Abstract
Protein arginine methyltransferases (PRMT) catalyze protein arginine methylation and play an important role in many biological processes. Aberrant PRMT expression in tumor cells has been documented in several common cancer types; however, its precise contribution to hepatocellular carcinoma (HCC) cell invasion and metastasis is not fully understood. In this study, we identified a new oncogene, PRMT9, whose overexpression strongly promotes HCC invasion and metastasis. PRMT9 expression was detected more frequently in HCC tissues than in adjacent noncancerous tissues. PRMT9 overexpression was significantly correlated with hepatitis B virus antigen (HBsAg) status, vascular invasion, poor tumor differentiation and advanced TNM stage. Patients with higher PRMT9 expression had a shorter survival time and higher recurrence rate. PRMT9 expression was an independent and significant risk factor for survival after curative resection. Functional studies demonstrated that PRMT9 increased HCC cell invasion and lung metastasis. Knocking down PRMT9 with short hairpin RNA (shRNA) inhibited HCC cell invasion. Further investigations found that PRMT9 increased cell migration and invasion through epithelial‐mesenchymal transition (EMT) by regulating Snail expression via activation of the PI3K/Akt/GSK‐3β/Snail signaling pathway. In clinical HCC samples, PRMT9 expression was positively associated with Snail expression and was negatively associated with E‐cadherin expression. In conclusion, our study demonstrated that PRMT9 is an oncogene that plays an important role in HCC invasion and metastasis through EMT by regulating Snail expression via activation of the PI3K/Akt/GSK‐3β/Snail signaling pathway. Thus, PRMT9 may serve as a candidate prognostic biomarker and a potential therapeutic target.
Keywords: Akt, hepatocellular carcinoma, invasion, metastasis, PRMT9
1. INTRODUCTION
Hepatocellular carcinoma (HCC) is the fifth most common cancer and the third leading cause of cancer‐related mortality worldwide.1 Curative surgical resection is the most common treatment for HCC, but the 5‐year postoperative survival rate is unsatisfactory. This poor prognosis is mainly due to the high frequency of tumor recurrence and distant metastasis after curative surgical resection.2 Therefore, a better understanding of the molecular mechanisms underlying HCC metastasis will help prevent HCC recurrence and metastasis.
Epithelial‐mesenchymal transition (EMT) is known to be involved in cancer metastasis and is a process in which epithelial cancer cells lose their typical epithelial characteristics and acquire mesenchymal traits.3 During EMT, proteins involved in cell junctions such as E‐cadherin and ZO‐1 are downregulated, while mesenchymal markers such as Fibronectin and vimentin are upregulated.4 This conversion is believed to be triggered by multiple zinc finger proteins, including Snail, Slug, ZEB1, ZEB2 and Twist.5, 6, 7 As the regulators of EMT, zinc finger proteins directly promote the repression of E‐cadherin at the transcriptional level by binding to E‐box motifs, resulting in a loss of cell adhesion, which allows cells to migrate and metastasize.8 A large number of studies show that EMT are regulated by numerous signaling pathways, including phosphoinositide 3‐kinase (PI3K)/Akt‐, Hedgehog‐, Notch‐, Wnt‐ and nuclear factor‐κB (NF‐κB)‐dependent pathways.9
Cancer cells undergoing EMT become mobile and invasive, enabling metastasis and chemotherapy resistance.10 Recently, it was shown that HCC cells undergoing EMT play a vital role in the dissemination of malignant hepatocytes and, thus, influence HCC recurrence and prognosis.11, 12 Therefore, identifying the underlying molecular mechanism during HCC progression may ultimately provide innovative therapeutic strategies against HCC.
Protein arginine methylation is a widespread posttranslational modification that impacts numerous cellular processes, including transcription, RNA splicing, DNA repair, cell signaling and cell fate decisions.13, 14 This posttranslational modification is catalyzed by the 9 protein arginine methyltransferases (PRMT).14 According to their catalytic activity, PRMT are categorized into 3 groups. Type I enzymes, including PRMT1, PRMT2, PRMT3, PRMT4, PRMT6 and PRMT8, catalyze the synthesis of monomethylarginine (MMA) and asymmetric dimethylarginine (aDMA), whereas type II enzymes (PRMT5 and PRMT9) carry out the formation of MMA and sDMA.14, 15 PRMT7 is the only type III enzyme, and it catalyzes the formation of MMA.16 Numerous studies have shown that the aberrant expression PRMT is associated with cancer. These dysregulated proteins can impact multiple pathways that contribute to tumorigenesis, including the proliferation, cell cycle, apoptosis, and invasion and metastatic capacity of tumor cells.14, 17 Several studies have shown that PRMT1 and PRMT5 are overexpressed in HCC, and their high expression indicates a poor prognosis in HCC patients.18, 19 This evidence implies that PRMT may play a crucial role in HCC progression. However, the roles of PRMT, especially PRMT9, in the regulation of HCC invasion and metastasis have not been fully elucidated.
In this study, we identified that PRMT9 has an important role in HCC invasion and in the EMT process of HCC cells. Further investigation showed that PRMT9 was upregulated in HCC tissues and cell lines. High expression of PRMT9 indicated a poor prognosis in HCC patients. Moreover, both in vitro and in vivo experiments indicated that PRMT9 had a strong effect on the invasion and metastasis of HCC. The molecular mechanism assays demonstrated that PRMT9 promoted the invasion of HCC cells via PI3K/Akt/GSK‐3β/Snail pathway.
2. MATERIALS AND METHODS
2.1. Cell lines, culture conditions and reagents
In this study, the HCC cell lines Huh7, MHCC97H and Sk‐Hep1 and an immortalized hepatocyte cell line, LO2, were obtained from the Cell Bank of Type Culture Collection of Chinese Academy of Sciences. The human HCC cell line Bel7402 was purchased from the Shanghai Institute of Cell Biology (Shanghai, China). All cell lines were maintained in high‐glucose DMEM supplemented with 10% heat‐inactivated FBS at 37°C with 5% CO2. The inhibitor 2‐morpholin‐4‐yl‐8‐phenylchromen‐4‐one (LY294002) was purchased from Selleckchem (Houston, TX, USA).
2.2. Patients and samples
Paired HCC tissues and adjacent non‐tumor liver tissues were obtained from HCC patients undergoing curative resection between 2000 and 2005 at Sun Yat‐Sen Memorial Hospital. A total of 139 HCC patients who had distinctive pathological diagnoses, no preoperative systemic or local treatments, and complete follow‐up data were enrolled in this study. All patients were monitored after surgery until 12 May 2013. The histological grade of tumor differentiation was determined according to the method of Edmondson and Steiner. The pTNM classification for HCC was determined according to The American Joint Committee on Cancer/International Union Against Cancer Staging System (7th edition, 2010). This study was approved by the research ethics committee of Sun Yat‐Sen University, and written informed consent was obtained from each patient. In addition, 20 pairs of HCC tissues with corresponding portal vein tumor thrombosis were collected from archived paraffin‐embedded samples.
2.3. Immunohistochemistry
A tissue microarray (TMA) containing 139 cases of paired HCC samples and corresponding non‐tumor tissues was analyzed. The immunochemistry staining process was strictly performed using the Dako Envision Plus System (Dako, Glostrup, Denmark) according to the manufacturer's protocol. The TMA was stained for PRMT9, Snail and E‐cadherin expression. Immunohistochemistry (IHC) staining was scored based on both staining intensity and the extent of protein expression by 2 independent pathologists who were blind to all patient clinical data. The percentage of positive cells was classified as follows: 0% ≤ 5%, 1 = 5%‐25%, 2 = 25%‐50%, 3 = 50%‐75% and 4 ≥ 75% positive cells. The intensity of staining was defined according to the following: 0 (negative), 1 (weak), 2 (moderate) or 3 (strong). The final protein expression score was calculated by multiplying the intensity score by the positive‐staining score. Each sample was defined as either negative expression if the final score was 0‐4 or positive expression if the final score was >4. The primary antibodies and dilutions used for IHC are listed in Table S1.
2.4. Plasmids and stable overexpression and knockdown of PRMT9 in hepatocellular carcinoma cells
The pLV‐Puro‐GFP‐PRMT9 lentiviral vector was purchased from Guangzhou Cyagen Biosciences. The GV248‐GFP‐shRNA‐PRMT9 lentiviral vectors containing shRNA targeting PRMT9 were purchased from Shanghai GeneChem and the hairpin shRNA sequences are described in Document S1. The lentiviruses produced from pLV‐Puro‐GFP‐PRMT9 lentiviral vectors were used to infect Huh7 and Bel7402 cells, and the shRNA‐PRMT9 lentiviruses were used to infect SK‐Hep1 cells. Cells infected with pLV‐Puro‐GFP or PGCSIL‐GFP were used as controls. Infected cells were selected with 2 μg/mL puromycin for 2 weeks.
2.5. Wound healing assay
Cells were grown to 100% confluence in 6‐well plates. The confluent cell monolayer was scratched with a 200‐μL sterile plastic pipette tip to produce an artificial wound. Cell debris was removed by washing with PBS, and cells were cultured in fresh medium without FBS for 48 hours. Then, the rate of wound closure was examined and photographed.
2.6. Cell migration and invasion assay
Cell migration and invasion ability were measured using Corning Transwell chambers (Corning, Corning, NY, USA) precoated with or without Matrigel (BD Biosciences, Franklin Lake, NJ, USA). DMEM medium (600 μL) containing 10% FBS was placed in the lower chambers. Next, 1 × 105 cells were suspended in 200 μL of serum‐free medium, added to the upper chambers and incubated at 37°C for 24 hours. Migrating and invading cells were fixed with 4% paraformaldehyde solution for 30 minutes, stained with 0.2% crystal violet for 30 min and counted under a microscope.
2.7. In vivo metastasis assays
BALB/C nude mice (5 weeks old) were bred under standard conditions and maintained according to the institutional guidelines for animal care. All animal research procedures were approved by the Animal Care and Use Ethics Committee, Sun Yat‐Sen University. For the in vivo lung metastasis assay, Bel7402‐control and Bel7402‐PRMT9 (2 × 106 in 25 μL PBS) cells were injected orthotopically into the left liver lobe of BALB/C nude mice (10 in each group). Six weeks later, the mice were killed, and the lung tissues were harvested. Liver and lung tissues were paraffin‐embedded and stained with H&E for histological validation.
2.8. RNA extraction and quantitative real‐time PCR
Total RNA from the HCC cell lines and snap‐frozen clinical specimens were extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) and reverse transcribed into cDNA using the Superscript III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer's instructions. The mRNA expression levels were determined by real‐time RT‐PCR analysis, which was performed by using a Light Cycler 480 system (Roche Applied Science, Penzberg, Germany) and SYBR Green Master Mix (Takara Bio, Otsu, Japan). The primer sequences used in this study are listed in Document S1.
2.9. Western blot analysis
Western blotting was performed in accordance with the standard methods. Briefly, cells were lysed in RIPA buffer containing protease inhibitor cocktail (FDbio Science, Shanghai, China). Protein extracts were separated by SDS‐PAGE and transferred to PVDF membranes (Millipore, Bedford, MA, USA). After blocking with 5% non‐fat dry milk, the membranes were incubated with primary antibody at 4°C overnight, followed by an incubation with HRP‐conjugated secondary antibody for 1 hours at room temperature. Immunoreactive bands were detected by enhanced chemiluminescence (ECL; Millipore, Billerica, MA, USA). The primary antibodies and dilutions used for western blot analysis are listed in xTable S1.
2.10. Immunofluorescence assay
Cells were fixed with 4% paraformaldehyde for 15 minutes at 4°C on glass coverslips and permeabilized with 0.1% Triton‐X 100 for 10 minutes at 4°C. Then, the cells were blocked with 1% BSA for 1 hours at 4°C and incubated with primary antibody at 4°C overnight. After rinsing with PBS, the cells were incubated with AlexaFluor 555‐conjugated goat‐anti‐rabbit IgG for 1 hours at room temperature. Cell nuclei were subsequently stained with DAPI for 3 minutes. Finally, the cells were examined using a confocal laser‐scanning microscope (LSM710, Carl Zeiss GmbH, Oberkochen, Germany). The primary antibodies and dilutions used for immunofluorescence assays are listed in Table S1.
2.11. Statistical analysis
Statistical analyses were performed using SPSS 17.0 software. Data are expressed as the means ± SD of 3 independent experiments. Quantitative data between groups were compared using Student's t test. Categorical data were analyzed using Pearson's χ2‐test or Fisher's exact test. Correlation analysis was performed for PRMT9, Snail and E‐cadherin. Survival analyses and cumulative recurrence rates were assessed using the Kaplan–Meier method and the log‐rank test. Univariate and multivariate survival analyses were performed using the Cox proportional hazards regression model. Results were considered statistically significant when P < .05.
3. Results
3.1. The expression of PRMT9 in hepatocellular carcinoma tissues and its correlation with patient clinicopathological features and survival
To explore whether PRMT9 is an important factor in determining the clinical outcomes of HCC patients, the expression of PRMT9 in a TMA containing 139 pairs of primary HCC was detected. In HCC tissues, positive PRMT9 staining was observed primarily in the nucleus, whereas in adjacent liver tissue, weak positive staining of PRMT9 was observed in the cytoplasm (Figure 1A1). Approximately half of the HCC samples (69/139, 49.6%) were positive for PRMT9 expression. In contrast, only 10.8% (15/139, 10.8%) of the adjacent liver samples expressed PRMT9 (P < .01, Fisher's exact test; Figure 1A2). However, the mRNA levels of PRMT9 were not changed between tumor and adjacent non‐tumor tissues in an additional 33 HCC samples, which suggested that the overexpression of PRMT9 was only at the post‐transcriptional level (data not shown).
Figure 1.
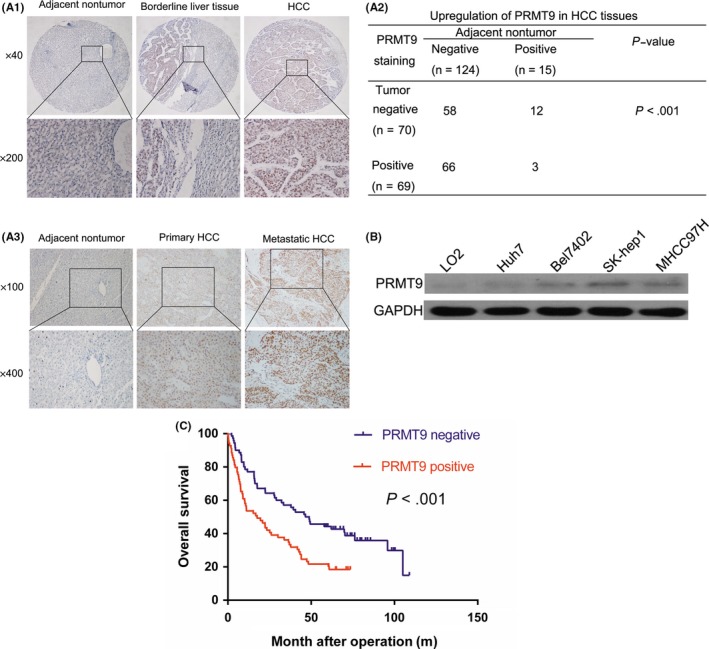
Overexpression of PRMT9 in hepatocellular carcinoma (HCC) tissues and its correlation with patient clinicopathological features and survival. A1, Representative immunohistochemical staining of PRMT9 expression in matched primary HCC samples (n = 139), borderline liver tissue (n = 139) and the corresponding non‐tumor tissue (n = 139). A2, Statistical analysis of PRMT9 expression in HCC. A3, Representative PRMT9 expression in adjacent non‐tumor tissues (n = 20), primary HCC tissues (n = 20) and portal vein tumor thrombosis tissues (n = 20) detected by immunohistochemistry (IHC). B, Western blotting analysis of PRMT9 expression in different HCC cell lines. C, Kaplan‐Meier analysis of the correlation between PRMT9 expression and overall survival (OS) in HCC patients
To investigate the role of PRMT9 in HCC metastasis, PRMT9 expression was compared in 20 pairs of matched primary and portal vein tumor thrombosis samples. A representative example of PRMT9 immunohistochemical staining is shown in Figure 1A3. Among the HCC specimens, 12 pairs (60%) had higher PRMT9 expression levels in the portal vein tumor thrombosis samples than in the primary HCC tissues (Figure 1A3).
PRMT9 expression levels were compared between the noncancerous LO2 liver cell line and the 4 HCC cell lines. Western blot assays showed that PRMT9 expression was increased in highly metastatic HCC cell lines (SK‐Hep1 and MHCC97H; Figure 1B).
Upregulation of PRMT9 was significantly associated with hepatitis B virus antigen (HBsAg, P = .011), vascular invasion (P = .02), tumor differentiation (P = .02) and TNM stage (P = .016; Table 1). Kaplan–Meier analysis showed that HCC patients with positive PRMT9 staining had shorter overall survival times than those with negative PRMT9 expression (Figure 1C). Cox's multivariate proportional hazards model indicated that PRMT9‐positive staining was an independent predictor of survival in HCC patients after curative resection (Table 2). Taken together, these observations indicated that the overexpression of PRMT9 may contribute to HCC metastasis.
Table 1.
Correlation between PRMT9 expression and patient's clinicopathologic features in hepatocellular carcinoma
| Clinicopathological variables | Case number (n = 139) | Tumor PRMT9 expression | ||
|---|---|---|---|---|
| Negative (n = 70) | Positive (n = 69) | P value | ||
| Age | ||||
| ≤60 years | 101 | 52 | 49 | .404 |
| ≥60 years | 38 | 18 | 20 | |
| Sex | ||||
| Male | 117 | 59 | 58 | .577 |
| Female | 22 | 11 | 11 | |
| Serum AFP | ||||
| ≤20 ng/mL | 42 | 17 | 25 | .079 |
| >20 ng/mL | 94 | 52 | 42 | |
| HBsAg | ||||
| Negative | 18 | 14 | 4 | .011* |
| Positive | 121 | 56 | 65 | |
| Cirrhosis | ||||
| Absent | 33 | 16 | 17 | .481 |
| Present | 106 | 54 | 52 | |
| Tumor size | ||||
| ≤5 cm | 54 | 31 | 23 | .125 |
| >5 cm | 85 | 39 | 46 | |
| Vascular invasion | ||||
| Absent | 93 | 53 | 40 | .020* |
| Present | 46 | 17 | 29 | |
| Tumor encapsulation | ||||
| Absent | 63 | 28 | 35 | .136 |
| Present | 76 | 42 | 56 | |
| Tumor differentiation | ||||
| I‐II | 101 | 25 | 13 | .020* |
| III‐IV | 38 | 33 | 6 | |
| TNM stage | ||||
| I‐II | 66 | 40 | 26 | .016* |
| III‐IV | 73 | 30 | 43 | |
*indicates P < 0.05.
Table 2.
Univariate and multivariate analysis of factors associated with survival and recurrence of 139 hepatocellular carcinoma
| Variables | Survival | Recurrence | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Univariate analysis | Multivariate analysis | Univariate analysis | Multivariate analysis | |||||||||
| HR | 95% CI | P‐value | HR | 95% CI | P‐value | HR | 95% CI | P‐value | HR | 95% CI | P‐value | |
| Age (≤60.0 vs >60.0) | 0.700 | 0.461‐1.062 | .093 | 1.573 | 1.042‐2.373 | .031* | ||||||
| Sex (male vs female) | 0.811 | 0.461‐1.429 | .469 | 1.218 | 0.744‐1.994 | .432 | ||||||
| Serum AFP (≤20 vs >20 ng/mL) | 1.294 | 0.837‐2.000 | .246 | 1.374 | 0.928‐2.034 | .112 | ||||||
| HBsAg (negative vs positive) | 1.182 | 0.669‐2.087 | .564 | 1.138 | 0.671‐1.931 | .631 | ||||||
| Tumor encapsulation (absent vs present) | 0.666 | 0.449‐0.989 | .044* | 0.559 | 0.387‐0.808 | .002* | ||||||
| Cirrhosis (absent vs present) | 1.408 | 0.877‐2.261 | .157 | 1.089 | 0.715‐1.659 | .692 | ||||||
| Tumor size (≤5 vs >5 cm) | 1.840 | 1.211‐2.795 | .004* | 1.898 | 1.290‐2.795 | .001* | ||||||
| Vascular invasion (absent vs present) | 4.527 | 2.968‐6.904 | <.001* | 4.426 | 2.205‐8.817 | <.001* | 2.196 | 1.460‐3.301 | <.001* | |||
| Tumor differentiation (I‐II vs III‐IV) | 1.314 | 0.847‐2.039 | .223 | 1.433 | 0.939‐2.186 | .095 | ||||||
| TNM stage (I‐II vs III‐IV) | 3.261 | 2.150‐4.945 | <.001* | 1.916 | 1.080‐3.401 | .026* | 2.401 | 1.632‐3.531 | <.001* | 1.720 | 1.046‐2.282 | .032* |
| Tumor PRMT9 expression (negative vs positive) | 1.968 | 1.315‐2.945 | .001* | 1.284 | 1.020‐1.616 | .033* | 1.528 | 1.055‐2.212 | .025* | |||
*indicates P < 0.05.
3.2. PRMT9 promotes hepatocellular carcinoma cell migration and tumor metastasis
Because our clinicopathological correlation analysis indicated that PRMT9 upregulation was significantly associated with vascular invasion, we further investigated the role of PRMT9 in HCC cell migration and invasion. Two stable cell lines, Huh7‐PRMT9 and Bel7402‐PRMT9, were established for this purpose. Wound healing assays showed that the upregulation of PRMT9 expression significantly increased the capacity of Huh7 and Bel7402 cells to close the scratched “wound” (Figure 2A,B and Figure S1A,B). Transwell assays further revealed that the upregulation of PRMT9 enhanced the migration and invasion of Huh7 and Bel7402 cells (Figure 2C,D). In contrast, when PRMT9 was silenced by targeted shRNA, cell migration and invasion were significantly inhibited (Figure 2E,F and Figure S1C).
Figure 2.
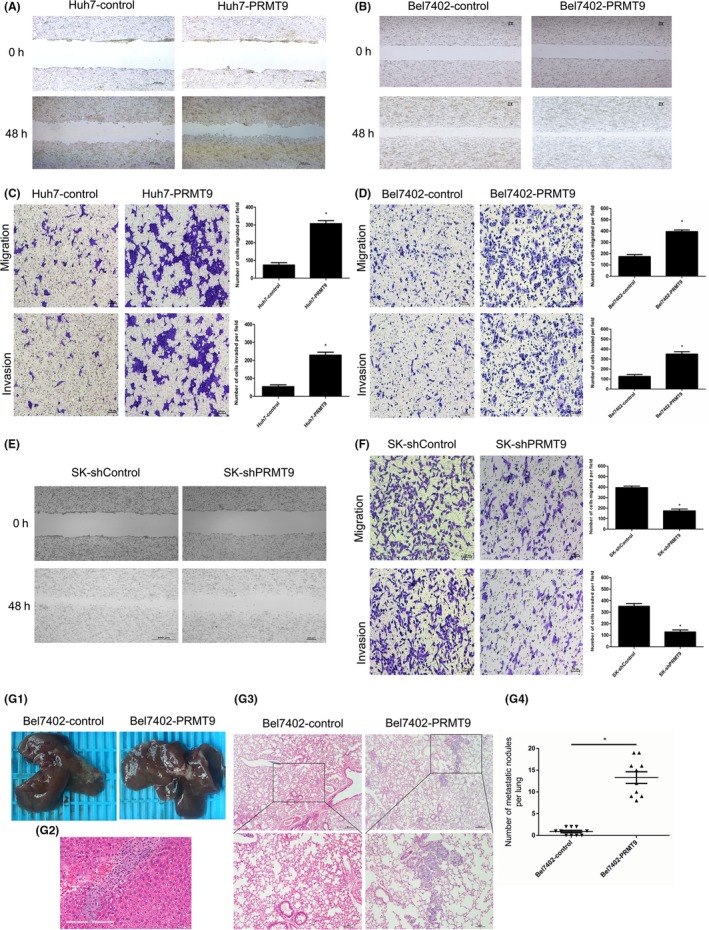
PRMT9 promotes hepatocellular carcinoma (HCC) cell migration and tumor metastasis. A,B, Wound healing assays showing PRMT9‐promoted cell migration. Representative images of wound healing assays were taken at 0 and 48 h after the scratch was made. C,D, Migration and invasion assay analysis showing that the upregulation of PRMT9 enhanced cell migration and invasion in vitro. E, Wound healing assays showing that silencing PRMT9 inhibited cell migration. Representative images of wound healing assays were taken at 0 and 48 h after scratching. F, Migration and invasion assay analysis showing that the silencing of PRMT9 inhibited cell migration and invasion in vitro. G, In vivo metastasis assay. Bel7402 cells with either the empty or PRMT9 overexpression vector (n = 10 per group) were transplanted into the livers of nude mice. G1, Representative views of tumor nodules at six weeks following orthotopic implantation. G2, Representative images of microscopic tumor nodes in liver stained with H&E. G3, Incidence of metastasis in the Bel7402 group of nude mice. G4, Representative images of microscopic tumor nodes in lungs stained with H&E. (All the experiments were repeated 3 times; *P < .05 indicates significant difference in independent Student's t test.)
To further evaluate the effect of PRMT9 on tumor metastasis in vivo, cells were injected into the livers of nude mice. The PRMT9‐expressing group showed an increased incidence of lung metastasis compared with that of the control group (Figure 2G). These data indicated that PRMT9 had an important role in promoting HCC invasion and metastasis.
3.3. PRMT9 induces epithelial‐mesenchymal transition in hepatocellular carcinoma
As PRMT9 was involved in cell migration and invasion, we speculated that PRMT9 may affect the EMT process. The relationship among PRMT9 and EMT markers as well as EMT‐related transcription factors was investigated. Real‐time PCR showed that the upregulation of PRMT9 in Huh7 and Bel7402 cells led to a decreased expression of epithelial markers (E‐cadherin and ZO‐1) and an increased expression of mesenchymal markers (vimentin and fibronectin; Figure 3A,B). Moreover, by screening 5 EMT‐related transcription factors, we found that PRMT9 markedly increased Snail protein expression (Figure 3D). In contrast, silencing PRMT9 in SK‐Hep1 cells caused the opposite expression pattern of EMT markers and Snail (Figure 4A,C).
Figure 3.
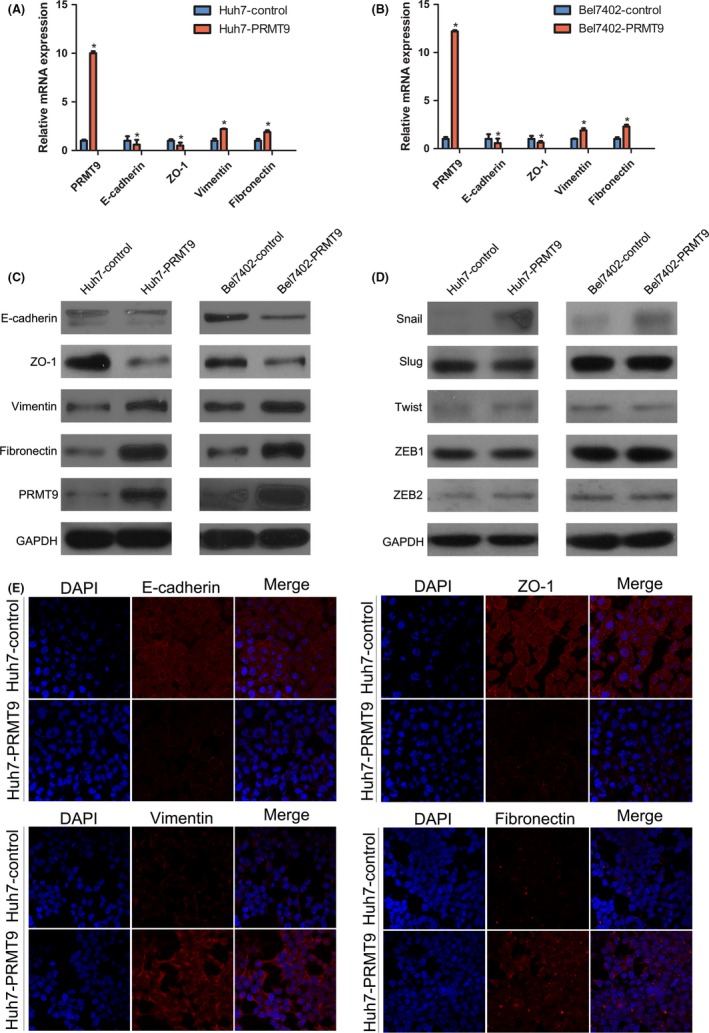
PRMT9 induces epithelial‐mesenchymal transition in hepatocellular carcinoma (HCC). A,B, The relative expression levels of E‐cadherin, ZO‐1, vimentin and fibronectin, as measured by qPCR, were compared between PRMT9‐expressing and control cells. C,D, Western blot analysis showed changes in the expression of EMT markers (E‐cadherin, ZO‐1, vimentin and fibronectin) snail, slug, twist, ZEB1 and ZEB2 in HCC cell lines. E, Representative immunofluorescent staining (IF) images showing the increased expression of vimentin and fibronectin and decreased expression of E‐cadherin and ZO‐1 in Huh7‐PRMT9 cells compared with the expression levels of these proteins in Huh7‐control cells. Nuclei were counterstained with DAPI. (All the experiments were repeated 3 times; *P < .05 indicates significant difference in independent Student's t test.)
Figure 4.
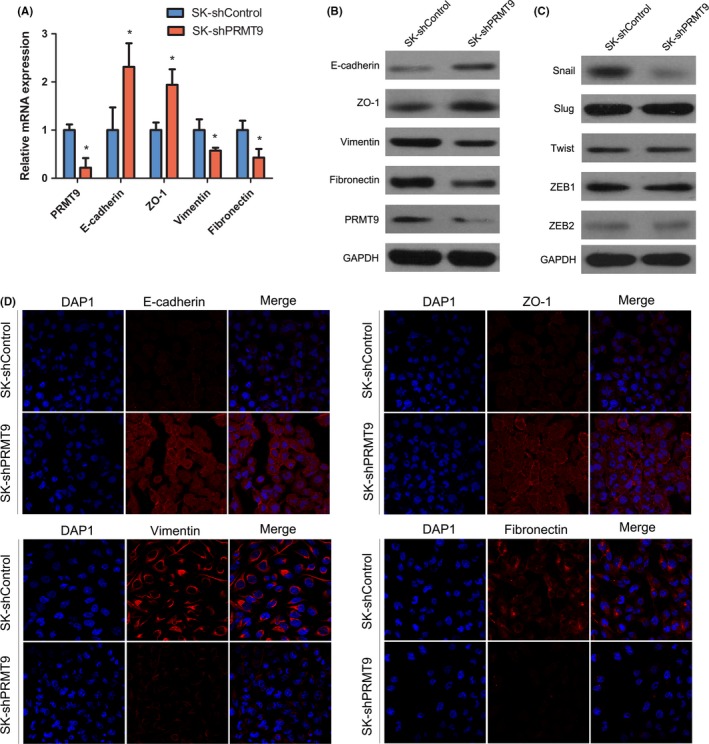
Silencing of PRMT9 reverses epithelial‐mesenchymal transition in hepatocellular carcinoma (HCC). A, The RT‐qPCR results showing changes in the expression of epithelial‐mesenchymal transition (EMT) markers (E‐cadherin, ZO‐1, vimentin and fibronectin) in the indicated HCC cell lines. B, Western blot analysis showing changes in EMT marker expression (E‐cadherin, ZO‐1, vimentin and fibronectin) in the indicated HCC cell lines. C, Western blots showing changes in snail, slug, twist, ZEB1 and ZEB2 expression. D, Representative immunofluorescent staining (IF) images showing the decreased expression of vimentin and fibronectin and increased expression of E‐cadherin and ZO‐1 in SK‐shPRMT9 cells compared with these expression levels in SK‐shControl cells. Nuclei were counterstained with DAPI. (All the experiments were repeated 3 times; *P < .05 indicates significant difference in independent Student's t test.)
Western blotting analysis and immunofluorescent staining further showed that PRMT9 decreased epithelial markers (E‐cadherin and ZO‐1) but increased the expression of mesenchymal markers (vimentin and fibronectin) and Snail (Figure 3C, E). The results from PRMT9 knockdown in SK‐Hep1 cells were consistent with these findings (Figure 4B‐D). These results indicated that PRMT9 induced EMT in HCC cells.
3.4. PRMT9 induces epithelial‐mesenchymal transition by activating the PI3K/Akt/GSK‐3β/Snail pathway in hepatocellular carcinoma
It is well known that AKT activation plays a crucial role in inducing EMT by inhibiting GSK‐3β activation and subsequently leading to Snail stabilization. In addition, it has been reported that the Akt signaling pathway is constitutively active in HCC.20 We therefore hypothesized that PRMT9 might promote HCC metastasis through the PI3K/Akt/GSK‐3β/Snail pathway. Western blot assays showed increased expression of phosphorylated Akt, phosphorylated GSK‐3β and Snail in PRMT9‐transfected cells (Figure S2A). In contrast, in PRMT9‐silenced cells, the expression of phosphorylated Akt, phosphorylated GSK‐3β and Snail were significantly reduced compared with the expression levels in SK‐shControl cells (Figure S2B). These results indicated that PRMT9 probably enhanced Snail expression via the PI3K/Akt/GSK‐3β/Snail pathway. To further elucidate whether PRMT9 induced the expression of Snail by activating Akt signaling, PRMT9‐infected cells were treated with LY294002 (a PI3K‐specific inhibitor). The results showed that the expression levels of phosphorylated Akt, phosphorylated GSK‐3β, Snail and EMT markers were reversed by LY294002 treatment in PRMT9‐infected cells (Figure 5A,B). These results indicated that the PRMT9‐induced expression of Snail was probably dependent on the Akt pathway. Next, we further determined if the oncogenic effect of PRMT9 was through the activation of the Akt/GSK‐3β/Snail pathway. The migration ability of PRMT9‐infected cells was inhibited by LY294002, as indicated by Transwell and wound healing assays (Figure 5C,D and Figure S3).
Figure 5.
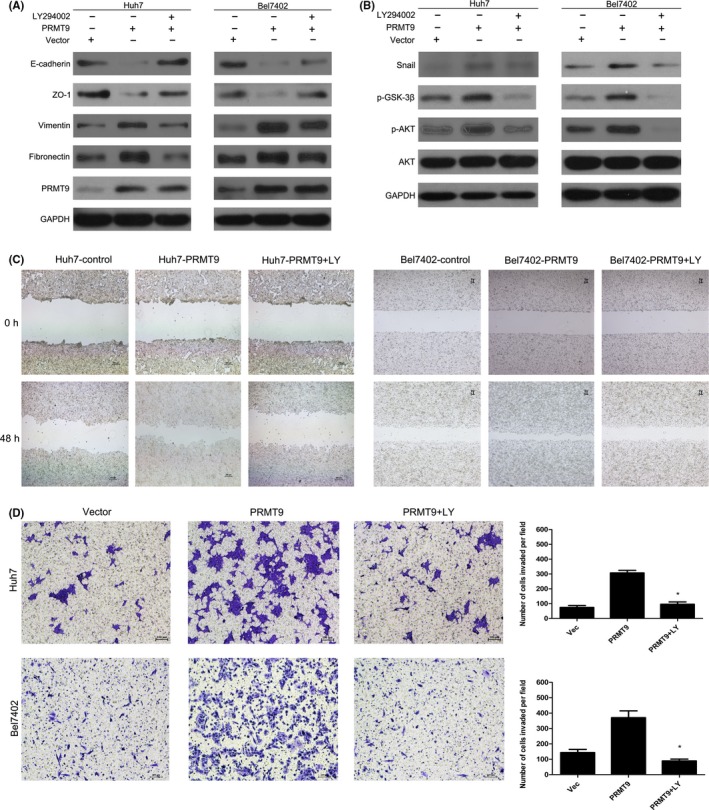
PRMT9 induces epithelial‐mesenchymal transition (EMT) via activating the PI3K/Akt/GSK‐3β/Snail pathway in hepatocellular carcinoma (HCC). A,B, Western blot analysis showing that the Akt inhibitor LY294002 effectively inhibits the activation of the PI3K/Akt signaling induced by PRMT9 and EMT. C, Wound healing assay showing that LY294002 inhibits PRMT9‐induced cell migration. D, Invasion assays showing that LY294002 inhibits PRMT9‐induced cell invasion. (All the experiments were repeated 3 times; *P < .05 indicates significant difference in independent Student's t test.)
3.5. High PRMT9 expression correlates with Snail and E‐cadherin expression in hepatocellular carcinoma tissues
We further evaluated the expression of PRMT9, Snail, and the EMT marker E‐cadherin using IHC in a TMA consisting of primary tumors from 139 HCC patients. IHC showed that PRMT9 expression was positively correlated with Snail expression and was negatively correlated with E‐cadherin expression (Figure 6A,B). To further explore the relationship between PRMT9 and Snail or E‐cadherin during HCC metastasis, 20 paired primary and portal vein tumor thrombosis tissues were investigated (Figure 6C,D). Our results showed that higher levels of PRMT9 and Snail expression were detected in metastatic HCC tissues than in primary HCC tissues. For the same patients, a lower level of E‐cadherin expression was detected in portal vein tumor thrombosis tissues than in primary HCC tissues. These data further support the crucial role of PRMT9 in HCC metastasis.
Figure 6.
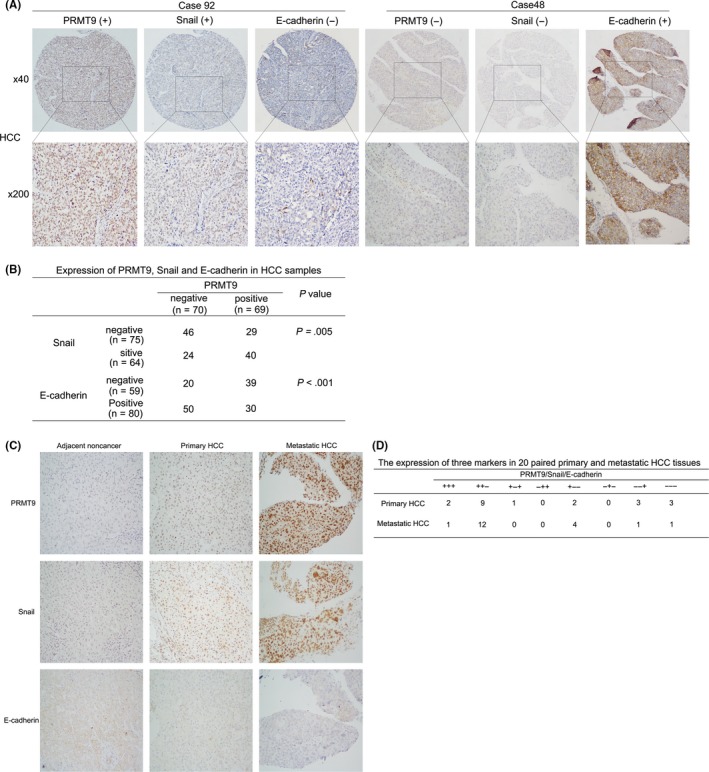
High expression of PRMT9 correlates with Snail and E‐cadherin in hepatocellular carcinoma (HCC) tissues. A, Expression of PRMT9, Snail and E‐cadherin in HCC samples (n = 139); Case 92: high expression of both PRMT9 and Snail and low expression of E‐cadherin; Case 48: low expression of both PRMT9 and Snail and high expression of E‐cadherin. B, Statistical analyses of the expression of 3 markers in HCC. C, Immunohistochemical analysis of PRMT9, Snail and E‐cadherin expression in 20 paired primary and metastatic HCC tissues. Representative immunohistochemical staining of all 3 markers is shown. D, Comprehensive correlation analysis of the expression of the 3 markers in HCC
4. DISCUSSION
In this study, we first identified a novel gene, PRMT9, whose overexpression promotes the invasion and EMT of HCC cells. Second, we determined that the PRMT9‐related enhancement HCC cell invasion and metastasis may depend on the activation of the PI3K/Akt/GSK‐3β/Snail pathway. Finally, in clinical HCC tissues, we observed that PRMT9 expression was positively associated with Snail expression and was negatively associated with E‐cadherin expression. In summary, our findings indicate that PRMT9 has a crucial role in facilitating HCC EMT and metastasis. PRMT9 may serve as a candidate prognostic biomarker and a potential therapeutic target in HCC patients.
PRMT9 was recently identified as a new type II protein arginine methyltransferases.15 It contains a conserved core methyltransferase domain, consisting of S‐adenosyl‐L‐methionine binding sequences and substrate binding sequences, and a THW loop. Like PRMT5, another type II PRMT, PRMT9, transfers methyl groups on different terminal guanidino nitrogen atoms to form MMA and ADMA residues.15, 21 For example, Yang et al and Hadjikyriacou et al showed that PRMT9 symmetrically demethylates arginine residues on SAP145 and SF3B2.15, 21 Because SF3B2 is involved in cell cycle progression and is often hijacked by HIV, SF3B2 methylation may be related to disease progression. This indicates that PRMT9 may play an important role in disease progression. Furthermore, Uhlén et al report that the high expression of PRMT9 has been detected in several types of cancer, including melanoma, and testicular, pancreatic and lymphoma cancer.21 These studies imply that PRMT9 plays a role in cancer development. However, until now, no study has reported the role of PRMT9 in HCC. Our findings showed that, compared with normal liver tissues, the protein levels of PRMT9 were significantly increased in HCC tissues. In addition, 12 pairs of HCC tissue (60%) showed higher levels of PRMT9 expression in the portal vein tumor thrombosis samples than in the paired primary HCC samples. Furthermore, we observed an inverse correlation between PRMT9 expression and prognosis in a large cohort of HCC patients. A clinicopathological correlation study showed that PRMT9 overexpression was correlated with HBsAg status, vascular invasion, poor tumor differentiation and advanced TNM stage. These results imply that PRMT9 may play an important role during HCC progression, especially in HCC metastasis. In fact, our in vitro and in vivo studies confirmed that PRMT9 plays an important role specifically in HCC invasion and metastasis.
Epithelial‐mesenchymal transition plays an important role in cancer cell invasiveness and metastasis.22, 23 The EMT process is believed to be involved in the progression of HCC and correlates with HCC patient prognosis.24 Although EMT is triggered by numerous factors, whether PRMT9 promotes EMT was undetermined. To explore the exact function of PRMT9 in HCC progression, we created cells lines with PRMT9 overexpression and knockdown. As expected, decreased epithelial marker expression and increased mesenchymal marker expression were observed in HCC cells when PRMT9 was overexpressed. In contrast, knocking down PRMT9 increased the expression of epithelial markers and decreased the expression of mesenchymal markers. In addition, the key regulator of EMT, Snail, was significantly altered by PRMT9. The protein levels of Snail were significantly downregulated when PRMT9 was silenced, whereas the level of Snail was increased when PRMT9 was overexpressed. These results indicated that PRMT9 may induce EMT in HCC cells and play an important role in HCC invasion and metastasis.
The activation of Akt inhibits GSK‐3β activity, subsequently suppressing the phosphorylation of Snail, inducing Snail protein stabilization and nuclear localization, which ultimately promotes EMT.25, 26 Our data showed that the expression levels of phosphorylated Akt (active form), phosphorylated GSK‐3β (inactive form) and Snail were increased or decreased when PRMT9 was overexpressed or silenced, respectively. In addition, the activation of Akt, overexpression of Snail and motility induced by PRMT9 overexpression in HCC cells were reversed by LY294002 (a PI3K inhibitor). The crucial role of Akt/GSK‐3β/Snail pathway during EMT in HCC has been previously reported.27, 28 Taken together, these findings strongly suggest that PRMT9‐induced EMT in HCC is through the activation of the Akt/GSK‐3β/Snail signaling pathway. However, it remains to be determined how PRMT9 activates Akt. Many studies have shown that PTEN can negatively regulate Akt activity.29, 30 In addition, one recent study showed that PRMT5 regulates the self‐renewal capacity of primary glioblastoma neurosphere cells through modulation of the PTEN‐Akt axis.31 We speculated that PRMT9, like PRMT5, which also belongs to the type II enzymes, may regulate Akt activity via modulation of PTEN. Our western blot results showed that PRMT9 can repress PTEN expression (Figure S4). Further studies are needed to elucidate how PRMT9 induces Akt activation.
Using in vitro and in vivo studies, we measured the relative expression of PRMT9, Snail and E‐cadherin in HCC tissues. We found that a high expression of PRMT9 was significantly correlated with a high expression of Snail and a low expression of E‐cadherin in 139 specimens of primary HCC tissue. In 20 pairs of primary and metastatic HCC tissues, the expression levels of PRMT9 and Snail were frequently higher in portal vein tumor thrombosis samples than in primary HCC samples. However, E‐cadherin expression was often lower in portal vein tumor thrombosis tissues than in primary HCC tissues. Clinical data further indicated that PRMT9 may play an important role in HCC metastasis.
Protein arginine methylation is thought to be a reversible phenomenon, and the dysregulation of protein arginine methyltransferases has been observed in various types of cancer.32 According to these characteristics, small‐molecule inhibitors targeting protein arginine methyltransferases have been developed as anticancer drugs.33 In this study, we showed that PTMT9 was frequently overexpressed in liver cancer, and PRMT9 overexpression significantly promoted cancer cell migration and invasion in vivo and in vitro. Thus, targeting PRMT9 may be an effective therapeutic strategy to prevent HCC metastasis. However, for the development of effective small‐molecule inhibitors targeting PRMT9, it is necessary to clarify the biological and physiological significance of other unidentified substrates by PRMT9. In addition, to better understand the biological roles of PRMT9, generating mice in which PRMT9 is overexpressed in a defined organ is essential.
In summary, this study revealed that the overexpression of PRMT9 is a strong indicator of more aggressive tumors and a poor prognosis in HCC. PRMT9 promotes HCC metastasis by inducing EMT via the PI3K/Akt/GSK‐3β/Snail signaling pathway. Thus, PRMT9 may be a candidate biomarker for HCC prognosis and a potential therapeutic target.
CONFLICT OF INTEREST
The authors have no conflict of interest to declare.
Supporting information
ACKNOWLEDGMENTS
We thank all members of Professor Jie Wang's laboratory for helpful discussion and technical support.
Jiang H, Zhou Z, Jin S, et al. PRMT9 promotes hepatocellular carcinoma invasion and metastasis via activating PI3K/Akt/GSK‐3β/Snail signaling. Cancer Sci. 2018;109:1414–1427. 10.1111/cas.13598
Funding information
Sun Yat‐Sen University (No. 15ykpy20), National Natural Science Foundation of China (No. 30700803, No. 81372565, No. 81672401, No. 81672403 and No. 81672405); Natural science Foundation of Guangdong, China (2016A030311051); Science and Technology Program of Guangzhou, China (201607010225); Foundation from the Key Laboratory of Malignant Tumor Gene Regulation and Target Therapy of Guangdong Higher Education Institutes (No. KLB 09001); Grant [2013] 163 from the Key Laboratory of Malignant Tumor Mechanism and Translational Medicine of Guangzhou Bureau of Science and Information Technology
Hai Jiang, Zhenyu Zhou and Shaowen Jin contributed equally to this work.
Contributor Information
Jie Wang, Email: jiewsysu@163.com.
Junyao Xu, Email: xuyuny@mail.sysu.edu.cn.
REFERENCES
- 1. Farazi PA, DePinho RA. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer. 2006;6:674‐687. [DOI] [PubMed] [Google Scholar]
- 2. Sherman M. Recurrence of hepatocellular carcinoma. N Engl J Med. 2008;359:2045‐2047. [DOI] [PubMed] [Google Scholar]
- 3. Brabletz T, Jung A, Spaderna S, Hlubek F, Kirchner T. Opinion: migrating cancer stem cells ‐ An integrated concept of malignant tumour progression. Nat Rev Cancer. 2005;5:744‐749. [DOI] [PubMed] [Google Scholar]
- 4. Klymkowsky MW, Savagner P. Epithelial‐mesenchymal transition: a cancer researcher's conceptual friend and foe. Am J Pathol. 2009;174:1588‐1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7:415‐428. [DOI] [PubMed] [Google Scholar]
- 6. Ell B, Kang Y. Transcriptional control of cancer metastasis. Trends Cell Biol. 2013;23:603‐611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chaffer CL, Weinberg RA. A perspective on cancer cell metastasis. Science. 2011;331:1559‐1564. [DOI] [PubMed] [Google Scholar]
- 8. Batlle E, Sancho E, Franci C, et al. The transcription factor snail is a repressor of E‐cadherin gene expression in epithelial tumour cells. Nat Cell Biol. 2000;2:84‐89. [DOI] [PubMed] [Google Scholar]
- 9. Iwatsuki M, Mimori K, Yokobori T, et al. Epithelial‐mesenchymal transition in cancer development and its clinical significance. Cancer Sci. 2010;10:293‐299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fischer KR, Durrans A, Lee S, et al. Epithelial‐to‐mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature. 2015;527:472‐476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chang L, Yuan Y, Li C, et al. Upregulation of SNHG6 regulates ZEB1 expression by competitively binding miR‐101‐3p and interacting with UPF1 in hepatocellular carcinoma. Cancer Lett. 2016;383:183‐194. [DOI] [PubMed] [Google Scholar]
- 12. Xu Q, Liu X, Liu Z, et al. MicroRNA‐1296 inhibits metastasis and epithelial‐mesenchymal transition of hepatocellular carcinoma by targeting SRPK1‐mediated PI3K/AKT pathway. Mol Cancer. 2017;16:103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bedford MT, Clarke SG. Protein arginine methylation in mammals: who, what, and why. Mol Cell. 2009;33:1‐13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang Y, Bedford MT. Protein arginine methyltransferases and cancer. Nat Rev Cancer. 2013;13:37‐50. [DOI] [PubMed] [Google Scholar]
- 15. Yang Y, Hadjikyriacou A, Xia Z, et al. PRMT9 is a type II methyltransferase that methylates the splicing factor SAP145. Nat Commun. 2015;6:6428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feng Y, Maity R, Whitelegge JP, et al. Mammalian protein arginine methyltransferase 7 (PRMT7) specifically targets RXR sites in lysine‐ and arginine‐rich regions. J Biol Chem. 2013;288:37010‐37025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Blanc RS, Richard S. Arginine methylation. The coming of age. Mol Cell. 2017;65:8‐24. [DOI] [PubMed] [Google Scholar]
- 18. Li B, Liu L, Li X, Wu L. miR‐503 suppresses metastasis of hepatocellular carcinoma cell by targeting PRMT1. Biochem Biophys Res Commun. 2015;464:982‐987. [DOI] [PubMed] [Google Scholar]
- 19. Liu L, Zhao X, Zhao L, et al. Arginine methylation of SREBP1a via PRMT5 promotes de novo lipogenesis and tumor growth. Cancer Res. 2016;76:1260‐1272. [DOI] [PubMed] [Google Scholar]
- 20. Wu J, Ru NY, Zhang Y, et al. HAb18G/CD147 promotes epithelial‐mesenchymal transition through TGF‐beta signaling and is transcriptionally regulated by Slug. Oncogene. 2011;30:4410‐4427. [DOI] [PubMed] [Google Scholar]
- 21. Hadjikyriacou A, Yang Y, Espejo A, Bedford MT, Clarke SG. Unique features of human protein arginine methyltransferase 9 (PRMT9) and its substrate RNA splicing factor SF3B2. J Biol Chem. 2015;290:16723‐16743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thiery JP. Epithelial‐mesenchymal transitions in tumour progression. Nat Rev Cancer. 2002;2:442‐454. [DOI] [PubMed] [Google Scholar]
- 23. Kang Y, Massague J. Epithelial‐mesenchymal transitions: twist in development and metastasis. Cell. 2004;118:277‐279. [DOI] [PubMed] [Google Scholar]
- 24. Yang MH, Chen CL, Chau GY, et al. Comprehensive analysis of the independent effect of twist and snail in promoting metastasis of hepatocellular carcinoma. Hepatology. 2009;50:1464‐1474. [DOI] [PubMed] [Google Scholar]
- 25. Zhou BP, Deng J, Xia W, et al. Dual regulation of Snail by GSK‐3beta‐mediated phosphorylation in control of epithelial‐mesenchymal transition. Nat Cell Biol. 2004;6:931‐940. [DOI] [PubMed] [Google Scholar]
- 26. Cohen P, Frame S. The renaissance of GSK3. Nat Rev Mol Cell Biol. 2001;2:769‐776. [DOI] [PubMed] [Google Scholar]
- 27. Zhou SL, Zhou ZJ, Hu ZQ, et al. CXCR2/CXCL5 axis contributes to epithelial‐mesenchymal transition of HCC cells through activating PI3K/Akt/GSK‐3beta/Snail signaling. Cancer Lett. 2015;358:124‐135. [DOI] [PubMed] [Google Scholar]
- 28. Liu L, Dai Y, Chen J, et al. Maelstrom promotes hepatocellular carcinoma metastasis by inducing epithelial‐mesenchymal transition by way of Akt/GSK‐3beta/Snail signaling. Hepatology. 2014;59:531‐543. [DOI] [PubMed] [Google Scholar]
- 29. Manning BD, Logsdon MN, Lipovsky AI. Feedback inhibition of Akt signaling limits the growth of tumors lacking Tsc2. Genes Dev. 2005;19:1773‐1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Aoki M, Fujishita T. Oncogenic roles of the PI3K/AKT/mTOR axis. Curr Top Microbiol Immunol. 2017;407:153‐189. [DOI] [PubMed] [Google Scholar]
- 31. Banasavadi‐Siddegowda YK, Russell L, Frair E, et al. PRMT5‐PTEN molecular pathway regulates senescence and self‐renewal of primary glioblastoma neurosphere cells. Oncogene. 2017;36:263‐274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hamamoto R, Nakamura Y. Dysregulation of protein methyltransferases in human cancer: an emerging target class for anticancer therapy. Cancer Sci. 2016;107:377‐384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hu H, Qian K, Ho MC, Zheng YG. Small molecule inhibitors of protein arginine methyltransferases. Expert Opin Investig Drugs. 2016;25:335‐358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


