Summary
The family of non‐classical major histocompatibility complex (MHC) class‐I like CD1 molecules has an emerging role in human disease. Group 1 CD1 includes CD1a, CD1b and CD1c, which function to display lipids on the cell surface of antigen‐presenting cells for direct recognition by T‐cells. The recent advent of CD1 tetramers and the identification of novel lipid ligands has contributed towards the increasing number of CD1‐restricted T‐cell clones captured. These advances have helped to identify novel donor unrestricted and semi‐invariant T‐cell populations in humans and new mechanisms of T‐cell recognition. However, although there is an opportunity to design broadly acting lipids and harness the therapeutic potential of conserved T‐cells, knowledge of their role in health and disease is lacking. We briefly summarize the current evidence implicating group 1 CD1 molecules in infection, cancer and autoimmunity and show that although CD1 are not as diverse as MHC, recent discoveries highlight their versatility as they exhibit intricate mechanisms of antigen presentation.
Keywords: CD1, cholesteryl‐esters, lipids, Mycobacterium tuberculosis, mycolic acid, T‐cells
Introduction
Cellular adaptive immunity in higher vertebrates is critically dependent on T‐cells and their specific interactions with antigen‐presenting molecules on the surface of antigen‐presenting cells (APCs), including dendritic cells, macrophages and B cells. The major families of antigen‐presenting molecules in mammals include the peptide‐binding major histocompatibility complex (MHC) class I and II molecules, which are highly polymorphic, butyrophillin 3A1 molecules, which are thought to mediate phosphoantigen‐sensing by γδ T‐cells,1 the non‐polymorphic MR1, which presents small metabolites,2, 3 and the lipid‐binding CD1 proteins.4 Of the five CD1 isotypes, CD1a, CD1b, CD1c and CD1d function to present lipid antigen at the cell surface to both αβ and γδ T‐cells (Fig. 1). CD1e is not expressed on the cell surface and functions as a lipid transfer protein.5 Group 1 CD1 comprise CD1a, CD1b and CD1c, which are not present in mice, whereas group 2 CD1 comprise CD1d, which is found in all mammals.4 Upon recognition of the CD1 ligand complex by the T‐cell receptor (TCR), CD1‐dependent T‐cells are activated in a variety of immunological contexts.
Figure 1.
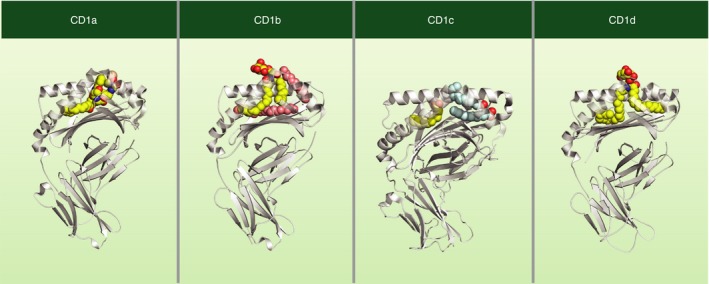
Structure of human CD1–ligand complexes. Each human CD1 protein has a unique lipid‐binding pocket architecture. CD1a is shown bound to a mycobacterial lipopeptide,60 CD1b is bound to a self‐phospholipid, phosphatidylinositol (PI) and two spacer lipids,61 CD1c is shown bound to spacer lipids (stearic and lauric acids),12 and CD1d is bound to α‐galactosylceramide.8
CD1‐restricted T‐cells can be activated by a range of self and non‐self lipids.6 Two broad mechanisms of CD1–lipid/TCR complex interactions have emerged as models of T‐cell activation. These include highly specific interactions of the antigen surface‐exposed polar head‐group with the TCR, as is the case for many of the CD1‐bound foreign bacterial lipids defined so far.7, 8 Another mechanism is governed by dominant interactions between the TCR and the CD1 protein, as is the case for the limited number of autoreactive group 1 CD1‐restricted TCRs described,9 and for high‐affinity human CD1d‐restricted TCRs.10 Two mechanisms exist for generating CD1 conformations that induce protein–protein interactions with the TCR. The first is absence of interference, as is the case for CD1a.11 The second is conformational remodelling, which is driven by ligand binding to CD1c,12 but this model requires further validation. These probably add to the many mechanisms driving CD1 autoreactive T‐cell activation that include increased CD1 expression, de novo synthesis of endogenous CD1 lipid antigens, altered cytokine secretion as well as the absence of steric and electrostatic TCR hindrance.13, 14, 15, 16, 17, 18 Understanding the basic mechanisms of CD1‐mediated T‐cell activation is essential to harness their potential in disease treatment. Here, we summarize evidence implicating CD1 molecules in a variety of disease states and review their unique features and emerging concepts of CD1–lipid presentation to T‐cells, highlighting the versatility of the CD1 system.
Group 1 CD1 elicit T‐cell responses in a variety of disease states
The presentation of lipid antigens by CD1 molecules on the cell surface represents a snapshot of the cell condition. Whether the cell is at homeostasis, infected or under stress; CD1 molecules can report these changes in the lipidome to T‐cells, and have increasingly recognized roles in many disease states.
Infection
The majority of foreign antigens that bind CD1a, CD1b and CD1c have so far been identified as mycobacterial cell‐wall‐associated lipids such as: mycolic acid,19 glucose monomycolate (GMM),7 glycerol monomycolate,20 isoprenoids,21 sulphoglycolipids,22 phosphmycoketides,23, 24 lipoarabinomannan,25 phosphatidyliniositol mannosides26 and dideoxymycobactin.27
CD1‐restricted T‐cells specific for mycobacterial antigens are well described, and some are being investigated for their role in host immunity against infection. For example, peripheral blood lymphocytes of Mycobacterium tuberculosis‐infected patients, but not those from healthy subjects, proliferate in response to CD1c‐presented mycobacterial phosphodolichols.21 T‐cell responses toward CD1b presented mycolates have been broadly characterized and are arguably the current focus of lipid specific T‐cell investigation in M. tuberculosis infection.7, 19, 28, 29, 30, 31, 32 Importantly, local expression of CD1b in M. tuberculosis‐infected lung granulomas from patients with tuberculosis implies the presentation of CD1b‐presented lipid antigens, including mycolates, within infected lesions.33 Furthermore, insights from lepromatous dermal lesions showed that lower CD1 expression correlated with a lack of effective cell‐mediated immunity.34 Once activated, it is possible that lipid‐specific T‐cells mediate protection, as they are capable of secreting the anti‐microbial cytokines interferon‐γ and tumour necrosis factor‐α, important for APC activation and granuloma formation, respectively, among other roles.28, 29, 35, 36 Additionally, polycytotoxic mycolic‐acid‐specific T‐cells isolated from bronchoalveolar fluid in tuberculosis patients are capable of limiting mycobacterial growth ex vivo.37 In a humanized CD1 mouse model expressing the mycolic‐acid‐specific TCR DN1, T‐cells were capable of reducing bacterial growth both in vitro and in vivo.31 Moreover, CD1b tetramers treated with mycobacterial GMM defined two populations of GMM‐specific T‐cells within the human CD1b‐restricted repertoire, the germline‐encoded mycolyl‐lipid‐reactive T‐cells (GEMs), and the LDN5‐like T‐cells. These T‐cells produce anti‐microbial cytokines and increase in numbers post infection with M. tuberculosis.28, 32 Indeed, the presence of conserved GEM T‐cells in humans with such exquisite specificity, represents possible selective pressure for lipid‐reactive T‐cells in mycobacterial infection.28
Other antigens displayed by CD1 include phospholipids, which are shared by both host and pathogens. T‐cells derived against Staphylococcus and Salmonella antigens were shown to prominently target CD1b presentation of phosphatidylglycerol and phosphatidylethanolamine.38 Although abundant in eukaryotic mitochondrial membranes, phospholipids such as phosphatidylglycerol and phosphatidylethanolamine are also highly expressed by bacterial membranes. As they are only available for loading onto CD1 under conditions of cell stress, damage and infection, the circumstances of their presentation represent a new mechanism of distinguishing between self and non‐self.38 Furthermore, solved co‐crystal structures prompted speculation that more abundant phospholipids sterically or electrostatically prevented CD1b‐restricted autoreactive TCRs from binding, expanding our knowledge regarding regulation of autoreactivity.39 In addition, lipopeptides are also targets for CD1‐restricted T‐cells. Dideoxymycobactin is a well‐characterized mycobacterial CD1a‐presented antigen that is specifically recognized by T‐cells.40 Lipo‐12, an N‐terminally acylated protein similar to the myrisolyated negative regulatory factor protein from human immunodeficiency virus, is also presented by CD1c.41 The recognition of such a ligand by T‐cells highlights the possibility that viral, ribosomal and mammalian acyl‐peptides could be presented by CD1 and demonstrates the flexibility of the CD1 system in reporting infectious disease to T‐cells.
Due to the tight regulation of group 1 CD1 expression on APC, involvement of CD1a, CD1b and CD1c in infection is confined to their expression. Central to regulating CD1 expression on professional APCs is Toll‐like receptors.18 Bacterial lipids and products trigger group 1 CD1 expression on dendritic cells through Toll‐like receptor 2 ligation.13, 42, 43, 44, 45 However, regulation of CD1 expression is clearly more complex due to the inconsistencies of expression on various cell types. Macrophages rarely express detectable levels of group 1 CD1; however, their expression in macrophages derived from human decidua, and on lipid‐laden foam cell macrophages in atherosclerotic tissue, suggest that other, as yet unknown, mechanisms determine CD1 expression.46, 47
Cancer
A role for CD1b‐restricted T‐cells in cancer has only recently been shown, which is highlighted in the double transgenic mouse model that expresses human group 1 CD1 molecules and the CD1b autoreactive HJ1 TCR. The CD1b autoreactive HJ1 T‐cells recognized several self phospholipids that are known to accumulate in tumours including phosphatidylethanolamine, phosphatidylglycerol, ether‐linked phosphatidylethanolamine, phosphatidylcholine and phosphatidylinositol.48, 49 HJ1 T‐cells were more potently activated by tumour‐derived self‐lipids than those isolated from healthy cells and were able to kill CD1b‐expressing tumours. They also exhibited anti‐tumour immunity in vivo, and their response was enhanced by various Toll‐like recptor agonists and by dendritic cell‐derived IL‐12 cytokine secretion. Given that there is little data on the CD1b autoreactive TCR repertoire, and the possible range of self‐antigens presented by CD1b, there is potential for CD1b to demonstrate further functions in cancer.
Recently, CD1c‐restricted T‐cell clones isolated from different donors recognized several CD1c expressing leukaemic cell lines in a CD1c‐dependent manner.50 Biochemical studies combined with T‐cell activity data identified methyl‐lysophosphatidic acids (mLPAs) as the CD1c‐presented antigen, which was specifically recognized by CD1c autoreactive T‐cells. mLPA is a self‐lipid antigen that accumulates in leukaemic cells including primary human leukaemia but not in healthy monocytes or B cells. It potently activated CD1c autoreactive T‐cell clones in vivo, limiting the growth of leukaemic cells in mice with human tumour cell xenografts.50 Therefore, CD1c‐mLPA‐specific TCRs are candidates for adoptive T‐cell therapy in acute myeloid leukaemia.
Autoimmunity and allergy
Presentation of self‐lipids to T‐cells represents a major function of the group 1 CD1 system. CD1 autoreactive T‐cells, particularly CD1a and CD1c, are abundant among circulating T‐cells from healthy human adults and neonates.51 In conditions manifesting in hyperthyroidism such as Graves’ disease and the autoimmune condition Hashimoto's thyroiditis, CD1 has a potential role. B cells positive for CD1c and dendritic cells expressing CD1a, CD1b and CD1c infiltrate afflicted thyroid glands, resulting in major tissue destruction and loss of structure. CD1 autoreactive T‐cells present within the thyroid are capable of lysing thyroid target cells in a CD1a‐ and CD1c‐dependent manner.52 In systemic lupus erythematosus, a systemic autoimmune condition of unknown origin, CD1c autoreactive T‐cells induce auto‐antibody isotype switching on CD1c‐expressing B cells, leading to an increase in IgM antibody production.53 Potential antigens such as the so‐called ‘headless antigens’ derived from self and presented by CD1a have been described, which are present in skin oils.51, 54 In psoriasis, an autoimmune skin condition, neolipid antigens are generated by phospholipase A2 (PLA2), which is found in active psoriasis lesions, implying a pathogenic role in inflammation through CD1a antigen presentation.55 Allergic skin conditions such as atopic dermatitis may also become exacerbated by house dust mite allergen PLA2 through generation of CD1a‐presented neolipid antigens that cause inflammation.56 Interestingly, filaggrin, a skin barrier protein, inhibits PLA2 and therefore prevents the development of atopic dermatitis in healthy individuals.56 Furthermore, PLA2 is also found in bee and wasp venom that, once delivered subcutaneously, generates a local inflammatory response.57 The recent understanding of CD1a presentation of headless antigens that mediate inflammatory skin disorders led to the hypothesis that urushiol, found in poison ivy, can elicit a T‐cell response in a CD1a‐dependent manner.58 These studies also demonstrated a role for CD1a in driving psoriatic skin inflammation, and provided a potential therapy for inflammatory skin conditions through targeting CD1a.58
CD1 lipid antigen presentation therefore, is associated with a wide variety of diseases. New data are consistent with an important role for CD1a‐, CD1b‐ and CD1c‐restricted T‐cells in human immunity to infection, cancer and autoimmune disease. The small number of CD1‐restricted T‐cell clones and their biased selection in culture have contributed to our limited knowledge of their overall function in disease. In addition, little attention has been placed upon the translational potential of CD1 and the creation of novel therapeutic treatments. Greater use of humanized mouse models and tetramers for direct ex vivo T‐cell analysis will aid our understanding of the function of the CD1 system in health and disease.
Unique features allow diverse lipid sampling
CD1 structure
CD1 molecules consist of a heavy chain comprising the α1–α3 domains, which non‐covalently bind the light chain of β 2‐microglubulin59 (Fig. 1). Structural insights of CD1 proteins displaying glycolipids reveal that the antigen‐binding function of CD1 is embedded within the α1 and α2 domains, which harbour hydrophobic channels that are suited for binding the lipidic parts of CD1 antigens. Polar moieties of lipid antigens are positioned above the binding pocket at the TCR interface for direct recognition by T‐cells. CD1 isotypes have evolved antigen‐binding pockets of differing shapes and sizes, presumably to allow them to bind and present a large number of structurally diverse lipids.8, 12, 60, 61 Insights from crystal structures and molecular modelling have greatly contributed to our current understanding of how CD1 proteins bind to a broad range of lipids and glycolipids.
All CD1 proteins have an A’ and an F’ channel, whereas CD1b has an additional T’ tunnel and a C’ channel.61 In CD1b, the interconnected A’, T’ and F’ channels form a superchannel, which is a unique feature of human CD1b that enables the presentation of the very long mycolic acids of mycobacteria and nocardia.62 The C’ channel has a C’ portal under the α2 helix, which may allow exit of longer hydrocarbon chains. CD1a in comparison, has the smallest groove comprising a short A’ channel that limits the length of alkyl chains able to bind, and it has a less restrictive F’ channel, which accommodates larger lipid moieties.60, 63 The A’ and F’ channels of CD1d are ideally suited for binding ceramide lipids such as α‐galactosylceramide (α‐GC), a potent agonist of invariant natural killer T‐cells.8 Lastly, CD1c features an A’ and an F’ channel12, 64 which can present diverse lipid antigens, including cholesteryl‐esters to T‐cells. The F’ channel of CD1c adopts dramatically different ‘open’ and ‘closed’ conformations depending on ligand occupation.12 Hence, each CD1 isotype has a set of unique antigen‐binding channels highlighting their ability to bind diverse lipid structures.
CD1 localization
CD1 ensures presentation of a broad range of lipids by surveying distinct cellular subcompartments. Upon assembly in the endoplasmic reticulum, CD1 molecules initially bind polar and non‐polar self‐lipids, believed to function as stabilizing chaperones.65 These include sphingomyelins and phospholipids,66, 67, 68 and in the case of CD1b, multiple scaffold ligands are bound.65 Upon correct folding in the endoplasmic reticulum, CD1 complexes travel through the secretory pathway directly to the plasma membrane. Once expressed, they are internalized into lysosomal compartments where they can encounter self and bacterial lipid antigens before recycling back to the plasma membrane.69 The continuous recycling of CD1 molecules from the plasma membrane to lysosomal compartments is regulated by tyrosine‐containing motifs in their cytoplasmic tails, with the exception of CD1a, which lacks such a motif.69, 70 CD1a localizes predominantly in the early endosomes, and does not require the low pH conditions for lipid loading. CD1b primarily locates to the late endosomes and lysosomes, where the acidic conditions regulate lipid loading by altering ionic tethers in the F’ portal.70, 71, 72, 73 CD1c and CD1d recycle to early and late endosomes, as a result of binding adaptor protein 2 (AP‐2) via their cytoplasmic tails.74, 75 Additionally, while CD1a, CD1b, CD1c and CD1d are expressed by myeloid dendritic cells,76 they all have niche cellular and tissue expression patterns. CD1d is constitutively expressed on many haematopoietic and non‐haematopoietic cells including epithelial cells.76, 77 Whereas CD1a, CD1b and CD1c expression is restricted to professional APCs. CD1c and CD1d are expressed by B cells including leukaemic lymphoma cells.50, 78 CD1a and to a lesser extent CD1c are expressed by skin‐resident Langerhans cells.74, 77, 79 The unique intracellular trafficking routes and cellular expression patterns within different tissues is consistent with a specialized role for each CD1 isotype.
Diverse mechanisms of antigen display to T‐cells
CD1‐restricted TCRs recognize a broad range of lipids when bound to CD1 proteins through diverse mechanisms. The TCRs of invariant natural killer T‐cells for example, predominantly bind CD1d, whereas the polar head group of the lipid modulates the interaction. Moreover, recent reports suggest that ligands bound to CD1 can modulate TCR interaction even when they are not contacted directly by the TCR.38, 48, 80 Here we describe the possible modes of recognition of CD1 by T‐cells and mechanisms behind the range of activity that T‐cells display to CD1 antigens.
The recognition of CD1–lipid complexes by TCRs is regulated by a number of mechanisms. The first and most typical is through T‐cell specificity for the large lipid head‐group moieties protruding above the CD1 protein, visible to the TCR.7, 81, 82, 83, 84 The best characterized example is recognition of GMMs by CD1b‐restricted TCRs.7, 85 Here, even the smallest alteration in hydroxyl group orientation on the carbohydrate moiety abrogates TCR binding. This specificity for the head group was recently demonstrated at the molecular level by the structure determination of a GEM TCR in complex with CD1b‐GMM. The GEM TCR employs an elegant ‘tweezer’‐like mechanism that grips the glucose head‐group moiety of GMM.85 Another example includes the presentation of mannosyl‐β1‐phosphomycoketide (MPM) by CD1c. The presence of the polar mannosyl moiety is recognized by the T‐cell line CD8‐1, whose activity is lost upon cleavage of the sugar.21, 86 The discrimination of different head groups is a classical mechanism for differentiating between different lipid ligands by TCRs.
CD1 can also communicate changes in lipid repertoires to T‐cells through a mechanism coined ‘absence of interference’. Identification of hundreds of small polar and non‐polar CD1a lipid antigens revealed the so‐called ‘permissive’ or ‘non‐permissive’ ligands for the CD1a autoreactive BC2 T‐cells.11 Generally, permissive ligands were headless antigens as they lacked a polar head‐group moiety, and were derived from skin oils including squalene and wax esters. CD1a autoreactive T‐cells were activated in the presence of CD1a bound to headless lipids and not those with large polar head groups.54 Subsequent co‐crystal structures revealed that the autoreactive BK6 TCR binds the A’ roof without directly contacting the bound lipid cargo.11
Conformational remodelling is also employed by CD1 as a mechanism thought to facilitate autoreactive TCR binding.77 Initial CD1c structures were solved, bound to MPM and subsequently phosphomycoketide, which are lipids derived from the cell wall of M. tuberculosis.24, 64 These highly methylated and branched lipid molecules sat within the A’ channel, leaving the F’ groove unoccupied, containing only a small spacer lipid likely to have originated from the insect cells from which CD1c was produced. Key features of these CD1c structures included their open F’ groove, reminiscent of the peptide‐binding grooves of MHC molecules, and two portals, designated D’ and E’, connecting the A’ and F’ channels to the exterior, respectively.24, 64 A more recent study reported an entirely different conformation of CD1c, whereby a lipid‐saturated F’ channel induced extensive remodelling of the protein forming an F’ roof above the F’ channel and a G’ portal, open on the right side of the F’ channel with the apparent loss of the E’ portal.12 Furthermore, aromatic self‐ligands including cholesteryl‐esters, were able to induce this remodelling, which is apparently critical for the binding of some autoreactive TCRs.12 Structural insights of CD1c in complex with autoreactive TCRs will shed further light onto this novel mechanism.
Buried hydrophobic moieties translate distinct T‐cell activities
Identifying mechanisms of ligand recognition by crystal structures are indispensable, but only describe a snapshot of the interaction. However, to activate T‐cells, the engagement of the TCR with the CD1–lipid complex must occur over time. Therefore, activation is not only a function of binding affinity and avidity, but also of complex dynamics.87
T‐cell activity can be influenced by architecture of the CD1 groove and subtle changes in the structural composition of lipid ligands including the fine structure of lipid tails. This concept was first implied in 2002 with crudely separated classes of mycolates. Stimulation of the mycolic‐acid‐specific and CD1b‐restricted DN1 TCR with α‐keto‐ and methoxy classes of mycolate revealed the sensitivity of the TCR towards the fine structure of the long meromycolate tails of mycolic acids.88 Furthermore, MPMs with differing methyl‐branching patterns influence the activation levels of CD8‐1 T‐cells when presented by CD1c.84 Another class of lipid, mycobacterial sulphoglycolipids, were shown to alter the potency of the T‐cell response when the aliphatic hydrocarbon chains were altered.89 This effect was evident when the number of C‐methyl substituents within the fatty acid, the positioning and the stereochemistry of the functional groups were changed.89 The invariant natural killer T‐cell TCR also differentiated between CD1d bound α‐GC, when the length of the phytosphingosine chain and the extent of saturation was changed.90 These studies suggest that CD1‐restricted TCRs are not only influenced by lipid polar head‐group moieties, but also the fine structure of hydrophobic moieties deeply buried within the CD1 proteins, which are in all cases assumed to be not in direct contact with the TCR.
Until recently, a role for the distal or proximal functional groups of the meromycolate tails of mycolic acids as antigenic determinants were not well understood.88 By treating soluble CD1b molecules with extremely insoluble mycolic acids, Van Rhijn et al.19 showed definitively, a direct impact of distal meromycolate functional group alteration on TCR recognition. Synthetic representatives of the α‐, keto‐ and methoxy‐mycolic acids that appear on the cell wall of M. tuberculosis show clear differential activity on a variety of CD1b‐mycolic acid‐specific T‐cell clones and lines when presented by CD1b. Furthermore, CD1b tetramers were treated with two forms of mycolic acid classes, the keto‐ and methoxy‐ and tetramer staining revealed differing patterns of binding to T‐cells. Interestingly, each of the T‐cell clones recognized distinct classes of mycolates leading to the conclusion that each class of mycolic acid should be considered as separate antigens. Most recently, work investigating GEM TCR activity toward an extensive panel of synthetic mycolates based on natural structures, corroborated this, also showing that both proximal and distal meromycolate functional groups impact T‐cell responses.33
A putative mechanism for the differential recognition of hydrocarbon chains, which are expected to lie outside direct contact by the TCR, was alluded to by computational modelling simulations of lipid–protein interactions.33 These data indicated that meromycolate chain dynamics within the CD1b groove appear to be directly linked to the activity of GEM18 TCR (Fig. 2). The result is a range of T‐cell activities from one TCR against a number of subtly altered mycolic acids of the same class. This model suggests that weakly stimulatory lipid tails are immobile due to the position and nature of their chain substituents caused by functional group ‘trapping’ in hydrophobic crevices within the binding channels of CD1b. This may restrict the head group from adopting positions that facilitate TCR binding. In contrast, strongly stimulatory lipid chain substituents do not ‘catch’ on pocket features and therefore remain fluid. The apparent combination of greater chain fluidity and reduced head‐group mobility may allow conformations that are more productive for GEM18 TCR binding. The differing activity of diverse CD1b‐restricted mycolate‐specific TCRs may be because they have different modes of engagement and interaction with the CD1b‐mycolic acid complex, and therefore preferentially bind to different classes of mycolic acid that adopt different head‐group positions.19 However, this model does not account for the ‘combinatorial epitope’ hypothesis, which suggests apparently different conformations of CD1b protein and lipid such that the meromycolate functional groups are displayed directly to the TCR.88 However, given the similarity of lipid structure of mycolic acid to GMM, whose crystal structure has been resolved when bound to CD1b, the required major conformational change is hard to envisage.62 Mutational and structural studies as well as specifically designed mycolic acid analogues will reveal the mechanism behind such enigmatic TCR binding requirements. Elucidation of these will help contribute to the design of lipid vaccines and potential therapies.
Figure 2.
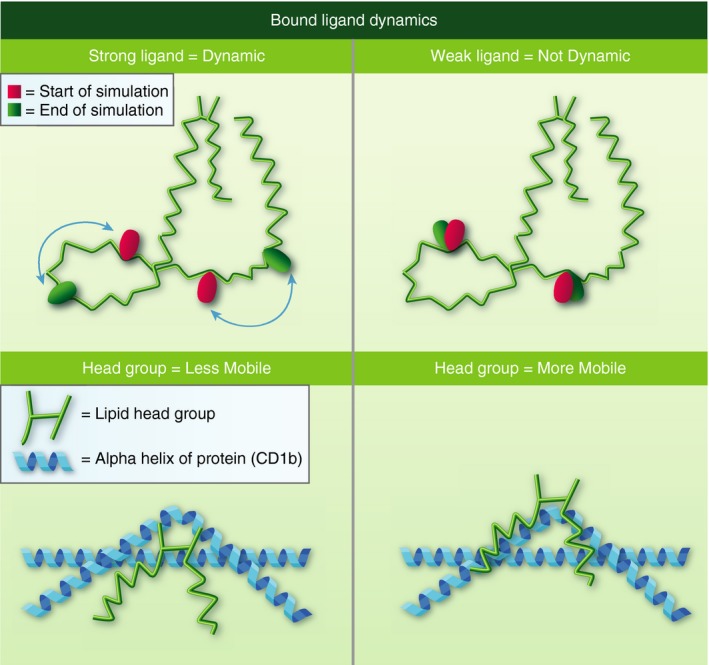
Putative dynamics model of CD1b‐bound mycolic acids (MAs). Representation of the molecular simulation of mycolic acids when bound to CD1b showing the chain functional groups at the start of simulation (red spheres) and at the end of simulation (green spheres). The model predicts that MAs immunogenic for GEM18 TCR have meromycolate chains that are highly fluid within the CD1b pocket whereas less immunogenic chains are not fluid due to functional group trapping within crevices found in the CD1b pocket (upper panels). Immunogenic lipids have head groups that are less mobile and ‘sink’ into position to allow GEM18 TCR recognition, whereas less immunogenic lipids have head groups that are mobile or ‘unstable’, which do not position correctly to allow productive GEM18 TCR recognition (lower panels).33
Summary
The discovery that CD1 is a TCR binding epitope,44 was shortly followed by the recognition that lipid antigens are capable of eliciting specific T‐cell responses, when presented by CD1.81 MHC molecules are highly polymorphic and have the capacity to bind and present a vast number of peptide sequences. This results in the generation of an almost infinite number of binding epitopes that can be recognized specifically by TCRs. On the other hand, CD1 molecules are non‐polymorphic, therefore CD1‐restricted TCRs do not have the luxury of such diverse binding sites or variety of bound lipids. Despite this, CD1 molecules prove highly versatile in lipid presentation and play a role in a number of disease conditions, employing diverse mechanisms to elicit a variety of specific T‐cell responses. Recent insights suggest multiple modes of TCR engagement to CD1–lipid complexes – head‐group discrimination, absence of interference and protein remodelling. Moreover, the hydrophobic groove architecture can communicate subtle differences in lipid structure to T‐cells, modulating their responses. These features highlight the plasticity of CD1 antigen presentation and supports evolved mechanisms that paint a complete picture of the cellular lipid environment to T‐cells. Ongoing emphasis on translational research within CD1 will contribute towards the effort to derive new therapeutic targets in cancer, autoimmunity and infectious disease.
Disclosures
The authors declare no conflict of interest.
Acknowledgements
The authors thank Paul Elkington and Marc Tebruegge for advice. This work was supported by Cancer Research UK (23562).
References
- 1. Karunakaran MM, Herrmann T. The Vγ9Vδ2 T cell antigen receptor and Butyrophilin‐3 A1: models of interaction, the possibility of co‐evolution, and the case of dendritic epidermal T cells. Front Immunol 2014; 5:648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lepore M, Kalinichenko A, Calogero S, Kumar P, Paleja B, Schmaler M et al Functionally diverse human T cells recognize non‐microbial antigens presented by MR1. Elife 2017; 6:pii: e24476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kjer‐Nielsen L, Patel O, Corbett AJ, Le Nours J, Meehan B, Liu L et al MR1 presents microbial vitamin B metabolites to MAIT cells. Nature 2012; 491:717–23. [DOI] [PubMed] [Google Scholar]
- 4. Mori L, Lepore M, De Libero G. The immunology of CD1‐ and MR1‐restricted T cells. Annu Rev Immunol 2016; 34:479–510. [DOI] [PubMed] [Google Scholar]
- 5. Garcia‐Alles LF, Giacometti G, Versluis C, Maveyraud L, de Paepe D, Guiard J et al Crystal structure of human CD1e reveals a groove suited for lipid‐exchange processes. Proc Natl Acad Sci U S A 2011; 108:13230–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Van Rhijn I, Moody DB. CD1 and mycobacterial lipids activate human T cells. Immunol Rev 2015; 264:138–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Moody DB, Reinhold BB, Guy MR, Beckman EM, Frederique DE, Furlong ST et al Structural requirements for glycolipid antigen recognition by CD1b‐restricted T cells. Science 1997; 278:283–6. [DOI] [PubMed] [Google Scholar]
- 8. Koch M, Stronge VS, Shepherd D, Gadola SD, Mathew B, Ritter G et al The crystal structure of human CD1d with and without α‐galactosylceramide. Nat Immunol 2005; 6:819–26. [DOI] [PubMed] [Google Scholar]
- 9. Van Rhijn I, Godfrey DI, Rossjohn J, Moody DB. Lipid and small‐molecule display by CD1 and MR1. Nat Rev Immunol 2015; 15:643–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Matulis G, Sanderson JP, Lissin NM, Asparuhova MB, Bommineni GR, Schümperli D et al Innate‐like control of human iNKT cell autoreactivity via the hypervariable CDR3β loop. PLoS Biol 2010; 8:e1000402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Birkinshaw RW, Pellicci DG, Cheng TY, Keller AN, Sandoval‐Romero M, Gras S et al αβ T cell antigen receptor recognition of CD1a presenting self lipid ligands. Nat Immunol 2015; 16:258–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mansour S, Tocheva AS, Cave‐Ayland C, Machelett MM, Sander B, Lissin NM et al Cholesteryl esters stabilize human CD1c conformations for recognition by self‐reactive T cells. Proc Natl Acad Sci USA 2016; 113:E1266–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roura‐Mir C, Wang L, Cheng TY, Matsunaga I, Dascher CC, Peng SL et al Mycobacterium tuberculosis regulates CD1 antigen presentation pathways through TLR‐2. J Immunol 2005; 175:1758–66. [DOI] [PubMed] [Google Scholar]
- 14. Ishikawa E, Ishikawa T, Morita YS, Toyonaga K, Yamada H, Takeuchi O et al Direct recognition of the mycobacterial glycolipid, trehalose dimycolate, by C‐type lectin Mincle. J Exp Med 2009; 206:2879–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Paget C, Mallevaey T, Speak AO, Torres D, Fontaine J, Sheehan KC et al Activation of invariant NKT cells by toll‐like receptor 9‐stimulated dendritic cells requires type I interferon and charged glycosphingolipids. Immunity 2007; 27:597–609. [DOI] [PubMed] [Google Scholar]
- 16. Brigl M, Bry L, Kent SC, Gumperz JE, Brenner MB. Mechanism of CD1d‐restricted natural killer T cell activation during microbial infection. Nat Immunol 2003; 4:1230–7. [DOI] [PubMed] [Google Scholar]
- 17. Brennan PJ, Tatituri RV, Brigl M, Kim EY, Tuli A, Sanderson JP et al Invariant natural killer T cells recognize lipid self antigen induced by microbial danger signals. Nat Immunol 2011; 12:1202–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Moody DB. TLR gateways to CD1 function. Nat Immunol 2006; 7:811–7. [DOI] [PubMed] [Google Scholar]
- 19. Van Rhijn I, Iwany SK, Fodran P, Cheng TY, Gapin L, Minnaard AJ et al CD1b‐mycolic acid tetramers demonstrate T‐cell fine specificity for mycobacterial lipid tails. Eur J Immunol 2017; 47:1525–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Layre E, Collmann A, Bastian M, Mariotti S, Czaplicki J, Prandi J et al Mycolic acids constitute a scaffold for mycobacterial lipid antigens stimulating CD1‐restricted T cells. Chem Biol 2009; 16:82–92. [DOI] [PubMed] [Google Scholar]
- 21. Moody DB, Ulrichs T, Mühlecker W, Young DC, Gurcha SS, Grant E et al CD1c‐mediated T‐cell recognition of isoprenoid glycolipids in Mycobacterium tuberculosis infection. Nature 2000; 404:884–8. [DOI] [PubMed] [Google Scholar]
- 22. Garcia‐Alles LF, Collmann A, Versluis C, Lindner B, Guiard J, Maveyraud L et al Structural reorganization of the antigen‐binding groove of human CD1b for presentation of mycobacterial sulfoglycolipids. Proc Natl Acad Sci USA 2011; 108:17755–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Matsunaga I, Bhatt A, Young DC, Cheng TY, Eyles SJ, Besra GS et al Mycobacterium tuberculosis pks12 produces a novel polyketide presented by CD1c to T cells. J Exp Med 2004; 200:1559–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roy S, Ly D, Li NS, Altman JD, Piccirilli JA, Moody DB et al Molecular basis of mycobacterial lipid antigen presentation by CD1c and its recognition by αβ T cells. Proc Natl Acad Sci U S A 2014; 111:E4648–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sieling PA, Chatterjee D, Porcelli SA, Prigozy TI, Mazzaccaro RJ, Soriano T et al CD1‐restricted T cell recognition of microbial lipoglycan antigens. Science 1995; 269:227–30. [DOI] [PubMed] [Google Scholar]
- 26. Fischer K, Scotet E, Niemeyer M, Koebernick H, Zerrahn J, Maillet S et al Mycobacterial phosphatidylinositol mannoside is a natural antigen for CD1d‐restricted T cells. Proc Natl Acad Sci USA 2004; 101:10685–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Young DC, Kasmar A, Moraski G, Cheng TY, Walz AJ, Hu J et al Synthesis of dideoxymycobactin antigens presented by CD1a reveals T cell fine specificity for natural lipopeptide structures. J Biol Chem 2009; 284:25087–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Van Rhijn I, Kasmar A, de Jong A, Gras S, Bhati M, Doorenspleet ME et al A conserved human T cell population targets mycobacterial antigens presented by CD1b. Nat Immunol 2013; 14:706–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Montamat‐Sicotte DJ, Millington KA, Willcox CR, Hingley‐Wilson S, Hackforth S, Innes J et al A mycolic acid‐specific CD1‐restricted T cell population contributes to acute and memory immune responses in human tuberculosis infection. J Clin Invest 2011; 121:2493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moody DB, Briken V, Cheng TY, Roura‐Mir C, Guy MR, Geho DH et al Lipid length controls antigen entry into endosomal and nonendosomal pathways for CD1b presentation. Nat Immunol 2002; 3:435–42. [DOI] [PubMed] [Google Scholar]
- 31. Zhao J, Siddiqui S, Shang S, Bian Y, Bagchi S, He Y et al Mycolic acid‐specific T cells protect against Mycobacterium tuberculosis infection in a humanized transgenic mouse model. Elife 2015; 4:pii: e08525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Van Rhijn I, Gherardin NA, Kasmar A, de Jager W, Pellicci DG, Kostenko L et al TCR bias and affinity define two compartments of the CD1b‐glycolipid‐specific T Cell repertoire. J Immunol 2014; 192:4054–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chancellor A, Tocheva AS, Cave‐Ayland C, Tezera L, White A, Al Dulayymi JR et al CD1b‐restricted GEM T cell responses are modulated by Mycobacterium tuberculosis mycolic acid meromycolate chains. Proc Natl Acad Sci U S A 2017; 114:E10956–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sieling PA, Jullien D, Dahlem M, Tedder TF, Rea TH, Modlin RL et al CD1 expression by dendritic cells in human leprosy lesions: correlation with effective host immunity. J Immunol 1999; 162:1851–8. [PubMed] [Google Scholar]
- 35. Flynn JL, Chan J, Triebold KJ, Dalton DK, Stewart TA, Bloom BR. An essential role for interferon γ in resistance to Mycobacterium tuberculosis infection. J Exp Med 1993; 178:2249–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kindler V, Sappino AP, Grau GE, Piguet PF, Vassalli P. The inducing role of tumor necrosis factor in the development of bactericidal granulomas during BCG infection. Cell 1989; 56:731–40. [DOI] [PubMed] [Google Scholar]
- 37. Busch M, Herzmann C, Kallert S, Zimmermann A, Höfer C, Mayer D et al Lipoarabinomannan‐responsive polycytotoxic T cells are associated with protection in human tuberculosis. Am J Respir Crit Care Med 2016; 194:345–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Van Rhijn I, van Berlo T, Hilmenyuk T, Cheng TY, Wolf BJ, Tatituri RV et al Human autoreactive T cells recognize CD1b and phospholipids. Proc Natl Acad Sci USA 2016; 113:380–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shahine A, Van Rhijn I, Cheng TY, Iwany S, Gras S, Moody DB et al A molecular basis of human T cell receptor autoreactivity toward self‐phospholipids. Sci Immunol 2017; 2:pii: eaao1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cheng JM, Liu L, Pellicci DG, Reddiex SJ, Cotton RN, Cheng TY et al Total synthesis of Mycobacterium tuberculosis Dideoxymycobactin‐838 and Stereoisomers: diverse CD1a‐restricted T cells display a common hierarchy of lipopeptide recognition. Chemistry 2017; 23:1694–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Van Rhijn I, Young DC, De Jong A, Vazquez J, Cheng TY, Talekar R et al CD1c bypasses lysosomes to present a lipopeptide antigen with 12 amino acids. J Exp Med 2009; 206:1409–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Krutzik SR, Tan B, Li H, Ochoa MT, Liu PT, Sharfstein SE et al TLR activation triggers the rapid differentiation of monocytes into macrophages and dendritic cells. Nat Med 2005; 11:653–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Yakimchuk K, Roura‐Mir C, Magalhaes KG, de Jong A, Kasmar AG, Granter SR et al Borrelia burgdorferi infection regulates CD1 expression in human cells and tissues via IL1‐β . Eur J Immunol 2011; 41:694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Porcelli S, Morita CT, Brenner MB. CD1b restricts the response of human CD4–8– T lymphocytes to a microbial antigen. Nature 1992; 360:593–7. [DOI] [PubMed] [Google Scholar]
- 45. Inkeles MS, Teles RM, Pouldar D, Andrade PR, Madigan CA, Lopez D et al Cell‐type deconvolution with immune pathways identifies gene networks of host defense and immunopathology in leprosy. JCI Insight 2016; 1:e88843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guerin L, Wu V, Houser B, Tilburgs T, de Jong A, Moody DB et al CD1 antigen presentation and autoreactivity in the pregnant human uterus. Am J Reprod Immunol 2015; 74:126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Melian A, Geng YJ, Sukhova GK, Libby P, Porcelli SA. CD1 expression in human atherosclerosis. A potential mechanism for T cell activation by foam cells. Am J Pathol 1999; 155:775–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bagchi S, Li S, Wang CR. CD1b‐autoreactive T cells recognize phospholipid antigens and contribute to antitumor immunity against a CD1b+ T cell lymphoma. Oncoimmunology 2016; 5:e1213932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Daly PF, Zugmaier G, Sandler D, Carpen M, Myers CE, Cohen JS. Regulation of the cytidine phospholipid pathways in human cancer cells and effects of 1‐β‐d‐arabinofuranosylcytosine: a noninvasive 31P nuclear magnetic resonance study. Cancer Res 1990; 50:552–7. [PubMed] [Google Scholar]
- 50. Lepore M, de Lalla C, Gundimeda SR, Gsellinger H, Consonni M, Garavaglia C et al A novel self‐lipid antigen targets human T cells against CD1c+ leukemias. J Exp Med 2014; 211:1363–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. de Jong A, Peña‐Cruz V, Cheng TY, Clark RA, Van Rhijn I, Moody DB. CD1a‐autoreactive T cells are a normal component of the human αβ T cell repertoire. Nat Immunol 2010; 11:1102–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Roura‐Mir C, Catálfamo M, Cheng TY, Marqusee E, Besra GS, Jaraquemada D et al CD1a and CD1c activate intrathyroidal T cells during Graves’ disease and Hashimoto's thyroiditis. J Immunol 2005; 174:3773–80. [DOI] [PubMed] [Google Scholar]
- 53. Sieling PA, Porcelli SA, Duong BT, Spada F, Bloom BR, Diamond B et al Human double‐negative T cells in systemic lupus erythematosus provide help for IgG and are restricted by CD1c. J Immunol 2000; 165:5338–44. [DOI] [PubMed] [Google Scholar]
- 54. de Jong A, Cheng TY, Huang S, Gras S, Birkinshaw RW, Kasmar AG et al CD1a‐autoreactive T cells recognize natural skin oils that function as headless antigens. Nat Immunol 2014; 15:177–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Cheung KL, Jarrett R, Subramaniam S, Salimi M, Gutowska‐Owsiak D, Chen YL et al Psoriatic T cells recognize neolipid antigens generated by mast cell phospholipase delivered by exosomes and presented by CD1a. J Exp Med 2016; 213:2399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Jarrett R, Salio M, Lloyd‐Lavery A, Subramaniam S, Bourgeois E, Archer C et al Filaggrin inhibits generation of CD1a neolipid antigens by house dust mite‐derived phospholipase. Sci Transl Med 2016; 8:325ra318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bourgeois EA, Subramaniam S, Cheng TY, De Jong A, Layre E, Ly D et al Bee venom processes human skin lipids for presentation by CD1a. J Exp Med 2015; 212:149–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kim JH, Hu Y, Yongqing T, Kim J, Hughes VA, Le Nours J et al CD1a on Langerhans cells controls inflammatory skin disease. Nat Immunol 2016; 17:1159–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zeng Z, Castaño AR, Segelke BW, Stura EA, Peterson PA, Wilson IA. Crystal structure of mouse CD1: an MHC‐like fold with a large hydrophobic binding groove. Science 1997; 277:339–45. [DOI] [PubMed] [Google Scholar]
- 60. Zajonc DM, Crispin MD, Bowden TA, Young DC, Cheng TY, Hu J et al Molecular mechanism of lipopeptide presentation by CD1a. Immunity 2005; 22:209–19. [DOI] [PubMed] [Google Scholar]
- 61. Gadola SD, Zaccai NR, Harlos K, Shepherd D, Castro‐Palomino JC, Ritter G et al Structure of human CD1b with bound ligands at 2.3 A, a maze for alkyl chains. Nat Immunol 2002; 3:721–6. [DOI] [PubMed] [Google Scholar]
- 62. Batuwangala T, Shepherd D, Gadola SD, Gibson KJ, Zaccai NR, Fersht AR et al The crystal structure of human CD1b with a bound bacterial glycolipid. J Immunol 2004; 172:2382–8. [DOI] [PubMed] [Google Scholar]
- 63. Zajonc DM, Elsliger MA, Teyton L, Wilson IA. Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 A. Nat Immunol 2003; 4:808–15. [DOI] [PubMed] [Google Scholar]
- 64. Scharf L, Li NS, Hawk AJ, Garzón D, Zhang T, Fox LM et al The 2.5 a structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation. Immunity 2010; 33:853–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Huang S, Cheng TY, Young DC, Layre E, Madigan CA, Shires J et al Discovery of deoxyceramides and diacylglycerols as CD1b scaffold lipids among diverse groove‐blocking lipids of the human CD1 system. Proc Natl Acad Sci U S A 2011; 108:19335–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Cox D, Fox L, Tian R, Bardet W, Skaley M, Mojsilovic D et al Determination of cellular lipids bound to human CD1d molecules. PLoS ONE 2009; 4:e5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fox LM, Cox DG, Lockridge JL, Wang X, Chen X, Scharf L et al Recognition of lyso‐phospholipids by human natural killer T lymphocytes. PLoS Biol 2009; 7:e1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Haig NA, Guan Z, Li D, McMichael A, Raetz CR, Xu XN. Identification of self‐lipids presented by CD1c and CD1d proteins. J Biol Chem 2011; 286:37692–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Jayawardena‐Wolf J, Benlagha K, Chiu YH, Mehr R, Bendelac A. CD1d endosomal trafficking is independently regulated by an intrinsic CD1d‐encoded tyrosine motif and by the invariant chain. Immunity 2001; 15:897–908. [DOI] [PubMed] [Google Scholar]
- 70. Cernadas M, Cavallari M, Watts G, Mori L, De Libero G, Brenner MB. Early recycling compartment trafficking of CD1a is essential for its intersection and presentation of lipid antigens. J Immunol 2010; 184:1235–41. [DOI] [PubMed] [Google Scholar]
- 71. Relloso M, Cheng TY, Im JS, Parisini E, Roura‐Mir C, DeBono C et al pH‐dependent interdomain tethers of CD1b regulate its antigen capture. Immunity 2008; 28:774–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Sugita M, Jackman RM, van Donselaar E, Behar SM, Rogers RA, Peters PJ et al Cytoplasmic tail‐dependent localization of CD1b antigen‐presenting molecules to MIICs. Science 1996; 273:349–52. [DOI] [PubMed] [Google Scholar]
- 73. Jackman RM, Stenger S, Lee A, Moody DB, Rogers RA, Niazi KR et al The tyrosine‐containing cytoplasmic tail of CD1b is essential for its efficient presentation of bacterial lipid antigens. Immunity 1998; 8:341–51. [DOI] [PubMed] [Google Scholar]
- 74. Sugita M, van Der Wel N, Rogers RA, Peters PJ, Brenner MB. CD1c molecules broadly survey the endocytic system. Proc Natl Acad Sci USA 2000; 97:8445–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Briken V, Jackman RM, Watts GF, Rogers RA, Porcelli SA. Human CD1b and CD1c isoforms survey different intracellular compartments for the presentation of microbial lipid antigens. J Exp Med 2000; 192:281–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Dougan SK, Kaser A, Blumberg RS. CD1 expression on antigen‐presenting cells. Curr Top Microbiol Immunol 2007; 314:113–41. [DOI] [PubMed] [Google Scholar]
- 77. Moody DB, Cotton RN. Four pathways of CD1 antigen presentation to T cells. Curr Opin Immunol 2017; 46:127–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Allan LL, Stax AM, Zheng DJ, Chung BK, Kozak FK, Tan R et al CD1d and CD1c expression in human B cells is regulated by activation and retinoic acid receptor signaling. J Immunol 2011; 186:5261–72. [DOI] [PubMed] [Google Scholar]
- 79. Milne P, Bigley V, Gunawan M, Haniffa M, Collin M. CD1c+ blood dendritic cells have Langerhans cell potential. Blood 2015; 125:470–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Roy S, Ly D, Castro CD, Li NS, Hawk AJ, Altman JD et al Molecular analysis of lipid‐reactive Vδ1 γδ T cells identified by CD1c tetramers. J Immunol 2016; 196:1933–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Beckman EM, Porcelli SA, Morita CT, Behar SM, Furlong ST, Brenner MB. Recognition of a lipid antigen by CD1‐restricted αβ + T cells. Nature 1994; 372:691–4. [DOI] [PubMed] [Google Scholar]
- 82. Gilleron M, Stenger S, Mazorra Z, Wittke F, Mariotti S, Böhmer G et al Diacylated sulfoglycolipids are novel mycobacterial antigens stimulating CD1‐restricted T cells during infection with Mycobacterium tuberculosis . J Exp Med 2004; 199:649–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Moody DB, Young DC, Cheng TY, Rosat JP, Roura‐Mir C et al T cell activation by lipopeptide antigens. Science 2004; 303:527–31. [DOI] [PubMed] [Google Scholar]
- 84. de Jong A et al CD1c presentation of synthetic glycolipid antigens with foreign alkyl branching motifs. Chem Biol 2007; 14:1232–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Gras S, Arce EC, Cheng TY, van Summeren RP, Feringa BL, Dudkin V et al T cell receptor recognition of CD1b presenting a mycobacterial glycolipid. Nat Commun 2016; 7:13257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Ly D, Kasmar AG, Cheng TY, de Jong A, Huang S, Roy S et al CD1c tetramers detect ex vivo T cell responses to processed phosphomycoketide antigens. J Exp Med 2013; 210:729–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Baker BM, Scott DR, Blevins SJ, Hawse WF. Structural and dynamic control of T‐cell receptor specificity, cross‐reactivity, and binding mechanism. Immunol Rev 2012; 250:10–31. [DOI] [PubMed] [Google Scholar]
- 88. Grant EP, Beckman EM, Behar SM, Degano M, Frederique D, Besra GS et al Fine specificity of TCR complementarity‐determining region residues and lipid antigen hydrophilic moieties in the recognition of a CD1–lipid complex. J Immunol 2002; 168:3933–40. [DOI] [PubMed] [Google Scholar]
- 89. Guiard J, Collmann A, Garcia‐Alles LF, Mourey L, Brando T, Mori L et al Fatty acyl structures of mycobacterium tuberculosis sulfoglycolipid govern T cell response. J Immunol 2009; 182:7030–7. [DOI] [PubMed] [Google Scholar]
- 90. McCarthy C, Shepherd D, Fleire S, Stronge VS, Koch M, Illarionov PA et al The length of lipids bound to human CD1d molecules modulates the affinity of NKT cell TCR and the threshold of NKT cell activation. J Exp Med 2007; 204:1131–44. [DOI] [PMC free article] [PubMed] [Google Scholar]


