Abstract
Interpretations of steroid hormone actions as slow, nuclear, transcriptional events have frequently been seen as competing against inferences of rapid membrane actions. We have discovered conditions where membrane-limited effects potentiate later transcriptional actions in a nerve cell line. Making use of a two-pulse hormonal schedule in a transfection system, early and brief administration of conjugated, membrane-limited estradiol was necessary but not sufficient for full transcriptional potency of the second estrogen pulse. Efficacy of the first pulse depended on intact signal transduction pathways. Surprisingly, the actions of both pulses were blocked by a classical estrogen receptor (ER) antagonist. Thus, two different modes of steroid hormone action can synergize.
The report of prolonged hormone retention in cell nuclei (1) began an era of research on nuclear hormone receptors as ligand-activated transcription factors and also started an enduring controversy. Fast, membrane-mediated hormone effects have frequently been argued to be alternatives to nuclear hormone actions. Rapid effects of estrogens certainly exist and are especially prominent in the neurophysiological literature (2–11). In turn, estrogen receptor (ER) genomic actions in neurons triggering transcriptional events governing reproductive behavior are also well established (12). What is the relation of the early steroid hormone actions to the slow? Based on new findings, we suggest that, far from the two types of estrogenic actions being “opposed,” the former can amplify the latter.
Methods
Cell Culture.
SK-N-BE2C, a human neuroblastoma cell line, was maintained in Ham's F-12:minimal essential media (MEM) (1:1) supplemented with 15% FBS (Bioreclamation, New York), 100 units/ml penicillin, and 50 μg/ml streptomycin. For transfections, the cells were grown in phenol-red free Ham's F-12:MEM (1:1), supplemented with 10% charcoal dextran-stripped FBS (Gemini Biotech, Alachua, FL) and 100 units/ml penicillin and 50 μg/ml streptomycin.
Plasmids/Constructs.
The pGL2-TATA-Inr-Luc construct was a kind gift from Donald McDonnell and has three consensus estrogen response elements (EREs) arranged in tandem upstream of the luciferase reporter gene. The pSG-hERα is a kind gift of Pierre Chambon and has been described (13).
Cell Culture and Transfections.
SK-N-BE2C cells, a human neuroblastoma cell line, were plated in Ham's F-12:MEM (1:1) supplemented with 15% FBS (Bioreclamation), 100 units/ml penicillin, and 50 μg/ml streptomycin in 6-well plates (Falcon) at a density of 0.3 × 106 cells per well and transfected by using the Effectene reagent (Qiagen) according to the manufacturer's instructions. Forty-eight hours after plating, the cells were cotransfected with pGL2-TATA-Inr-Luc (200 ng), pSG-hERα (80 ng), pSV-βgal (80 ng), and pBSKII+ to a total of 400 ng per well. The plasmid pSV-βgal was used to control for the efficiency of transfection. Twenty-four hours after transfection, the cells were washed free of the media and phenol-red free Ham's F-12:MEM (1:1), supplemented with 10% charcoal dextran-stripped FBS (Gemini Biotech), and 100 units/ml penicillin and 50 μg/ml streptomycin was added to the cells. Unless otherwise mentioned, a two-pulse regimen consisting of two 2-h pulses separated by 4 h was then initiated. In Figs. 2B and 4A, the first pulse consisted only of 20 min and was followed by a second 2-h pulse administered 4 h after the first pulse. Hormones and inhibitors were added as detailed in the figure captions. The 10−9 M estrogen conjugated to BSA (E-BSA) used in this study refers to the concentration of BSA. At the end of the first pulse (either 2 h or 20 min as mentioned in figure captions), the media containing hormones/inhibitors was removed from the cells and the cells were washed with PBS (D-PBS). Fresh media was added, free of hormones, for the duration of the 4 h, after which the second pulse of 2 h was initiated. At the end of the second pulse, the media containing the hormone was removed, the cells washed with D-PBS, and fresh phenol-red free media added. Incubation was continued for another 16 h. The cells were then lysed by using the Reporter Lysis buffer (Promega) according to the manufacturer's instructions and lysate from each sample was used for both luciferase assays and β-galactosidase (β-gal) assays. Luciferase and β-gal assays were performed according to the manufacturer's instructions (Promega). The β-gal assays were used to normalize both lysate preparation and transfection efficiency. The luciferase activity from each sample was normalized to the β-gal activity. Results are plotted (PRISM software, GraphPad, San Diego) as fold induction over that obtained in the vehicle-treated group. Results (bars in graphs) represent mean ± SEM and are obtained from replicate experiments (n = at least 4 per treatment group in each experiment). Statistical analysis was done by using one-way ANOVA followed by Student Newman Keuls post hoc test (PRISM software) to compare between treatment groups. A P value < 0.05 was taken as significant.
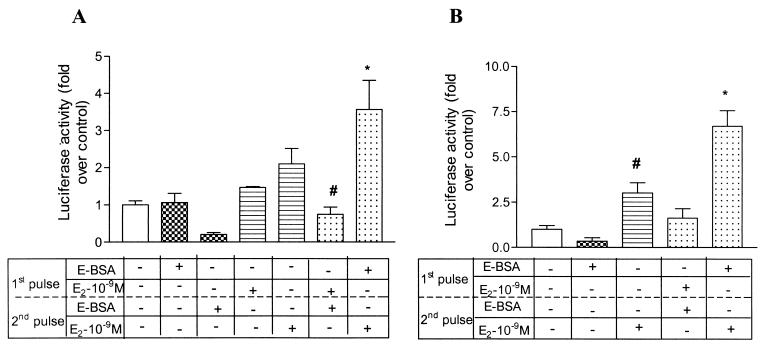
Figure 2.
Membrane-impermeable E-BSA in the first pulse can potentiate transactivation by 17β-estradiol in the second pulse. A brief description of the protocol and analysis is outlined in the legend for Fig. 1A. (A) E-BSA (checkered bars) or 17β-estradiol (horizontal bars) at 10−9 M were given in the first or second pulse alone. Control condition: 17β-estradiol (10−9 M) in the first pulse was followed by E-BSA (10−9 M) in the second pulse (sixth bar from left). Test of “potentiation” hypothesis: E-BSA given in the first pulse was followed by 17β-estradiol in the second pulse (far right bar). Results represent mean + SEM (n = 7 per treatment group from replicate experiments). One-way ANOVA followed by Student Newman Keuls post hoc test was used to compare between treatment groups. *, P < 0.001 compared with vehicle-treated group. #, P < 0.01 compared with the group administered E-BSA in the first pulse followed by 17β-estradiol in the second pulse (far right bar). (B) E-BSA (10−9 M), given for only 20 min in the first pulse, can potentiate transcription mediated by 17β-estradiol (10−9 M) in the second pulse. E-BSA (10−9 M) was given for 20 min in the first pulse, and 17β-estradiol (10−9 M) was given in the second pulse for a duration of 2 h. The two pulses were separated by 4 h. All other details are as in Fig. 1A. Control condition: 17β-estradiol (10−9 M) in the first pulse was followed by E-BSA (10−9 M) in the second pulse (fourth bar from left). Test of “potentiation” hypothesis: E-BSA given in the first pulse was followed by 17β-estradiol in the second pulse (far right bar). Results (n = 4 per treatment group from replicate experiments) are represented as mean + SEM. Statistical analysis was done by using one-way ANOVA, using Student Newman Keuls post hoc test to compare between treatment groups. *, P < 0.05 compared with all other treatment groups. #, P < 0.05 compared with the vehicle group.
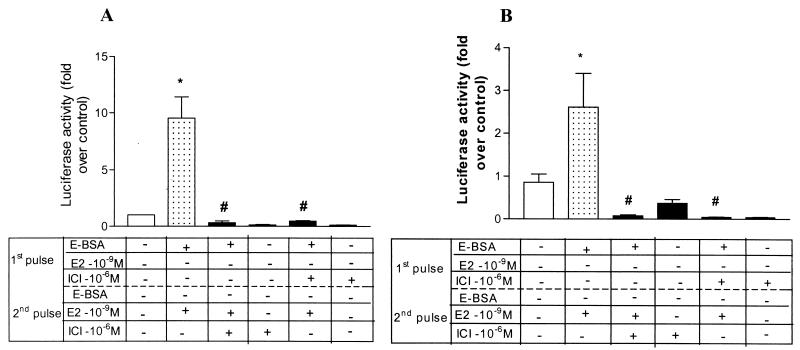
Figure 4.
ICI 182,780, an ER antagonist, blocks the potentiating effect of E-BSA from a brief 20-min first pulse (A) or a longer 2-h first pulse (B), as well as the transactivation of estrogen in the second pulse.(A) ICI 182,780 (10−6 M) was added either during the second 2-h pulse with 10−9 M 17β-estradiol (third bar from left) or during the first 20-min pulse with 10−9 M E-BSA (fifth bar from left). (B) ICI 182,780 (10−6 M) was added during the first 2-h pulse with 10−9 M E-BSA (fifth bar from left) and later in the second 2-h pulse with 10−9 M 17β-estradiol (third bar from left). Results are represented as mean + SEM. Statistical analysis was done by using one way ANOVA, using Student Newman Keuls post hoc test to compare between treatment groups. n = 5 and 8 per treatment group for A and B, respectively. *, P < 0.001 compared with vehicle-treated group. #, P < 0.001 compared with E-BSA given in the first pulse followed by 17β-estradiol in the second pulse.
Results
Use was made of a two-pulse paradigm of hormone administration shown effective for mediating estradiol's effects on uterine cell division (14) and reproductive behavior (15–17). Estrogen was given in two discrete pulses (Fig. 1), geared to initiate nongenomic actions with the first pulse, and then (Fig. 2) to limit direct genomic actions to the second pulse.
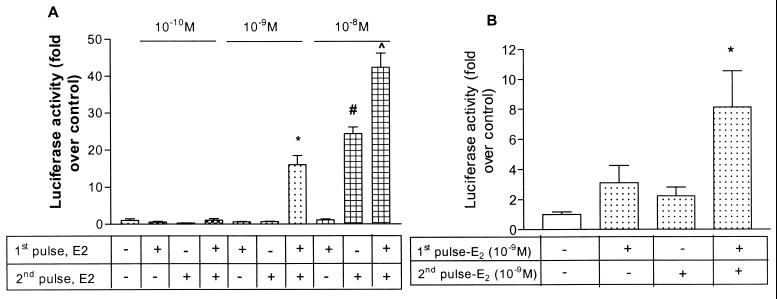
Figure 1.
(A) Different doses of 17β-estradiol administered in a two-pulse paradigm potentiate transactivation of a consensus ERE-driven construct. SK-N-BE2C, a human neuroblastoma cell line, was cotransfected with ERE-luciferase, pSG-hERα, and pSVβ-gal plasmids, 48 h after plating, using the Effectene reagent (Qiagen) according to the manufacturer's protocol. The day following the transfection, 17β-estradiol was added at the various concentrations detailed in the figure. Briefly, 17β-estradiol was added to the media twice for 2 h each, with these two pulses separated by an interval of 4 h. At the end of the second 2-h pulse of 17β-estradiol, the cells were washed free of hormones, fresh media was added, and incubation was continued for a further 16 h. Cell lysates were then prepared by using the Reporter Lysis buffer (Promega) as per the manufacturer's instructions (Promega) and the luciferase activity normalized to the β-galactosidase (β-gal) activity for every sample. The results (fold over control) are expressed as fold induction over that achieved by the vehicle, ethanol, alone. Results (n = 4 per treatment group) represent mean + SEM. Statistical analysis was done using one-way ANOVA followed by Student Newman Keuls post hoc test to compare between treatment groups. *, P < 0.001 compared with the vehicle-treated group or estradiol given in either of the two pulses at 10−9 M concentration. ^, P < 0.001 compared with the vehicle-treated group or estradiol given in the first pulse at 10−10 M concentration. #, P < 0.001 compared with vehicle-treated group or the group administered two pulses of 17β-estradiol. (B) The first pulse of 17β-estradiol potentiates the transactivation mediated by the second pulse in the SK-N-BE2C cell line. A brief description of the protocol and analysis is outlined in the legend for Fig. 1A. Results from replicate experiments are depicted; the bars represent mean + SEM (n = 8 per treatment group). Statistical analysis was done by using one-way ANOVA followed by Student Newman Keuls post hoc test to compare between treatment groups. 17β-estradiol given in both pulses (fourth bar from left) was compared with estradiol given in either the first or second pulse (second and third bars from left). *, P < 0.01 compared with vehicle-treated group and P < 0.05 compared with a estradiol given in either pulse alone.
In the SK-N-BE2C neuroblastoma cell line, we demonstrated the ability of discrete pulses of different concentrations of 17β-estradiol (Fig. 1A) to transactivate a luciferase-based reporter construct driven by three tandem consensus EREs, via ERα (18). We then used 10−9 M 17β-estradiol, a physiological level, for all subsequent experiments using the two-pulse paradigm. Can the first pulse amplify the transcriptional responses to the second pulse? In experiments with two 2-h pulses of estradiol given 4 h apart, expression of the reporter gene following the second pulse was enhanced by the first pulse (Fig. 1B).
To demonstrate that estrogen in the first pulse is acting through nongenomic mechanisms at the cell membrane, we used a membrane-impermeable E-BSA in the first pulse, followed by 17β-estradiol in the second pulse. The duration of the first pulse was shortened from 2 h (Fig. 2A) to 20 min (Fig. 2B), and in both cases this led to significantly higher transactivation of the luciferase reporter gene than seen with either pulse alone (Fig. 2 A and B). Therefore, the membrane-impermeable E-BSA at 10−9 M could potentiate the transcriptional activation by 17β-estradiol, at 10−9 M, liganded to the classical nuclear ER, ERα. Moreover, reversing the order—a first pulse of 17β-estradiol followed by a pulse of E-BSA—did not support transcription; this genomic effect required preceding nongenomic events occurring at the membrane (Fig. 2 A and B).
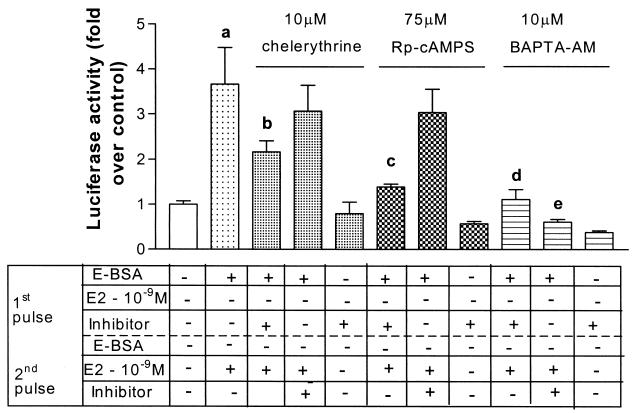
To further support the notion that the actions of the first pulse depend on early signal transduction, we investigated whether protein kinase A (PKA), protein kinase C (PKC), and/or calcium pathways play a role in E-BSA potentiation of ERα-mediated transcription. We used specific inhibitors coadministered in the first pulse along with E-BSA for 2 h (Fig. 3). The PKA-specific inhibitor (Rp-cAMPs) and the PKC-specific inhibitor (chelerythrine) were both capable of reducing the nongenomic E-BSA-mediated potentiation. These data demonstrate that both PKA and PKC play roles in the nongenomic mechanisms by which E-BSA potentiated the ERα-mediated genomic effect. As a control, administration of the inhibitors in the second pulse along with the 17β-estradiol, acting genomically, had no effect on the transactivation.
Figure 3.
Inhibitors of PKA, PKC, and Ca2+ release, block the ability of E-BSA in the first pulse to potentiate transactivation. SK-N-BE2C cells were treated as described in Fig. 1A. Chelerythrine (PKC inhibitor), Rp-cAMPS (PKA inhibitor), and BAPTA-AM (a calcium chelator) were coadministered with 10−9 M E-BSA in the first 2-h pulse (third, sixth, and ninth bars from the left, respectively) at the concentrations detailed above. They were also coadministered with 10−9 M 17β-estradiol in the second 2-h pulse (fourth, seventh, and tenth bars from the left) or given alone in the first 2-h pulse (fifth, eight, and eleventh bars from the left). Results, plotted as fold over control, represent mean + SEM (n = 8 per treatment group from replicate experiments). Statistical analysis was done by using one-way ANOVA, using Student Newman Keuls post hoc test to compare between treatment groups. a, P < 0.001 compared with vehicle treatment. b, P < 0.01 compared with E-BSA given in the first pulse followed by 17β-estradiol in the second pulse. c, d, and e, P < 0.001 compared with E-BSA given in the first pulse followed by 17β-estradiol in the second pulse.
The Ca2+ chelator, 1,2-bis(2-aminophenoxy)ethane-N,N,N′,N′-tetraacetate–acetoxymethyl ester (BAPTA-AM), blocked transcriptional activation when administered in either the first or second pulse (Fig. 3), implicating calcium release as a necessary event in both the nongenomic and genomic effects of estradiol. Phosphorylation of the ER and subsequent activation depends on a calcium-activated kinase (19, 20) and hence, calcium may also be essential for the genomic actions of the ER. In sum, early, membrane-limited effects of estradiol fostered later transcriptional facilitation in this neuroblastoma cell line.
Does this potentiation rely on the classical nuclear ER? Surprisingly, ICI 182,780 abolished the ability of E-BSA even in the first brief 20-min pulse (Fig. 4A), and in a first longer 2-h pulse (Fig. 4B), to activate the nuclear ER. This result suggests that this potentiation is mediated by classical ERα acting at the membrane or by another receptor with a highly homologous estrogen binding site. As expected, ICI 182,780 given in the second pulse, coadministered with 17β-estradiol for 2 h, blocked the genomic effect of ERα. ICI, a pure ER antagonist, blocks the ability of E-BSA to activate mitogen-activated protein kinase (MAPK) in Rat-2 fibroblasts (21), as well as the interaction between ERα and Gαi (22).
Discussion
Although nongenomic and genomic mechanisms have been widely viewed as alternate, discrete modes of steroid hormone action, we have shown in this study that they can synergize to potentiate transcription. The nongenomic mode of action in this study is demonstrated by using membrane-limited E-BSA. Although there may be concern that E can dissociate from the BSA to produce an effect on the nuclear receptor, other studies have reported a very small (<0.03%) dissociation of E-BSA (23, 24). These data may also explain a surprising result of Roy et al. (25, 26) in which anesthetization of a rat at the very time of estrogen administration, 48 h before behavioral test, contravened the ability of the estradiol to facilitate mating behavior. Interfering with the early membrane actions of the hormone elucidated here by disrupting normal neuronal membrane activity through anesthesia could easily account for their result.
These findings open as many questions as they answer. Through which pathways can estrogens signal the nerve cell nucleus, thus setting the stage for later genomic actions? Nongenomic actions of estradiol have been proposed to involve several signal transduction pathways, dependent on cell type. McDonnell and colleagues have provided evidence for MAP kinase pathway components and intracellular calcium as important players (27), whereas the molecular pharmacological results of O'Malley and his coworkers make a strong case for dopamine and subsequent cAMP signaling in the CNS (28–30). Mitogen-activated protein kinase (MAPK) has been implicated in estrogen's rapid action at the membrane in the related cell line SK-N-SH (31), as have PKA (32) and PKC (33). In a cell line with endogenous ERα, nongenomic mechanisms have been shown to activate MAPK (27). In addition, the rapid rise in intracellular free Ca2+ has been thought to be mediated by membrane estrogen receptors (34–36). In turn, the presence of ERα at membrane sites has been explored in several studies (37, 38).
The ability of nongenomic modes of steroid hormone action to potentiate nuclear transcriptional effects provides a unified view of estrogen action and has potential physiological relevance. Dopamine agonists have been shown to activate ER in the SK-N-SH cell line, an effect blocked by PKA antagonists (39). The presence of D1 receptor in the ventromedial hypothalamus (VMH), a brain region rich in ERα, and the ability of dopamine to induce lordosis (29) suggest that dopamine effects involving intracellular second messengers can activate ER and consequently result in an ER-controlled behavior.
Did one of the mechanisms of hormone action evolve first, or did they evolve in parallel? With respect to therapeutic uses of estrogens related to women's health, is one type of mechanism more important than the other? In any case, we can see that in the CNS, early responses to estrogens may be important for the eventual genomic responses that govern reproductive behaviors and other neuroendocrine functions.
Acknowledgments
We wish to thank Dr. Donald McDonnell for providing the ERE-luciferase construct and Dr. Pierre Chambon for the pSG-hER α construct. We also thank Ms. Elena Won and Ms. Elena Drekovic for technical assistance. This work was supported by National Institutes of Health Grant 05751.
Abbreviations
- ER
estrogen receptor
- PKA
protein kinase A
- PKC
protein kinase C
- ERE
estrogen response elements
- E-BSA
estrogen conjugated to BSA
References
- 1.Jensen E V, Jacobson H I. Recent Prog Horm Res. 1962;18:387–414. [Google Scholar]
- 2.Kelly M J, Levin E R. Trends Endocrinol Metab. 2001;12:152–156. doi: 10.1016/s1043-2760(01)00377-0. [DOI] [PubMed] [Google Scholar]
- 3.Kelly M J, Moss R L, Dudley C A. Brain Res. 1976;114:152–157. doi: 10.1016/0006-8993(76)91017-9. [DOI] [PubMed] [Google Scholar]
- 4.Kelly M J, Moss R L, Dudley C A. Exp Brain Res. 1977;30:53–64. doi: 10.1007/BF00237858. [DOI] [PubMed] [Google Scholar]
- 5.Kelly M J, Moss R L, Dudley C A, Fawcett C P. Exp Brain Res. 1977b;30:43–52. doi: 10.1007/BF00237857. [DOI] [PubMed] [Google Scholar]
- 6.Moss R L, Gu Q, Wong M. Recent Prog Horm Res. 1997;52:1–37. [PubMed] [Google Scholar]
- 7.Kousteni S, Bellido T, Plotkin L I, O'Brien C A, Bodenner D L, Han L, Han K, DiGregorio G B, Katzenellenbogen J A, Katzenellenbogen B S, et al. Cell. 2001;104:719–730. [PubMed] [Google Scholar]
- 8.Becker J B, Rudick C N. Pharmacol Biochem Behav. 1999;64:53–57. doi: 10.1016/s0091-3057(99)00091-x. [DOI] [PubMed] [Google Scholar]
- 9.Becker J B, Beer M E. Behav Brain Res. 1986;19:27–33. doi: 10.1016/0166-4328(86)90044-6. [DOI] [PubMed] [Google Scholar]
- 10.Becker J B, Rudick C N, Jenkins W J. J Neurosci. 2001;21:3236–3241. doi: 10.1523/JNEUROSCI.21-09-03236.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Becker J B. In: Behavioral Endocrinology. Becker J B, Breedlove M S, Crews D, editors. Cambridge, MA: MIT Press; 1992. pp. 325–356. [Google Scholar]
- 12.Pfaff D W. Drive: Neural and Molecular Mechanisms for Sexual Motivation. Cambridge, MA: MIT Press; 1999. [Google Scholar]
- 13.Zhu Y S, Yen P M, Chin W W, Pfaff D W. Proc Natl Acad Sci USA. 1996;93:12587–12592. doi: 10.1073/pnas.93.22.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harris J, Gorski J. Endocrinology. 1978;103:240–245. doi: 10.1210/endo-103-1-240. [DOI] [PubMed] [Google Scholar]
- 15.Parsons B, Rainbow T, Pfaff D, McEwen B. Nature (London) 1981;292:58–59. doi: 10.1038/292058a0. [DOI] [PubMed] [Google Scholar]
- 16.Clark A S, Roy E J. Physiol Behav. 1983;4:561–565. doi: 10.1016/0031-9384(83)90221-4. [DOI] [PubMed] [Google Scholar]
- 17.Sodersten P, Pettersson A, Eneroth P. Endocrinology. 1983;112:1883–1885. doi: 10.1210/endo-112-5-1883. [DOI] [PubMed] [Google Scholar]
- 18.Hall J M, McDonnell D P. Endocrinology. 1999;140:5566–5578. doi: 10.1210/endo.140.12.7179. [DOI] [PubMed] [Google Scholar]
- 19.Auricchio F, Migliaccio A, Castoria G, Rotondi A, Lastoria S. J Steroid Biochem. 1984;20:31–35. doi: 10.1016/0022-4731(84)90185-7. [DOI] [PubMed] [Google Scholar]
- 20.Auricchio F, Migliaccio A, Castoria G, Rotondi A, Di Domenico M, Pagano M. J Steroid Biochem. 1986;24:39–43. doi: 10.1016/0022-4731(86)90029-4. [DOI] [PubMed] [Google Scholar]
- 21.Wade C B, Robinson S, Shapiro R A, Dorsa D M. Endocrinology. 2001;142:2336–2342. doi: 10.1210/endo.142.6.8071. [DOI] [PubMed] [Google Scholar]
- 22.Wyckoff M H, Chambliss K L, Mineo C, Yuhanna I S, Mendelsohn M E, Mumby S M, Shaul P W. J Biol Chem. 2001;276:27071–27076. doi: 10.1074/jbc.M100312200. [DOI] [PubMed] [Google Scholar]
- 23.Caldwell J D, Moe B D. Horm Behav. 1999;35:38–46. doi: 10.1006/hbeh.1998.1494. [DOI] [PubMed] [Google Scholar]
- 24.Stevis P E, Deecher D C, Suhadolnik L, Mallis L M, Frail D E. Endocrinology. 1999;140:5455–5458. doi: 10.1210/endo.140.11.7247. [DOI] [PubMed] [Google Scholar]
- 25.Roy E J, Lynn D M, Clark A S. Brain Res. 1985;337:163–166. doi: 10.1016/0006-8993(85)91624-5. [DOI] [PubMed] [Google Scholar]
- 26.Quinones-Jenab V, Zhang C, Jenab S, Brown H, Pfaff D. Mol Brain Res. 1996;35:297–303. doi: 10.1016/0169-328x(95)00232-h. [DOI] [PubMed] [Google Scholar]
- 27.Improta-Brears T, Whorton A R, Codazzi F, York J D, Meyer T, McDonnell D P. Proc Natl Acad Sci USA. 1999;96:4686–4691. doi: 10.1073/pnas.96.8.4686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Apostolakis E M, Garai J, Fox C, Smith C L, Watson S J, Clark J H, O'Malley B W. J Neurosci. 1996;16:4823–4834. doi: 10.1523/JNEUROSCI.16-16-04823.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou D, Apostolakis E M, O'Malley B W. Biochem Biophys Res Commun. 1999;266:556–559. doi: 10.1006/bbrc.1999.1851. [DOI] [PubMed] [Google Scholar]
- 30.Mani S K, Allen J M, Clark J H, Blaustein J D, O'Malley B W. Science. 1994;265:1246–1249. doi: 10.1126/science.7915049. [DOI] [PubMed] [Google Scholar]
- 31.Watters J J, Campbell J S, Cunningham M J, Krebs E G, Dorsa D M. Endocrinology. 1997;138:4030–4033. doi: 10.1210/endo.138.9.5489. [DOI] [PubMed] [Google Scholar]
- 32.Watters J J, Dorsa D M. J Neurosci. 1998;18:6672–6680. doi: 10.1523/JNEUROSCI.18-17-06672.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sylvia V L, Walton J, Lopez D, Dean D D, Boyan B D, Schwartz Z. J Cell Biochem. 2001;81:413–429. doi: 10.1002/1097-4644(20010601)81:3<413::aid-jcb1055>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 34.Morley P, Whitfiled J F, Vanderhyden B C, Tsang B K, Schwartz J L. Endocrinology. 1992;131:1305–1312. doi: 10.1210/endo.131.3.1505465. [DOI] [PubMed] [Google Scholar]
- 35.Audy M C, Vacher P, Dufy B. Eur J Endocrinol. 1996;135:367–373. doi: 10.1530/eje.0.1350367. [DOI] [PubMed] [Google Scholar]
- 36.Stefano G B, Prevot V, Beauvillain J C, Fimiani C, Welters I, Cadet P, Breton C, Pestel J, Salzet M, Bilfinger T V. J Immunol. 1999;163:3758–3763. [PubMed] [Google Scholar]
- 37.Razandi M, Pedram A, Greene G L, Levin E R. Mol Endocrinol. 1999;13:307–319. doi: 10.1210/mend.13.2.0239. [DOI] [PubMed] [Google Scholar]
- 38.Clarke C H, Norfleet A M, Clarke M S, Watson C S, Cunningham K A, Thomas M L. Neuroendocrinology. 2000;71:34–42. doi: 10.1159/000054518. [DOI] [PubMed] [Google Scholar]
- 39.Gangolli E A, Conneely O M, O'Malley B W. J Steroid Biochem Mol Biol. 1997;61:1–9. doi: 10.1016/s0960-0760(97)00003-4. [DOI] [PubMed] [Google Scholar]