Abstract
Objective
To identify the most appropriate imputation method for missing data in the HCUP State Inpatient Databases (SID) and assess the impact of different missing data methods on racial disparities research.
Data Sources/Study Setting
HCUP SID.
Study Design
A novel simulation study compared four imputation methods (random draw, hot deck, joint multiple imputation [MI], conditional MI) for missing values for multiple variables, including race, gender, admission source, median household income, and total charges. The simulation was built on real data from the SID to retain their hierarchical data structures and missing data patterns. Additional predictive information from the U.S. Census and American Hospital Association (AHA) database was incorporated into the imputation.
Principal Findings
Conditional MI prediction was equivalent or superior to the best performing alternatives for all missing data structures and substantially outperformed each of the alternatives in various scenarios.
Conclusions
Conditional MI substantially improved statistical inferences for racial health disparities research with the SID.
Keywords: Missing data, multiple imputation, racial disparities
The Healthcare Cost and Utilization Project (HCUP) is a family of health care databases, related software tools, and products sponsored by the Agency for Healthcare Research and Quality (AHRQ). The State Inpatient Databases (SID), a member of the HCUP family since 1990, contain inpatient discharge abstracts from community hospitals in 28 participating states as of 2014. With such comprehensive information, the SID provide a unique and powerful platform for a broad range of health care and medical research (Hellinger 2004; Hamlat et al. 2012; Vosseller, Karl, and Greisberg 2014; Chan et al. 2016).
As with any large‐scale data collection effort, the SID have a moderate amount of missing data from several patient‐level variables. These variables are continuous (e.g., total hospital charges), ordered categorical (e.g., median household income), and unordered categorical (e.g., race, admission source). Patient race has a particularly high proportion of missingness across multiple states and years. Figure 1 shows race data missingness patterns from 2004 to 2010 for five racially diverse states in the SID. Although rates of incomplete race data decreased over time in some states (e.g., North Carolina and Colorado), a large amount of race data was still missing (>5 percent) in 2010. The availability of high‐quality, valid, and reliable data is critically important for identifying the causes of racial health disparities and, ultimately, developing solutions. Due to the lack of knowledge or tools to address missing data issues in public‐use databases, researchers often rely on crude strategies that either conduct complete case analysis (CCA) by eliminating incomplete cases or impute the missing data with a single set of replacement values. Unfortunately, these outdated methods, which not only affect the quality of research, but also lead to inconsistent results across studies, are still widely used in practice. In a systematic review of missing race/ethnicity data in Veterans Health Administration (VHA) based health disparities research, CCA was found in over half of 114 health disparities articles using administrative data (Long et al. 2006). Although simple to implement, CCA has two fundamental problems. First, it produces biased parameter estimates when subjects with missing values are systematically different from those with complete observations. Second, it can discard a large number of cases, resulting in inefficient estimation. If CCA was applied to the SID, it would cause a significant loss of sample due to substantial race data missingness.
Figure 1.
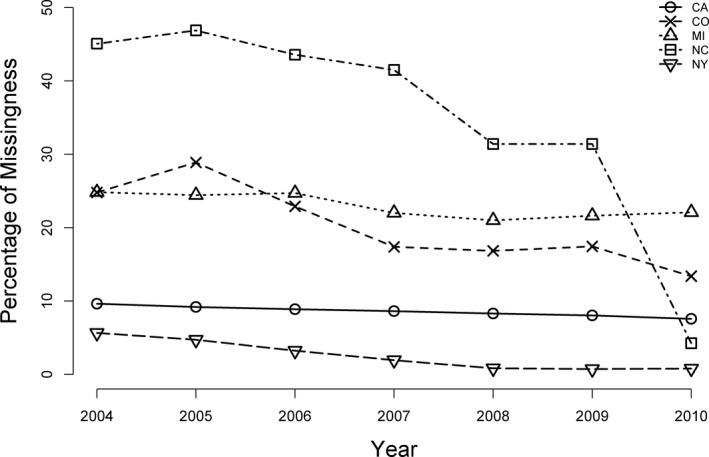
Missing Race Data in Selected SID Participating States from 2004 to 2010
Imputation methods were developed to address the issue of incomplete data by filling in missing observations. A naive imputation method is the random draw, which imputes missing data with a randomly drawn nonmissing value from the same variable. A more sophisticated method is “hot deck” imputation. This approach involves replacing missing values from a nonrespondent (recipient) with observed values from a respondent (donor) that is similar to the nonrespondent with respect to a subset of other fully observed (“deck”) variables (Andridge and Little 2009). Although the hot deck method has been used by federal agencies to impute missing data in public‐use datasets (Coffey et al. 2008; National Hospital Ambulatory Medical Care Survey 2008), it has limitations. In principle, the more deck variables considered, the more predictive information can be used to increase the accuracy of the imputation. However, dimensionality problems can quickly arise, dramatically limiting the number of deck variables that can be included (Andridge and Little 2009). In addition, both random draw and hot deck are single imputation methods, which treat imputed values as known. As a result, the variability in imputed values is underestimated. For this reason, single imputation has been criticized for more than two decades (Madow, Olkin, and Rubin 1983; Rubin 1987; Rubin and Little 2002; Scheuren 2005).
Multiple imputation (MI) is an advanced statistical method that handles missing data by replacing each missing value with a set of plausible values, producing a set of imputed datasets. As opposed to single imputation methods, multiple imputation methods account for the uncertainty of the imputed data. Each imputed dataset is analyzed using standard methods for complete data, and results are pooled using Rubin's rule (Rubin 1987). MI is particularly effective in large datasets and has been used to impute missing data in several well‐respected nationwide health studies, including the National Health and Nutritional Examination Survey III (Schafer et al. 1996), the National Health Interview Survey (Schenker et al. 2006), Cardiovascular Health Survey (Arnold and Kronmal 2003), Cancer Care Outcomes Research and Surveillance (CanCORS) Consortium (He et al. 2010), and several others. However, there are no imputed datasets available to SID users. Improved imputation methodology for the SID is crucial given the anticipated impact of national databases on health policy and medical practice. In 2013, we began a project to impute missing data for the SID with funding provided by the AHRQ. Our goal is to generate imputed companion datasets to the SID that will allow public users to perform analysis on complete data using existing software. To identify the appropriate imputation methods for the SID, we compared the performance of two MI methods (joint MI, conditional MI) with three other methods (complete case analysis, random draw method, hot deck imputation) through a novel simulation study. We also assessed the impact of different missing data methods on health disparities research.
Methods
To understand which imputation methods are most appropriate and how best to apply them, one must first determine the patterns and mechanisms of the missing data. In this section, we will first describe the characteristics of the missing data in the SID. This will be followed by a review of the two MI approaches (conditional and joint). For illustrative purposes, we will focus on the 2005 SID Colorado (SID‐CO) data. This dataset contains patient baseline characteristics (e.g., age, gender, race, median household income for patient zip code), admission information (e.g., admission type and source, insurance type, length of hospital stay), comorbidities, diagnosis, procedure, and discharge status (e.g., mortality, disposition of patient at discharge) for 474,057 admissions from 79 hospitals in CO.
Missing Data in SID‐CO
The majority of missing data were found in five variables across 30.5 percent (n = 144,337) of the admissions in the 2005 SID‐CO (Table 1). In particular, patient race had the highest rate of missingness. This missingness could be caused by a variety of factors. Registration staff are usually not trained in interviewing and may feel uncomfortable soliciting these data from patients. Patients, meanwhile, may refuse to provide race data because of concerns about privacy and their own uncertainty as to why these data are needed. Under these circumstances, missing data are inevitable. Logistic regression also revealed that race missingness was related to many observed patient characteristics (e.g., age, gender, admission source, mortality, admission type, insurance type, disposition of patient, diagnosis and procedure type, comorbidities, length of hospital stay).
Table 1.
Summary of Missing Data in the 2005 SID‐CO
| Variable Name | Variable Type | Total Number of Missing Observations (%) | Description |
|---|---|---|---|
| Total charges | Continuous | 9,156 (1.93) | Total hospital charges |
| Gender | Binary | 161 (0.034) | Male/female |
| Median income quartile | Ordinal | 14,191 (2.99) |
|
| Admission source | Unordered categorical | 13,755 (2.90) |
|
| Race | Unordered categorical | 136,955 (28.9) |
|
The statistical literature has defined three types of missingness mechanisms: missing completely at random (MCAR), missing at random (MAR), and missing not at random (MNAR). In MCAR, data missingness is independent of both unobserved and observed data. In MAR, data missingness depends only on observed information. In MNAR, data missingness depends on unobserved information. Data in the SID‐CO were clearly not MCAR as the probability of missingness depended on observed data. However, it is impossible to determine whether data are MAR or MNAR solely based on observed data. The SID are de‐identified to prevent tracking patients for follow‐up or prior information, so the MNAR assumption cannot be tested. Because of this de‐identification, it is impossible to impute missing race data based on name and census tract (Elliott et al. 2013). Nevertheless, the MAR assumption is generally considered to be realistic for well‐conducted surveys (Rubin, Stern, and Vehovar 1995) and has been recommended for practical applications (Verbeke and Molenberghs 2000). The assumption of MAR becomes more reasonable as more predictors are included in the imputation model (Little and Rubin 2003; Gelman and Hill 2006). As the SID contain high‐quality data with a large amount of predictive information, the MAR assumption can be justified. Additional predictive information was obtained from the U.S. Census (e.g., racial and socioeconomic status distributions) and the American Hospital Association (AHA) database (e.g., hospital level characteristics) and taken into account in our imputation.
The SID contain missing data not only on race, but also on gender, total charge, admission source, and median income. The issue is complicated by the fact that these variables have different distributions. There are two general MI methods for handling multivariate missing data: joint modeling and conditional specification. Next, we discuss these two methods in detail.
Multiple Imputation Methods
The joint MI assumes that data follow a joint distribution, typically a multivariate normal (MVN), and draws imputed values from this distribution. Unless stated otherwise, the joint MI in this paper refers to the joint MVN MI. Under this assumption, discrete variables must be treated as if they were continuous. Continuous imputed values are then often converted back to discrete values by rounding to the nearest category. However, this rounding approach has been criticized for reduced accuracy (Horton, Lipsitz, and Parzen 2003; Horton and Kleinman 2007; Yucel, He, and Zaslavsky 2008; Honaker, King, and Blackwell 2011). The disadvantages of rounding are even more salient for the imputation of binary or ordinal variables with asymmetric distributions (Yucel, He, and Zaslavsky 2008). An alternative approach is to transform the continuous imputed values to probabilities from Bernoulli, binomial, and multinomial distributions for binary, ordinal, and nominal variables, respectively (Honaker, King, and Blackwell 2011). For binary and ordinal variables, the imputed values that are outside the range of the categories are first rounded to the nearest (either the lowest or the highest) category. The continuous values imputed for binary variables are within [0, 1] and are treated as probabilities from independent Bernoulli distributions, where an imputed value of 0 or 1 is randomly generated from each distribution. The continuous values for ordinal variables are scaled into [0, 1] and then used as probabilities from independent binomial distributions. A categorical imputed value can then be drawn from each distribution. To retain the unordered nature of nominal variables, a binary indicator is created for each category. Continuous values from MVN are assigned to these binary variables and constrained so that they are within [0, 1] and can sum to 1. The transformed values are then treated as probabilities from independent multinomial distributions. A value is then drawn from each distribution for each nominal variable. For example, the six‐category race variable in the SID can be imputed using the following steps. First, the variable is broken into five dummy variables (i.e., one for each racial category besides the reference category). These dummy variables are treated as continuous variables and imputed simultaneously using draws from a MVN distribution. The continuous imputed values are then appropriately scaled into probabilities for each of the six race categories from a multinomial distribution. The missing race data are then replaced with draws from the multinomial distribution. This procedure can be implemented using the Amelia package in R.
Conditional MI imputes data in a variable‐by‐variable fashion rather than relying on a joint distribution. Raghunathan et al. (2001) formalized this concept using a sequential regression multivariate imputation approach. This method provides substantial flexibility for handling complex data structures (e.g., bounds, skip patterns) where it is difficult to formulate a joint distribution. Conditional MI also allows variables of different types to be modeled separately. For example, continuous, binary, ordinal, and nominal variables are modeled using linear, logistic, ordinal logistic, and multinomial regression, respectively. Incomplete variables are imputed consecutively and iteratively from their respective conditional distributions. In each iteration of the procedure, one incomplete variable is regressed on the observed and imputed values for the other variables in the data. Incomplete predictors in each regression model are replaced with imputed values from the last iteration. The missing outcome values are drawn from the corresponding predictive distribution given the observed values. Conditional MI can be implemented in R by the mice (Van Buuren and Oudshoorn 2011) and mi (Su et al. 2011) packages. The mi package uses Bayesian versions of linear, logistic, and ordered logistic regression as imputation models for continuous, binary, and ordered categorical variables, respectively, while mice runs standard regressions. In our study, we compared imputation results from these two packages. Throughout the paper, “conditional MI (mi)” and “conditional MI (mice)” refer to conditional multiple imputation implemented in mi and mice, respectively.
Joint MI and conditional MI are the most accessible MI methods because they can be implemented in almost all standard statistical software including R, SAS, and Stata (Royston 2004; Yuan 2010; Honaker, King, and Blackwell 2011). Prior comparisons of these methods were limited by multiple factors. First, previous comparisons used less complex data structures than the SID and tested fewer variable types (Yu, Burton, and Rivero‐Arias 2007; Lee and John 2010). For example, Raghunathan et al. (2001) compared conditional MI with a general location model (Little and Schluchter 1985; Belin et al. 1999). The latter model is a more plausible alternative to multivariate normal imputation for mixed categorical and continuous data. However, the comparison in Raghunathan's study was limited to imputation of missing continuous and binary data. Second, prior comparisons were not comprehensive as they focused solely on either the accuracy of the imputed values (Yu, Burton, and Rivero‐Arias 2007) or the impact of imputation on regression analysis (Yu, Burton, and Rivero‐Arias 2007; Lee and John 2010). Third, MI methods were only compared with CCA. Other methods such as hot deck, a widely used imputation procedure for survey data, were not included in prior comparisons. Therefore, the existing literature does not provide adequate guidance on the use of MI methods, and the most appropriate method for imputing the SID is unknown. To fill the gap, we performed a comprehensive simulation study.
Simulation Study
Statistical simulation has become popular for comparing the performance of competing methods. However, classic simulations are built entirely on fake data. Given the large sample size and complex data structure, the SID would be almost impossible to simulate. Therefore, we developed a novel simulation using real data from the 2005 SID‐CO.
Missing Data Generation
Missing data were generated with the following steps:
Step 1. Missing data patterns were identified. Missing observations were found in 28.9 percent of race and less than 5 percent on all other variables (e.g., total charges, admission source, median household income, gender).
Step 2. A complete subset of the 2005 SID‐CO was left after removing the partially observed cases that had missing data on the aforementioned variables in Step 1. This complete dataset, denoted by Z, includes a total of n = 329,720 admissions, a 69.5 percent sample of the original dataset. The values of the data in Z were referred to as true values (i.e., gold standard) when assessing the accuracy of the imputed values in the simulation study. The dataset Z was further split into two datasets, X and Y, where X is a (n × p) matrix containing a set of p variables and Y a (n × 5) matrix containing five variables. For ease of computation, categorical variables in X were broken into dummy variables. In this simulation, there were a total of p = 155 variables in X, including patient age, admission type, insurance type, disposition of patient, weekend admission status, length of hospital stay, comorbidities, mortality, and major diagnostic and procedure categories. The variables in Y were those found to have missing data in the original dataset, including race, total charges, admission source, median household income, and gender. Variables in X and Y were all from the SID, not generated by statistical models. Hence, the data structure and correlations between these variables remained intact.
Step 3. To mimic the missing data patterns identified in Step 1, missingness was generated for the variables in Y under the assumption of MAR: 25 percent missingness on race and 5 percent on total charge, gender, median household income, and admission source. As the true values of these missing data were known from Y, the accuracy of imputed values could be assessed. The missingness was generated through a logistic regression on the p predictors in X. Let θ =Xβ, where β denotes the (155 × 5) matrix of regression coefficients associated with the predictors in X. As the correlations between predictors and missingness indicators are unknown, elements in the matrix β were independently generated from the standard normal distribution. The rows of β correspond to the 155 predictors in X, and the columns of β correspond to the aforementioned five variables that have missing data.
Step 4. A matrix Ω of the same dimensions as θ was randomly generated from a multivariate normal distribution MVN(μ,Σ), where μ was a vector of 0 and Σ a random correlation matrix (Lewandowski, Kurowicka, and Joe 2009). The columns of Ω could be correlated as Σ was not constrained to be diagonal in our study. Let W = θ + Ω. By adding Ω to θ, these correlations were integrated into missing data generation such that observations missing on one variable would be likely to be missing on another variable—a phenomenon that occurs in many real samples. Then, the elements of W were transformed to probabilities π using the logistic distribution function.
Step 5. Let U be a matrix of the same dimensions as π. The elements of U were independently drawn from the uniform distribution on [0,1]. Let Π = π−U. In the first column of Π, the highest 25 percent of observations were selected to be missing in race. In the other four columns of Π, the highest 5 percent were selected to be missing in total charge, gender, median income quartile, and admission source, respectively. Following this procedure, we obtained a dataset with simulated missing observations. This dataset is denoted by , where contains the same variables in Y with missing observations.
Imputation Procedure
Missing data were imputed using four statistical methods, including random draw, hot deck, joint MI, and conditional MI. Each MI generated five imputed datasets. Predictive variables used for the imputation included patient age, mortality, weekend admission status, disposition of patient, admission type, insurance type, length of hospital stay, comorbidities, and major diagnostic and procedure categories. The original dataset contains a total of 231 procedure categories. For analytic simplicity, we collapsed the categories into 33 groups based on guidance from clinicians (detailed information can be found in Data S1 in Appendix SA2). To further improve the prediction of missing data, additional predictive information was incorporated in the imputation from the U.S. Census and the AHA databases. This information included zip code racial distribution, median household income, poverty and education levels, and hospital characteristics such as bed size, service type, hospital teaching status, and hospital location. These additional data were linked to the SID via patient zip code or hospital ID.
To reduce the computational burden caused by the large sample size, we split the population into 10 age groups: newborn, 1–17 (years old), 18–25, 26–34, 35–44, 45–54, 55–64, 65–74, 75–84, and age greater or equal to 85. Imputation was performed for each age group separately. This approach also captures the characteristics of age‐dependent variables such as admission type, insurance type, and comorbidities. Sample R code of all imputation strategies can be found in Data S2 in Appendix SA2.
Evaluation of Imputation Performance
Upon the completion of imputation, the following procedures were carried out to evaluate the performance of imputation methods over 100 Monte Carlo replications.
Hellinger Distance
We calculated Hellinger distances (HDs; Escofier 1978; Pollard 2002) for all variables to assess the similarity between the marginal distributions of the imputed and true values. Smaller values of HD imply more similarity between the distributions. The HD between two continuous distributions is given by , where f (.) and g (.) are density functions. For two discrete distributions P and Q taking k values with probabilities and , the HD can be expressed as .
Brier Score
We also calculated squared errors of prediction, also called Brier scores, for categorical variables. Brier scores have been used to measure the accuracy of probabilistic predictions for discrete outcomes in MI (Heltshe et al. 2012; Held et al. 2016). Let X be a K‐level (K > 1) categorical variable with M missing observations and denote the predicted probability of a specific level of X. For a MI with five iterations, , where is the imputed value of from the lth iteration, . The Brier score is given by , where , if the true value of is (not) j. A smaller Brier score implies a more precise imputation. The average Brier score was obtained across all simulation iterations. As the predicted probability is calculated based on multiply imputed data, Brier scores were used to compare multiple imputation methods only.
Post‐Imputation Analysis
We assessed the impact of each imputation method on inferences from regression analysis. For illustrative purposes, we chose three outcomes of interest in total knee arthroplasty (TKA) racial disparities research: length of hospital stay (a continuous outcome), any surgical complications (a binary outcome), and utilization of high TKA volume hospitals (an ordinal outcome: low [<200], medium [200–400], high annual TKA volume [400+]). Linear regression, logistic regression, and multinomial logistic regression were conducted for length of hospital stay, complications, and TKA volume, respectively. In addition to race, regression model covariates included age, gender, comorbidity index (Deyo, Cherkin, and Ciol 1992), median household income, admission type, admission source, insurance type, hospital bed size, teaching status, and location. Each regression was performed using the true dataset Z, complete cases in , and the imputed datasets. Regression coefficient estimates from regression analyses of complete cases and imputed datasets were compared with those calculated from the true dataset Z. Let denote the coefficient estimates from the analyses of true data and (i = 1, 2,…,100) the coefficient estimates from the analyses of imputed data or complete cases in the ith simulation, respectively. We computed the root mean square difference (RMSD) of each coefficient estimate using .
Simulation Results
The ratios of HDs for each method versus conditional MI (mi) are summarized in Figure 2. In the following discussion, unless otherwise stated, “conditional MI” refers to conditional multiple imputation implemented in both mice and mi. Among different imputation methods, conditional MI was associated with the smallest HD for all variables (Figure 2), implying that the marginal distributions of imputed data from conditional MI were most similar to the marginal distributions of true data. Results from conditional MI implemented in mice and mi were very close or equivalent. Random draw had consistently inferior performance for all variable types. Joint and conditional MI performed similarly for binary and continuous variables, but joint MI had significantly larger HD than conditional MI for nominal and ordinal variables. For example, the HDs for race were 43 percent greater for joint MI than for conditional MI (mi). The HDs for income were 70 percent greater for joint MI than for conditional MI (mi). Hot deck had significantly larger HDs for continuous, ordinal, and nominal variables than both MI methods. Specifically, hot deck had nearly 200 percent greater HD for total charge than joint or conditional MI, and 100 percent greater HD for income than conditional MI. Hot deck had 15 percent and 73 percent larger HDs than conditional MI for race and admission source, respectively.
Figure 2.

Evaluation of Imputation Performance: Ratio of Hellinger Distances for Each Method versus Conditional MI (mi)
The ratios of Brier scores for joint MI and conditional MI (mice) versus conditional MI (mi) are summarized in Figure 3 for all categorical variables. Conditional MI methods had smaller Brier scores than joint MI in all cases, implying that conditional MI was associated with more accurate predicted probabilities than joint MI. Such differences were pronounced for nominal variables. For example, conditional MI had 22 and 60 percent lower Brier score than joint MI for race and admission source, respectively. Results from conditional MI implemented in mice and mi were nearly equivalent for all variables.
Figure 3.
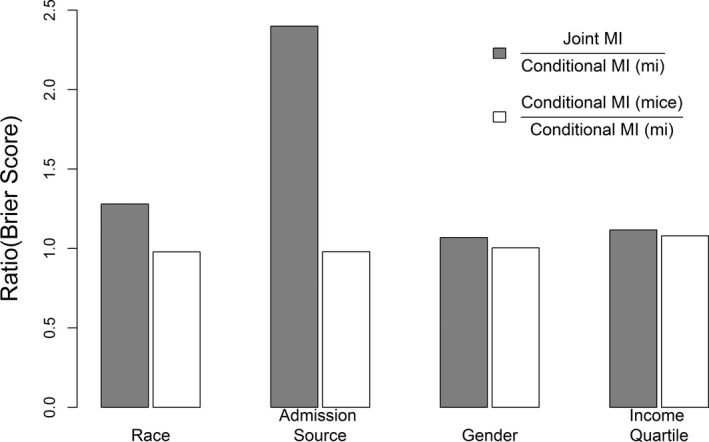
Evaluation of Predicted Probability for Categorical Variable: Ratio of Brier Scores for Joint MI and Conditional MI (mice) versus Conditional MI (mi)
The RMSDs of regression coefficient estimates are summarized in Figures 4, 5, 6. Among the covariates, admission source, admission type, and race had comparatively larger RMSDs for all methods in all models, but the magnitudes of these RMSDs were highly heterogeneous between different methods. Random draw, joint MI, and hot deck had significantly inflated RMSDs for admission source, admission type, and race in all models. The performance of CCA varied by the type of regression. Logistic regression based on CCA tended to have the highest RMSDs for all covariates (the RMSDs for all covariates can be found in Figures S1–S3 in Data S3 in Appendix SA2). For linear regression and multinomial logistic regression, CCA had higher RMSDs than other methods for income, comorbidity index, age, hospital bed size, gender, and insurance type. Conditional MI using mice or mi tended to have the smallest RMSDs for nearly all coefficient estimates in all three regression models (Figures S1–S3).
Figure 4.

Evaluation of Post‐Imputation Performance: Root Mean Square Difference (RMSD) of Coefficient Estimates for Linear Regression for Length of Stay
Figure 5.

Evaluation of Post‐Imputation Performance: Root Mean Square Difference (RMSD) of Coefficient Estimates for Logistic Regression for Any Complications
Figure 6.
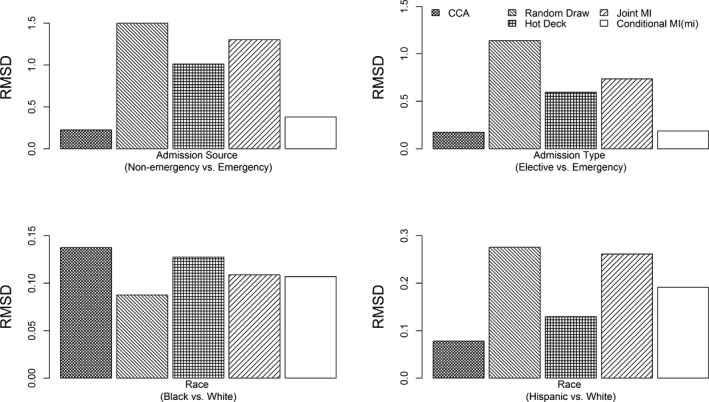
Evaluation of Post‐Imputation Performance: Root Mean Square Difference (RMSD) of Coefficient Estimates for Multinomial Logistic Regression for Hospital TKA Volume
Discussion
Eliminating racial disparities in health care continues to be an important goal for our nation. Large hospital administrative datasets have been used widely to study racial health disparities. However, incomplete race data are a serious and persistent problem that hampers the progress of research in health disparities. Fortunately, multiple procedures have been developed to address this issue both retrospectively and prospectively. Some efforts aim to improve future race/ethnicity data collection and reporting. For example, the AHRQ and the Institute of Medicine have been collaborating to identify standardized categories for race and ethnicity (Institute of Medicine 2009). Other efforts, such as imputation, aim to reduce the impact of existing missing data on disparities research (Mulugeta et al. 2012). This project was funded by AHRQ to impute missing data, including race, in the SID. Upon completion of the study, an imputed version of the SID will be available for public use.
To find the most appropriate imputation method for the SID, we systematically compared multiple approaches through a novel simulation study built on real data from the 2005 SID‐CO. Among the tested imputation approaches, conditional MI provided the most accurate imputed data for all types of variables. Joint MI, which is built on the assumption of the multivariate normal distribution, generated severe bias when imputing categorical data. Hot deck provided suboptimal imputation for continuous, nominal, and ordinal data, which affected its statistical inferences in regression analysis. Further, we assessed the impact of imputed data on analysis of racial disparities among TKA patients. Regression coefficient estimates from analyses of datasets imputed with conditional MI were most similar to those from analyses of true datasets. In contrast, the popular, naive approaches (CCA and random draw) gave substantially different coefficient estimates.
Collection of race data can be influenced by a variety of factors, including patient perceptions, culture, staff discomfort, legal concerns, or lack of appropriate categories. These factors could induce complicated missingness mechanisms. Nevertheless, MI can provide reasonably stable results under the assumption of MAR (Schafer et al. 1996; Schenker et al. 2006). Our study is limited by a number of issues inherent to secondary data analysis of large administrative databases. The SID lack patient‐level clinical and socioeconomic status data, so we were unable to incorporate such information into our imputation. To address this limitation, we used zip code–level information (e.g., racial distribution, income, education, and poverty level) from the U.S. Census. In addition to the simulation study presented in this paper, we conducted the same set of regression analyses using real data from 2005 SID‐CO to study racial disparities in TKA. The results from this real data analysis using different imputation methods are available in Data S4 in Appendix SA2).
Conclusions
Conditional MI prediction was uniformly equivalent or superior to the best performing alternatives for all missing data structures, while substantially outperforming each of the alternatives in various scenarios. The validated imputed datasets generated from this study will improve the value of the SID as a data source for a variety of future studies. The approaches used in this study can be applied to other large datasets and tested in other priority populations and health conditions, yielding benefits that extend beyond the current study.
Supporting information
Appendix SA1: Author Matrix.
Appendix SA2:
Data S1. SAS Code for Collapsing Categories in Procedure Types.
Data S2. Sample R Code for Performing Imputation Using Random Draw, Hotdeck, Joint MI, Conditional MI.
Data S3. Evaluation of Post‐Imputation Performance.
Data S4. Real Data Analysis.
Acknowledgments
Joint Acknowledgment/Disclosure Statement: This research was supported by grant funding from the Agency for Healthcare Research and Quality (R01HS21734). We sincerely thank Dr. Andrew Gelman at Columbia University for his substantial contributions to the success of the project.
Disclosures: None.
Disclaimers: None.
References
- Andridge, R. H. , and Little R. J. A.. 2009. “The Use of Sample Weights in Hot Deck Imputation.” Journal of Official Statistics 25: 21–36. [PMC free article] [PubMed] [Google Scholar]
- Arnold, A. M. , and Kronmal R. A.. 2003. “Multiple Imputation of Baseline Data in the Cardiovascular Health Study.” American Journal of Epidemiology 157: 74–84. [DOI] [PubMed] [Google Scholar]
- Belin, T. R. , Hu M. Y., Young A. S., and Grusky O.. 1999. “Performance of a General Location Model with an Ignorable Missing‐Data Assumption in a Multivariate Mental Health Services Study.” Statistics in Medicine 18: 3123–35. [DOI] [PubMed] [Google Scholar]
- Chan, M. Y. , Malik S., Hallstrom B. R., and Hughes R. E.. 2016. “Factors Affecting Readmission Cost after Primary Total Knee Arthroplasty in Michigan.” Journal of Arthroplasty 31 (6): 1179–81. doi:10.1016/j.arth.2015.11.037 [DOI] [PubMed] [Google Scholar]
- Coffey, R. , Barrett M., Houchens R., Ho K., Moy E., Brady J., and Andrews R.. 2008. Methods Applying AHRQ Quality Indicators to Healthcare Cost and Utilization Project (HCUP) Data for the Sixth (2008) National Healthcare Disparities Report. HCUP Methods Series Report # 2008‐06. Online October 23, 2008. U.S. Agency for Healthcare Research and Quality. Available at http://www.hcup-us.ahrq.gov/reports/methods.jsp
- Deyo, R. A. , Cherkin D. C., and Ciol M. A.. 1992. “Adapting a Clinical Comorbidity Index for Use with ICD‐9‐CM Administrative Databases.” Journal of Clinical Epidemiology 45: 613–9. [DOI] [PubMed] [Google Scholar]
- Elliott, M. N. , Becker K., Beckett M. K., Hambarsoomian K., Pantoja P., and Karney B.. 2013. “Using Indirect Estimates Based on Name and Census Tract to Improve the Efficiency of Sampling Matched Ethnic Couples from Marriage License Data.” Public Opinion Quarterly 77 (1): 375–84. [Google Scholar]
- Escofier, B. 1978. “Analyse Factorielle et Distances Répondant au Principe D’équivalence Distributionnelle.” Revue de Statistiques Appliquées 26: 29–37. [Google Scholar]
- Gelman, A. , and Hill J.. 2006. Data Analysis Using Regression and Multilevel/Hierarchical Models. New York: Cambridge University Press. [Google Scholar]
- Hamlat, C. A. , Arbabi S., Koepsell T. D., Maier R. V., Jurkovich G. J., and Rivara F. P.. 2012. “National Variation in Outcomes and Costs for Splenic Injury and the Impact of Trauma Systems: A Population‐Based Cohort Study.” Annals of Surgery 255 (1): 165–70. [DOI] [PubMed] [Google Scholar]
- He, Y. , Zaslavsky A. M., Landrum M. B., Harrington D. P., and Catalano P.. 2010. “Multiple Imputation in a Large‐Scale Complex Survey: A Practical Guide.” Statistical Methods in Medical Research 19: 653–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held, U. , Kessels A., Aymerich J. A., Basagaña X., Riet G., Moons K. G. M., Puhan M. A., and International COPD Cohorts Collaboration Working Group . 2016. “Methods for Handling Missing Variables in Risk Prediction Models.” American Journal of Epidemiology 184 (7): 545–51. doi:10.1093/aje/kwv346 [DOI] [PubMed] [Google Scholar]
- Hellinger, F. J. 2004. “HIV Patients in the HCUP Database: A Study of Hospital Utilization and Costs.” Inquiry 41 (1): 95–105. [DOI] [PubMed] [Google Scholar]
- Heltshe, S. L. , Lubin J. H., Koutros S., Coble J. B., Ji B. T., Alavanja M. C., Blair A., Sandler D. P., Hines C. J., Thomas K. W., Barker J., Andreotti G., Hoppin J. A., and Beane Freeman L. E.. 2012. “Using Multiple Imputation to Assign Pesticide Use for Non‐Responders in the Follow‐up Questionnaire in the Agricultural Health Study.” Journal of Exposure Science and Environmental Epidemiology 22: 409–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honaker, J. , King G., and Blackwell M.. 2011. “Amelia II: A Program for Missing Data.” Journal of Statistical Software 45 (7): 1–47. [Google Scholar]
- Horton, N. J. , and Kleinman K. P.. 2007. “Much Ado about Nothing: A Comparison of Missing Data Methods and Software to Fit Incomplete Data Regression Models.” American Statistician 61: 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton, N. J. , Lipsitz S. R., and Parzen M.. 2003. “A Potential for Bias When Rounding in Multiple Imputation.” American Statistician 57: 229–32. [Google Scholar]
- Institute of Medicine (IOM). 2009. Race, Ethnicity, and Language Data: Standardization for Health Care Quality Improvement. Washington, DC: The National Academies Press. [PubMed] [Google Scholar]
- Lee, K. J. , and John B. C.. 2010. “Multiple Imputation for Missing Data: Fully Conditional Specification Versus Multivariate Normal Imputation.” American Journal of Epidemiology 171 (5): 624–32. [DOI] [PubMed] [Google Scholar]
- Lewandowski, D. , Kurowicka D., and Joe H.. 2009. “Generating Random Correlation Matrices Based on Vines and Extended Onion Method.” Journal of Multivariate Analysis 100 (9): 1989–2001. [Google Scholar]
- Little, R. J. A. , and Rubin D. B.. 2003. Statistical Analysis with Missing Data. Hoboken, NJ: John Wiley & Son. [Google Scholar]
- Little, R. J. , and Schluchter M. D.. 1985. “Maximum Likelihood Estimation for Mixed Continuous and Categorical Data with Missing Values.” Biometrika 72: 497–512. [Google Scholar]
- Long, J. A. , Bamba M. I., Ling B., and Shea J. A.. 2006. “Missing Race/Ethnicity Data in Veterans Health Administration Based Disparities Research: A Systematic Review.” Journal of Health Care for the Poor and Underserved 17: 128–40. [DOI] [PubMed] [Google Scholar]
- Madow, W. G. , Olkin I., and Rubin D. B.. 1983. Incomplete Data in Sample Surveys (Volume 2): Theory and Bibliographies. New York: Academic Press. [Google Scholar]
- Mulugeta, G. , Yumin Z., Neal A., Gregory E. G., Carrae E., and Leonard E. E.. 2012. “Lessons Learned in Dealing with Missing Race Data: An Empirical Investigation.” Journal of Biometrics & Biostatistics 3: 138. [Google Scholar]
- Pollard, D. 2002. A User's Guide to Measure Theoretic Probability. New York: Cambridge University Press. [Google Scholar]
- Raghunathan, T. E. , Lepkowski J. M., Hoewyk J. V., and Solenberger P.. 2001. “A Multivariate Technique for Multiply Imputing Missing Values Using a Sequence of Regression Models.” Survey Methodology 27 (1): 85–96. [Google Scholar]
- Royston, P. 2004. “Multiple Imputation of Missing Values.” Stata Journal 4 (3): 227–41. [Google Scholar]
- Rubin, D. 1987. Multiple Imputation for Nonresponse in Surveys. New York: John Wiley & Sons. [Google Scholar]
- Rubin, D. B. , and Little R. J. A.. 2002. Statistical Analysis With Missing Data, 2d Edition New York: John Wiley and Sons. [Google Scholar]
- Rubin, D. B. , Stern H. S., and Vehovar V.. 1995. “Handling ‘Don't Know’ Survey Responses: The Case of the Slovenian Plebiscite.” Journal of the American Statistical Association 90 (431): 822–8. [Google Scholar]
- Schafer, J. L. , Ezzati‐Rice T. M., Johnson W., Khare M., Little R. J. A., and Rubin D. B.. 1996. The NHANES III Multiple Imputation Project. Available at http://ftp.cdc.gov/pub/health_Statistics/nchs/nhanes/nhanes3/7a/doc/jsm96.pdf
- Schenker, N. , Raghunathan T. E., Chiu P. L., Makuc D. M., Zhang G., and Cohen A. J.. 2006. “Multiple Imputation of Missing Income Data in the National Health Interview Survey.” Journal of the American Statistical Association 101 (475): 924–33. [Google Scholar]
- Scheuren, F. 2005. “Multiple Imputation: How It Began and Continues.” American Statistician 59: 315–9. [Google Scholar]
- Su, Y. S. , Yajima M., Gelman A. E., and Hill J.. 2011. “Multiple Imputation with Diagnostics (mi) in R: Opening Windows into the Black box.” Journal of Statistical Software 45 (2): 1–31. [Google Scholar]
- United States Department of Health and Human Services. Centers for Disease Control and Prevention. National Center for Health Statistics . 2008. National Hospital Ambulatory Medical Care Survey. ICPSR29922‐v1. Available at http://doi.org/10.3886/ICPSR29922.v1
- Van Buuren, S. , and Oudshoorn C. G.. 2011. “Multivariate Imputation by Chained Equations in R.” Journal of Statistical Software 45 (3): 1–67. [Google Scholar]
- Verbeke, G. , and Molenberghs G.. 2000. Linear Mixed Models for Longitudinal Data. New York: Springer. [Google Scholar]
- Vosseller, J. T. , Karl J. W., and Greisberg J. K.. 2014. “Incidence of Syndesmotic Injury.” Orthopedics 37 (3): e226–9. [DOI] [PubMed] [Google Scholar]
- Yu, L. M. , Burton A., and Rivero‐Arias O.. 2007. “Evaluation of Software for Multiple Imputation of Semi‐Continuous Data.” Statistical Methods in Medical Research 16 (3): 243–58. [DOI] [PubMed] [Google Scholar]
- Yuan, Y. C. . 2010. Multiple Imputation for Missing Data: Concepts and New Development (Version 9.0). Rockville, MD: SAS Institute Inc. [Google Scholar]
- Yucel, R. M. , He Y., and Zaslavsky A. M.. 2008. “Using Calibration to Improve Rounding in Imputation.” American Statistician 62: 125–9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix SA1: Author Matrix.
Appendix SA2:
Data S1. SAS Code for Collapsing Categories in Procedure Types.
Data S2. Sample R Code for Performing Imputation Using Random Draw, Hotdeck, Joint MI, Conditional MI.
Data S3. Evaluation of Post‐Imputation Performance.
Data S4. Real Data Analysis.


