Summary
Commensal microbes and the host immune system have been co‐evolved for mutual regulation. Microbes regulate the host immune system, in part, by producing metabolites. A mounting body of evidence indicates that diverse microbial metabolites profoundly regulate the immune system via host receptors and other target molecules. Immune cells express metabolite‐specific receptors such as P2X7, GPR41, GPR43, GPR109A, aryl hydrocarbon receptor precursor (AhR), pregnane X receptor (PXR), farnesoid X receptor (FXR), TGR5 and other molecular targets. Microbial metabolites and their receptors form an extensive array of signals to respond to changes in nutrition, health and immunological status. As a consequence, microbial metabolite signals contribute to nutrient harvest from diet, and regulate host metabolism and the immune system. Importantly, microbial metabolites bidirectionally function to promote both tolerance and immunity to effectively fight infection without developing inflammatory diseases. In pathogenic conditions, adverse effects of microbial metabolites have been observed as well. Key immune‐regulatory functions of the metabolites, generated from carbohydrates, proteins and bile acids, are reviewed in this article.
Keywords: barrier function, bile acids, immunity, indole, inflammation, metabolites, microbiome, short‐chain fatty acids
Introduction
Commensal microbiota functionally maturate the host immune system. This effect is mediated by microbial factors that stimulate host cells. Microbial factors work through a variety of host receptors and molecular targets on or within host cells. Host receptors for microbial factors include pattern recognition receptors such as Toll‐like receptors, nucleotide oligomerization receptors (NLRs), C‐type lectin receptors and RIG‐1‐like receptors, which sense major microbial constituents such as DNA, RNA, proteins and cell wall components. However, these microbial constituents are just a fraction of what commensal microbes produce to regulate the host immune system. In the alimentary tract, host enzymes process dietary materials, such as starch, dietary fibres, proteins/peptides and lipids for nutrients harvest in the small intestine. However, some dietary materials inevitably reach the colon for microbial fermentation due to incomplete digestion and absorption. These materials feed the microbiota for production of a variety of microbial metabolites, some of which have immune regulatory roles. It is thought that there are 500–1000 microbial operating taxonomic units in the mammalian gut. While each microbial species has a finite number of genes that code for nutrient transporters and metabolizing enzymes, the combined number of these genes for the whole microbiota would be enormous.1 Moreover, the microbiota employ a wide variety of nutrient‐utilizing enzymes in numbers that far exceed that of the human enzyme repertoire. Because the products of microbial enzymes are absorbed in the colon and utilized by the host, the gut microbiota increase nutrient or energy recovery from consumed diets. Microbial metabolites have significant effects on the host metabolism.2, 3, 4 Moreover, some of the metabolites also condition and activate the immune system to increase the immune function but decrease harmful inflammatory responses.5, 6, 7, 8
The concentrations of certain microbial metabolites, best exemplified by short‐chain fatty acids (SCFAs), reach very high concentrations in the colon,9 lowering pH, fulfilling nutritional needs, regulating microbial function and composition, and conditioning the immune system. Host cells express various receptors to sense microbial metabolites, which include purinergic receptors such as P2X7 to detect microbial and host‐derived nucleotides [e.g. adenosine triphosphate (ATP) and nicotinamide adenine dinucleotide (NAD)];10 GPR43 and GPR41 to detect SCFAs; membrane bile acid receptor (M‐BAR/TGR5) and farnesoid X receptor (FXR) to detect bile acid and xenobiotic metabolites;11, 12 and aryl hydrocarbon receptor precursor (AhR) and pregnane X receptor (PXR) to detect tryptophan, indole, bile acid and toxicant metabolites.13, 14, 15 Moreover, the roles of dietary factors that activate immune cells, including certain phytochemicals, even without microbial modifications should not be ignored. This article is to review our current understanding of the origin, host receptors and target cells of major microbial metabolites in the immune system. Detailed impacts of these metabolites on the innate and adaptive arms of the immune system are discussed.
Major groups of microbial metabolites and their receptors
Carbohydrate metabolites
Bacteria produce SCFAs as the result of carbohydrate fermentation in the colon (Fig. 1). Bacteria express carbohydrate‐active enzymes, such as glycoside hydrolases and polysaccharide lyases.16, 17 These enzymes often form enzyme complexes (e.g. cellulosomes) to process long carbohydrate fibres into simple sugars.18 Released component sugars, such as glucose and xylose, are transported into bacteria via the phosphotransferase system for uptake and phosphorylation of carbohydrates for fermentation.19 While the human genome encodes 17 enzymes to breakdown carbohydrate nutrients, some gut bacteria such as Bacteroides thetaiotaomicron encode over 260 glycoside hydrolases.20 Because there are a few hundred bacterial and yeast species in the gut, the total number of carbohydrate‐active enzymes and their overall combined capacity to handle different dietary fibres are expected to be sufficient to handle most of the consumed polysaccharides. While both soluble and insoluble dietary fibres can be processed by bacteria, soluble fibres such as arabinoxylan, pectin, inulin and hemicellulose are preferentially utilized to produce SCFAs in the gut over insoluble fibres such as cellulose and chitin.21 Also, digestion‐resistant oligosaccharides, such as fructooligosaccharide and xylooligosaccharide, and resistant starches along with host glycoproteins, such as mucins, can be processed to produce SCFAs. Microbes greatly differ in their ability to ferment dietary fibres and sugars to produce different SCFAs. In general, members of the Bacteroidetes phylum are good producers of acetate (C2) and propionate (C3), whereas bacteria in the Firmicutes phylum are efficient butyrate (C4) producers.22 More specifically, Akkermansia municiphilla produces C3 from mucin.23 Roseburia inulinivorans and Coprococcus catus produce both C3 and C4.24, 25, 26 Faecalibacterium prausnitzii, Eubacterium rectale, Eubacterium hallii and Ruminococcus bromii are good producers of C4.27 Also, Roseburia intestinalis, Eubacterium rectale and Clostridium symbiosum are good C4‐producers and are increased in numbers with a high‐fibre diet.28 Ruminococcus bromii produces C4 from resistant starch.29 SCFAs are absorbed by colonocytes and other cells via solute transporters and simple diffusion. SLC16a1 and SLC5a8 are major transporters for SCFAs.8 C4 is mainly used by colonocytes, whereas C2 and C3 are transported out to the portal circulation. C2 and C3 are transported to the liver, muscle, brain and other organs. C2 is converted into acetyl‐CoA for lipogenesis or oxidation in peripheral muscles. Most C3 is metabolized in the liver and contributes to gluconeogenesis. SCFAs affect the metabolism of host cells, activating multiple metabolic pathways to produce energy and building blocks and regulating host metabolism.30, 31 Also produced by microbes are lactate and succinate, which can be converted to C3 by many bacterial species.32 SCFAs activate several cell surface G‐protein‐coupled receptors (GPCRs), such as GPR43, GPR41, GPR109A and Olfr78.33, 34, 35, 36 GPR43 and GPR41 are highly expressed by intestinal epithelial cells.37 T‐ and B‐cells do not express these SCFA receptors, but certain myeloid cells, such as neutrophils, macrophages and dendritic cells (DCs), express GPR43 and GPR109A at variable levels to sense the concentration of SCFAs in tissue environments.33, 34, 38, 39, 40
Figure 1.
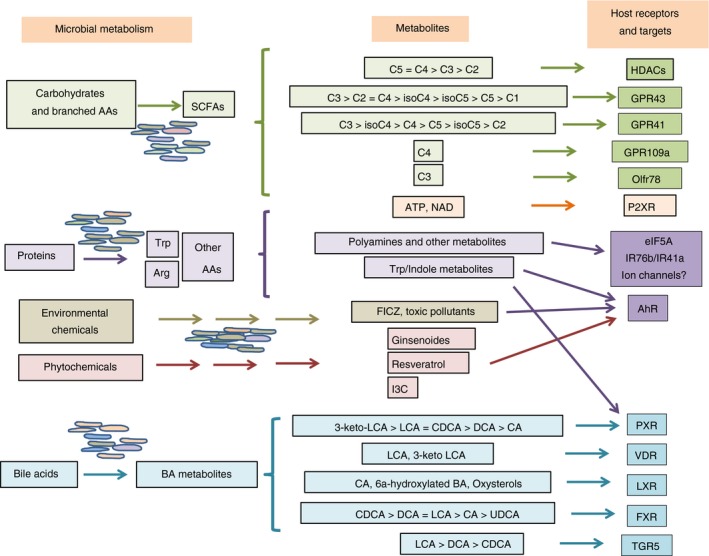
Production of microbial metabolites and their major receptors in the immune system. The gut microbiota can metabolize a variety of dietary materials, which include carbohydrates, proteins, lipids, plant‐derived molecules, bile acids and environmental contaminants. These materials are metabolized into short‐chain fatty acids (SCFAs), polyamines, ATP, indoles, phytochemical metabolites and bile acid metabolites. SCFAs function as histone deacetylase (HDAC) inhibitors to regulate gene expression and activate G‐protein‐coupled receptors (GPCRs) such as GPR43, GPR41, GPR109A (C4) and Olfr78 (C3). Other metabolites collectively activate nuclear receptors [aryl hydrocarbon receptor precursor (AhR), pregnane X receptor (PXR), VDR, LXR and farnesoid X receptor (FXR)], TGR5 and P2XRs. These receptors are expressed by various cells in the innate and adaptive immune systems to sense the presence of the gut microbial metabolites.
Amino acid and related metabolites
Another abundant group of dietary materials includes proteins. Proteins are digested into oligopeptides and amino acids, which are largely absorbed in the small intestine. Some proteins, oligopeptides and amino acids, not processed or absorbed in the small intestine, reach the colon for bacterial catabolism and utilization. Many bacterial groups, including certain Clostridium, Bacillus, Lactobacillus, Streptococcus and Proteobacteria groups, are effective metabolizers of proteins, causing ‘protein putrefaction’.41 Among amino acids, glycine, lysine, arginine, leucine, isoleucine and valine are preferred amino acid substrates for gut bacteria.42 Microbial catabolism of these amino acids generates ammonia, biogenic amines (monoamines and polyamines) and other metabolites. Polyamines are produced from ornithine, arginine, lysine and methionine.43 Decarboxylation of amino acids generates histamine (from histidine), agmatine (from arginine) and cadaverine (from lysine).44 For example, Lactobacillus plantarum decarboxylates ornithine to produce putrescine, a major polyamine.45 Moreover, branched‐chain SCFAs, such as isobutyrate, valerate and isovalerate, are produced from respective branched amino acids (i.e. leucine, valine and isoleucine).46 Similar to C4, branched‐chain SCFAs are potent histone deacetylase (HDAC) inhibitors and, therefore, their functions in regulating host cells are expected to be similar to that of C4. The luminal concentrations of branched‐chain SCFAs are relatively lower than those of the major SCFAs (C2–C4).
Indole is produced from tryptophan and metabolized into kynurenine, indole‐3‐acetic acid and tryptamine. Bacterial trytophanase produces indole, and host‐produced indoleamine‐pyrrole 2,3‐dioxygenase (IDO) generates kynurenine. These indole metabolites activate AhR and PXR.15, 47, 48 Non‐protein materials such as glucobrassicin, a compound found in cruciferous vegetables, are metabolized to indole‐3‐carbinole, which also activates AhR.49 AhR acts as a transcription factor to induce expression of genes such as CYP4501A1, which detoxifies chemicals and toxins.50
Lipid and bile acid metabolites
Gut bacteria produce many lipid‐modifying and metabolizing enzymes. For example, hydroxy fatty acids are generated from polyunsaturated fatty acids by gastrointestinal microorganisms such as Lactobacillus plantarum, which encodes polyunsaturated fatty acid‐saturating enzymes.51, 52 Roseburia species are active in metabolizing linoleic acid (cis‐9,cis‐12‐18:2).53 They produce vaccenic acid, a precursor of health‐promoting linoleic acids. These fatty acid‐modifying functions of the microbiota can change the lipid profile in the gut lumen. Bile acids are produced from the liver and secreted into the small intestine to emulsify triglyceride and fatty acids. Primary bile acids, such as cholic acid and chenodeoxycholic acid, are produced in the gall bladder and secreted into the duodenum as conjugated forms to glycine and taurine. Most of the secreted bile acids are reabsorbed in the terminal ileum. Gut microbiota alter bile acids through various modifications, including hydrolysis of the C24N‐acyl amide bond, oxidation and epimerization of hydroxyl groups, 7α‐dehydroxylation, esterification and desulphatation.54 These processes generate more than 20 different secondary bile acids, including deoxycholic acid (DCA) and lithocholic acid (LCA). Also, phosphatidylcholine is metabolized to produce important metabolites such as trimethylamine‐N‐oxide (TMAO). Gut microbes convert choline to trimethylamine, which is absorbed and then converted by the flavin monooxygenase system in the liver to TMAO.55 Other substrates of gut bacteria to produce trimethylamin include betaine (trimethylglycine) and carnitine (a lysine derivative). Bile acids activate many cell types through cell receptors, such as FXR, VDR, PXR and TGR5 (also called GP‐BAR1 or M‐BAR).12, 56 FXR, VDR and PXR are nuclear receptor family members, whereas TGR5 is a G‐protein‐coupled receptor. FXR is expressed by many cell types, including epithelial and endothelial cells in the liver, gastrointestinal tract and kidney.57, 58 PXR is expressed by cells in the liver, small intestine and colon.14, 59 TGR5 is widely expressed in the body. Its expression is particularly high in the intestines, pancreas, lungs, lymphoid tissues and brain.60
Altered microbial metabolites in diseases
Metabolites are produced by the microbiota and, thus, changes in the microbiota or dysbiosis can alter metabolite profiles. Particularly, metabolites are altered in many pathogenic conditions such as obesity, type I diabetes, alcoholic‐ and hepatitis B virus‐induced liver diseases, inflammatory bowel diseases (IBD), cancer and other chronic diseases. Alteration of SCFA levels and/or microbial producers has been observed in many diseases. It has been observed that C2 and C3 are decreased in some patients with type I diabetes or colitis.61, 62 Similarly, C2 and C4 levels were decreased in patients with colitis, colon cancer or other intestinal disorders.63, 64 Also, low dietary fibre intake and decreased levels of SCFAs and some other longer‐chain fatty acids have been observed in allergy patients.65, 66, 67 Increased levels of C2 and C4 along with the expansion of Firmicutes species are associated with obesity.68 In addition, indole derivatives are also altered in some diseases. For example, tryptophan metabolism seems to be increased in patients with IBD, and the levels of tryptophan metabolites such as kynurenine and indole‐3‐acetate were altered in some colitis patients.69, 70 However, more data are needed to firmly connect the changes of specific metabolites to diseases. Assuming that these metabolites regulate many cell types in most parts of the body, changes in the metabolites could have comprehensive effects, potentially altering immune and inflammatory responses, metabolism, neuronal functions, cancer development and other processes.
Immune regulatory functions of SCFAs
As the most abundant microbial metabolites among all in the colonic lumen, SCFAs play multi‐faceted regulatory roles in the immune system (Fig. 2). First, SCFAs are used by colonocytes as a major energy source, and they influence gene expression necessary for epithelial barrier and defense functions. Second, SCFAs regulate innate immune cells such as macrophages, neutrophils and DCs. Third, SCFAs bidirectionally regulate antigen‐specific adaptive immunity mediated by T‐cells and B‐cells.
Figure 2.
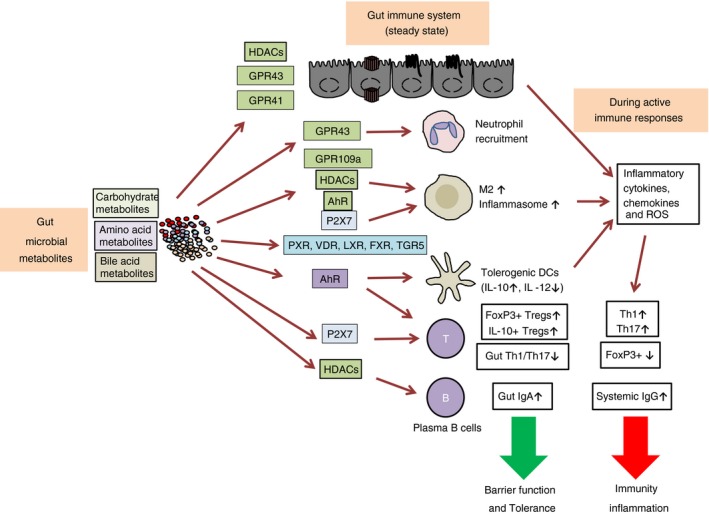
Impacts of microbial metabolites on the immune system. Gut microbial metabolites exert far‐reaching influences on the host. They regulate the immune system through histone deacetylases (HDACs), receptors and/or metabolic integration. Short‐chain fatty acids (SCFAs) fuel and fortify epithelial cells for promoting barrier functions. SCFAs also function as a neutrophil chemotaxin, and regulate macrophages and dendritic cells (DCs) through G‐protein‐coupled receptors (GPCRs) and HDACs. Bile acid metabolites also induce tolerogenic DCs and type 2 macrophages (M2), effects mediated by multiple receptors. Aryl hydrocarbon receptor precursor (AhR) activation by gut microbial metabolites induces regulatory T‐cells that express FoxP3 and IL‐10. Many gut microbial metabolites support the generation of induced Tregs for immune tolerance. SCFAs also fuel B‐cells and promote their differentiation into IgA‐ or IgG‐producing plasma B‐cells, an effect mediated by HDAC inhibition and metabolic regulation by SCFAs. However, during infection, gut microbial metabolites promote the generation of effector T‐cells such as Th1 and Th17 cells to fight pathogens. P2X7 activation by adenosine triphosphate (ATP) and nicotinamide adenine dinucleotide (NAD) exert both positive and negative regulatory roles in the immune system. Both tolerogenic and inflammatory functions of gut microbial metabolites have been reported, indicating that the overall functions of gut microbial metabolites are determined by the immune status of the host.
Dietary fibre and SCFAs support intestinal epithelial proliferation.71, 72 SCFAs are converted to acetyl‐CoA for energy production through the tricarboxylic acid cycle and lipid synthesis.73 SCFAs also inhibit HDACs to promote gene expression in epithelial cells.74 In the colonic lumen, SCFAs decrease intracellular pH and facilitate sodium ion absorption through multiple mechanisms, mediated in part through the Na+‐H+ and Cl−‐HCO3 − exchange pumps.71, 75, 76 SCFAs are weak acids and play important roles in lowering pH in the gut lumen. SCFAs induce cancer cell differentiation and apoptosis and, therefore, are generally perceived as tumour suppresors.77 This function is closely associated with the HDAC inhibition and the histone hyperacetylation effects of SCFAs to regulate genes that control key cellular processes.78 A caveat for this role is that C4 promotes hyperproliferation of mutated (MSH2−/)− colon epithelial cells, and thus has the potential to even promote certain types of tumours.79 A function important for barrier immunity is the positive effects of SCFAs on epithelial cell production of certain cytokines such as IL‐18 and antimicrobial peptides.80, 81 SCFAs also enhance the expression of epithelial barrier‐forming molecules and mucin production, which are mediated, in part, by HDAC inhibition and AMP‐activated protein kinase (AMPK) activation.82, 83, 84, 85
Short‐chain fatty acids regulate myeloid cells. Neutrophils are the prototype myeloid cells that express the SCFA receptor GPR43. GPR43 activation by SCFAs induces chemotaxis and functional activation of neutrophils.32, 33, 37, 86 SCFAs also regulate macrophages. C4 conditions intestinal macrophages through GPR109A to induce IL‐10‐producing T‐cells.87 While GPCR activation by SCFAs is involved in regulating macrophages, the HDAC inhibitor function of SCFAs is also important. For example, C4 downregulates lipopolysaccharide‐induced production of nitric oxide and inflammatory cytokines such as IL‐6 and IL‐12 in a GPCR‐independent manner presumably through HDAC inhibition.88 C4 suppresses production of inflammatory cytokines, such as TNF‐α, MCP‐1 and IL‐6 in macrophages.89 SCFAs also regulate DCs. Human monocyte‐derived DCs cultured with C3 or C4 were less inflammatory with decreased production of pro‐inflammatory cytokines and chemokines.90 It was documented that SCFAs also indirectly affect DCs by inducing retinaldehyde dehydrogenase 1 (RALDH1) expression in intestinal epithelial cells, which leads to retinoic acid (RA) production and subsequent generation of RA‐regulated DCs with a tolerogenic phenotype.91
Short‐chain fatty acids can also directly and indirectly regulate T‐cell differentiation into functionally specialized cells.8 This occurs when T‐cells undergo antigen priming by antigen presenting cells in the presence of SCFAs. It was initially reported that C4 suppresses CD4+ T‐cell proliferation but does not induce FoxP3+ T‐cells.92 However, several groups reported that C4 and C3 can increase colonic FoxP3+ Tregs in vivo and/or in vitro settings, potentially through their HDAC inhibitor activity.93, 94, 95 In our own study, all major SCFAs (i.e. C2, C3 and C4) enhanced the generation of effector T‐cells such as Th17 and Th1 as well as IL‐10+ CD4+ T‐cells in both in vitro and in vivo settings.38 However, the number of FoxP3+ T‐cells was not necessarily increased in vivo and in vitro, whereas the number of IL‐10+ T‐cells was increased by SCFAs. However, the IL‐10+ T‐cell‐inducing effect of SCFAs is lost during active immune responses to C. rodentium.38 During active immune responses, the numbers of Th17 cells and Th1 cells were increased by SCFAs. Overall, SCFAs have significant impacts on regulatory T‐cells and effector T‐cells depending on immunological context. This is perhaps due to the booster effect of SCFAs on gene expression during lymphocyte activation. HDAC inhibition increases histone acetylation but it does not have selectivity toward Treg versus effector T‐cells. Rather, SCFAs have the tendency to boost the polarization effects set by cytokine milieus present at the time of T‐cell priming and differentiation. Because SCFA levels are high in the colonic tissues and gut‐associated lymphoid tissues, intestinal T‐cells are likely to be a major target of SCFAs.
It has long been recognized that consumption of dietary fibre increases host antibody responses in animals.96 In vitro, C4 increased B‐cell production of antibodies, which was associated with increased histone acetylation.97 Our laboratory systematically studied the impact of dietary fibre and SCFAs on antibody responses to commensal bacteria and pathogens.98 SCFAs exert strong epigenetic regulatory effects on B‐cells through their HDAC inhibitory activity, and promote B‐cell differentiation into plasma B‐cells. SCFAs can boost the production of both IgA and IgG isotypes. They increase IgA‐coated intestinal bacteria and promote IgA and IgG production in response to C. rodentium infection. This indicates that both mucosal and systemic antibody responses are boosted by SCFAs. SCFAs upregulate three important metabolic processes, such as glycolysis, oxidative phosphorylation and lipogenesis, in B‐cells, which are necessary to produce cellular building blocks and energy to support plasma B‐cell differentiation. It is believed that HDAC inhibition by SCFAs promotes histone acetylation to enhance gene expression necessary for plasma B‐cell differentiation. Another potentially important regulatory mechanism of SCFAs is mediated by their impact on cellular metabolism. SCFAs increase the levels of acetyl‐CoA and ATP but decrease AMP level and AMPK activity, leading to increased mTOR activity that sustains the high metabolic demand of differentiating B‐cells.98 High metabolic activity is necessary to fully support plasma B‐cell differentiation.
Through the effects on many cell types, SCFAs exert comprehensive regulatory effects on inflammatory diseases. Compared with wild‐type (WT) mice, GPR43−/− mice suffer more from dextran sulphate sodium (DSS)‐induced colitis.99 However, GPR43 signalling promotes also acute inflammatory responses required to mount normal immune responses to microbes, which is delayed in GPR43−/− mice.100 C4 enema has been reported to suppress chronic colitis in mice and humans.101, 102 However, C4 was not effective in suppressing acute colitis responses such as 2,4,6‐trinitrobenzene sulphonic acid (TNBS)‐induced inflammation.103 SCFAs and dietary fibre also suppress inflammatory responses associated with type I diabetes and food allergy responses in animal models.104, 105 SCFAs can also suppress acute kidney injury in an animal model.106 However, chronic administration of SCFAs induced urethritis in mice characterized by smooth muscle cell and epithelial hyperplasia leading to hydronephrosis.107 Therefore, SCFAs can promote also inflammatory responses depending on their concentrations, organs and host condition.
Immune regulatory functions of amino acid and indole‐related metabolites
Major indole derivatives, generated either from tryptophan or plant‐derived glucobrassicin, activate AhR. AhR is activated by many natural and synthetic chemicals such as indole‐3‐acetate, indole‐3‐carbinol, kynurenine, resveratrol, indirubin, flavonoids, omeprazole and dioxin (2,3,7,8‐tetrachlorodibenzo‐p‐dioxin, TCDD). Therefore, AhR is important not only for dietary factor‐regulated but also for environmental pollutant‐modulated immune responses. Upon ligand binding, cytoplasmic AhR translocates to the nucleus and heterodimerizes with AhR nuclear translocator. Dimerized AhR binds the consensus xenobiotic responsive element (XRE, 5′‐GCGTG‐3′) in many genes encoding toxicant‐modifying enzymes or immune‐regulatory molecules. The XRE is thought to be created by insertion of transposable elements such as LINE‐1 and Alu.108 AhR regulates many genes, including the genes coding for CYP450 1A1, IDO, IL‐10, Aiolos, FoxP3, IL‐21 and CD39. For T‐cells, AhR activation supports Th17 but suppresses Treg differentiation.109 AhR works together with other transcription factors, such as STAT3, to turn on Aiolos or regulates STAT1.110, 111 Interestingly, AhR activation by TCDD had an opposite effect, boosting Tregs but suppressing Th17 cell differentiation. In this case, AhR works with another transcription factor c‐Maf, which promotes IL‐27‐induced IL‐10+ T‐cells.112 AhR activation promotes monocyte differentiation into DCs over macrophages.113 In vivo, dietary indoles suppress Th17 but increase Treg responses.114 Thus, the effects of AhR ligands on T‐cell responses are complex, mediated by many factors and cell types. AhR function is not necessarily equal to that of AhR ligands in overall immune responses. This is probably because AhR functions as a transcription regulator regardless of the presence of ligands, and its function is further regulated by ligands. Moreover, different ligands and cooperating transcription factors could lead to heterogenous outcomes.
Polyamines regulate transcription, protein translation, stress protein responses and cellular metabolism. They have the potential to exert regulatory functions on immune cells. Polyamines have anti‐inflammatory effects, in part, by suppressing inflammatory T‐cells and the production of cytokines and nitric oxide (NO).115 Blocking of polyamine synthesizing enzymes in host cells can break tolerogenic tumour microenvironments.116 Spermine can regulate autophagy and apoptosis and suppress the production of IL‐12 but increase that of IL‐10.117, 118 Polyamines also exert repair functions following tissue damages. However, the functions and mechanisms of actions of various polyamines in regulating the immune system remain unclear at the molecular level.42 Particularly it remains to be determined how gut microbiota‐derived polyamines are transported through the gut barrier and regulate immune cells.
Immune regulatory functions of bile acid and related cholesterol metabolites
Bile acids play important anti‐inflammatory roles in cholestatic and metabolically driven inflammatory diseases. Low bile acid levels correlate with increased susceptibility to infection.119 However, chronic exposure to high levels of bile acids can induce inflammation and cancer.120 Secondary bile acids, such as DCA and LCA, regulate the immune system, in part, through their receptors, such as TGR5 (also called GPBAR1 or M‐BAR) and two nuclear receptors, FXR and PXR. Animals deficient in TGR5 develop more severe colitis induced by T‐cell‐activating haptens (TNBS and oxazolone).121 Similarly, FXR−/− mice were more susceptible to TNBS‐induced colitis than WT mice.122 Also, DSS‐induced colitis was more severe in PXR−/− mice.123 These data suggest that bile acid receptors promote immune tolerance. One caveat is that not all the phenotype of FXR−/− mice can be attributed to the function of bile acid metabolites, because these receptors, particularly PXR and FXR, are activated also by non‐bile acid metabolite ligands. Moreover, these receptors, particularly those receptors that function as transcription factors, can have ligand‐independent regulatory functions. TGR5 ligands, such as LCA and DCA, suppress TNF‐α production by macrophages.124 TGR5 activation induces cAMP production and subsequent phosphorylation of c‐Fos, leading to NF‐κB inhibition. In this regard, TGR5 ligands generate DCs that have decreased production of IL‐12.125 Consistently, instillation of DCA into the colon exacerbated colitis responses.126, 127 Bile acid metabolites regulate NLRP3 inflammasome activation, but their exact roles in this regard are controversial.128, 129 While the functions of bile acid metabolites appear to be complex and are probably determined by receptors, cell types, tissue sites and immunological context, these metabolites play important roles in regulating the immune system.
Concluding remarks
Gut commensal bacteria produce a myriad of microbial metabolites. These metabolites function as nutrients and/or activate host receptors, including GPR43, GPR41, GPR109A, PXR, FXR and TGR5. While not discussed in detail, P2X receptors (P2X1–7) are expressed by many cell types, including tissue, myeloid, mast and T‐cells to sense ATP,130 which is produced by both host and microbial cells. Activation of P2X7, for example, leads to Ca2+ influx and subsequent inflammasome activation or cell death for positive and negative regulation of the immune system.131, 132, 133 The host receptors for microbial metabolites are expressed by diverse cell types in barrier and systemic tissues. Therefore, gut microbial metabolites and their receptors create an extensive array of signalling to sense and respond to nutritional status and host conditions reflected in microbial activity. It is apparent that gut microbial metabolites can promote both immunity and tolerance, both of which are required to maintain health by preventing chronic infection and inflammatory diseases. Microbial metabolites strengthen barrier tissues and train the immune system to prevent and prepare for possible infection by pathogens. Beyond the immune system, gut metabolites and their receptors maintain homeostasis of metabolism, which is important to maintain host health by balancing nutrients intake and utilization. Diseases and pathological conditions cause gut dysbiosis and altered production of microbial metabolites, leading to dysregulation of the immune system and metabolism. More research is required to identify the immune regulatory functions of individual metabolites in health and disease. More importantly, how these microbial metabolites in combination regulate the host immune system remains to be elucidated. Furthermore, how altered composition of microbial metabolites is associated with specific diseases should be studied in an effort to identify biomarkers for pathological conditions.
Funding
Grant support: this study was supported, in part, from grants from NIH (R01AI121302 and R01AI080769) to CHK.
Disclosure
The author has no competing interests.
Acknowledgements
The author thanks all current and past students and research fellows in former and current labs at Purdue University and University of Michigan for providing invaluable insights into the functions of metabolites in regulating the immune system.
References
- 1. Rowland I, Gibson G, Heinken A, Scott K, Swann J, Thiele I et al Gut microbiota functions: metabolism of nutrients and other food components. Eur J Nutr 2017; 57:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Koh A, De Vadder F, Kovatcheva‐Datchary P, Backhed F. From dietary fiber to host physiology: short‐chain fatty acids as key bacterial metabolites. Cell 2016; 165:1332–45. [DOI] [PubMed] [Google Scholar]
- 3. Wahlstrom A, Sayin SI, Marschall HU, Backhed F. Intestinal crosstalk between bile acids and microbiota and its impact on host metabolism. Cell Metab 2016; 24:41–50. [DOI] [PubMed] [Google Scholar]
- 4. den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. The role of short‐chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 2013; 54:2325–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol 2016; 16:341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blacher E, Levy M, Tatirovsky E, Elinav E. Microbiome‐modulated metabolites at the interface of host immunity. J Immunol 2017; 198:572–80. [DOI] [PubMed] [Google Scholar]
- 7. Postler TS, Ghosh S. Understanding the holobiont: how microbial metabolites affect human health and shape the immune system. Cell Metab 2017; 26:110–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kim CH, Park J, Kim M. Gut microbiota‐derived short‐chain fatty acids, T cells, and inflammation. Immune Netw 2014; 14:277–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Louis P, Flint HJ. Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 2017; 19:29–41. [DOI] [PubMed] [Google Scholar]
- 10. Surprenant A, Rassendren F, Kawashima E, North RA, Buell G. The cytolytic P2Z receptor for extracellular ATP identified as a P2X receptor (P2X7). Science 1996; 272:735–8. [DOI] [PubMed] [Google Scholar]
- 11. Wang H, Chen J, Hollister K, Sowers LC, Forman BM. Endogenous bile acids are ligands for the nuclear receptor FXR/BAR. Mol Cell 1999; 3:543–53. [DOI] [PubMed] [Google Scholar]
- 12. Maruyama T, Miyamoto Y, Nakamura T, Tamai Y, Okada H, Sugiyama E et al Identification of membrane‐type receptor for bile acids (M‐BAR). Biochem Biophys Res Commun 2002; 298:714–9. [DOI] [PubMed] [Google Scholar]
- 13. Roberts EA, Golas CL, Okey AB. Ah receptor mediating induction of aryl hydrocarbon hydroxylase: detection in human lung by binding of 2,3,7,8‐[3H]tetrachlorodibenzo‐p‐dioxin. Cancer Res 1986; 46:3739–43. [PubMed] [Google Scholar]
- 14. Kliewer SA, Moore JT, Wade L, Staudinger JL, Watson MA, Jones SA et al An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 1998; 92:73–82. [DOI] [PubMed] [Google Scholar]
- 15. Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP et al Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll‐like receptor 4. Immunity 2014; 41:296–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J 1991; 280:309–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Henrissat B, Callebaut I, Fabrega S, Lehn P, Mornon JP, Davies G. Conserved catalytic machinery and the prediction of a common fold for several families of glycosyl hydrolases. Proc Natl Acad Sci USA 1995; 92:7090–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shoham Y, Lamed R, Bayer EA. The cellulosome concept as an efficient microbial strategy for the degradation of insoluble polysaccharides. Trends Microbiol 1999; 7:275–81. [DOI] [PubMed] [Google Scholar]
- 19. Meadow ND, Fox DK, Roseman S. The bacterial phosphoenolpyruvate: glycose phosphotransferase system. Annu Rev Biochem 1990; 59:497–542. [DOI] [PubMed] [Google Scholar]
- 20. Cantarel BL, Lombard V, Henrissat B. Complex carbohydrate utilization by the healthy human microbiome. PLoS One 2012; 7:e28742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stark AH, Madar Z. In vitro production of short‐chain fatty acids by bacterial fermentation of dietary fiber compared with effects of those fibers on hepatic sterol synthesis in rats. J Nutr 1993; 123:2166–73. [DOI] [PubMed] [Google Scholar]
- 22. Macfarlane S, Macfarlane GT. Regulation of short‐chain fatty acid production. Proc Nutr Soc 2003; 62:67–72. [DOI] [PubMed] [Google Scholar]
- 23. Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin‐degrading bacterium. Int J Syst Evol Microbiol 2004; 54:1469–76. [DOI] [PubMed] [Google Scholar]
- 24. Louis P, Hold GL, Flint HJ. The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol 2014; 12:661–72. [DOI] [PubMed] [Google Scholar]
- 25. Li H, Myeroff L, Smiraglia D, Romero MF, Pretlow TP, Kasturi L et al SLC5A8, a sodium transporter, is a tumor suppressor gene silenced by methylation in human colon aberrant crypt foci and cancers. Proc Natl Acad Sci USA 2003; 100:8412–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yanase H, Takebe K, Nio‐Kobayashi J, Takahashi‐Iwanaga H, Iwanaga T. Cellular expression of a sodium‐dependent monocarboxylate transporter (Slc5a8) and the MCT family in the mouse kidney. Histochem Cell Biol 2008; 130:957–66. [DOI] [PubMed] [Google Scholar]
- 27. Louis P, Young P, Holtrop G, Flint HJ. Diversity of human colonic butyrate‐producing bacteria revealed by analysis of the butyryl‐CoA:acetate CoA‐transferase gene. Environ Microbiol 2010; 12:304–14. [DOI] [PubMed] [Google Scholar]
- 28. Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016; 7:189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ze X, Duncan SH, Louis P, Flint HJ. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. ISME J 2012; 6:1535–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim M, Kim CH. Regulation of humoral immunity by gut microbial products. Gut Microbes 2017; 8:392–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kasubuchi M, Hasegawa S, Hiramatsu T, Ichimura A, Kimura I. Dietary gut microbial metabolites, short‐chain fatty acids, and host metabolic regulation. Nutrients 2015; 7:2839–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Miller TL, Wolin MJ. Pathways of acetate, propionate, and butyrate formation by the human fecal microbial flora. Appl Environ Microbiol 1996; 62:1589–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brown AJ, Goldsworthy SM, Barnes AA, Eilert MM, Tcheang L, Daniels D et al The Orphan G protein‐coupled receptors GPR41 and GPR43 are activated by propionate and other short chain carboxylic acids. J Biol Chem 2003; 278:11 312–9. [DOI] [PubMed] [Google Scholar]
- 34. Nilsson NE, Kotarsky K, Owman C, Olde B. Identification of a free fatty acid receptor, FFA2R, expressed on leukocytes and activated by short‐chain fatty acids. Biochem Biophys Res Commun 2003; 303:1047–52. [DOI] [PubMed] [Google Scholar]
- 35. Thangaraju M, Cresci GA, Liu K, Ananth S, Gnanaprakasam JP, Browning DD et al GPR109A is a G‐protein‐coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer Res 2009; 69:2826–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pluznick JL, Protzko RJ, Gevorgyan H, Peterlin Z, Sipos A, Han J et al Olfactory receptor responding to gut microbiota‐derived signals plays a role in renin secretion and blood pressure regulation. Proc Natl Acad Sci USA 2013; 110:4410–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim MH, Kang SG, Park JH, Yanagisawa M, Kim CH. Short‐chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 2013; 145:e1–10. [DOI] [PubMed] [Google Scholar]
- 38. Le Poul E, Loison C, Struyf S, Springael JY, Lannoy V, Decobecq ME et al Functional characterization of human receptors for short chain fatty acids and their role in polymorphonuclear cell activation. J Biol Chem 2003; 278:25 481–9. [DOI] [PubMed] [Google Scholar]
- 39. Park J, Kim M, Kang SG, Jannasch AH, Cooper B, Patterson J et al Short‐chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR‐S6K pathway. Mucosal Immunol 2015; 8:80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nakajima A, Nakatani A, Hasegawa S, Irie J, Ozawa K, Tsujimoto G et al The short chain fatty acid receptor GPR43 regulates inflammatory signals in adipose tissue M2‐type macrophages. PLoS One 2017; 12:e0179696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dai ZL, Wu G, Zhu WY. Amino acid metabolism in intestinal bacteria: links between gut ecology and host health. Front Biosci (Landmark Ed) 2011; 16:1768–86. [DOI] [PubMed] [Google Scholar]
- 42. Macfarlane GT, Allison C, Gibson SA, Cummings JH. Contribution of the microflora to proteolysis in the human large intestine. J Appl Bacteriol 1988; 64:37–46. [DOI] [PubMed] [Google Scholar]
- 43. Miller‐Fleming L, Olin‐Sandoval V, Campbell K, Ralser M. Remaining mysteries of molecular biology: the role of polyamines in the cell. J Mol Biol 2015; 427:3389–406. [DOI] [PubMed] [Google Scholar]
- 44. Sanchez‐Jimenez F, Ruiz‐Perez MV, Urdiales JL, Medina MA. Pharmacological potential of biogenic amine‐polyamine interactions beyond neurotransmission. Br J Pharmacol 2013; 170:4–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Arena ME, Manca de Nadra MC. Biogenic amine production by Lactobacillus. J Appl Microbiol 2001; 90:158–62. [DOI] [PubMed] [Google Scholar]
- 46. Blachier F, Mariotti F, Huneau JF, Tome D. Effects of amino acid‐derived luminal metabolites on the colonic epithelium and physiopathological consequences. Amino Acids 2007; 33:547–62. [DOI] [PubMed] [Google Scholar]
- 47. Miller CA 3rd. Expression of the human aryl hydrocarbon receptor complex in yeast. Activation of transcription by indole compounds. J Biol Chem 1997; 272:32 824–9. [DOI] [PubMed] [Google Scholar]
- 48. Heath‐Pagliuso S, Rogers WJ, Tullis K, Seidel SD, Cenijn PH, Brouwer A et al Activation of the Ah receptor by tryptophan and tryptophan metabolites. Biochemistry 1998; 37:11 508–15. [DOI] [PubMed] [Google Scholar]
- 49. Bjeldanes LF, Kim JY, Grose KR, Bartholomew JC, Bradfield CA. Aromatic hydrocarbon responsiveness‐receptor agonists generated from indole‐3‐carbinol in vitro and in vivo: comparisons with 2,3,7,8‐tetrachlorodibenzo‐p‐dioxin. Proc Natl Acad Sci USA 1991; 88:9543–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ito S, Chen C, Satoh J, Yim S, Gonzalez FJ. Dietary phytochemicals regulate whole‐body CYP1A1 expression through an arylhydrocarbon receptor nuclear translocator‐dependent system in gut. J Clin Invest 2007; 117:1940–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Polan CE, McNeill JJ, Tove SB. Biohydrogenation of unsaturated fatty acids by rumen bacteria. J Bacteriol 1964; 88:1056–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kishino S, Takeuchi M, Park SB, Hirata A, Kitamura N, Kunisawa J et al Polyunsaturated fatty acid saturation by gut lactic acid bacteria affecting host lipid composition. Proc Natl Acad Sci USA 2013; 110:17 808–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Devillard E, McIntosh FM, Duncan SH, Wallace RJ. Metabolism of linoleic acid by human gut bacteria: different routes for biosynthesis of conjugated linoleic acid. J Bacteriol 2007; 189:2566–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gerard P. Metabolism of cholesterol and bile acids by the gut microbiota. Pathogens 2013; 3:14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Fennema D, Phillips IR, Shephard EA. Trimethylamine and trimethylamine N‐Oxide, a Flavin‐Containing Monooxygenase 3 (FMO3)‐mediated host‐microbiome metabolic axis implicated in health and disease. Drug Metab Dispos 2016; 44:1839–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M et al A G protein‐coupled receptor responsive to bile acids. J Biol Chem 2003; 278:9435–40. [DOI] [PubMed] [Google Scholar]
- 57. Forman BM, Goode E, Chen J, Oro AE, Bradley DJ, Perlmann T et al Identification of a nuclear receptor that is activated by farnesol metabolites. Cell 1995; 81:687–93. [DOI] [PubMed] [Google Scholar]
- 58. Bishop‐Bailey D, Walsh DT, Warner TD. Expression and activation of the farnesoid X receptor in the vasculature. Proc Natl Acad Sci USA 2004; 101:3668–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bertilsson G, Heidrich J, Svensson K, Asman M, Jendeberg L, Sydow‐Backman M et al Identification of a human nuclear receptor defines a new signaling pathway for CYP3A induction. Proc Natl Acad Sci USA 1998; 95:12 208–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Duboc H, Tache Y, Hofmann AF. The bile acid TGR5 membrane receptor: from basic research to clinical application. Dig Liver Dis 2014; 46:302–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. de Groot PF, Belzer C, Aydin O, Levin E, Levels JH, Aalvink S et al Distinct fecal and oral microbiota composition in human type 1 diabetes, an observational study. PLoS One 2017; 12:e0188475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Murri M, Leiva I, Gomez‐Zumaquero JM, Tinahones FJ, Cardona F, Soriguer F et al Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case‐control study. BMC Med 2013; 11:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mortensen PB, Clausen MR. Short‐chain fatty acids in the human colon: relation to gastrointestinal health and disease. Scand J Gastroenterol Suppl 1996; 216:132–48. [DOI] [PubMed] [Google Scholar]
- 64. Chen HM, Yu YN, Wang JL, Lin YW, Kong X, Yang CQ et al Decreased dietary fiber intake and structural alteration of gut microbiota in patients with advanced colorectal adenoma. Am J Clin Nutr 2013; 97:1044–52. [DOI] [PubMed] [Google Scholar]
- 65. Sandin A, Braback L, Norin E, Bjorksten B. Faecal short chain fatty acid pattern and allergy in early childhood. Acta Paediatr 2009; 98:823–7. [DOI] [PubMed] [Google Scholar]
- 66. Bottcher MF, Nordin EK, Sandin A, Midtvedt T, Bjorksten B. Microflora‐associated characteristics in faeces from allergic and nonallergic infants. Clin Exp Allergy 2000; 30:1590–6. [DOI] [PubMed] [Google Scholar]
- 67. De Preter V, Machiels K, Joossens M, Arijs I, Matthys C, Vermeire S et al Faecal metabolite profiling identifies medium‐chain fatty acids as discriminating compounds in IBD. Gut 2015; 64:447–58. [DOI] [PubMed] [Google Scholar]
- 68. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity‐associated gut microbiome with increased capacity for energy harvest. Nature 2006; 444:1027–31. [DOI] [PubMed] [Google Scholar]
- 69. Lamas B, Richard ML, Leducq V, Pham HP, Michel ML, Da Costa G et al CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat Med 2016; 22:598–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nikolaus S, Schulte B, Al‐Massad N, Thieme F, Schulte DM, Bethge J et al Increased tryptophan metabolism is associated with activity of inflammatory bowel diseases. Gastroenterology 2017; 153:e2. [DOI] [PubMed] [Google Scholar]
- 71. Sakata T, von Engelhardt W. Stimulatory effect of short chain fatty acids on the epithelial cell proliferation in rat large intestine. Comp Biochem Physiol A Comp Physiol 1983; 74:459–62. [DOI] [PubMed] [Google Scholar]
- 72. Zhang J, Lupton JR. Dietary fibers stimulate colonic cell proliferation by different mechanisms at different sites. Nutr Cancer 1994; 22:267–76. [DOI] [PubMed] [Google Scholar]
- 73. Zambell KL, Fitch MD, Fleming SE. Acetate and butyrate are the major substrates for de novo lipogenesis in rat colonic epithelial cells. J Nutr 2003; 133:3509–15. [DOI] [PubMed] [Google Scholar]
- 74. Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther 2008; 27:104–19. [DOI] [PubMed] [Google Scholar]
- 75. DeSoignie R, Sellin JH. Propionate‐initiated changes in intracellular pH in rabbit colonocytes. Gastroenterology 1994; 107:347–56. [DOI] [PubMed] [Google Scholar]
- 76. Gonda T, Maouyo D, Rees SE, Montrose MH. Regulation of intracellular pH gradients by identified Na/H exchanger isoforms and a short‐chain fatty acid. Am J Physiol 1999; 276:G259–70. [DOI] [PubMed] [Google Scholar]
- 77. Heerdt BG, Houston MA, Augenlicht LH. Potentiation by specific short‐chain fatty acids of differentiation and apoptosis in human colonic carcinoma cell lines. Cancer Res 1994; 54:3288–93. [PubMed] [Google Scholar]
- 78. Hinnebusch BF, Meng S, Wu JT, Archer SY, Hodin RA. The effects of short‐chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J Nutr 2002; 132:1012–7. [DOI] [PubMed] [Google Scholar]
- 79. Belcheva A, Irrazabal T, Robertson SJ, Streutker C, Maughan H, Rubino S et al Gut microbial metabolism drives transformation of MSH2‐deficient colon epithelial cells. Cell 2014; 158:288–99. [DOI] [PubMed] [Google Scholar]
- 80. Kalina U, Koyama N, Hosoda T, Nuernberger H, Sato K, Hoelzer D et al Enhanced production of IL‐18 in butyrate‐treated intestinal epithelium by stimulation of the proximal promoter region. Eur J Immunol 2002; 32:2635–43. [DOI] [PubMed] [Google Scholar]
- 81. Schauber J, Svanholm C, Termen S, Iffland K, Menzel T, Scheppach W et al Expression of the cathelicidin LL‐37 is modulated by short chain fatty acids in colonocytes: relevance of signalling pathways. Gut 2003; 52:735–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Peng L, Li ZR, Green RS, Holzman IR, Lin J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP‐activated protein kinase in Caco‐2 cell monolayers. J Nutr 2009; 139:1619–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Krishnan M, Singh AB, Smith JJ, Sharma A, Chen X, Eschrich S et al HDAC inhibitors regulate claudin‐1 expression in colon cancer cells through modulation of mRNA stability. Oncogene 2010; 29:305–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Burger‐van Paassen N, Vincent A, Puiman PJ, van der Sluis M, Bouma J, Boehm G et al The regulation of intestinal mucin MUC2 expression by short‐chain fatty acids: implications for epithelial protection. Biochem J 2009; 420:211–9. [DOI] [PubMed] [Google Scholar]
- 85. Zheng L, Kelly CJ, Battista KD, Schaefer R, Lanis JM, Alexeev EE et al Microbial‐derived butyrate promotes epithelial barrier function through IL‐10 receptor‐dependent repression of Claudin‐2. J Immunol 2017; 199: 2976–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Vinolo MA, Ferguson GJ, Kulkarni S, Damoulakis G, Anderson K, Bohlooly YM et al SCFAs induce mouse neutrophil chemotaxis through the GPR43 receptor. PLoS One 2011; 6:e21205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Singh N, Gurav A, Sivaprakasam S, Brady E, Padia R, Shi H et al Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014; 40:128–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chang PV, Hao L, Offermanns S, Medzhitov R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc Natl Acad Sci USA 2014; 111:2247–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ohira H, Fujioka Y, Katagiri C, Mamoto R, Aoyama‐Ishikawa M, Amako K et al Butyrate attenuates inflammation and lipolysis generated by the interaction of adipocytes and macrophages. J Atheroscler Thromb 2013; 20:425–42. [DOI] [PubMed] [Google Scholar]
- 90. Nastasi C, Candela M, Bonefeld CM, Geisler C, Hansen M, Krejsgaard T et al The effect of short‐chain fatty acids on human monocyte‐derived dendritic cells. Sci Rep 2015; 5:16 148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Goverse G, Molenaar R, Macia L, Tan J, Erkelens MN, Konijn T et al Diet‐derived short chain fatty acids stimulate intestinal epithelial cells to induce mucosal tolerogenic dendritic cells. J Immunol 2017; 198:2172–81. [DOI] [PubMed] [Google Scholar]
- 92. Fontenelle B, Gilbert KM. n‐Butyrate anergized effector CD4 + T cells independent of regulatory T cell generation or activity. Scand J Immunol 2012; 76:457–63. [DOI] [PubMed] [Google Scholar]
- 93. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly YM et al The microbial metabolites, short‐chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013; 341:569–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P et al Metabolites produced by commensal bacteria promote peripheral regulatory T‐cell generation. Nature 2013; 504:451–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D et al Commensal microbe‐derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013; 504:446–50. [DOI] [PubMed] [Google Scholar]
- 96. Nakamura Y, Nosaka S, Suzuki M, Nagafuchi S, Takahashi T, Yajima T et al Dietary fructooligosaccharides up‐regulate immunoglobulin A response and polymeric immunoglobulin receptor expression in intestines of infant mice. Clin Exp Immunol 2004; 137:52–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Okamura T, Gohda E, Kohge T, Yamamoto I. An increase in histone acetylation and IL‐2 antagonizing the immunoinhibitory effect are necessary for augmentation by butyrate of in vitro anti‐TNP antibody production. Biol Pharm Bull 1999; 22:1288–92. [DOI] [PubMed] [Google Scholar]
- 98. Kim M, Qie Y, Park J, Kim CH. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe 2016; 20:202–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D et al Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009; 461:1282–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Sina C, Gavrilova O, Forster M, Till A, Derer S, Hildebrand F et al G protein‐coupled receptor 43 is essential for neutrophil recruitment during intestinal inflammation. J Immunol 2009; 183:7514–22. [DOI] [PubMed] [Google Scholar]
- 101. Venkatraman A, Ramakrishna BS, Shaji RV, Kumar NS, Pulimood A, Patra S. Amelioration of dextran sulfate colitis by butyrate: role of heat shock protein 70 and NF‐kappaB. Am J Physiol Gastrointest Liver Physiol 2003; 285:G177–84. [DOI] [PubMed] [Google Scholar]
- 102. Hamer HM, Jonkers DM, Vanhoutvin SA, Troost FJ, Rijkers G, de Bruine A et al Effect of butyrate enemas on inflammation and antioxidant status in the colonic mucosa of patients with ulcerative colitis in remission. Clin Nutr 2010; 29:738–44. [DOI] [PubMed] [Google Scholar]
- 103. Tarrerias AL, Millecamps M, Alloui A, Beaughard C, Kemeny JL, Bourdu S et al Short‐chain fatty acid enemas fail to decrease colonic hypersensitivity and inflammation in TNBS‐induced colonic inflammation in rats. Pain 2002; 100:91–7. [DOI] [PubMed] [Google Scholar]
- 104. Tan J, McKenzie C, Vuillermin PJ, Goverse G, Vinuesa CG, Mebius RE et al Dietary fiber and bacterial SCFA enhance oral tolerance and protect against food allergy through diverse cellular pathways. Cell Rep 2016; 15:2809–24. [DOI] [PubMed] [Google Scholar]
- 105. Marino E, Richards JL, McLeod KH, Stanley D, Yap YA, Knight J et al Gut microbial metabolites limit the frequency of autoimmune T cells and protect against type 1 diabetes. Nat Immunol 2017; 18:552–62. [DOI] [PubMed] [Google Scholar]
- 106. Andrade‐Oliveira V, Amano MT, Correa‐Costa M, Castoldi A, Felizardo RJ, de Almeida DC et al Gut bacteria products prevent AKI induced by ischemia‐reperfusion. J Am Soc Nephrol 2015; 26:1877–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Park J, Goergen CJ, HogenEsch H, Kim CH. Chronically elevated levels of short‐chain fatty acids induce T cell‐mediated ureteritis and hydronephrosis. J Immunol 2016; 196:2388–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Mulero‐Navarro S, Fernandez‐Salguero PM. New trends in aryl hydrocarbon receptor biology. Front Cell Dev Biol 2016; 4:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Quintana FJ, Basso AS, Iglesias AH, Korn T, Farez MF, Bettelli E et al Control of T(reg) and T(H)17 cell differentiation by the aryl hydrocarbon receptor. Nature 2008; 453:65–71. [DOI] [PubMed] [Google Scholar]
- 110. Quintana FJ, Jin H, Burns EJ, Nadeau M, Yeste A, Kumar D et al Aiolos promotes TH17 differentiation by directly silencing Il2 expression. Nat Immunol 2012; 13:770–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Kimura A, Naka T, Nohara K, Fujii‐Kuriyama Y, Kishimoto T. Aryl hydrocarbon receptor regulates Stat1 activation and participates in the development of Th17 cells. Proc Natl Acad Sci USA 2008; 105:9721–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Apetoh L, Quintana FJ, Pot C, Joller N, Xiao S, Kumar D et al The aryl hydrocarbon receptor interacts with c‐Maf to promote the differentiation of type 1 regulatory T cells induced by IL‐27. Nat Immunol 2010; 11:854–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Goudot C, Coillard A, Villani AC, Gueguen P, Cros A, Sarkizova S et al Aryl hydrocarbon receptor controls monocyte differentiation into dendritic cells versus macrophages. Immunity 2017; 47:e6. [DOI] [PubMed] [Google Scholar]
- 114. Singh NP, Singh UP, Rouse M, Zhang J, Chatterjee S, Nagarkatti PS et al Dietary indoles suppress delayed‐type hypersensitivity by inducing a switch from proinflammatory Th17 cells to anti‐inflammatory regulatory T cells through regulation of MicroRNA. J Immunol 2016; 196:1108–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Keough MP, Hayes CS, DeFeo K, Gilmour SK. Elevated epidermal ornithine decarboxylase activity suppresses contact hypersensitivity. J Invest Dermatol 2011; 131:158–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Hayes CS, Shicora AC, Keough MP, Snook AE, Burns MR, Gilmour SK. Polyamine‐blocking therapy reverses immunosuppression in the tumor microenvironment. Cancer Immunol Res 2014; 2:274–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Hasko G, Kuhel DG, Marton A, Nemeth ZH, Deitch EA, Szabo C. Spermine differentially regulates the production of interleukin‐12 p40 and interleukin‐10 and suppresses the release of the T helper 1 cytokine interferon‐gamma. Shock 2000; 14:144–9. [DOI] [PubMed] [Google Scholar]
- 118. Ilmarinen P, Moilanen E, Erjefalt JS, Kankaanranta H. The polyamine spermine promotes survival and activation of human eosinophils. J Allergy Clin Immunol 2015; 136:e11. [DOI] [PubMed] [Google Scholar]
- 119. Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A et al Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 2015; 517:205–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Yoshimoto S, Loo TM, Atarashi K, Kanda H, Sato S, Oyadomari S et al Obesity‐induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 2013; 499:97–101. [DOI] [PubMed] [Google Scholar]
- 121. Biagioli M, Carino A, Cipriani S, Francisci D, Marchiano S, Scarpelli P et al The bile acid receptor GPBAR1 regulates the M1/M2 phenotype of intestinal macrophages and activation of GPBAR1 rescues mice from murine colitis. J Immunol 2017; 199:718–33. [DOI] [PubMed] [Google Scholar]
- 122. Renga B, Mencarelli A, Cipriani S, D'Amore C, Carino A, Bruno A et al The bile acid sensor FXR is required for immune‐regulatory activities of TLR‐9 in intestinal inflammation. PLoS One 2013; 8:e54472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Shah YM, Ma X, Morimura K, Kim I, Gonzalez FJ. Pregnane X receptor activation ameliorates DSS‐induced inflammatory bowel disease via inhibition of NF‐kappaB target gene expression. Am J Physiol Gastrointest Liver Physiol 2007; 292:G1114–22. [DOI] [PubMed] [Google Scholar]
- 124. Yoneno K, Hisamatsu T, Shimamura K, Kamada N, Ichikawa R, Kitazume MT et al TGR5 signalling inhibits the production of pro‐inflammatory cytokines by in vitro differentiated inflammatory and intestinal macrophages in Crohn's disease. Immunology 2013; 139:19–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Ichikawa R, Takayama T, Yoneno K, Kamada N, Kitazume MT, Higuchi H et al Bile acids induce monocyte differentiation toward interleukin‐12 hypo‐producing dendritic cells via a TGR5‐dependent pathway. Immunology 2012; 136:153–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Traub RJ, Tang B, Ji Y, Pandya S, Yfantis H, Sun Y. A rat model of chronic postinflammatory visceral pain induced by deoxycholic acid. Gastroenterology 2008; 135:2075–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Bernstein H, Holubec H, Bernstein C, Ignatenko N, Gerner E, Dvorak K et al Unique dietary‐related mouse model of colitis. Inflamm Bowel Dis 2006; 12:278–93. [DOI] [PubMed] [Google Scholar]
- 128. Zhao S, Gong Z, Zhou J, Tian C, Gao Y, Xu C et al Deoxycholic acid triggers NLRP3 inflammasome activation and aggravates DSS‐induced colitis in mice. Front Immunol 2016; 7:536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Guo C, Xie S, Chi Z, Zhang J, Liu Y, Zhang L et al Bile acids control inflammation and metabolic disorder through inhibition of NLRP3 inflammasome. Immunity 2016; 45:944. [DOI] [PubMed] [Google Scholar]
- 130. Khakh BS, North RA. P2X receptors as cell‐surface ATP sensors in health and disease. Nature 2006; 442:527–32. [DOI] [PubMed] [Google Scholar]
- 131. Di Virgilio F, Dal Ben D, Sarti AC, Giuliani AL, Falzoni S. The P2X7 receptor in infection and inflammation. Immunity 2017; 47:15–31. [DOI] [PubMed] [Google Scholar]
- 132. Yip L, Woehrle T, Corriden R, Hirsh M, Chen Y, Inoue Y et al Autocrine regulation of T‐cell activation by ATP release and P2X7 receptors. FASEB J 2009; 23:1685–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Hashimoto‐Hill S, Friesen L, Kim M, Kim CH. Contraction of intestinal effector T cells by retinoic acid‐induced purinergic receptor P2X7. Mucosal Immunol 2017; 10:912–23. [DOI] [PMC free article] [PubMed] [Google Scholar]


