinin chlorophyll protein
- TCM
central memory T cells
- TE
effector T cells
- TEM
effector memory T cells
- TTM
transitional memory T cells
- TCR‐γδ
T‐cell receptor‐γδ
Introduction
The immune response to pathogens, alloantigens and tumours is mainly investigated in peripheral blood (PB). There is limited information concerning the identification and physiological mechanism of immunological participating factors in peripheral tissues and organs. The liver‐specific immune response depends on an array of non‐parenchymal cells that include liver sinusoidal endothelial cells, Kupffer cells, hepatic stellate cells and dendritic cells, which interact with liver‐associated lymphocytes.1 Previous studies have attempted to characterize the phenotype of non‐parenchymal liver cells isolated after enzymatic digestion of liver fragments,2 and several studies3, 4, 5 have analysed the phenotypic and functional characteristics of human‐liver mononuclear cells (LMCs). However, extended phenotypic and functional studies may be restricted by the limited amount of cells available from tissues or organs of patients who underwent surgery for an underlying disease or during liver transplantation. Recently, LMCs were isolated during the perfusion of the living donor liver lobe before transplantation or through perfusion of livers from deceased donors.6, 7 The advantage of these approaches is that they avoid tissue destruction and do not require enzymatic treatment. Previous studies7, 8 have demonstrated that liver natural killer (NK) cells, CD3+ CD56+ and CD3+ CD56− T cells, differ in their phenotypes with peripheral blood mononuclear cells (PBMCs) but are immersed in a tolerogenic microenvironment.9 NK cells are innate immune cells that are specialized to eliminate virus‐infected cells. These cells can be divided into functionally distinct subsets based on their level of CD56 surface expression as primarily cytotoxic CD56dim NK cells and more immunoregulatory cytokine‐producing CD56bright NK cells. The functions of both NK cell groups are modulated through inhibitory and activating signals from distinct classes of receptors. Inhibitory receptors include the polymorphic system, killer cell immunoglobulin‐like receptors (KIR)10 and members of the C‐type lectin‐like receptor CD94/NKG2A family, which recognize HLA‐E.11 Activating receptors include natural cytotoxicity receptors (NKp30, NKp44 and NKp46), NKG2C expressed as a dimer with CD94, signalling lymphocyte activation molecule family receptors,12 lectin‐like receptor NKG2D and FcγRIIIa receptor (CD16), which mediates antibody‐dependent cytotoxicity.13 KIR expression on NK cells is largely random and determined based on the KIR gene contents, polymorphisms and stochastic epigenetic regulation at the promoter level.14, 15 The interactions between KIRs and HLA ligands ensure the maintenance of self‐tolerance. Expression of inhibitory receptors is genetically determined, and in the absence of infection, inflammation and other diseases, NK cells primarily receive inhibitory signals. Alternatively, NK cell activation increases the expression of activating receptors,16 enabling NK cells to reach a more responsive state in the context of infection and inflammation.
At least for some KIRs, the expression frequencies at the cell surface level are higher in individuals with two copies of the respective KIR gene compared with those with only one copy, suggesting a gene‐dose effect.14, 17
These receptors modulate NK cell function for the rapid targeting and destruction of virus‐infected cells without the need for previous sensitization, making them an important first line of defence against viral pathogens.
Liver CD3+ cells are characterized by a high frequency of CD8+ cells and a subset of NKT cells that are represented by their expression of CD3 and CD56.2, 7 CD8+ T cells can exist in at least three states of reactivity: naive CD8+ T cells with low reactivity, activated (effector) CD8+ T cells with high reactivity and memory CD8+ T cells with intermediate reactivity. The overall memory CD8+ T‐cell compartment is represented by central and memory subsets, which can be recognized based on their phenotype and function.18, 19 However, the state of activation and cell differentiation of liver CD8+ T cells remains controversial. A previous report using liver perfusion described a relatively high level of CD8+ T cells with a naive phenotype, suggesting that in addition to their role as a graveyard for antigen‐specific activated CD8+ T cells, naive CD8+ T cells may enter the liver without previous activation.20 By contrast, other studies have demonstrated that the healthy human liver is a site of intense immunological activity7 and contains a liver‐homing/retention marker‐expressing CD8+ T‐cell pool identified as tissue‐resident memory T cells, which are not observed within circulating memory CD8+ T cells.4
Recently, we showed that expression of KIR genes on T cells is primarily restricted to liver T cells,21 potentially representing highly differentiated T cells.
In the present study, using LMCs isolated from liver perfusion, we performed extensive analyses of NK cells and T‐cell subsets to obtain novel insights into the prevalence of various lymphocyte subsets and the putative use of KIR genes by intrahepatic NK and T cells.
Patients and methods
Obtaining LMCs from healthy liver donors
Liver wedge samples
Liver samples (10–20 g) were obtained from hepatic transplant donors, collected in sterile physiological solution, stored at 4°, and processed within the following 12 hr. After washing with sterile PBS, the samples were mechanically disintegrated and collected in a Petri dish with 10 ml of RPMI‐1640 containing 0·5 mg/ml collagenase type II (clostridiopeptidase A), 0·02 mg/ml gentamycin, 2 mmol/l l‐glutamine (Sigma‐Aldrich, Darmstadt, Germany) and 10% fetal bovine serum (Natocor, Cordoba, Argentina) and incubated at 37° for 30 min. The samples were subsequently filtered through a 70‐mm metal mesh and washed with 1× PBS. Mononuclear cells were purified through a Ficoll–Hypaque gradient (Sigma‐Aldrich).
Liver perfusion and cell sample collection
Samples were collected from donor livers according to Kelly et al.6 Briefly, 23 samples were collected from donor livers during orthotropic liver transplantation: 14 donors at the Austral University Hospital and nine donors at the liver transplant unit of the Italiano Hospital. During retrieval (at the time of exsanguination), the donor aorta and superior mesenteric vein were flushed with University of Wisconsin (UW) solution (Bristol‐Myers Squibb, Uxbridge, UK) or with HTK (histidine‐tryptophan‐ketoglutarate) solution (Custodiol®; Essential Pharmaceuticals, Ewing, NJ). After excision of the organ, the liver was flushed again with UW solution until all blood was removed and the perfused solution appeared clear. At implantation, after completion of the upper inferior cava anastomosis, the livers were flushed with ringer lactate at 4° through the portal vein to wash out the UW solution before reperfusion. The perfused hepatic fluid was collected from the inferior vena cava (600–1200 ml). Within the following 12 hr, the samples were sent to the laboratory and centrifuged at 500 g for 5 min, and the supernatant was discarded. All pellets were collected, washed twice with 1× PBS at 500 g for 5 min, and mononuclear cells were subsequently isolated through a Ficoll–Hypaque (GE Healthcare Bio‐Sciences, Uppsala, Sweden) gradient.
All protocols were approved by the Hospital de Clínicas and Hospital Italiano Institutional Review Board in accordance with the ethical guidelines of the 1975 Declaration of Helsinki. All samples were identified using a transplant procedure number provided by the INCUCAI without the name of the donor. None of the transplant donors were recruited from a vulnerable population, and all donors or next of kin provided written informed consent that was freely given.
Isolation of PBMCs
Peripheral blood mononuclear cells were obtained from 5 to 20 ml of heparinized blood from 35 adult healthy controls and isolated through a Ficoll–Hypaque density gradient.
Monoclonal antibodies and flow cytometry
Both PBMCs and LMCs were stained with unconjugated antibodies against KIR 3DL1/3DL2 (clone 5.133, courtesy of Dr M. Colonna), KIR 2DL2/2DS2/2DL3 (clone CHL, courtesy of Dr S. Ferrini), KIR 2DL1/2DS1 (clone HPMA4), KIR 2DL1/2DS1/2DS3 (clone HP3E4), CD94 (clone 3D9) and NKG2A (clone Z199) courtesy of Dr M. Bottet and unconjugated isotypes against IgG1, IgG2a, IgG2b and IgM (Becton Dickinson, San Jose, CA). In addition, mouse monoclonal antibodies against 3DL1‐fluorescein isothiocyanate (FITC), 2DL3‐phycoerythrin (PE) (Becton Dickinson), 2DS4‐PE, NKG2C‐PE (R&D Systems, Minneapolis, MN) NKp44‐PE, NKp46‐PE, 2B4‐FITC, CD16‐FITC, CD57‐allophycocyanin (APC), CD8‐PE/APC, CD4‐FITC, CD45RA‐PE‐Cy7, CD28‐PE, CD27‐PE‐CF594, CCR7‐FITC, CD11b‐FITC, CD161‐APC/PE, HLA‐DR‐FITC, T‐cell receptor‐γδ (TCR‐γδ) ‐PE and TCR Vα7.2 MAIT‐PE (Biolegend, San Diego, CA), and labelled isotypes against IgG1, IgG2a (Biolegend) were used. The amount of antibody used was previously determined by a titration experiment performed on PBMCs and LMCs. For the characterization of T‐cell differentiation, anti‐CD27, anti‐CD28 and anti‐CD45RA were simultaneously labelled, and for NK differentiation analysis, anti‐CD27 and CD11b were used in conjunction. Incubation of cells with all of the above‐mentioned antibodies was performed at room temperature for 30 min, and subsequently, the cells were washed twice with 1 ml of 1× PBS and centrifuged for 5 min at 500 g. For unconjugated antibodies, a subsequent incubation with 3 μl of an FITC‐ or PE‐conjugated anti‐mouse immunoglobulin antibody (Dako, Glostrup, Denmark) was performed for 30 min at room temperature. After washing, 3 μl of normal mouse serum was added. To analyse the labelling corresponding to NK, NKT and T cells, an incubation was performed with 3 μl of anti‐CD3 (Becton Dickinson) conjugated to Peridinin chlorophyll protein (PerCP)/FITC and 5 μl of anti‐CD56 conjugated to PE/APC (Becton Dickinson) or the BV421 antibody (Biolegend) for 15 min at room temperature. A suitable fluorochrome combination was performed for each labelling scheme. In the liver samples, the anti‐CD45 pan‐leucocyte marker conjugated with APC‐H7 (Becton Dickinson) was also added. After washing again with 1× PBS, cells were fixed with 2% paraformaldehyde and subsequently analysed using a FACSAria II cell sorter (Becton Dickinson) flow cytometer. The results were analysed using flowjo 7.6.2 software (Tree Star, Inc., Ashland, OR). Analyses of a particular population of interest were based on a gate of at least 100 000 events. For KIR analysis, expression of three receptors was indirectly inferred: the antibody 3DL1/3DL2 showed two clouds with different mean fluorescence intensity values, where the lower cloud corresponded to 3DL2 and the upper cloud corresponded to 3DL1. As a control method, in all cases, expression of the upper cloud was compared with the monoclonal antibody against 3DL1 and there were no significant differences. Expression of 2DS3 was deduced from the subtraction of expression between 2DL1/2DS1/2DS3 – 2DL1/2DS1. Expression of 2DS1 was deduced from subtraction of expression between 2DL1/2DS1 – 2DL1. Finally, we inferred the expression of 2DL2/2DS2 from subtraction of expression between 2DL2/2DS2/2DL3 – 2DL3. In all cases, the presence of the KIR gene was verified.
We have previously assessed the quality of the results through dead cell staining, demonstrating that dead cell discrimination by forward/side scatter presented excellent correlation with the results obtained using the LIVE/DEAD® Fixable Aqua Dead Cell Stain Kit (Thermo Fisher Scientific, Waltham, MA), which specifically stains lymphocytes (see Supplementary material, Fig. S1). Hence, dead cells were excluded by forward/side scatter gating.
Functional studies
Mononuclear cells obtained from liver or PB samples were placed in a 24‐well plate at a concentration of 1 × 106 lymphocytes per well in 1 ml of complete RPMI medium. Three wells were used for each sample according to the following scheme: two wells were stimulated with phorbol 12‐myristate 13‐acetate (PMA; Sigma‐Aldrich) at a final concentration of 25 ng/ml and ionomycin (calcium salt of Streptomyces conglobatus; Sigma‐Aldrich) at a final concentration of 0·5 μg/ml and compared with a not stimulated negative control. An anti‐CD107a antibody (Becton Dickinson) conjugated to FITC was added (20 μl) to the positive and negative controls and 20 μl of the FITC‐conjugated IgG1 (Becton Dickinson) was included as isotype control. The culture plates were incubated for 1 hr at 37° in 5% CO2. Subsequently, 1 μl of monensin (BD‐Golgi Stop Protein Transport Inhibitor; Becton Dickinson) was added to all wells, and the plates were incubated for another 5 hr. Cells were washed with 1 ml of 1× PBS/A and centrifuged at 500 g for 5 min. Cell surface staining was performed on all samples using the following monoclonal antibodies (AtcMo): 3 μl of anti‐CD3 PerCP and 5 μl of anti‐CD56 APC (Becton Dickinson). Pan‐Leukocyte anti‐CD45 APC‐H7 (Becton Dickinson) was included in the liver samples. All samples were subsequently incubated for 20 min at room temperature and washed twice with 1× PBS/A. Intracytoplasmic labelling was performed for the detection of interferon (IFN‐γ). The cells were treated with 100 μl of 1× fixation and permeation solution (Cytofix/Cytoperm; Becton Dickinson) for 20 min at 4°, washed twice with 1 ml of 1× Perm/Wash buffer (Becton Dickinson), and centrifuged at 500 g for 5 min. Cells were also incubated with 20 μl of anti‐IFN‐γ PE (Becton Dickinson), and the isotype control was incubated with 20 μl of the IgG1 PE (Becton Dickinson). The tubes were incubated for 30 min at 4°, followed by two washes with 1× buffer Perm/Wash. The pellets suspended in 200 μl of 1× PBS/A and fixed with 2% paraformaldehyde were analysed using the FACSAria II flow cytometer (Becton Dickinson).
Statistical analysis
Statistical analyses were performed using graphpad prism 5 (GraphPad Software Inc., San Diego, CA). Comparisons between samples (PB and liver) were performed using the Mann–Whitney U‐test. The data are presented as the median and interquartile range (IQR). The reported P values are two‐tailed, and P < 0·05 was considered significant.
Results
Comparative analysis of liver and PB NK and T‐cell frequencies
In the initial experiments, we analysed the frequencies of NK and T cells in LMCs by comparing the frequencies of these cells in samples obtained from liver homogenate (n = 9) and samples obtained from liver perfusion (n = 14). We observed no significant differences in the frequencies of NK (CD56+ CD3−), NKT (CD3+ CD56+) and T (CD3+) cells collected using these two techniques. Therefore, because of the low number of cells recovered from liver homogenates, we collected these cell populations from liver perfusion eluates.
Compared with PB, the liver showed different frequencies of T cells, NKT cells and NK cells. Similar to previous reports,2, 22, 23 we observed more NK cells in the liver. In particular, CD56bright NK cells represented almost 50% of all liver NK cells (14·8% of liver lymphocytes versus 0·7% in PB lymphocytes, P < 0·001), with CD56dim NK cells representing almost the other 50% of NK cells (11·6% versus 8·3% in PB, P = 0·002). Similarly, NKT cells were also highly increased in the liver (13·4 versus 3·8% in PB, P < 0·001). As indicated in Fig. 1, NK and NKT populations accounted for approximately 50% of the liver lymphocyte population. The increased frequencies of liver NK and NKT cells occur at the expense of the relatively decreased frequencies of T cells (54·7% in the liver versus 73·2% in PB, P < 0·001).
Figure 1.
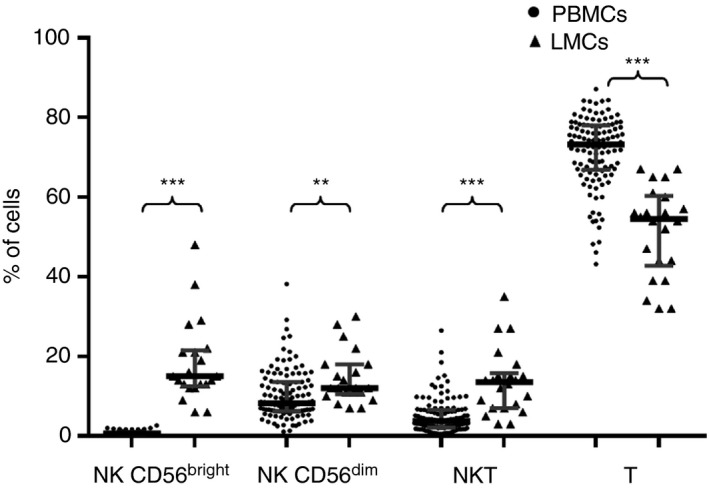
CD56bright natural killer (NK) cells, CD56dim NK cells, NKT and T‐cell frequencies detected in peripheral blood and liver. Data are expressed as the median and interquartile range. **P < 0·01, ***P < 0·001, Mann–Whitney U–test.
Comparative expression of T‐cell markers in the PB and liver of healthy individuals
The liver shows a higher frequency of CD8+ cells (66·1% versus 31·0% in PB, P < 0·001) and a reduced frequency of CD4+ cells (17·7% versus 61·5% in PB, P < 0·001), with an increased frequency of TCR‐γδ cells (10·8% versus 2·0% in PB, P = 0·01) (Fig. 2a). Notably, NKT cells from PB or liver samples showed similar frequencies of CD4, CD8 or TCR‐γδ T cells (Fig. 2b). These findings indicated that although NKT cells are a heterogeneous population, certain common characteristics are independently maintained in the tissue studied.
Figure 2.
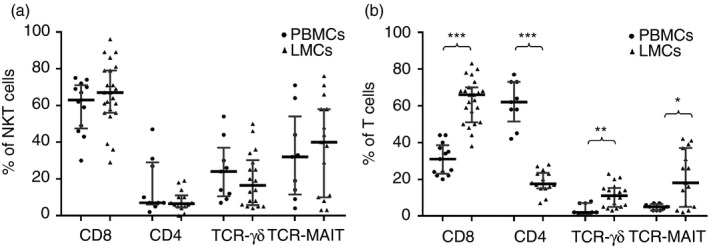
(a) CD4, CD8, T‐cell receptor‐γδ (TCR‐γδ) and TCR‐MAIT cell frequencies within CD3+ CD56+ natural killer T (NKT) cells from peripheral blood mononuclear cells (PBMCs) and liver mononculear cells (LMCs). (b) CD4+, CD8+, TCR‐γδ and TCR‐MAIT cell frequencies detected within CD3+ in PBMCs and LMCs. Data are expressed as the median and interquartile range. *P < 0·05, **P < 0·01, ***P < 0·001, Mann–Whitney U‐test.
In addition, we analysed the invariant receptor (TCR Vα7.2) of MAIT cells (mucosal‐associated invariant T‐cell antigen). In the liver, 18·4% of T cells expressed TCR‐MAIT versus 5·0% in PB, P < 0·01. This observation was expected because most MAIT cells are CD8+, consistent with the increased amount of liver CD8+ T cells, indicating that the composition of liver TCRs differs with respect to PB (Fig. 2a).
T‐cell markers from liver and PB samples identified different maturation stages
The drastically decreased CCR7 expression in liver T cells (11·1% versus 74·0% in PB, P < 0·01) was used to identify cells that lost their capacity to migrate to lymph nodes. Although the PB expression of CCR7 in NKT cells was low, its expression in liver NKT cells was even lower (0·7% versus 6·4% in PB, P < 0·001). The expression of HLA‐DR on CD8 T lymphocytes identified a particular subpopulation with a regulatory capacity.24 We observed significantly increased expression of HLA‐DR in both NKT liver cells (15·9% versus 4·0% in PB, P = 0·05) and T liver cells (19·7% versus 3·2% in PB, P = 0·01) (Fig. 3).
Figure 3.
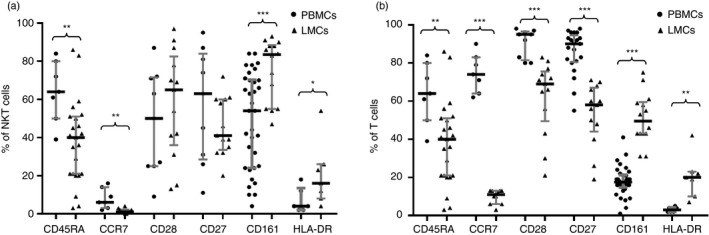
CD45RA, CCR7, CD28, CD27, CD161 and HLA‐DR cell frequencies within CD3+ CD56+ natural killer T (NKT) cells from peripheral blood mononuclear cells (PBMCs) and liver mononculear cells (LMCs) (a) and within CD3+ cells from PBMCs and LMCs. (b) Data are expressed as the median and interquartile range. *P < 0·05, **P < 0·01, ***P < 0·001, Mann–Whitney U‐test.
Additionally, liver T cells revealed decreased expression of CD45RA (31·3% versus 53·7% in PB, P < 0·01) and the co‐stimulatory molecules CD28 (68·7% versus 94·5% in PB, P < 0·001) and CD27 (58·0% versus 89·7% in PB, P < 0·001). Notably, CD161, a molecule with ascribed homing properties, was increased within liver NKT cells (83·7% versus 53·9% in PB, P < 0·001) and liver T cells (49·5% versus 17·5% in PB, P < 0·001) (Fig. 3). Expression of CD45RA, CD27, CD28 and CCR7 enabled characterization of different subsets of T cells: naive, effector cells (TE), central memory (TCM), transitional memory (TTM), effector memory (TEM) and terminally differentiated T cells. The strategy used to analyse T‐cell differentiation is depicted in Fig. 4, and the results are summarized in Table 1.
Figure 4.
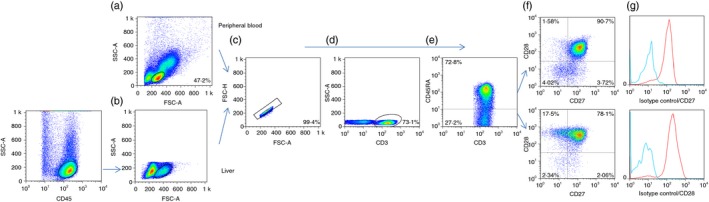
Dot plots showing the strategy used to analyse the T‐cell differentiation pattern. (a) Peripheral blood deduced from forward/side scatter. (b) The pan‐leucocyte CD45 marker was used to select the liver lymphocyte cloud. Illustrations in the followings dot plots correspond to analysis of peripheral blood. (c) Doublets were discarded. (d) The CD3+ population was selected, and (e) T cells were subdivided into CD45RA + and CD45RA − populations. (f) The combined presence of CD27 and CD28. The percentage of positive cells was calculated after subtracting the isotype control (g) and referred to the total CD3+ population. [Colour figure can be viewed at http://www.wileyonlinelibrary.com
Table 1.
T‐cell differentiation pattern
| T lymphocytes | T lymphocytes | P | |
|---|---|---|---|
| Peripheral blood | Liver | ||
| n = 7 | n = 7 | ||
| Median, % (range) | Median, % (range) | ||
| CCR7− | |||
| 45RA− 27− 28− | 0·6 (0·1–3·8) | 10·5 (4·6–16·2) | < 0·001 |
| 45RA+ 27+ 28+ | 3·1 (1·3–5·7) | 4·9 (0·9–8·0) | NS |
| 45RA− 27+ 28+ | 10·4 (5·5–15·2) | 38·7 (4·1–52·2) | < 0·05 |
| 45RA− 27− 28+ | 2·0 (0·7–3·0) | 13·8 (3·7–26·1) | < 0·001 |
| 45RA− 27+ 28− | 0·5 (0·1–2·5) | 3·3 (0·5–8·4) | < 0·01 |
| 45RA+ 27− 28+ | 0·1 (0·0–0·9) | 0·6 (0·4–2·0) | NS |
| 45RA+ 27+ 28− | 2·6 (0·5–5·0) | 6·2 (2·7–8·8) | < 0·05 |
| 45RA+ 27− 28− | 1·5 (0·5–13·6) | 14·6 (3·1–61·9) | < 0·05 |
| CCR7+ | |||
| 45RA− 27− 28− | 0·0 (0·0–0·1) | 0·0 (0·0–0·2) | NS |
| 45RA+ 27+ 28+ | 43·7 (29·1–67·0) | 3·2 (1·5–10·7) | < 0·001 |
| 45RA− 27+ 28+ | 33·7 (22·4–39·3) | 3·8 (1·7–5·9) | < 0·001 |
| 45RA− 27− 28+ | 1·6 (0·5–2·6) | 0·2 (0·1–3·9) | NS |
| 45RA− 27+ 28− | 0·0 (0·0–0·1) | 0·0 (0·0–0·6) | NS |
| 45RA+ 27− 28+ | 0·0 (0·0–0·1) | 0·0 (0·0–0·2) | NS |
| 45RA+ 27+ 28− | 0·2 (0·2–0·7) | 0·2 (0·1–1·3) | NS |
| 45RA+ 27− 28− | 0·0 (0·0–0·1) | 0·0 (0·0–2·6) | NS |
Comparison of the T‐cell differentiation pattern between peripheral blood (n = 7) and liver (n = 7) CD3+ cells. Phenotypes were separated into CCR7− (upper table) and CCR7+ (lower table) populations. Data are expressed as the median (range) of cells positive for the different T‐cell subpopulations according to the combined expression of CD45RA, CD27 and CD28 referred to as the total number of T lymphocytes. Mann–Whitney U‐test.
The liver showed a low frequency of CD45RA+ CD27+ CD28+ CCR7+ naive cells (3·2% versus 43·7% in PB, P < 0·001). The phenotype CD45RA− CD27+ CD28+ CCR7−, which represents a transitional stage between TCM and TEM cells, was increased in the liver (38·7% versus 10·4% in PB, P < 0·05). Interestingly, the TCM CD45RA− CD27+ CD28+ CCR7+, which represented 33·7% of PB T cells, was almost undetectable in the liver (3·8%, P < 0·001). Notably, all TEM cells were increased in the liver: CD45RA− CD27+ CD28− CCR7− (3·3% versus 0·5% in PB, P < 0·01); CD45RA− CD27− CD28+ CCR7− (13·8% versus 2·0% in PB, P < 0·001); and CD45RA− CD27− CD28− CCR7− (10·5% versus 0·6% in PB, P < 0·001). Finally, terminally differentiated T cells were also increased in the liver: CD45RA+ CD27+ CD28− CCR7− (6·2% versus 2·6% in PB, P < 0·05) and CD45RA+ CD27− CD28− CCR7− (14·6% versus 1·5% in PB, P < 0·05).
Different phenotype of liver and PB NK cells
NKG2A, one of the strongest NK inhibitory receptors, was detected in 89·4% of CD56bright NK cells in PB versus 56·3% of CD56bright NK cells in the liver (P < 0·001), suggesting a lower activation threshold in the liver (Fig. 5). Additionally, liver CD56bright NK cells showed a slight decrease in cell surface expression of their NKG2C‐activating counterparts (4·1% versus 16·6% in PB, P < 0·05), whereas liver CD56dim NK cells showed a significant increase of the activator receptor NKp44 (1·1% versus 0·1% in PB, P < 0·01) and decrease of the NK differentiation marker CD57 (8·9% versus 22·5% in PB, P < 0·01). Occasionally, it is difficult to differentiate liver CD56bright NK cells from CD56dim NK cells. The comparison between liver and blood CD56bright NK cells showed that CD16 expression was not differentially expressed in both subsets. By contrast, this difference was significant when we analysed CD56dim NK cells obtained from blood or liver samples (31·7% versus 92·0% in PB, P < 0·001; Fig. 6). Notably, the significantly decreased expression of CD16 on liver CD56dim NK cells supports the concept of the potentially decreased capacity of liver NK cells to perform antibody‐dependent cellular cytotoxicity (ADCC). In an attempt to further characterize the differences between PB and liver NK cells, we analysed the expression of CD11b and CD27, which identifies distinct stages of human NK cells from different sources.
Figure 5.
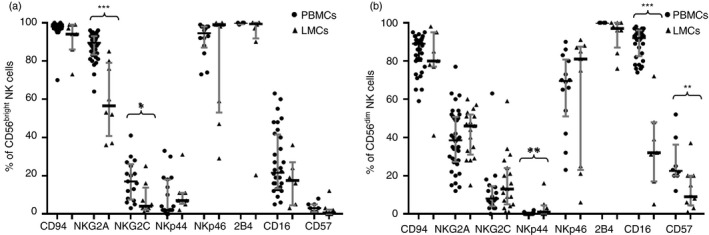
CD94, NKG2A, NKG2C, NKp44, NKp46, 2B4, CD16 and CD57 cell frequencies within CD56bright natural killer (NK) cells from peripheral blood mononuclear cells (PBMCs) and liver mononculear cells (LMCs) (a) and within CD56dim cells from PBMCs and LMCs (b). Data are expressed as the median and interquartile range. *P < 0·05, **P < 0·01, ***P < 0·001, Mann–Whitney U‐test.
Figure 6.
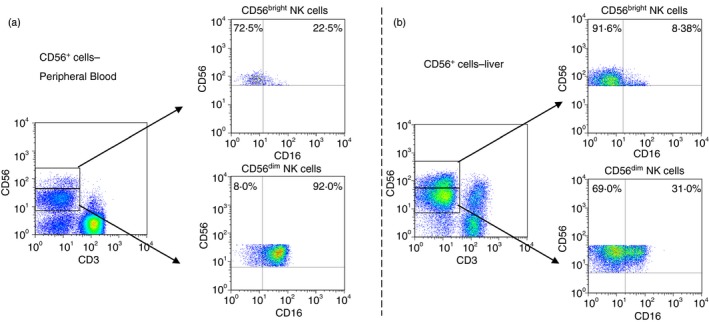
(a) Expression of CD16 on peripheral blood CD56bright and CD56dim natural killer (NK) cells, (b) shows the expression of CD16 on liver CD56bright and CD56dim NK cells. [Colour figure can be viewed at http://www.wileyonlinelibrary.com
As shown in Fig. 7, most liver and PB NK cells are CD11b+. However, liver CD56bright and CD56dim NK cells showed increased CD27+ expression (71·3% versus 30·0% in PB, P < 0·001 and 8·4% versus 4·6%, P < 0·05, respectively), suggesting that the liver has increased amounts of CD11b and CD27 double‐positive NK cells, which are associated with a higher secretory capacity.25 However, under a non‐specific stimulus, such as PMA‐ionomycin, we did not detect differences in the secretory or cytotoxic capacity between PB and liver NK cells (Fig. 9). Additionally, the lowest expression of CD45RA in liver CD56dim NK cells (96·8% versus 99·6%, P < 0·001) may indicate a previous antigenic experience.25
Figure 7.
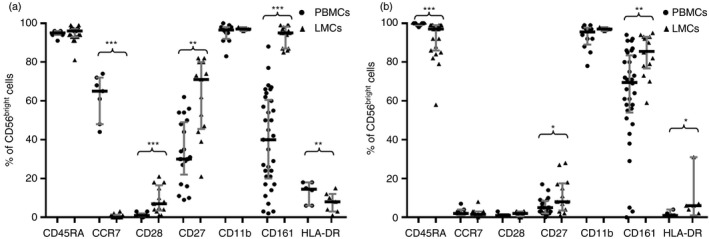
CD45RA, CCR7, CD28, CD27, CD11b, CD161 and HLA‐DR cell frequencies within CD56bright cells from peripheral blood mononuclear cells (PBMCs) (a) and within CD56dim NK cells from liver mononculear cells (LMCs) (b). Data are expressed as the median and interquartile range. *P < 0·05, **P < 0·01, ***P < 0·001, Mann–Whitney U‐test.
Figure 9.
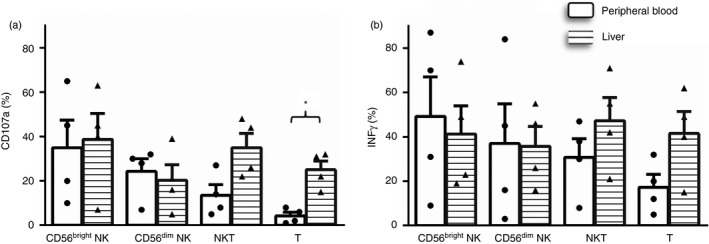
Comparative cytotoxic capacity measured as the expression of CD107a (a) and the secretory capacity (b) of liver (n = 4) and peripheral blood (n = 4) CD56bright, CD56dim, natural killer T (NKT) and total T cells. Figures show the percentage of positive cells. Data are presented as the means ± SEM. *P < 0·05, Mann–Whitney U test.
A feature of liver CD56bright NK cells is their dramatic loss of CCR7 (0·4% versus 65·1% in PB, P < 0·001), which may be responsible for their inability to migrate to the regional lymph node, as also recently described.26 However, a small but significant increase of CD28 was observed in liver CD56bright NK cells (6·8% versus 1·1% in PB, P = 0·001). HLA‐DR expression, also increased in liver CD56dim NK cells (5·8% versus 1·1% in PB, P < 0·05), may also be associated with their activated state.
Similar to liver NKT and T cells, the CD161 receptor showed high expression in liver CD56bright NK cells (95·2% versus 40·3% in PB, P < 0·001) and CD56dim NK cells: 85·8% versus 69·5%, P < 0·01 (Fig. 7, respectively. The functional role of CD161 is not clear because this protein has been described as both an activation marker and a homing marker and may also be expressed in cells with interleukin‐17 secretion capacity. The high expression of CD161 observed in liver NK, NKT and T cells may reflect its homing capacity.27
Expression of KIRs in PB and liver NK, NKT and T cells
We next compared expression of KIR genes in PB (n = 35), liver eluates (n = 7) and a few samples of mononuclear cells obtained from liver biopsies (the number of cells per sample enabled the analysis of only a few KIRs). The strategy used to analyse expression of KIRs is depicted in Fig. 8, and the results are summarized in Table 2. In PB, expression of the KIR genes in NK, NKT and T cells showed the following decreasing order CD56dim > NKT > CD56bright > T cells. In the liver, KIR expression was as follows: CD56dim > NKT > T > CD56bright cells (Table 2). In PB, the expression of KIRs on CD56bright NK cells never exceeded 10%, but liver CD56bright NK cells showed increased expression of KIR3DL2 (12·9% versus 4·1% in PB, P = 0·001) and KIR2DL3 (8·2% versus 1·7%, P < 0·01). As expected, expression of KIR genes was higher in CD56dim cells, with KIR2DS4 showing the highest expression in PB CD56dim cells (44·1% versus 22·2% in the liver, P < 0·01). Similar comparative analyses between liver and PB NKT cells revealed increased frequencies in the liver NKT cells of KIR2DL3 (15·9% versus 3·1%, P < 0·01) and KIR2DS1 (2·0% versus 0·0%, P < 0·01). Changes were obvious when we compared KIR expression on liver and PB T cells. Additionally, KIR expression on PB T cells was almost negligible, as most KIRs were detected in liver T cells: KIR2DL3 (12·9% versus 0·2%, P < 0·001), KIR3DL2 (8·6 versus 1·6%, P < 0·001), KIR2DS1 (1·0% versus 0·0%, P < 0·01), KIR2DS3 (3·1% versus 0·5%, P < 0·05) and KIR2DS4 (11·3% versus 0·4%, P < 0·001). A trend towards significance was also observed in KIR2DL2/2DS2. Interestingly, three of these increased KIR genes are activators, and KIR2DS4 together with the weaker inhibitory KIR2DL3 gene showed the highest expression (Table 2).
Figure 8.
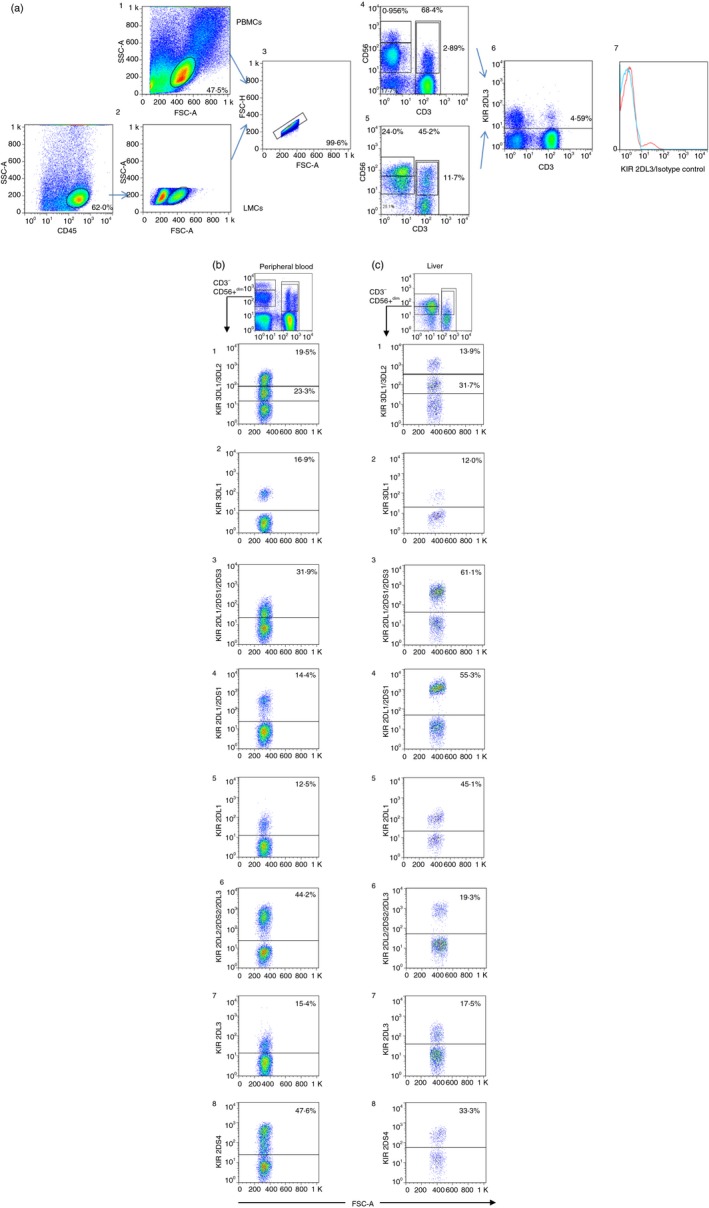
(a) Dot plots showing the strategy used to analyse the expression of killer cell immunoglobulin‐like receptors (KIRs) in peripheral blood (1) and liver CD45+ cells (2) deduced from FSC/SSC, Doublets were discarded (3). CD56bright, CD56dim, natural killer T (NKT) and T‐cell populations were selected in peripheral blood mononuclear cells (PBMCs) (4) and liver mononculear cells (LMCs) (5). (6) Illustrates an example of KIR2DL3 expression on total PBMCs and (7) after the subtraction of its isotype control. (b,c) The gating strategy for peripheral blood and liver NK, NKT and T cells. The following figures illustrate the KIR staining pattern of the different antibodies on CD56+ CD3− NK cells obtained from a single individual (one sample for peripheral blood and another sample for liver). b1 and c1 illustrate the staining pattern of the anti‐KIR3DL1/3DL2 antibody. The antibody showed two clouds with different mean fluorescence intensity values, where the lower cloud corresponds to 3DL2 and the upper cloud corresponds to 3DL1. The staining with anti‐KIR3DL1 shown in b2 and c2 confirmed that the frequency in the expression of the KIR 3DL1 antibody correlated with the upper cloud detected by the anti‐KIR3DL1/3DL2 antibody. The expression of 2DS3 was deduced from subtraction of expression between 2DL1/2DS1/2DS3, as shown in b3 and c3, and frequency of the expression of the anti‐2DL1/2DS1 illustrated in b4 and c4. Notably, expression of 2DL1/2DS1 in the liver sample showed a small difference with 2DL1/2DS1/2DS3 because the genotyping of this sample revealed the absence of the 2DS3 gene. b5 and c5 illustrates the expression of 2DL1 in peripheral blood and liver, respectively. b6 and c6 shows the expression of 2DL2/2DS2/2DL3, which can be compared with the expression of 2DL2/2DS2 obtained from subtraction of the signal detected using the anti‐2DL3 antibody (b7 and c7). b8 and c8 shows the expression of KIR2DS4. In all cases, the presence of the KIR genes was verified. [Colour figure can be viewed at http://www.wileyonlinelibrary.com
Table 2.
Expression of killer cell immunoglobulin‐like receptors
| NK CD56bright | NK CD56dim | NKT | T | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PB | Liver | PB | Liver | PB | Liver | PB | Liver | PB | Liver | |
| Median (range) | Median (range) | Median (range) | Median (range) | Median (range) | Median (range) | Median (range) | Median (range) | n | n | |
| 2DL1 | 1·7 (0·0–17·3) | 3·0 (1·0–20·1) | 16·1 (4·1–70·4) | 10·5 (5·5–45·1) | 3·8 (0·2–30·4) | 1·2 (0·3–7·7) | 0·4 (0·0–8·8) | 1·0 (0·2–3·7) | 34 | 7 |
| 2DL1/2DS1 | 2·5 (0·0–20·7) | 1·9 (0·4–33·8) | 17·5 (2·9–65·5) | 13·2 (8·6–55·3) | 1·1 (0·0–26·8) | 2·7 (1·2–11·4) | 0·0 (0·0–4·1) | 1·2*** (0·7–5·2) | 35 | 7 |
| 2DL1/2DS1/2DS3 | 4·0 (0·6–22·5) | 5·7 (2·0–27·0) | 19·9 (8·9–70·5) | 37·1 (9·7–61·1) | 2·7 (0·0–35·3) | 3·9 (1·1–53·0) | 0·2 (0·0–6·1) | 3·7*** (0·6–42·6) | 35 | 7 |
| 2DL2/2DS2/2DL3 | 3·7 (0·5–14·2) | 7·5 (2·8–9·4) | 31·4 (7·5–67·7) | 21·0 (12–36·8) | 12·8 (0·3–34·2) | 10·6 (1·9–75·0) | 1·6 (0·0–15·9) | 5·7*** (1·4–57·9) | 25 | 7 |
| 2DL3 | 1·7 (0·0–28·8) | 8·2** (1·4–15·9) | 11·9 (4·3–44·9) | 14·1 (2·6–46·2) | 3·1 (0·0–27·8) | 15·9** (2·5–50·4) | 0·2 (0·0–15·6) | 12·9*** (1·3–36·9) | 25 | 11 |
| 3DL1 | 2·2 (0·0–11·0) | 2·1 (0·7–18·4) | 16·4 (4·2–59·8) | 7·8 (1·7–36·4) | 5·9 (0·0–24·7) | 1·5 (0·2–15·6) | 0·7 (0·0–8·0) | 1·2 (0·2–8·1) | 31 | 6 |
| 3DL2a | 4·1 (0·0–18·1) | 12·9** (4·4–33·8) | 15·3 (4·9–44·3) | 31·2 (8·0–58·8) | 6·6 (0·2–45·3) | 15·6 (3·8–39·1) | 1·6 (0·0–12·6) | 8·6*** (4·2–36·2) | 35 | 7 |
| 2DS1a | 1·2 (0·0–15·4) | 0·0 (0·0–13·7) | 6·1 (0·7–19·1) | 8·7 (2·1–10·2) | 0·0 (0·0–1·9) | 2·0** (0·6–6·8) | 0·0 (0·0–0·2) | 1·0** (0·1–3·7) | 19 | 5 |
| 2DL2/2DS2a | 1·8 (0·0–13·2) | 0·7 (0·0–3·9) | 20·6 (2·9–35·6) | 7·4 (2·0–19·8) | 5·9 (0·0–23·1) | 5·3 (0·0–24·6) | 0·8 (0·0–2·6) | 3·1 (0·0–22·9) | 22 | 7 |
| 2DS3a | 3·2 (0·0–17·6) | 4·7 (0·0–8·0) | 14·5 (0·7–53·1) | 28·1 (4·0–28·5) | 3·6 (0·2–25·6) | 2·7 (0·0–44·9) | 0·5 (0·0–2·6) | 3·1* (0·1–38·6) | 15 | 4 |
| 2DS4 | 5·7 (0·1–21·1) | 7·3 (2·8–17·5) | 44·1 (5·9–69·5) | 22·2** (2·1–49·4) | 4·2 (1·2–32·5) | 12·6 (1·6–60·1) | 0·4 (0·0–6·3) | 11·3*** (1·2–45·5) | 16 | 12 |
Abbreviations: KIR, killer cell immunoglobulin‐like receptor; NK, natural killer; NKT, natural killer T; PB, peripheral blood.
Killer cell immunoglobulin‐like receptor expression in NK, NKT and T cells in peripheral blood and liver samples obtained from healthy individuals. Data are expressed as the median (range) of a specific marker within NK CD56bright, NK CD56dim, NKT and T populations. *P < 0·05, **P < 0·01, ***P < 0·001, Mann–Whitney U‐test.
Inferred KIR expression (see text).
Comparative analysis of the cytotoxic and secretor capacity of PB and liver T and NK cells from healthy subjects
The cytotoxic and secretory capacities of liver and PB T cells were measured by flow cytometry on PMA and ionomycin‐stimulated T cells. Liver CD3+ cells showed an increased frequency of CD107a+ cells as a marker of degranulation (25·0% versus 4·5% in PB, P = 0·03), which are associated with the increased frequency of CD8+ T cells, effector memory T cells and TCR γδ effector cells, representing CD3+ cells. Increased IFN‐γ secretion was also detected in liver T and NKT cells, but did not reach statistical significance, most likely because of the small number of experiments performed (Fig. 9).
It was speculated that decreased expression of NKG2A conferred a higher cytotoxic capacity on these cells. This pattern was not reflected in experiments in which PB and liver CD56bright and CD56dim NK cells showed similar levels of cytotoxicity and IFN‐γ secretor capacity (Fig. 9).
Discussion
The present study aimed to characterize and compare the frequencies, phenotypes and functions of NK, NKT and different subsets of T cells in PB and liver samples. Consistent with previous reports2, 22 we confirmed the increased frequency of intrahepatic NK cells, particularly CD56bright NK cells, which represent almost half of liver NK cells. These cells, as immediate precursors of CD56dim NK cells, are rare in PB, but constitute the majority of NK cells in secondary lymphoid tissues. Peripheral blood CD56bright NK cells are abundant cytokine producers, but are only weakly cytotoxic before activation.28 There are limited data regarding the recirculation of NK cells among human organs. Currently, the available data confirmed that human NK cells populate the blood, lymphoid organs, lung, liver, uterus (during pregnancy) and gut, following an NK cell homing pattern that appears to be subset‐specific.29 These data indicate that liver NK cells lack CCR7 expression, which could be necessary to migrate back to the lymph nodes, although these cells are able to interact with liver antigen‐presenting cells. Together with the loss of CCR7, liver CD56bright NK cells showed increased expression of CD27 and CD28. Most PB NK cells were CD56dim CD11b+ CD27− populations that exhibited a cytotoxic capacity, whereas NK cells from cord blood were CD11b+ CD27− and CD11b+ CD27+ populations. The most immature CD11b− CD27− NK subset represents the prevalent NK cells present in the decidua. Previous studies have reported that CD11b− CD27+ and CD11b+ CD27+ NK cells secrete cytokines.25 Data from the present and previous studies suggest that once NK cells migrate to the liver, these cells become tissue‐resident cells with particular features and activities. A previous study showed that some NK cells can be retained in the liver for up to 2 years.30 A recent study showed that some NK cells are retained for up to 13 years,3 and these long‐term resident NK cells represent an Eomeshi population,3, 31 which are unable to re‐enter the circulation and are long‐lived in the liver. In addition to the differences that we detected between liver and PB NK cells, the distribution and phenotype of NK cells across different human organs and tissues was markedly different. In the normal intestinal mucosa, NK cells show a phenotype similar to blood CD56bright NK cells.32, 33 NK cells represent ~ 10% of lymphocytes present in human normal lung and belong to the CD56dim CD16+ subset.34 As described above, the most immature CD11b− CD27−, CD56bright CD16neg subset is observed during the first trimester of pregnancy, representing 50–90% of the lymphoid cells that infiltrate this tissue.35 Increased expression of HLA‐DR in liver CD56dim NK cells could also be associated with the activation state of liver NK cells. At least in mice, activated NK cells expressing MHC class II have an antigenic presentation.36 Consistently, liver CD56bright NK cells also showed an increased frequency of NKp44, associated with decreased expression of NKG2A, which is one of the strongest NK inhibitory receptors. These results suggest that this subpopulation (or at least part of it) may represent activated NK cells, in coincidence with a previous study showing that liver‐resident CD56bright NK cells with reduced pro‐inflammatory potential had also enhanced degranulation activity.31 As previously discussed, it is occasionally difficult to discriminate between CD56bright and CD56dim NK cells in the liver. Although we achieved some discrimination between these two subsets based on differences in CD16 expression, we should be cautious with the results obtained in liver CD56bright cells, considering that some contamination with CD56dim cells may occur. In addition to the contamination of CD56dim into CD56bright cells, there can also be a contamination of CD56bright into CD56dim cells.
Differentiation of PB CD56dim NK cells was reported to involve acquisition of CD57, loss of NKG2A and gain of KIRs.37 In the present study, we observed that in comparison with PB, the NK differentiation marker CD57 is decreased in liver CD56dim NK cells, but the expression of NKG2A and KIR genes (with the exception of KIR2DS4) did not differ between PB and liver CD56dim NK cells. These results indicate that in contrast to CD56bright NK cells, the maturation state of liver and PB CD56dim NK cells was similar. The decreased expression of CD57 in liver CD56dim NK cells could indicate fewer terminally differentiated cells. We cannot exclude that decreased CD57 could be associated with a diminished memory capacity, as previously suggested.38, 39 In addition, we detected a strong decrease in the differentiation marker CD16, indicating a potential decreased capacity of liver NK cells to perform ADCC. Hence, liver CD56bright NK cells could more likely be associated with activation, whereas the ADCC capacity and maturation stage of CD56dim NK cells may be decreased.
Activation of NK cells is governed by the complex interplay of activating and inhibitory receptors. As inferred above, increased KIR expression on NK cells is associated with their maturation, and PB CD56bright NK cells showed low KIR gene expression. However, liver CD56bright NK cells showed a small but significant increase in KIR3DL2 and KIR2DL3 expression. In PB, expression of KIR genes was higher in CD56dim NK cells, with KIR2DS4 showing the highest surface expression. KIR2DS4 also showed the highest expression at the surface of liver CD56dim NK cells.
T cells expressing the semi‐invariant TCR (TCR Vα7.2‐Jα33) are highly abundant in humans. Within the liver, these cells represent approximately 20% of T cells. Although this increment is consistent with the increase of CD8+ T cells, liver TCR‐MAIT cells showed a fourfold increase compared with PB, whereas the frequency of CD8+ cells (which include MAIT cells) only showed a twofold increase, indicating that the TCR repertoire in the liver is different from that in PB. A subset of TCR‐γδ T cells also increased in the livers of healthy subjects. In contrast, NKT from PB or liver tissues showed similar frequencies of CD4, CD8 or TCR‐γδ T cells.
A main feature of liver T cells is the low frequency of naive T cells, and most of these cells have been ascribed to the effector memory phenotype CD45RA+ CD27− CD28− and CD45RA− CD27− CD28− T cells, with a greater capacity for degranulation and higher secretion of IFN‐γ. CD107a, a degranulation marker, is also considered to be a cytotoxicity marker. However, we should be cautious as previous studies4, 5 have reported that liver‐resident NK and T cells have the ability to degranulate but lack cytotoxic mediators.
Expression of the C‐type lectin CD161 in the liver was remarkable because, in contrast with PB, CD161 was expressed in most liver NK, NKT and T cells, Within the adult circulation, CD161 expression is restricted to T cells with a memory phenotype. Similarly, CD244 (2B4) is expressed by memory‐phenotype CD8+ T cells and all NK cells.40 The increased expression of these two receptors is consistent with the memory phenotype of liver T cells.
In contrast with the negligible expression in PB, a unique pattern of expression of KIR genes was observed in liver T cells. Expression of the activating KIR genes KIR2DS1, KIR2DS3 and KIR2DS4 has been associated with weaker expression of the inhibitory KIR2DL3 gene. It was previously reported that KIR expression could be induced after in vitro activation of a substantial fraction of terminally differentiated effector CD8 T cells.41, 42, 43 Additionally, genetics largely determines the expression of inhibitory receptors; activation receptors are heavily environmentally influenced.16 As previously reported,4 liver T cells primarily comprised effector memory and highly differentiated T cells. Hence, differential expression of KIR genes in liver T cells was expected, indicating their activation state. Although NK cells are the main KIR‐expressing immune cells, the presence of KIR on CD8 T cells was reported to enhance their HLA class I‐restricted antiviral effect.
We conclude that the liver has a large amount of CD8+ cells with a prevalence of memory and terminally differentiated phenotypes. The unique expression of KIR‐activating receptors with a high cytotoxic and functional capacity and decreased amount of CCR7 that are unable to migrate to the regional lymph node support the previously reported concept3, 4, 5 that liver T cells and NK cells are differentiated and activated cells that remain and die in the liver.
Disclosures
The authors have no financial conflicts of interest to disclose.
Supporting information
Figure S1. Entirely viable cells were selected from the lymphocyte cloud that had the highest FSC‐A and SSC‐A values.
Author contribution
AP, AM and SB performed the experiments, LF designed the study, LL, OI, SM, SM, OG and LGP provided samples, and LF and AP drafted the manuscript.
Acknowledgements
This work was financially supported by grants from the ANPCYT PICT2014 0925 and UBA grant 200201330100001. The authors would like to thank M Fernandez Viña for critical reading of the manuscript.
References
- 1. Racanelli V, Rehermann B. The liver as an immunological organ. Hepatology 2006; 43:S54–62. [DOI] [PubMed] [Google Scholar]
- 2. Doherty DG, O'Farrelly C. Innate and adaptive lymphoid cells in the human liver. Immunol Rev 2000; 174:5–20. [DOI] [PubMed] [Google Scholar]
- 3. Cuff AO, Robertson FP, Stegmann KA, Pallett LJ, Maini MK, Davidson BR et al Eomeshi NK cells in human liver are long‐lived and do not recirculate but can be replenished from the circulation. J Immunol 2016; 197:4283–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pallett LJ, Davies J, Colbeck EJ, Robertson F, Hansi N, Easom NJW et al IL‐2high tissue‐resident T cells in the human liver: sentinels for hepatotropic infection. J Exp Med 2017; 214:1567–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stegmann KA, Robertson F, Hansi N, Gill U, Pallant C, Christophides T et al CXCR6 marks a novel subset of T‐betlo Eomeshi natural killer cells residing in human liver. Sci Rep 2016; 6:26157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kelly A, Fahey R, Fletcher JM, Keogh C, Carroll AG, Siddachari R et al CD141+ myeloid dendritic cells are enriched in healthy human liver. J Hepatol 2014; 60:135–42. [DOI] [PubMed] [Google Scholar]
- 7. Tu Z, Bozorgzadeh A, Crispe IN, Orloff MS. The activation state of human intrahepatic lymphocytes. Clin Exp Immunol 2007; 149:186–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tian Z, Chen Y, Gao B. Natural killer cells in liver disease. Hepatology 2013; 57:1654–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Horst AK, Neumann K, Diehl L, Tiegs G. Modulation of liver tolerance by conventional and nonconventional antigen‐presenting cells and regulatory immune cells. Cell Mol Immunol 2016; 13:277–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Campbell KS, Purdy AK. Structure/function of human killer cell immunoglobulin‐like receptors: lessons from polymorphisms, evolution, crystal structures and mutations. Immunology 2011; 132:315–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Braud VM, Allan DS, O'Callaghan CA, Soderstrom K, D'Andrea A, Ogg GS et al HLA‐E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 1998; 391:795–9. [DOI] [PubMed] [Google Scholar]
- 12. Bryceson YT, March ME, Ljunggren HG, Long EO. Activation, coactivation, and costimulation of resting human natural killer cells. Immunol Rev 2006; 214:73–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mandelboim O, Malik P, Davis DM, Jo CH, Boyson JE, Strominger JL. Human CD16 as a lysis receptor mediating direct natural killer cell cytotoxicity. Proc Natl Acad Sci U S A 1999; 96:5640–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li H, Pascal V, Martin MP, Carrington M, Anderson SK. Genetic control of variegated KIR gene expression: polymorphisms of the bi‐directional KIR3DL1 promoter are associated with distinct frequencies of gene expression. PLoS Genet 2008; 4:e1000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yawata M, Yawata N, Draghi M, Partheniou F, Little AM, Parham P. MHC class I‐specific inhibitory receptors and their ligands structure diverse human NK‐cell repertoires toward a balance of missing self‐response. Blood 2008; 112:2369–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Horowitz A, Strauss‐Albee DM, Leipold M, Kubo J, Nemat‐Gorgani N, Dogan OC et al Genetic and environmental determinants of human NK cell diversity revealed by mass cytometry. Sci Transl Med 2013; 5:208ra145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pelak K, Need AC, Fellay J, Shianna KV, Feng S, Urban TJ et al Copy number variation of KIR genes influences HIV‐1 control. PLoS Biol 2011; 9:e1001208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaech SM, Wherry EJ. Heterogeneity and cell‐fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity 2007; 27:393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sallusto F, Geginat J, Lanzavecchia A. Central memory and effector memory T cell subsets: function, generation, and maintenance. Annu Rev Immunol 2004; 22:745–63. [DOI] [PubMed] [Google Scholar]
- 20. Golden‐Mason L, Douek DC, Koup RA, Kelly J, Hegarty JE, O'Farrelly C. Adult human liver contains CD8pos T cells with naive phenotype, but is not a site for conventional αβ T cell development. J Immunol 2004; 172:5980–5. [DOI] [PubMed] [Google Scholar]
- 21. Podhorzer A, Paladino N, Cuarterolo ML, Fainboim HA, Paz S, Theiler G et al The early onset of type 1 autoimmune hepatitis has a strong genetic influence: role of HLA and KIR genes. Genes Immun 2016; 17:187–92. [DOI] [PubMed] [Google Scholar]
- 22. Cosgrove C, Berger CT, Kroy DC, Cheney PC, Ghebremichael M, Aneja J et al Chronic HCV infection affects the NK cell phenotype in the blood more than in the liver. PLoS One 2014; 9:e105950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gao B, Radaeva S, Park O. Liver natural killer and natural killer T cells: immunobiology and emerging roles in liver diseases. J Leukoc Biol 2009; 86:513–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arruvito L, Payaslian F, Baz P, Podhorzer A, Billordo A, Pandolfi J et al Identification and clinical relevance of naturally occurring human CD8+HLA‐DR+ regulatory T cells. J Immunol 2014; 193:4469–76. [DOI] [PubMed] [Google Scholar]
- 25. Fu B, Wang F, Sun R, Ling B, Tian Z, Wei H. CD11b and CD27 reflect distinct population and functional specialization in human natural killer cells. Immunology 2011; 133:350–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hudspeth K, Donadon M, Cimino M, Pontarini E, Tentorio P, Preti M et al Human liver‐resident CD56bright/CD16neg NK cells are retained within hepatic sinusoids via the engagement of CCR5 and CXCR6 pathways. J Autoimmun 2016; 66:40–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fergusson JR, Fleming VM, Klenerman P. CD161‐expressing human T cells. Front Immunol 2011; 2:36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Poli A, Michel T, Theresine M, Andres E, Hentges F, Zimmer J. CD56bright natural killer (NK) cells: an important NK cell subset. Immunology 2009; 126:458–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Carrega P, Ferlazzo G. Natural killer cell distribution and trafficking in human tissues. Front Immunol 2012; 3:347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moroso V, Famili F, Papazian N, Cupedo T, van der Laan LJ, Kazemier G et al NK cells can generate from precursors in the adult human liver. Eur J Immunol 2011; 41:3340–50. [DOI] [PubMed] [Google Scholar]
- 31. Harmon C, Robinson MW, Fahey R, Whelan S, Houlihan DD, Geoghegan J et al Tissue‐resident Eomeshi T‐betlo CD56bright NK cells with reduced proinflammatory potential are enriched in the adult human liver. Eur J Immunol 2016; 46:2111–20. [DOI] [PubMed] [Google Scholar]
- 32. Chinen H, Matsuoka K, Sato T, Kamada N, Okamoto S, Hisamatsu T et al Lamina propria c‐kit+ immune precursors reside in human adult intestine and differentiate into natural killer cells. Gastroenterology 2007; 133:559–73. [DOI] [PubMed] [Google Scholar]
- 33. Cella M, Fuchs A, Vermi W, Facchetti F, Otero K, Lennerz JK et al A human natural killer cell subset provides an innate source of IL‐22 for mucosal immunity. Nature 2009; 457:722–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Carrega P, Morandi B, Costa R, Frumento G, Forte G, Altavilla G et al Natural killer cells infiltrating human nonsmall‐cell lung cancer are enriched in CD56bright CD16– cells and display an impaired capability to kill tumor cells. Cancer 2008; 112:863–75. [DOI] [PubMed] [Google Scholar]
- 35. King A, Balendran N, Wooding P, Carter NP, Loke YW. CD3– leukocytes present in the human uterus during early placentation: phenotypic and morphologic characterization of the CD56++ population. Dev Immunol 1991; 1:169–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nakayama M, Takeda K, Kawano M, Takai T, Ishii N, Ogasawara K. Natural killer (NK)‐dendritic cell interactions generate MHC class II‐dressed NK cells that regulate CD4+ T cells. Proc Natl Acad Sci U S A 2011; 108:18360–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bjorkstrom NK, Riese P, Heuts F, Andersson S, Fauriat C, Ivarsson MA et al Expression patterns of NKG2A, KIR, and CD57 define a process of CD56dim NK‐cell differentiation uncoupled from NK‐cell education. Blood 2010; 116:3853–64. [DOI] [PubMed] [Google Scholar]
- 38. Lopez‐Verges S, Milush JM, Pandey S, York VA, Arakawa‐Hoyt J, Pircher H et al CD57 defines a functionally distinct population of mature NK cells in the human CD56dimCD16+ NK‐cell subset. Blood 2010; 116:3865–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lopez‐Verges S, Milush JM, Schwartz BS, Pando MJ, Jarjoura J, York VA et al Expansion of a unique CD57+NKG2Chi natural killer cell subset during acute human cytomegalovirus infection. Proc Natl Acad Sci U S A 2011; 108:14725–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. McNerney ME, Lee KM, Kumar V. 2B4 (CD244) is a non‐MHC binding receptor with multiple functions on natural killer cells and CD8+ T cells. Mol Immunol 2005; 42:489–94. [DOI] [PubMed] [Google Scholar]
- 41. Snyder MR, Muegge LO, Offord C, O'Fallon WM, Bajzer Z, Weyand CM et al Formation of the killer Ig‐like receptor repertoire on CD4+CD28null T cells. J Immunol 2002; 168:3839–46. [DOI] [PubMed] [Google Scholar]
- 42. Young NT, Uhrberg M, Phillips JH, Lanier LL, Parham P. Differential expression of leukocyte receptor complex‐encoded Ig‐like receptors correlates with the transition from effector to memory CTL. J Immunol 2001; 166:3933–41. [DOI] [PubMed] [Google Scholar]
- 43. Anfossi N, Doisne JM, Peyrat MA, Ugolini S, Bonnaud O, Bossy D et al Coordinated expression of Ig‐like inhibitory MHC class I receptors and acquisition of cytotoxic function in human CD8+ T cells. J Immunol 2004; 173:7223–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Entirely viable cells were selected from the lymphocyte cloud that had the highest FSC‐A and SSC‐A values.


