Abstract
Gallbladder cancer (GBC) is the most common malignant tumor of the biliary tract system. Epithelial–mesenchymal transition (EMT) plays a vital role in the process of tumor metastasis. Mesenchymal‐like cells can serve as a source of cancer stem cells, which can confer the EMT phenotype. Placental growth factor (PLGF) belongs to the vascular endothelial growth factor family and plays a vital role in cancer. However, the underlying molecular mechanisms about the influence of PLGF on EMT in GBC remain unknown. Here we show that PLGF expression levels were higher in GBC tissues than in normal adjacent tissues and were associated with poor prognosis in GBC patients. Exogenous PLGF enhanced the migration, invasion, and tumorsphere formation of GBC cells. Conversely, knockdown of PLGF decreased the aggressive phenotype of GBC cells. Mechanistically, exogenous PLGF upregulated microRNA‐19a (miR‐19a) expression through the activation of c‐MYC. Moreover, Spearman's correlation analysis showed a positive pairwise correlation among PLGF, c‐MYC, and miR‐19a expression in GBC tissues. Taken together, these results suggest that PLGF promotes EMT and tumorsphere formation through inducing miR‐19a expression by upregulating c‐MYC. Thus, PLGF could be a promising molecular therapeutic target for GBC.
Keywords: cancer stem cell, c‐MYC, gallbladder cancer, miR‐19a, placental growth factor
Abbreviations
- CSC
cancer stem cell
- CT
cycle threshold
- EMT
epithelial–mesenchymal transition
- GBC
gallbladder cancer
- miR‐19a
microRNA‐19a
- NAT
normal adjacent tissue
- NC
negative control
- OS
overall survival
- PLGF
placental growth factor
- qRT‐PCR
quantitative real‐time PCR
1. INTRODUCTION
Gallbladder cancer is the most common malignant tumor of the biliary tract system with an incidence of approximately 2.5 cases per 1 × 105 people.1, 2, 3 Despite the lower incidence of GBC compared with other gastrointestinal cancers, the survival rate for GBC is poor, due to the high degree of malignancy and high mortality.2, 4, 5, 6 More attention should be given to the exact mechanisms contributing to GBC initiation and development.
Metastasis is a multistage process that includes ECM remodeling, blood vessel recruitment, tumor cell entry and exit from the circulation, and survival at a distant organ.7 Epithelial–mesenchymal transition plays a vital role in the tumor metastatic process.8, 9 Previous studies have confirmed that EMT is dynamically controlled by various pro‐invasion signals from the tumor microenvironment.10 The tumor microenvironment comprises cells and factors, such as growth factors, inflammatory factors, and angiogenesis factors.11 Chronic inflammation represents an important risk factor in the formation of GBCs. Chronic inflammation causes DNA damage and releases cytokines and growth factors.12 Placental growth factor belongs to the vascular endothelial growth factor family. Placental growth factor is dispensable for development and health but plays a prominent role in pathology, including in cancer.13 In addition to its angiogenic effects, PLGF contributes to metastasis and is considered a useful prognostic marker of cancer progression. Placental growth factor is overexpressed, and its expression is correlated with tumor stage, disease progression, recurrence, metastasis, and patient survival.13, 14, 15, 16, 17, 184 However, the biological and pathological roles of PLGF in GBC are relatively less understood.
Here, we found that PLGF overexpression promoted GBC cell stemness and metastasis. Furthermore, miR‐19a upregulation in GBC was associated with the amplification of PLGF and additional downstream signaling pathways. Evidence for PLGF functionality in GBC was also extended to the clinicopathological features and prognosis of GBC patients and could be considered for the development of potential therapeutics against GBC.
2. MATERIALS AND METHODS
2.1. Tissue samples
Human GBC samples and the corresponding normal gallbladder tissues were obtained from the Department of General Surgery, Xinhua Hospital (Shanghai, China) between 2008 and 2013. The tissue samples were sent for histological analysis and diagnostic confirmation. This study was approved by the research ethics committee, and written informed consent was obtained from all participants. The clinical features of the patients are listed in Table 1.
Table 1.
Association of placental growth factor (PLGF) expression with the clinicopathological features of patients with gallbladder cancer
| Variable | Category | Relative PLGF expression | χ2 | P‐value | |
|---|---|---|---|---|---|
| Low (n = 19) | High (n = 20) | ||||
| Age, years | <60 | 7 | 8 | 0.410 | 1.000 |
| ≥60 | 12 | 12 | |||
| Gender | Male | 7 | 6 | 0.205 | .741 |
| Female | 12 | 14 | |||
| Tumor size, cm | <3 | 9 | 6 | 1.242 | .333 |
| ≥3 | 10 | 14 | |||
| Histological differentiation | Well | 3 | 3 | 0.005 | 1.000 |
| Moderate or poor | 16 | 17 | |||
| Tumor invasion (AJCC) | Tis‐T2 | 12 | 5 | 5.770 | .025 * |
| T3‐T4 | 7 | 15 | |||
| Lymph node metastasis (AJCC) | Present | 6 | 14 | 5.757 | .026 * |
| Absent | 13 | 6 | |||
Bold values indicate statistical significance, *P < .05.
2.2. Cell culture and reagents
GBC‐SD and 293T cells were purchased from the cell bank of the type culture collection of the Chinese Academy of Sciences (Shanghai, China). NOZ and H69 cells were obtained from the Health Science Research Bank (Osaka, Japan). All of the cells were maintained in high‐glucose DMEM (Gibco, USA) supplemented with 10% FBS (Gibco) and penicillin G/streptomycin (100 U/mL, 100 g/mL; Gibco). The cells were maintained as monolayer cultures at 37°C in humidified air with 5% CO2 and 95% air. Recombinant PLGF was purchased from R&D Systems (USA).
2.3. Protein isolation and immunoblot analysis
RIPA buffer (Sigma‐Aldrich, USA) with Protease Inhibitor Cocktail (Roche Applied Science, USA) was used to isolate proteins from cells or tissues. Antibodies against PLGF were purchased from Proteintech (10642‐1‐AP; USA). Antibodies against Sox2 and Oct4 were purchased from Abcam (ab97959 and ab19857, respectively; Abcam, USA). Antibodies against Snail, Slug, N‐cadherin, E‐cadherin, C‐MYC, PTEN, and β‐actin were purchased from Cell Signaling Technology (#3879, #9585, #13116, #14472, #13987, #9188, and #3700, respectively; USA). Horseradish peroxidase‐linked anti‐rabbit IgG (#7074; CST, USA) and HRP‐linked anti‐mouse IgG (#7076; CST, USA) were used as secondary antibodies to detect the proteins.
2.4. RNA extraction and qRT‐PCR
Total RNA was extracted from the tissue samples or cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocols. Quantitative real‐time PCR was carried out using SYBR Green (Takara, Japan) according to the manufacturer's instructions. The qRT‐PCR results were analyzed and examined as the relative miRNA or mRNA levels based on CT values, which were converted to fold changes. The primer sequences used are listed in Table S1.
2.5. Sphere‐formation assays
Sphere‐formation assays were carried out according to the methods described by Wang et al19 The numbers of spheres >50 μm in diameter were counted under a microscope (Leica, Germany).
2.6. Cell migration and invasion assays
The cell scratch, migration, and invasion assays were undertaken as previously described.1
2.7. Immunofluorescence
After incubation with PLGF (100 ng/mL for 0, 24, and 48 hours), GBC cells were seeded onto 24‐well plates. The cells were then washed with PBS and fixed in 4% paraformaldehyde for 30 minutes at room temperature. Then the cells were incubated with blocking buffer (1% BSA and 0.1% Triton X‐100) alone for 1 hour and with the N‐cadherin and Slug primary antibodies overnight at 4°C. The primary antibody was removed, and cells were incubated with the secondary fluorescent antibody (green) for 1 hour at room temperature. After several washes with PBS, the cells were stained with DAPI and imaged under a fluorescence microscope (Leica).
2.8. Cell transfection
Hsa‐miRNA mimics, hsa‐miRNA inhibitors, and their cognate control RNAs were purchased from RioBio (Guangzhou, China). The PLGF siRNA, c‐MYC siRNA, and si‐NC were purchased from Biotend (Shanghai, China). The miRNA mimics, miRNA inhibitors, and siRNA were transfected into NOZ or GBC‐SD cells using Lipofectamine 2000 transfection reagent (Invitrogen). Total RNA and protein were collected 48 hours after transfection. The siRNA sequences used are listed in Table S2.
2.9. Generation of stable cell lines with miR‐19a downregulation
To construct cell lines stably expressing miR‐19a, the miR‐19a antagomir and NC antagomir were synthesized and inserted into the PGMLV‐hU6‐MCS‐CMV‐ZsGreen1‐PGK‐puromycin‐WPRE lentiviral vector. Recombinant lentiviruses expressing the miR‐19a antagomir or NC antagomir (Lv‐antagomir‐19a and Lv‐antagomir‐NC, respectively) were produced by Genomeditech (Shanghai, China). NOZ cells were infected with concentrated virus and treated with 1 μg/mL puromycin for 2 weeks for the selection of stable cell lines. The expression of miR‐19a antagomir in the stable cell lines was validated by qRT‐PCR analysis.
2.10. Luciferase reporter assay
Luciferase reporter assays were carried out as described previously.20 The miR‐19a binding sites were determined according the investigation of Jia et al21
2.11. In vivo animal study
The animal experiments were approved by the Institutional Animal Care and Use Committee of Xinhua Hospital (2013‐0106) and were conducted humanely. The liver metastatic model was established using 4‐6‐week‐old male BALB/c athymic nude mice. The mice were housed under specific pathogen‐free conditions and fed a regular autoclaved chow diet with water ad libitum. A total of 1 × 106 treated NOZ cells (Lv‐antagomir‐19a and Lv‐antagomir‐NC) were resuspended in 100 μL PBS and injected into BALB/c nude mice through the spleen (n = 5 per group); the spleen was then removed. The nude mice were killed 8 weeks after spleen injection. The livers were fixed with 4% paraformaldehyde.
2.12. Immunohistochemistry
Immunohistochemistry was carried out using anti‐N‐cadherin (1:200; Proteintech) and anti‐Slug (1:200; Proteintech) antibodies. Immunohistochemical staining of the metastatic nodule sections was undertaken as previously described.1
2.13. Statistical analysis
All experiments were repeated three times unless otherwise noted. The data are presented as the mean ± SD. Two‐tailed Student's t‐test was used for single comparisons, and one‐way anova was used to analyze for three or more groups. Survival curves were estimated by the Kaplan–Meier method. All data were analyzed with GraphPad Prism 5 (GraphPad Software, La Jolla, CA, USA) and spss version 17.0 (IBM, Chicago, IL, USA). P < .05 was considered statistically significant.
3. RESULTS
3.1. Placental growth factor is upregulated in GBC tissues and is associated with metastasis and poor patient survival
First, we examined PLGF protein levels in 10 pairs of human GBC tissues and NATs. We found that PLGF protein levels were significantly higher in GBC tissues than in NATs (Figure 1A). We further examined the mRNA levels of PLGF in another 39 pairs of human GBC tissues and NATs. Consistent with the protein levels, the average PLGF mRNA level was significantly higher in GBC tissues than in NATs (Figure 1B). To determine whether PLGF levels correlated with the OS of GBC patients, these 39 patients were followed up for 5 years. Thirty‐nine GBC patients were divided into two groups (PLGF‐low, n = 19; PLGF‐high, n = 20) by setting the cut‐off value to the median PLGF expression level. Kaplan–Meier curves showed that GBC patients with higher PLGF expression had a significantly shorter OS (P = .0375, Figure 1C). The mean survival time in the PLGF‐high group was 13.78 months, whereas the mean survival time in the PLGF‐low group was 27.32 months. A clinicopathological association study of the 39 GBCs showed that PLGF was significantly associated with tumor invasion (P = .025) and lymph node metastasis (P = .026; Table 1).
Figure 1.
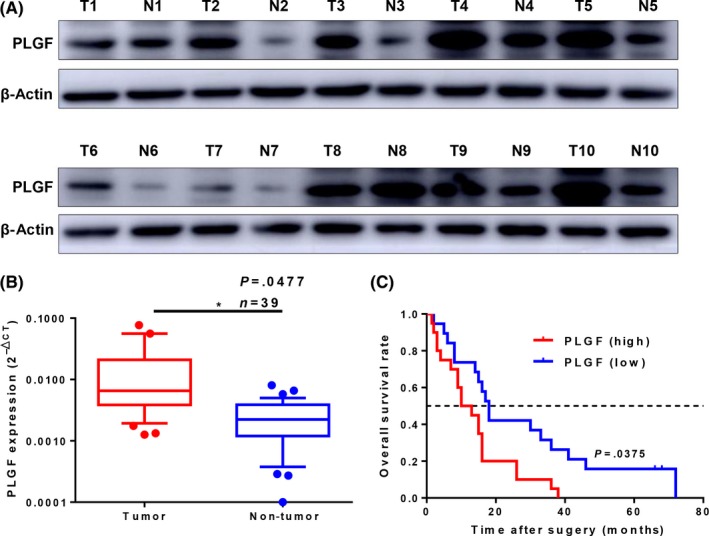
High placental growth factor (PLGF) expression correlates with poor clinical outcomes in patients with gallbladder cancer (GBC). A, Western blot analysis of PLGF protein levels in 10 paired GBC (T) and normal adjacent tissue (N) samples. β‐Actin was used as a loading control. B, Scatterplots of the relative expression levels of PLGF in another 39 paired GBC tissues and their corresponding normal adjacent tissues. PLGF expression was calculated and is expressed as the PLGF/GAPDH expression ratio (2−Δ CT); P = .0477. C, Kaplan–Meier overall survival curve of GBC patients based on PLGF expression; P = .0375
3.2. Placental growth factor promotes GBC cell migration, invasion, and stemness in vitro
The findings above revealed the prognostic value of PLGF in GBC, so we then addressed the role of PLGF in GBC cells. First, we examined the mRNA and protein expression levels in a series of GBC cell lines and normal cell lines. The H69 cell line was used as a representative of non‐tumorigenic biliary epithelial cells. Compared with H69, the expression level of PLGF in GBC cell lines was obviously increased. Among them, the PLGF expression level in NOZ was much higher than that in GBC‐SD (Figure 2A,B). Therefore, we selected NOZ for knockdown and GBC‐SD for overexpression.
Figure 2.
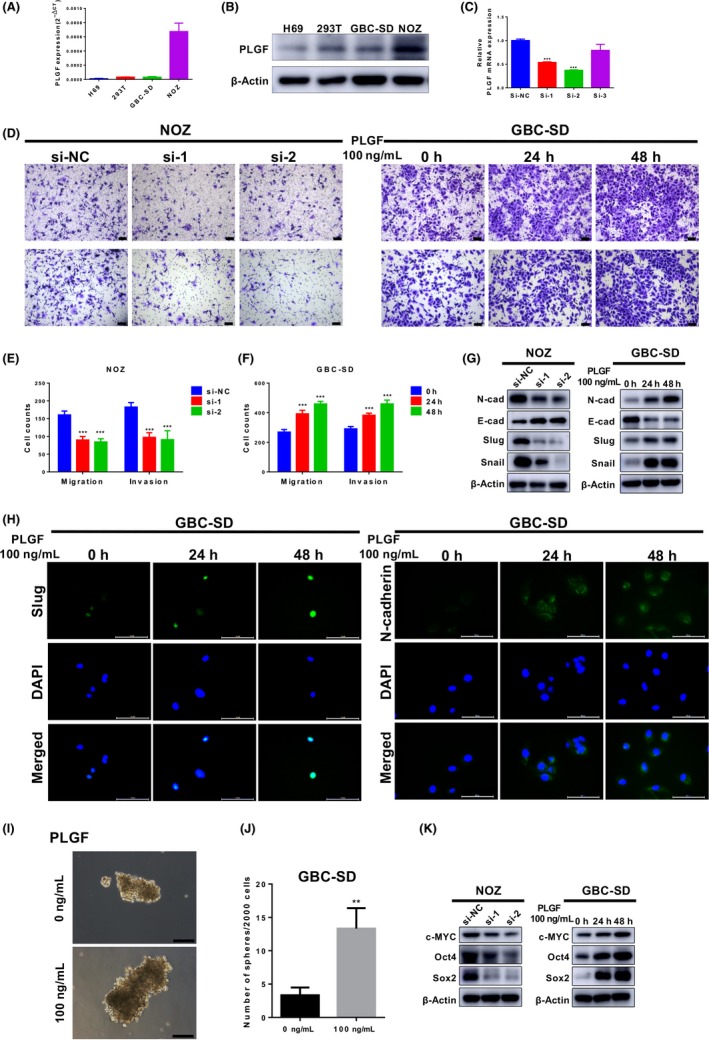
Placental growth factor (PLGF) promotes gallbladder cancer (GBC) cell metastasis and stemness in vitro. A, Quantitative RT‐PCR analysis of PLGF levels in H69, 293T, NOZ, and GBC‐SD cells. B, Immunoblotting analyses of the protein levels of PLGF in H69, 293T, NOZ, and GBC‐SD cells. C, Quantitative RT‐PCR analysis of PLGF levels in NOZ cells after transfection with PLGF siRNA (si‐1, si‐2, and si‐3) or negative control siRNA (si‐NC). ***P < .001. D‐F, Knockdown of PLGF suppresses NOZ cell migration and invasion. Exogenous PLGF reverses this process in GBC‐SD cells. Scale = 100 μm. D, Representative images. E, Quantitative analysis of the number of migrated and invaded NOZ cells after treatment with siRNA. ***P < .001. F, Quantitative analysis of the number of migrated and invaded GBC‐SD cells after treatment with exogenous PLGF. ***P < .001. Data are presented as the mean ± SD. G, Immunoblotting analyses of the levels of epithelial–mesenchymal transition marker proteins in treated GBC cells. E‐cad, E‐cadherin; N‐cad, N‐cadherin. H, Immunofluorescence analysis of Slug and N‐cadherin in treated GBC‐SD cells. Scale = 100 μm. I, Analysis of the sphere‐forming potential of GBC cells after treatment with or without 100 ng/mL PLGF. Shown are representative light microscopy images. Scale bar = 100 μm. J, Quantitative analysis of the numbers of spheres. Only spheres >50 μm in diameter were counted. **P < .01. K, Immunoblotting analyses of the levels of cancer stem cell marker proteins in treated GBC cells
We further investigated the functional role of PLGF in GBC progression. Epithelial–mesenchymal transition plays a key role in tumor progression and metastasis, in which epithelial cells obtain the migratory and invasive abilities of mesenchymal cells.8, 9 To investigate the effects of PLGF on GBC cells, we decreased PLGF expression by transfecting siRNA against PLGF and exogenously increased PLGF expression by treating GBC cells with recombinant PLGF (100 ng/mL).22
To gain insight into the role of PLGF in cell metastasis, Transwell migration and Matrigel invasion assays were carried out in vitro. After siRNA transfection, PLGF expression was significantly lower in the si‐PLGF‐1 (si‐1) and si‐PLGF‐2 (si‐2) groups than in the si‐NC group (Figure 2C). Following transfection with si‐1 and si‐2, NOZ cells showed decreased cell migration and invasion ability (Figure 2D, left, E). After treatment with PLGF for 24 hours, GBC‐SD migration and invasion were increased nearly 1.5‐fold compared with those of cells without PLGF. Moreover, after 48 hours, the increase was nearly 2‐fold (Figure 2D, right, F).
We also used the CCK8 assay to ascertain whether PLGF promotes the migration and invasion ability of GBC cells by promoting cell proliferation. There was no difference between the NC group and si‐PLGF group or PLGF‐treated group in terms of cell proliferation (Figure S1a,b), which means that PLGF promotes tumor migration and invasion ability without the effect of cell proliferation.
To determine whether PLGF induces EMT, the expression of EMT markers was evaluated. As shown in Figure 2(G), exogenous PLGF increased the levels of mesenchymal markers (including N‐cadherin, Slug, and Snail) while it decreased epithelial marker E‐cadherin expression in GBC‐SD cells. In contrast, PLGF knockdown decreased mesenchymal marker expression and increased epithelial marker expression in NOZ cells. Consistent with these findings, immunofluorescence staining showed that increased Slug and N‐cadherin expression was observed after PLGF treatment (Figure 2H). Thus, these results indicated that PLGF might promote GBC cell migration and invasion in vitro.
Numerous studies have reported that CSCs are responsible for heterogeneity, metastasis, drug resistance, and disease recurrence.23, 24 Therefore, we also undertook sphere‐formation assays to investigate the effect of PLGF on GBC CSC properties. Notably, spheres generated from exogenous PLGF‐treated GBC‐SD cells were larger in size than those generated from control cells (Figure 2I,J). The CSC marker c‐MYC, shown to be a key factor required for stem cell reprogramming,25, 26 was highly induced at the protein level in the PLGF‐treated group, unlike in the control group. Furthermore, another two CSC markers (Oct4 and Sox2) were significantly highly expressed in PLGF‐treated cells (Figure 2K, right). By contrast, after PLGF knockdown, the three markers were significantly downregulated compared to those in the si‐NC group (Figure 2K, left). Collectively, these data indicated that PLGF might play an important role in GBC EMT and tumorsphere formation.
3.3. Placental growth factor induces miR‐19a expression through c‐MYC
Because our results indicated that exogenous PLGF promoted c‐MYC protein levels, we used qRT‐PCR to determine the mRNA level of c‐MYC. The results were consistent with the protein level (Figure 3A,B). c‐MYC is one of the most commonly activated oncogenes implicated in the development of human cancers.26 Recent studies have shown that c‐MYC transcriptionally upregulates miR‐19a.27, 28, 29 Therefore, we speculated that PLGF influenced miR‐19a expression by modulating c‐MYC expression. To evaluate this hypothesis, we used siRNA to knock down c‐MYC expression in GBC cells (Figure 3C). We observed the anticipated decrease in the miR‐19a levels after treatment with c‐MYC siRNA (Figure 3D). To further investigate whether PLGF increased miR‐19a, we measured miR‐19a mRNA levels after treatment with exogenous PLGF or PLGF siRNA and detected miR‐19a levels (Figure 3E,F). In order to evaluate the clinical relevance of PLGF, c‐MYC, and miR‐19a in GBC specimens, we examined the mRNA levels of c‐MYC and miR‐19a in 39 pairs of human GBC tissues and NATs. The miR‐19a transcripts were expressed at higher levels in GBC tissues than in NATs (Figure 3G,H). More intriguingly, survival analysis showed that GBC patients with high miR‐19a expression displayed a shorter OS than patients with low miR‐19a expression (Figure 3I). Furthermore, the correlation among PLGF, c‐MYC, and miR‐19a in the GBC tissues was evaluated using Spearman's correlation analysis. Spearman's correlation analysis clearly showed a positive pairwise correlation among PLGF, c‐MYC, and miR‐19a expression in the GBC tissues (Figure 3J‐L).
Figure 3.
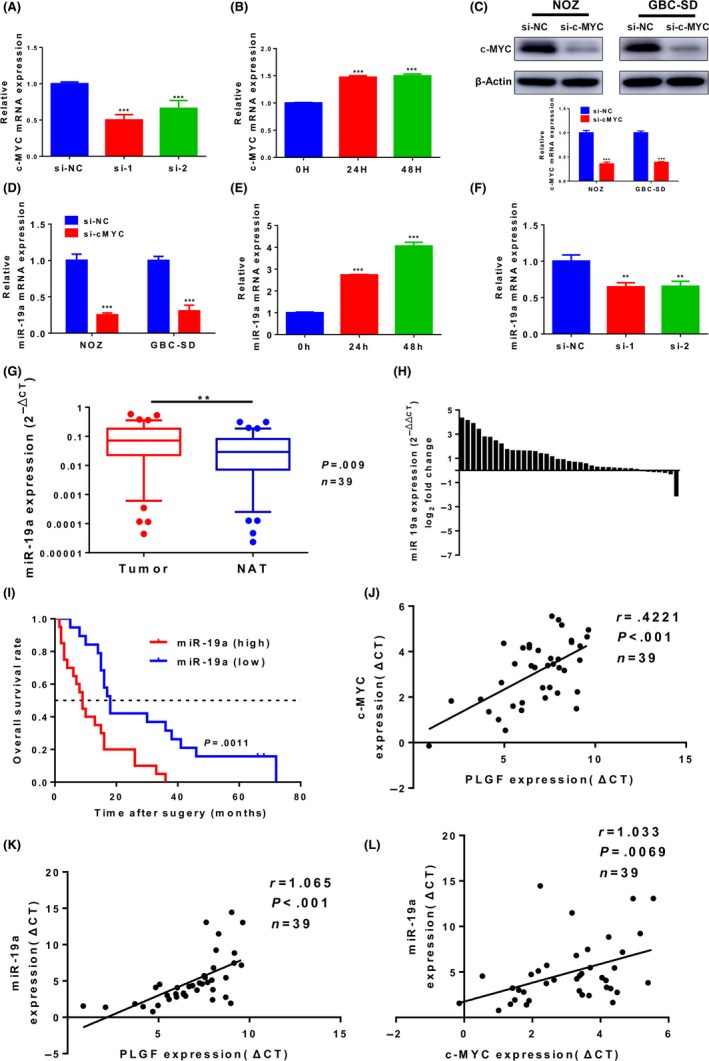
Placental growth factor (PLGF), c‐MYC, and microRNA‐19a (miR‐19a) form an axis in gallbladder cancer (GBC). A, Quantitative RT‐PCR analysis of c‐MYC levels in PLGF siRNA‐treated (si‐1 and si‐2) NOZ cells. ***P < .001. B, Quantitative RT‐PCR analysis of c‐MYC levels in exogenous PLGF‐treated GBC‐SD cells. ***P < .001. C, Quantitative RT‐PCR and Western blot analysis of c‐MYC in NOZ and GBC‐SD cells transfected with c‐MYC siRNA or negative control siRNA (si‐NC). ***P < .001. D, Quantitative RT‐PCR analysis of miR‐19a levels in NOZ and GBC cells transfected with c‐MYC siRNA or si‐NC. ***P < .001. E, Quantitative RT‐PCR analysis of miR‐19a levels in NOZ and GBC cells treated with 100 ng/mL PLGF for 0, 24, and 48 h. ***P < .001. F, Quantitative RT‐PCR analysis of miR‐19a levels in NOZ cells transfected with PLGF siRNA (si‐1 and si‐2) or si‐NC. **P < .01. G, Scatterplots of the relative expression levels of miR‐19a in 39 paired GBC tissues and their corresponding normal adjacent tissues (NAT). miR‐19a expression was calculated and expressed as the miR‐19a/U6 expression ratio (2−Δ CT); P = .009. H, miR‐19a expression in GBC tissues and NATs from the same 39 patients (quantitative RT‐PCR) (U6 as the internal control; Wilcoxon matched‐pairs test). I, Kaplan–Meier overall survival curve of GBC patients based on miR‐19a expression; P = .0011. J, Correlation between the expression levels of PLGF and c‐MYC was determined using linear regression analysis and paired t‐test with the same samples used (P < .001, r = .4221, n = 39; Pearson's correlation). K, Correlation between PLGF expression levels and miR‐19a (P < .001, r = 1.065, n = 39; Pearson's correlation). L, Correlation between the expression levels of c‐MYC and miR‐19a (P = .0069, r = 1.033, n = 39; Pearson's correlation)
3.4. MicroRNA‐19a phenocopies the effect of PLGF on metastasis and stemness in vitro
To determine whether the promotion of PLGF in the metastasis and stemness of GBC was indeed mediated by miR‐19a, we first examined the function of miR‐19a in GBC cells. We examined the miR‐19a expression level in GBC cell lines and normal cell lines. As shown in Figure S1(c), the expression levels of miR‐19a in GBC cells were higher than normal cells. Then we decreased miR‐19a expression by transfecting a miR‐19a inhibitor in NOZ cells and increased miR‐19a expression by transfecting a miR‐19a mimic in GBC‐SD cells. The levels of miR‐19a expression after transfection were confirmed by qRT‐PCR (Figure S1d). The downregulation of miR‐19a expression inhibited NOZ cell migration and invasion; by contrast, miR‐19a overexpression had the opposite effect on migration and invasion in GBC‐SD cells (Figure 4A‐C).
Figure 4.
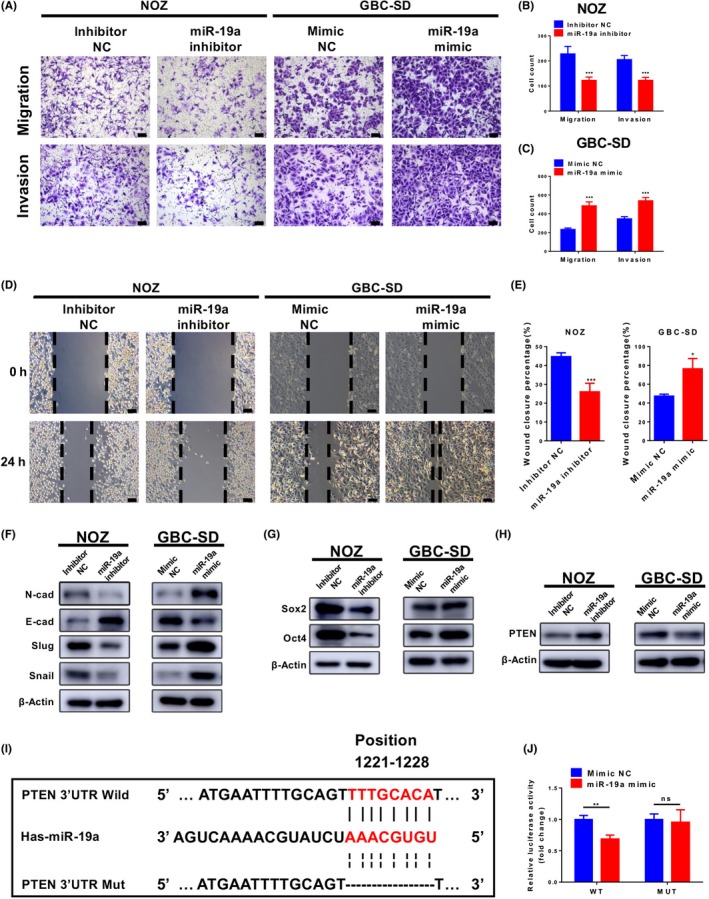
MicroRNA‐19a (miR‐19a) promotes gallbladder cancer (GBC) cell metastasis and stemness in vitro. A‐C, Cell migration and invasion assays were carried out in NOZ and GBC‐SD cells that were transfected with equal doses of mimic negative control (NC), miR‐19a mimic, inhibitor NC, or miR‐19a inhibitor. A, Representative images are shown. Scale = 100 μm. B, C, Quantitative analysis of the number of migrated and invaded cells. *P < .05; **P < .01. D, E, Wound‐healing assays were used to reveal the migration of treated NOZ cells. D, Representative images are shown. Scale = 100 μm. E, Quantitative analysis of wound closure. **P < .01; ***P < .001. F‐H, Immunoblotting analyses of the levels of epithelial–mesenchymal transition (EMT) marker proteins, cancer stem cell (CSC) marker protein, and PTEN in GBC cells after transfection with equal doses of mimic NC, miR‐19a mimic, inhibitor NC, or miR‐19a inhibitor. (F) Immunoblotting analyses of the levels of EMT marker proteins. E‐cad, E‐cadherin; N‐cad, N‐cadherin. G, Immunoblotting analyses of the levels of CSC marker proteins. H, Immunoblotting analyses of the levels of PTEN. I, Binding sites of miR‐19a in wild‐type (Wild) or mutant (Mut) PTEN 3ʹ‐UTRs. J, Relative luciferase activity of wild‐type or mutant PTEN 3ʹ‐UTR in GBC‐SD cells after transfection with miR‐19a mimic or mimic NC. **P < .01. ns, not significant
Wound‐healing assays also showed the same results (Figure 4D,E), indicating an oncogenic role for miR‐19a in GBC cells. Also, the proliferation ability of GBC cells was not affected by miR‐19a (Figure S2). Western blot analyses confirmed that the expression of mesenchymal markers (N‐cadherin, Slug, and Snail) and CSC markers (Sox2 and Oct4) were reduced by miR‐19a knockdown in NOZ cells and enhanced by miR‐19a overexpression in GBC‐SD cells, whereas epithelial marker E‐cadherin had the opposite change (Figure 4F, G).
MicroRNA‐19a, by binding to the 3ʹ‐UTR, regulates multiple target genes, including PTEN, which is associated with EMT and CSC formation.30, 31, 32 Thus, we detected PTEN protein levels after treatment with the miR‐19a mimic or inhibitor (Figure 4H). Furthermore, we constructed a dual‐luciferase reporter plasmid containing a fragment of the PTEN 3ʹ‐UTR (Figure 4I). The dual‐luciferase reporter assay showed that miR‐19a mimics significantly reduced the luciferase activity in the PTEN wild‐type group but had no effect on the PTEN mutated group (Fig. 4J). The results revealed that PTEN is the target of miR‐19a in GBC cells.
3.5. MicroRNA‐19a promotes metastasis in vivo
To determine whether miR‐19a could regulate the metastasis of GBC cells in vivo, we injected NOZ cells stably expressing antagomir‐19a or antagomir‐NC (Lv‐antagomir‐19a or Lv‐antagomir‐NC) into the spleens of nude mice. The mice injected with Lv‐antagomir‐19a NOZ cells showed a significant decrease in intrahepatic metastasis compared with the mice injected with Lv‐antagomir‐NC cells (Fig. 5A,B). Furthermore, we embedded the metastatic tumors in paraffin, carried out H&E staining, and examined N‐cadherin and Slug expression using immunohistochemical assays. Immunohistochemical staining revealed lower N‐cadherin and Slug levels in tumors from the Lv‐antagomir‐19a group (Fig. 5C). These findings confirmed that miR‐19a could induce EMT and increase cancer metastasis in vivo.
Figure 5.
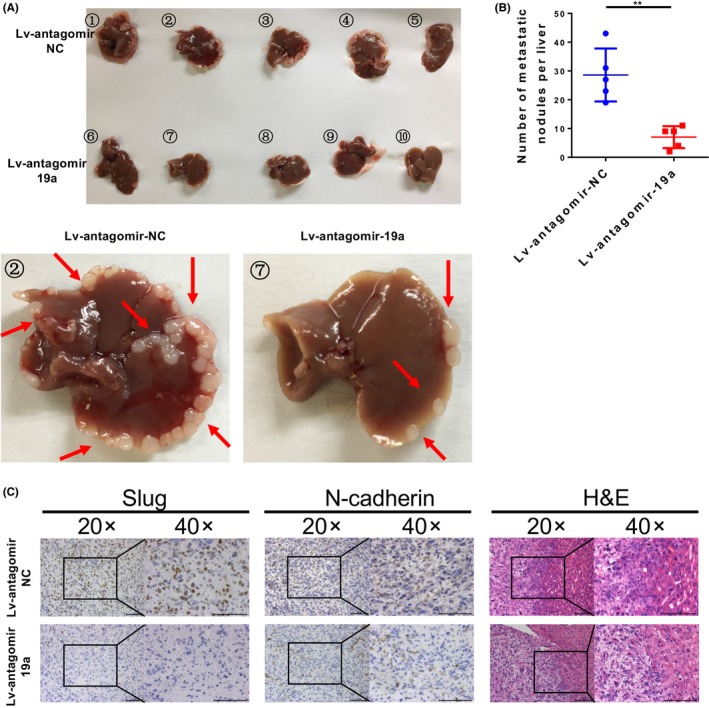
MicroRNA‐19a (miR‐19a) promotes gallbladder cancer cell metastasis in vivo. A, Representative images of liver metastases in nude mice injected with NOZ cells stably expressing antagomir‐19a or negative control (Lv‐antagomir‐19a or Lv‐antagomir‐NC). B, The number of metastatic nodules in the liver was calculated and compared in the diagrams. **P < .01. C, H&E, Slug, and N‐cadherin staining of the metastatic nodules. Scale bar = 100 μm
3.6. MicroRNA‐19a knockdown attenuates the enhancing effects of PLGF on GBC cells
To investigate whether PLGF affects the migration, invasion, and stemness of GBC cells through miR‐19a, we first treated GBC‐SD cells with or without PLGF and infected GBC‐SD cells with Lv‐antagomir‐19a or Lv‐antagomir‐NC. We then examined the effect of PLGF and Lv‐antagomir‐19a on GBC‐SD metastasis and stemness. The migration and invasion assay results showed that the PLGF‐mediated oncogenic phenotypes on GBC‐SD cells were weakened after transfection with Lv‐antagomir‐19a, indicating that PLGF could promote GBC‐SD cell metastasis by increasing miR‐19a (Fig. 6A,B). The sphere‐formation assay results confirmed that PLGF could enhance GBC‐SD cell stemness through miR‐19a (Figure 6C,D). Furthermore, we investigated the protein level of PTEN, EMT markers, and CSC markers after treatment with PLGF and infected them with antagomir‐19a lentivirus. As shown in Figure 6(E), when the antagomir‐19a lentivirus was used to neutralize the tumor‐promoting effects of PLGF, the levels of PTEN, a miR‐19a target gene, were also recovered. A similar effect was observed for the EMT and CSC markers (Figure 6E). However, the level of c‐MYC was not recovered. Taken together, these results revealed a PLGF/c‐MYC/miR‐19a/PTEN regulatory axis in GBC cells (Figure 6F).
Figure 6.
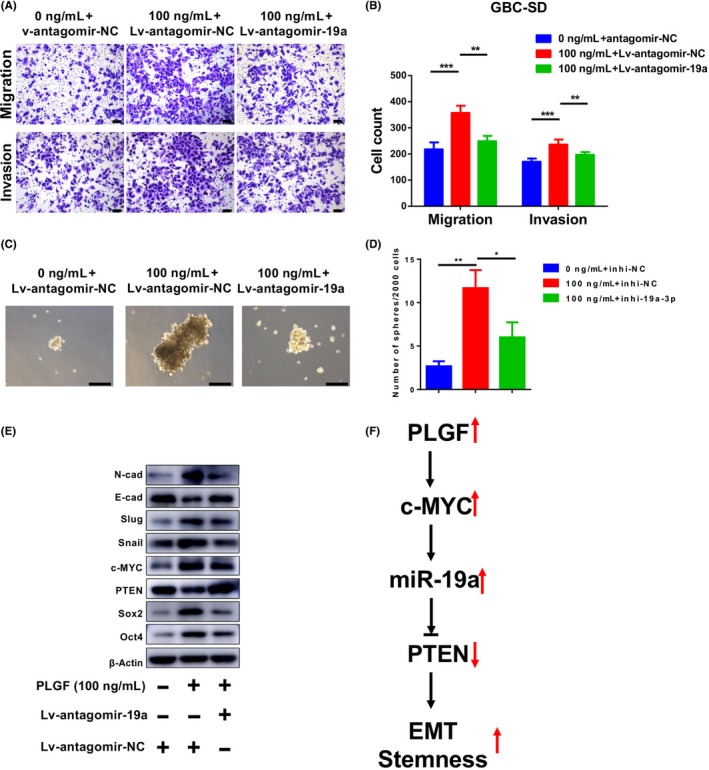
MicroRNA‐19a (miR‐19a) knockdown attenuates the promotion effects of placental growth factor (PLGF) on gallbladder cancer (GBC) cells. A, B, Migration and invasion assays of GBC‐SD cells infected with antagomir‐19a lentivirus or negative control (antagomir‐NC) and treated with or without 100 ng/mL PLGF. A, Representative images. B, Quantitative analysis of the numbers of migrated and invaded cells. C, D, Sphere‐formation assays of GBC‐SD cells infected with antagomir‐19a lentivirus or antagomir‐NC and treated with or without 100 ng/mL PLGF. (C) Representative images. D, Quantitative analysis of the numbers of spheres. *P < .05; **P < .01. E, Immunoblotting analyses of the levels of epithelial–mesenchymal transition (EMT) and cancer stem cell marker proteins in GBC‐SD cells after infection with antagomir‐19a lentivirus or antagomir‐NC and treatment with or without 100 ng/mL PLGF. E‐cad, E‐cadherin; N‐cad, N‐cadherin. F, Working model of the PLGF–c‐Myc–miR‐19a–PTEN regulatory axis in GBC
4. DISCUSSION
The tumor microenvironment promotes cancer progression and is used to predict the phenotypic characteristics of cancer.11 Most GBC patients have chronic cholecystitis, and gallstones are the main risk factor for the development of GBC.2 Placental growth factor, a member of the vascular endothelial growth factor family, was initially identified as an angiogenic factor.14 Interestingly, PLGF plays a vital role in tumor angiogenesis and metastasis and may be an unfavorable progression indicator for cancer patients.13, 14, 15, 16, 17, 33, 34 However, the precise molecular mechanisms by which PLGF influences various physiological processes of GBC remain unknown. In the present study, we found that PLGF is highly expressed in GBC tissues.
Tumor cell metastasis is a complex and multistage process that includes adhesion, migration, invasion, angiogenesis, and eventual unremitting proliferation in distant target organs.35 During tumor metastasis, EMT has been indicated to lead to the dissemination of single tumor cells from primary epithelial tumors. The EMT could be affected by various pro‐invasion signals from the tumor microenvironment. Growing evidence has indicated that EMT is stimulated by certain cytokines and growth factors, such as transforming growth factor‐β1. As an angiogenic factor, PLGF can induce EMT in human breast cancer cells.36 Mounting evidence has revealed that CSCs are associated with EMT, and EMT cells can serve as the source of CSCs, which also confer the EMT phenotype.37 The overlapping of these two properties suggests that they might be controlled by similar pathways. Therefore, it is important to identify whether PLGF could promote EMT and CSC formation and uncover the mechanistic role of PLGF for the development of targeted therapies. This study identified PLGF as a driver of EMT and CSC formation in GBC cells. Exogenous PLGF promoted migration and invasion, as well as sphere‐formation ability, in GBC cells. Knockdown of PLGF suppressed the EMT phenotype and CSC properties. In addition, PLGF significantly increased the protein levels of mesenchymal and CSC markers. Immunofluorescence analysis for N‐cadherin and Slug confirmed the results from Western blot analyses. These various observations showed that PLGF plays a vital role in EMT and CSC formation. Future studies could be applied to address the detailed regulation of CSC markers by PLGF as well as the exact effects of PLGF on EMT markers.
The c‐MYC oncogene is one of the most commonly activated oncogenes implicated in the development of human cancers, including GBC.38, 39 Recently, numerous studies have focused on the role of c‐MYC regulating miRNAs,40, 41 including the induction of miRNAs with oncogenic properties, such as the miR‐17~92 cluster.27, 42, 43 The miR‐17~92 cluster includes six mature miRNAs: miR‐17, miR‐18, miR‐19a, miR‐19b, miR‐20, and miR‐92a. As a direct transcriptional target of c‐MYC, the miR‐17~92 cluster was found to be highly expressed in various tumor cells and types of cancer.44, 45, 46, 47 Previous studies have revealed that miR‐19a triggers the EMT48 and regulates the activity of CSCs.46 Therefore, we proposed that PLGF promotes EMT and stemness through the c‐MYC/miR‐19a axis. In this study, we found that miR‐19a was dramatically upregulated after treatment with PLGF. Subsequently, we experimentally validated the oncogenic function of miR‐19a in two GBC cell lines. Furthermore, we revealed the important effects of the PLGF‐driven enhancement of miR‐19a in the promotion of GBC cell migration, invasion, and stemness. The positive correlation between PLGF and miR‐19a was also confirmed by Spearman's correlation analysis in 39 pairs of GBC tissues.
As an important oncogenic miRNA, miR‐19a regulates many target genes. Among them, PTEN is associated with EMT and CSC formation and is a target of miR‐19a,48, 49 It is easy to presume that, by miR‐19a upregulation, PLGF might also influence the downstream target genes of miR‐19a. Some oncogenic functions of PLGF may be indirectly executed by inhibiting its indirect target PTEN. Western blotting results of rescue assays proved that hypothesis.
In conclusion, our results show that PLGF and miR‐19a are significantly upregulated in GBC. The PLGF‐induced enhancement of miR‐19a expression plays vital roles in GBC metastasis and stemness in vitro and in vivo through the PLGF/c‐MYC/miR‐19a axis. These results provide new mechanistic insights into the molecular pathogenesis of PLGF‐mediated GBC metastasis and stemness and provide potential novel therapeutic targets for the treatment of GBC.
CONFLICT OF INTEREST
The authors have no conflict of interest.
Supporting information
ACKNOWLEDGMENTS
This study was supported by the National Natural Science Foundation of China (Nos. 91440203, 81502433, 31620103910, and 31601021) and the Shanghai Key Laboratory of Biliary Tract Disease Research Foundation (No. 17DZ2260200).
Li H, Jin Y, Hu Y, et al. The PLGF/c‐MYC/miR‐19a axis promotes metastasis and stemness in gallbladder cancer. Cancer Sci. 2018;109:1532–1544. https://doi.org/10.1111/cas.13585
Huaifeng Li and Yunpeng Jin contributed equally to this work.
Contributor Information
Xiangsong Wu, Email: wuxiangsong@xinhuamed.com.cn.
Yingbin Liu, Email: liuyingbin@xinhuamed.com.cn.
REFERENCES
- 1. Wu XS, Wang F, Li HF, et al. LncRNA‐PAGBC acts as a microRNA sponge and promotes gallbladder tumorigenesis. EMBO Rep. 2017;18:1837‐1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Li ML, Zhang Z, Li XG, et al. Whole‐exome and targeted gene sequencing of gallbladder carcinoma identifies recurrent mutations in the ErbB pathway. Nat Genet. 2014;46:872‐876. [DOI] [PubMed] [Google Scholar]
- 3. Shu YJ, Bao RF, Jiang L, et al. MicroRNA‐29c‐5p suppresses gallbladder carcinoma progression by directly targeting CPEB4 and inhibiting the MAPK pathway. Cell Death Differ. 2017;24:445‐457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Butte JM, Matsuo K, Gönen M, et al. Gallbladder cancer: differences in presentation, surgical treatment, and survival in patients treated at centers in three countries. J Am Coll Surg. 2011;212:50‐61. [DOI] [PubMed] [Google Scholar]
- 5. Wu XS, Shi LB, Li ML, et al. Evaluation of two inflammation‐based prognostic scores in patients with resectable gallbladder carcinoma. Ann Surg Oncol. 2014;21:449‐457. [DOI] [PubMed] [Google Scholar]
- 6. Liu Y, Wang Z, Li M, et al. Chloride intracellular channel 1 regulates the antineoplastic effects of metformin in gallbladder cancer cells. Cancer Sci. 2017;108:1240‐1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chou J, Lin JH, Brenot A, et al. GATA3 suppresses metastasis and modulates the tumour microenvironment by regulating microRNA‐29b expression. Nat Cell Biol. 2013;15:201‐213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Thiery JP, Acloque H, Huang RYJ, et al. Epithelial‐mesenchymal transitions in development and disease. Cell. 2009;139:871‐890. [DOI] [PubMed] [Google Scholar]
- 9. Nieto MA, Huang RYJ, Jackson RA, et al. EMT: 2016. Cell. 2016;166:21‐45. [DOI] [PubMed] [Google Scholar]
- 10. Song W, Mazzieri R, Yang T, et al. Translational significance for tumor metastasis of tumor‐associated macrophages and epithelial‐mesenchymal transition. Front Immunol. 2017;8:1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leonardi GC, Candido S, Cervello M, et al. The tumor microenvironment in hepatocellular carcinoma (review). Int J Oncol. 2012;40:1733‐1747. [DOI] [PubMed] [Google Scholar]
- 12. Goetze Thorsten O. Gallbladder carcinoma: prognostic factors and therapeutic options. World J Gastroenterol. 2015;21:12211‐12217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dewerchin M, Carmeliet P. Placental growth factor in cancer. Expert Opin Ther Targets. 2014;18:1339‐1354. [DOI] [PubMed] [Google Scholar]
- 14. Huang W, Zhu S, Liu Q, et al. Placenta growth factor promotes migration through regulating epithelial‐mesenchymal transition‐related protein expression in cervical cancer. Int J Clin Exp Pathol. 2014;7:8506‐8519. [PMC free article] [PubMed] [Google Scholar]
- 15. Chen CN, Hsieh FJ, Cheng YM, et al. The significance of placenta growth factor in angiogenesis and clinical outcome of human gastric cancer. Cancer Lett. 2004;213:73‐82. [DOI] [PubMed] [Google Scholar]
- 16. Parr C, Watkins G, Boulton M, et al. Placenta growth factor is over‐expressed and has prognostic value in human breast cancer. Eur J Cancer. 2005;41:2819‐2827. [DOI] [PubMed] [Google Scholar]
- 17. Matsumoto K, Suzuki K, Koike H, et al. Prognostic significance of plasma placental growth factor levels in renal cell cancer: an association with clinical characteristics and vascular endothelial growth factor levels. Anticancer Res. 2003;23:4953‐4958. [PubMed] [Google Scholar]
- 18. Zhang L, Chen J, Ke Y, et al. Expression of Placenta growth factor (PlGF) in non‐small cell lung cancer (NSCLC) and the clinical and prognostic significance. World J Surg Oncol. 2005;3:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Y, He L, Du Y, et al. The long noncoding RNA lncTCF7 promotes self‐renewal of human liver cancer stem cells through activation of Wnt signaling. Cell Stem Cell. 2015;16:413‐425. [DOI] [PubMed] [Google Scholar]
- 20. Jin YP, Hu YP, Wu XS, et al. miR‐143‐3p targeting of ITGA6 suppresses tumour growth and angiogenesis by downregulating PLGF expression via the PI3K/AKT pathway in gallbladder carcinoma. Cell Death Dis. 2018;9:182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jia Z, Wang K, Zhang A, et al. miR‐19a and miR‐19b overexpression in gliomas. Pathol Oncol Res. 2013;19:847‐853. [DOI] [PubMed] [Google Scholar]
- 22. Zhang L, Zhao S, Yuan L, et al. Placental growth factor triggers epithelial‐to‐mesenchymal transition‐like changes in rat type ii alveolar epithelial cells: activation of nuclear factor κB signalling pathway. Basic Clin Pharmacol Toxicol. 2016;119:498‐504. [DOI] [PubMed] [Google Scholar]
- 23. Zhao D, Pan C, Sun J, et al. VEGF drives cancer‐initiating stem cells through VEGFR‐2/Stat3 signaling to upregulate Myc and Sox2. Oncogene. 2015;34:3107‐3119. [DOI] [PubMed] [Google Scholar]
- 24. Lundin A, Driscoll B. Lung cancer stem cells: progress and prospects. Cancer Lett. 2013;338:89‐93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aimin Y, Shenghui Q, Schulte Bradley A, et al. MYC Inhibition depletes cancer stem‐like cells in triple‐negative breast cancer. Cancer Res. 2017;77:6641‐6650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Akita H, Marquardt JU, Durkin ME, et al. MYC activates stem‐like cell potential in hepatocarcinoma by a p53‐dependent mechanism. Cancer Res. 2014;74:5903‐5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu Y, Liu R, Yang F, et al. miR‐19a promotes colorectal cancer proliferation and migration by targeting TIA1. Mol Cancer. 2017;16:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mogilyansky E, Rigoutsos I. The miR‐17/92 cluster: a comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013;20:1603‐1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. O'Donnell KA, Wentzel EA, Zeller KI, et al. c‐Myc‐regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839‐843. [DOI] [PubMed] [Google Scholar]
- 30. Dou L, Meng X, Sui X, et al. MiR‐19a regulates PTEN expression to mediate glycogen synthesis in hepatocytes. Sci Rep. 2015;5:11602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ciuffreda L, Falcone I, Incani UC, et al. PTEN expression and function in adult cancer stem cells and prospects for therapeutic targeting. Adv Biol Regul. 2014;56:66‐80. [DOI] [PubMed] [Google Scholar]
- 32. Bahena‐Ocampo I, Espinosa M, Ceballos‐Cancino G, et al. miR‐10b expression in breast cancer stem cells supports self‐renewal through negative PTEN regulation and sustained AKT activation. EMBO Rep. 2016;17:648‐658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen J, Ye L, Zhang L, et al. Placenta growth factor, PLGF, influences the motility of lung cancer cells, the role of Rho associated kinase, Rock1. J Cell Biochem. 2008;105:313‐320. [DOI] [PubMed] [Google Scholar]
- 34. Li B, Wang C, Zhang Y, et al. Elevated PLGF contributes to small‐cell lung cancer brain metastasis. Oncogene. 2013;32:2952‐2962. [DOI] [PubMed] [Google Scholar]
- 35. Peng C, Zhao H, Song Y, et al. SHCBP1 promotes synovial sarcoma cell metastasis via targeting TGF‐β1/Smad signaling pathway and is associated with poor prognosis. J Exp Clin Cancer Res. 2017;36:141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ning Q, Liu C, Hou L, et al. Vascular endothelial growth factor receptor‐1 activation promotes migration and invasion of breast cancer cells through epithelial‐mesenchymal transition. PLoS One. 2013;8:e65217. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37. Gao Y, Zhang Z, Li K, et al. Linc‐DYNC2H1‐4 promotes EMT and CSC phenotypes by acting as a sponge of miR‐145 in pancreatic cancer cells. Cell Death Dis. 2017;8:e2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ishak G, Leal MF, Dos Santos NP, et al. Deregulation of MYC and TP53 through genetic and epigenetic alterations in gallbladder carcinomas. Clin Exp Med. 2015;15:421‐426. [DOI] [PubMed] [Google Scholar]
- 39. Dang CV. c‐Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1‐11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cairo S, Wang Y, de Reyniès A, et al. Stem cell‐like micro‐RNA signature driven by Myc in aggressive liver cancer. Proc Natl Acad Sci U S A. 2010;107:20471‐20476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Psathas JN, Thomas‐Tikhonenko A. MYC and the art of microRNA maintenance. Cold Spring Harb Perspect Med 2014;4:a014175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Frenzel A, Lovén J, Henriksson MA. Targeting MYC‐regulated miRNAs to combat cancer. Genes Cancer. 2010;1:660‐667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Olive V, Bennett MJ, Walker JC, et al. miR‐19 is a key oncogenic component of mir‐17‐92. Genes Dev. 2009;23:2839‐2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fang LL, Wang XH, Sun BF, et al. Expression, regulation and mechanism of action of the miR‐17‐92 cluster in tumor cells (Review). Int J Mol Med. 2017;40:1624‐1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Li X, Xie W, Xie C, et al. Curcumin modulates miR‐19/PTEN/AKT/p53 axis to suppress bisphenol A‐induced MCF‐7 breast cancer cell proliferation. Phytother Res. 2014;28:1553‐1560. [DOI] [PubMed] [Google Scholar]
- 46. Zhu J, Wang S, Chen Y, et al. miR‐19 targeting of GSK3β mediates sulforaphane suppression of lung cancer stem cells. J Nutr Biochem. 2017;44:80‐91. [DOI] [PubMed] [Google Scholar]
- 47. Wu Q, Yang Z, An Y, et al. MiR‐19a/b modulate the metastasis of gastric cancer cells by targeting the tumor suppressor MXD1. Cell Death Dis. 2014;5:e1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li J, Yang S, Yan W, et al. MicroRNA‐19 triggers epithelial‐mesenchymal transition of lung cancer cells accompanied by growth inhibition. Lab Invest. 2015;95:1056‐1070. [DOI] [PubMed] [Google Scholar]
- 49. Schubbert S, Jiao J, Ruscetti M, et al. Methods for PTEN in stem cells and cancer stem cells. Methods Mol Biol. 2016;1388:233‐285. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials


