ABSTRACT
Immunotherapies that target CD1d-restricted invariant NKT (iNKT) cells can prevent tumor growth in murine models but trials in humans have shown limited clinical efficacy. Here, we show that iNKT cells are depleted from blood and bronchial lavage samples from patients with non-small cell lung cancer (NSCLC) suggesting a role for these cells in immunity against NSCLC. We interrogated the Lung Cancer Explorer and Kaplan-Meier Plotter databases of NSCLC patients and found that pulmonary CD1d expression is reduced in patients with NSCLC and that low expression of CD1d mRNA is significantly associated with poor patient survival. We hypothesized that CD1d expression in NSCLC is epigenetically regulated and can be modulated using epigenetic targeting therapies. Treatment of the CD1d-negative NSCLC cell lines, A549 and SK-MES-1, with DNA methyltransferase inhibitors and histone deacetylase inhibitors resulted in a dose-dependent induction of CD1d mRNA and protein expression. Chromatin immunoprecipitation analysis indicated that this induction of CD1d expression directly involved chromatin remodelling. Induction of CD1d expression by A549 and SK-MES-1 cells using therapeutic low doses of DNA methyltransferase inhibitors and histone deacetylase inhibitors made them targets for iNKT cell-mediated cytolytic degranulation. Thus, epigenetic manipulation of CD1d expression may augment the efficacy of iNKT cell-based immunotherapies for NSCLC.
KEYWORDS: CD1d, cytolytic, epigenetic, degranulation, invariant NKT cells, lung cancer
Introduction
Lung cancer is the most common cause of cancer-related death worldwide, accounting for about 13% of all cancer diagnoses and 19% of cancer mortalities.1 Non-small cell lung cancer (NSCLC) accounts for approximately 85% of all lung cancer cases and can be classified into adenocarcinoma, squamous cell carcinoma, and large cell carcinoma. Mortality rates in NSCLC remain high because of difficulties in detection and resistance to current therapeutics. Surgical resection is a treatment for localized NSCLC, but only 15–40% of patients are considered candidates for surgery at diagnosis.2 Platinum-based therapy is the current gold standard of care for NSCLC patients,3 however, survival is only marginally improved in advanced NSCLC. The median survival for a patient with advanced NSCLC is approximately 1 year and only 3.5% of patients with advanced disease survive five years following diagnosis. In addition, many tumors develop resistance to current chemotherapies. Therefore, there is an urgent need to develop novel therapeutics for NSCLC.
We postulated that cellular therapies involving invariant natural killer T (iNKT) cells could benefit patients with NSCLC. iNKT cells are cytotoxic T lymphocytes that express NK cell markers and a TCR composed of an invariant α-chain (Vα24Jα18 in humans and Vα14Jα18 in mice) paired with one of a limited number of β-chains.4,5 iNKT cells recognise glycolipid antigens presented by the MHC class I-like molecule CD1d. They can recognise a number of self and bacterial glycolipids, however, most studies on murine and human iNKT cells have used the xenogeneic glycolipid α-galactosylceramide (α-GalCer).6 Upon activation by α-GalCer, iNKT cells can kill a wide range of tumor cell lines7,8 and rapidly secrete a diverse range of growth factors and cytokines that activate and polarize adaptive immune responses.9-11 Activated iNKT cells can also interact directly with other cells of the immune system, and can induce the maturation of DC into APC12,13 and of B cells into antibody secreting plasma cells.14,15 Therapeutic activation of iNKT cells in murine models can prevent and reverse tumor growth.7,16,17 Numerical and functional iNKT deficiencies have been reported in a number of human cancers including NSCLC,18-21 but clinical trials that have targeted iNKT cells in humans have to date shown limited efficacy.22-24 In the present study, we show that iNKT cells are depleted from the blood and lungs of NSCLC patients. We also interrogated the Lung Cancer Explorer and Kaplan-Meier Plotter (KMPlot) databases of patients with NSCLC and found that CD1d expression is lower in the lungs of patients with NSCLC compared to healthy lungs, and that low expression of CD1d mRNA is significantly associated with poor patient survival. We hypothesized that NSCLC cells may downregulate CD1d expression to evade recognition by iNKT cells.
Epigenetic alterations such as aberrant DNA methylation and histone acetylation are widespread in NSCLC. Hypermethylation of the promoter sequences of a number of tumor suppressor genes, resulting in loss of expression, occurs in most cases of NSCLC and is associated with tumor recurrence and shorter survival times following resection.25-27 DNA methyltransferases (DNMT) are overexpressed in NSCLC,28-30 leading to hypermethylation of tumor suppressor genes.30 Conversely, histone deacetylases (HDAC), which remove acetyl groups from ε-N-acetyl lysine amino acids on histones resulting in gene activation,31 are aberrantly expressed in NSCLC,32 which is associated with poor prognosis in NSCLC.33-35
Epigenetic modifications also play key roles in downregulating expression of molecules that are critical for immunogenicity and tumor recognition, leading to tumour evasion. Aberrant hypermethylation of MHC genes resulting in downregulated MHC class I protein expression has been demonstrated in gastric carcinoma and oesophageal squamous cell carcinoma.36-38 Treatment of oesophageal squamous cell and gastric carcinoma cell lines with decitabine (5-aza-2′-deoxycytidine or DAC), an inhibitor of DNA methylation, restored MHC class I expression.36,37 DNMT and HDAC inhibitors can also prevent the suppression of NKG2D ligand expression by tumor cells resulting in increased NKG2D-mediated activation of cytotoxic T cells and NK cells.39-41
A study by Yang and colleagues42 found that CD1d expression could be induced in lung adenocarcinoma cell lines from both mice and humans using HDAC inhibitors. This study showed that CD1d expression could be induced, either through knockdown of HDAC genes, or treatment with HDAC inhibitors trichostatin A (TSA) or suberoylanilide hydroxamic acid (SAHA). The reversible nature of epigenetic modifications makes them an attractive prospect as novel therapeutic targets for cancer. DNMT inhibitors such as DAC and HDAC inhibitors such as SAHA have been approved for the treatment of some cancers.43 In the present study we have investigated if DAC, TSA and SAHA can induce CD1d expression on NSCLC adenocarcinoma (A549) and squamous cell carcinoma (SK-MES-1) cell lines. We show that all epigenetic modulators tested induced CD1d mRNA and protein expression by both cell lines and that treatment with DAC sensitized NSCLC cells for killing by iNKT cells. Thus, epigenetic regulation of CD1d expression may benefit NSCLC patients by helping to overcome immune evasion by tumor cells.
Results
iNKT cells are depleted from the blood and lungs of patients with NSCLC
PBMCs were prepared from blood samples of 9 NSCLC patients and 13 healthy donors, and stained with mAbs specific for CD3 and Vα24Jα18, and analysed by flow cytometry (Fig. 1A). iNKT cell frequencies were determined as percentages of lymphocytes that expressed CD3 and Vα24Jα18. Absolute numbers of iNKT cells per millilitre of blood or BAL were calculated. Fig. 1B shows that the frequencies and absolute counts of circulating iNKT cells were significantly decreased in NSCLC patients compared to healthy controls.
Figure 1.
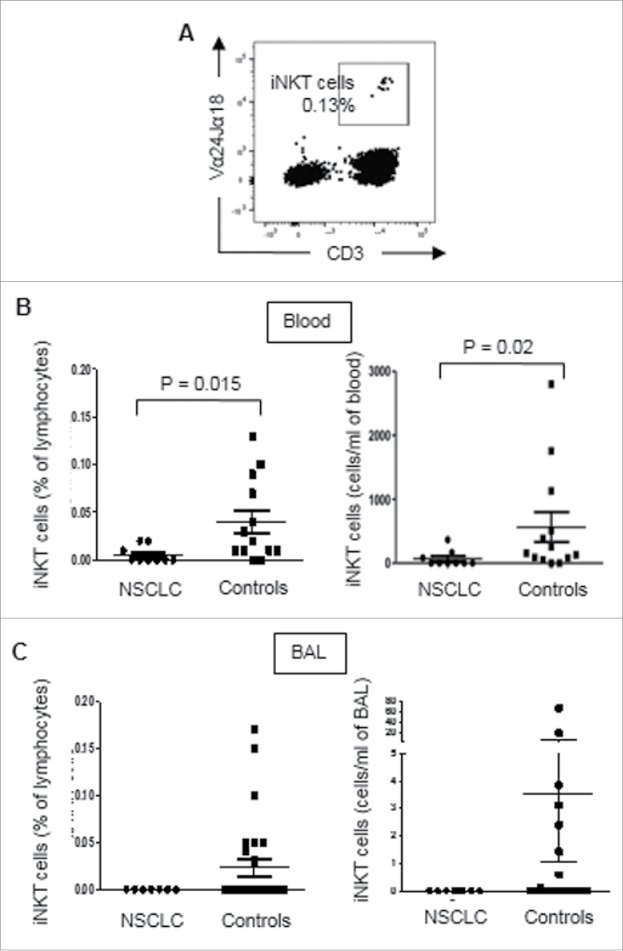
iNKT cells are depleted from the blood and BAL of patients with NSCLC. PBMCs were prepared from blood samples of 9 NSCLC patients and 13 healthy donors, and macrophage-depleted cells were prepared from BAL samples from 7 NSCLC patients and 26 non-cancer subjects. Cells were stained with mAbs specific for CD3 and Vα24Jα18 and analysed by flow cytometry. A, Flow cytometric dot plot showing CD3 and Vα24Jα18 expression by PBMCs after exclusion of doublets and dead cells. B, Scatter plots showing circulating iNKT cell frequencies, as percentages of lymphocytes (left) and absolute numbers of cells per ml of blood (right) in NSCLC patients and healthy control subjects. C, Scatter plot showing iNKT cell frequencies (left) and absolute numbers (right) in BAL samples from NSCLC patients and non-cancer subjects. Statistical analysis was performed using a two-tailed t test with Welch's correction. None of the NSCLC patients studied had detectable iNKT cells in their BAL samples.
The frequencies and numbers of iNKT cells in bronchoalveolar lavage (BAL) samples from 7 NSCLC patients and 26 non-cancer controls were determined after removal of macrophages by adherence purification. Cells were washed with PBS and stained with mAbs specific for CD3 and Vα24Jα18, and analysed by flow cytometry. Fig. 1C shows that iNKT cells were undetectable in BAL samples from NSCLC patients, but accounted for up to 0.15% of lymphocytes in BAL from non-cancer control subjects. These results show that iNKT cells are depleted from the blood and lungs of patients with lung cancer.
CD1d expression in lung tissue is reduced in patients with NSCLC
CD1d expression levels were compared between datasets on 498 samples of lung adenocarcinoma, 492 samples of lung squamous cell carcinoma and 59 samples of healthy lung by interrogating the Lung Cancer Explorer database. The relative levels of CD1d expression were significantly lower in the patients with adenocarcinoma (P = 3.3 × 10−5) and squamous cell carcinoma (P = 1.3 × 10−13) compared to controls (Fig. 2).
Figure 2.
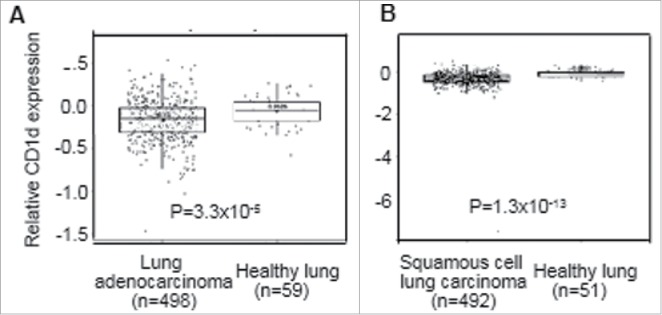
CD1d expression in lung tissue is reduced in patients with NSCLC. Comparative analysis of CD1d mRNA expression levels in resected lung tissues using datasets from 498 samples of lung adenocarcinoma (A), 492 samples of lung squamous cell carcinoma (B) and 59 and 51 samples of healthy lung in the Lung Cancer Explorer database. Statistical analysis was performed using a two-tailed t-test.
High expression of CD1d in NSCLC is associated with significantly improved overall survival
CD1d mRNA expression in tumors from NSCLC patients was analysed using the KMPlot database.44 Univariate analysis of mRNA expression in 1,926 NSCLC patients of all histologies determined that overall survival was significantly improved in NSCLC patients with high levels of CD1d mRNA expression (CD1d-high) compared with those who had low levels of CD1d mRNA expression (CD1d-low) (P = 0.0013), with a median overall survival of 56.8 months versus 76 months (Fig. 3A). Multivariate analysis confirmed that this benefit in overall survival based on CD1d-high and CD1d-low NSCLC patients was present regardless of grade, staging, treatment, gender or smoking status (P = 0.0014). When separated according to histological subtype, univariate analysis of survival for CD1d mRNA expression in 720 lung adenocarcinoma patients (Fig. 3B) and 524 lung squamous cell carcinoma patients (Fig. 3C), found that there was no significant difference in overall survival between CD1d-high and CD1d-low lung adenocarcinoma patients, however high CD1d expression in lung squamous cell carcinoma was associated with improved overall survival (P = 0.031; Fig. 3C).
Figure 3.
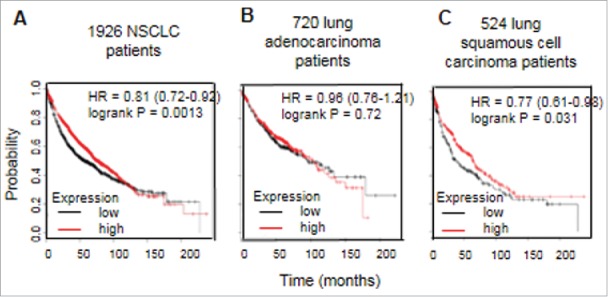
High expression of CD1d in NSCLC is associated with significantly improved overall survival. Kaplan-Meier plot analysis of CD1d mRNA expression in NSCLC patients. A, Univariate analysis on a dataset containing 1926 patients with NSCLC for all histologies demonstrating that high expression of CD1d mRNA is associated with significantly better overall survival (P = 0.0013). B, Univariate analysis of CD1d mRNA expression on a dataset containing 720 lung adenocarcinoma patients. C, Univariate analysis of CD1d mRNA expression on a dataset containing 524 lung squamous cell carcinomas. HR, hazard ratio. These data were generated using KMPlot44.
To determine whether gender or smoking status can affect overall survival in NSCLC patients, CD1d mRNA expression was assessed in the same dataset based on sex, smoking status, or in never smokers (Fig. 4). Using univariate analysis we found that high CD1d mRNA expression is associated with improved survival in males (P = 0.0057; Fig. 4A) and smokers (P = 0.037; Fig. 4C). There was no significant difference in overall survival between CD1d-high and CD1d-low females (Fig. 4B) and never smokers (Fig. 4D). These results show that low CD1d expression is associated with poorer survival rates in NSCLC.
Figure 4.
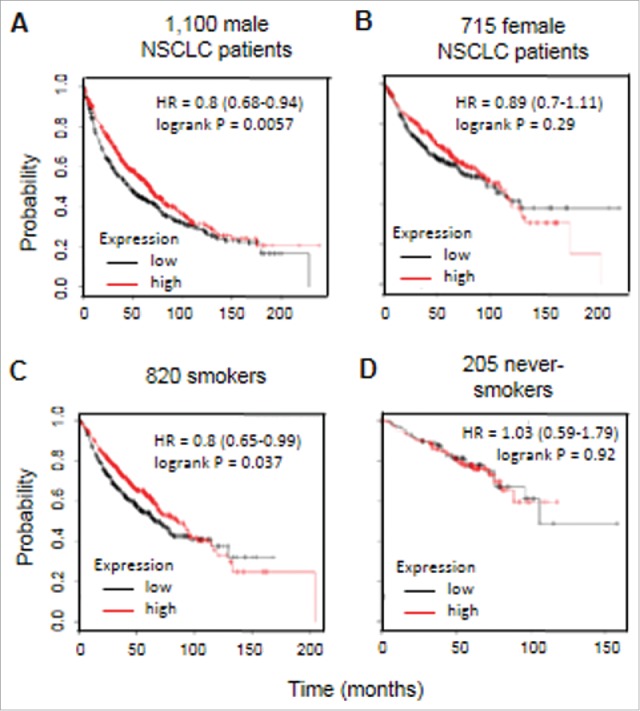
High expression of CD1d is associated with significantly improved survival in male patients and smokers with NSCLC. Kaplan-Meier plot analysis of CD1d mRNA expression in NSCLC patients divided according to gender and smoking status. A, Univariate analysis of CD1d mRNA expression on a dataset containing 1,100 male NSCLC patients. B, Univariate analysis of CD1d mRNA expression on a dataset containing 715 female NSCLC patients. C, Univariate analysis of CD1d mRNA expression on a dataset containing 820 smokers with NSCLC. D, Univariate analysis of CD1d mRNA expression on a dataset containing 205 never smokers with NSCLC. HR, hazard ratio. These data were generated using KMPlot.
We also compared survival rates of NSCLC patients with low and high levels of expression of a set of key inflammation-related genes identified by Loza and co-workers,45 using the KM Plot database. High expression of 8 out of 12 genes tested (IL-4R, CTLA4, PTPN22, IRF5, NOD2, TGFβ, MDA5 and CASP8) was significantly associated with improved survival (data not shown). Interrogation of the Lung Cancer Explorer database, revealed that the expression levels of all of these genes correlate positively with CD1d expression levels (not shown). Therefore, the high CD1d expression observed is associated with a generalized immune activation in the lungs.
Histone acetylation and DNA methylation inhibitors induce expression of CD1d mRNA in A549 and SK-MES-1 cells
The human adenocarcinoma alveolar basal epithelial cell line, A549, and the human lung squamous cell carcinoma cell line, SK-MES-1, were treated with HDAC inhibitor TSA at 250 ng/ml or vehicle control for a period of 24 h. Following treatment, RNA was extracted and quantified and CD1d mRNA expression was examined by RT-PCR (Fig. 5A). Changes in CD1d expression were expressed as ratios of CD1d to β-actin mRNA. Fig. 5B shows that a significant upregulation of CD1d was observed in A549 cells in response to TSA treatment (P = 0.0002). A similar upregulation of CD1d expression by SK-MES-1 cells was also observed (Fig. 5C; P = 0.0026).
Figure 5.
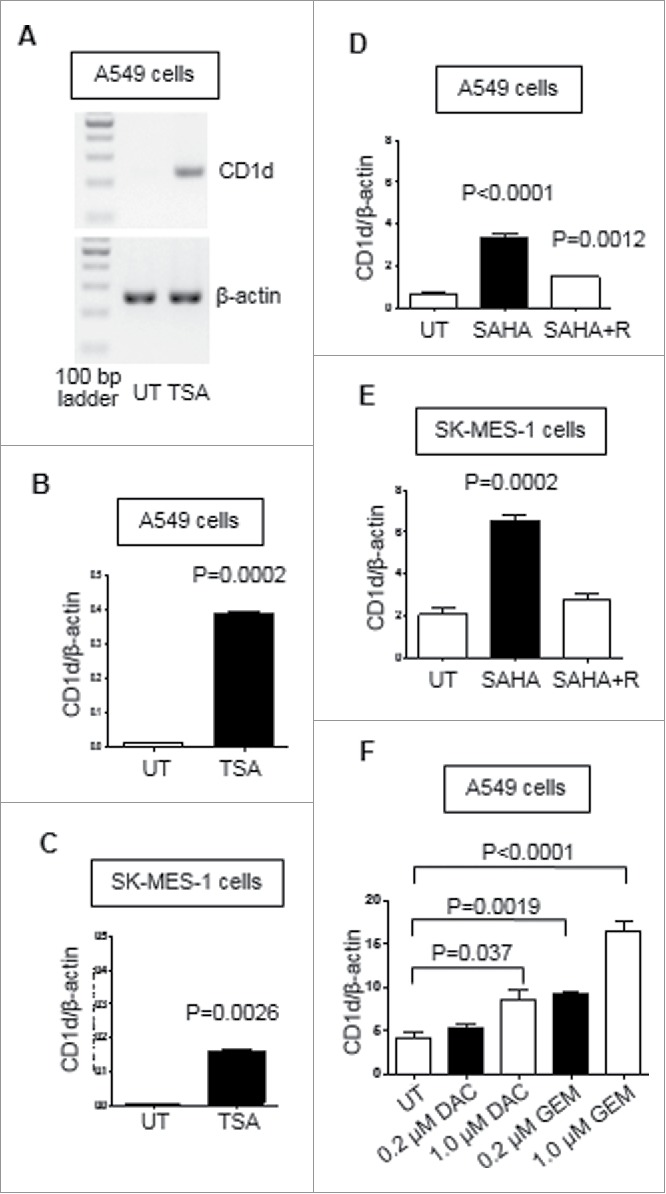
Histone deacetylase and DNA methylation inhibitors induce expression of CD1d in A549 and SK-MES-1 at the mRNA level. A, RT-PCR analysis of CD1d expression by untreated (UT) A549 cells and A549 cells treated for 24 hours with 250 ng/ml TSA. B, Mean (± SEM) ratios of CD1d to β-actin mRNA expression by untreated and TSA-treated A549 cells from 3 experiments. C, Mean (± SEM) ratios of expression of CD1d to β-actin by untreated and TSA-treated SK-MES-1 cells from 3 experiments. D, Mean (± SEM) ratios of expression of CD1d to β-actin by untreated, 5 μM SAHA-treated, and 5 μM SAHA-treated and rested (SAHA + R) A549 cells from 3 experiments. E, Mean (±SEM) ratios of expression of CD1d to β-actin by untreated and 5 μM SAHA-treated, and 5 μM SAHA-treated and rested SK-MES-1 cells from 3 experiments. F, Mean (± SEM) ratios of expression of CD1d to β-actin by untreated A549 cells and A549 cells treated for 48 hours with DAC, at 0.2 μM and 1.0 μM, and GEM, at 0.2 μM and 1.0 μM, from 3 experiments.
A549 and SK-MES-1 cells were also treated with the HDAC inhibitor SAHA at 5 μM or vehicle control for a period of 24 h. In order to determine whether the effects of this treatment could be maintained over time, the cells were treated with SAHA as before and then washed with PBS to remove SAHA and left for 24 hours. RNA was extracted and CD1d mRNA was quantified by RT-PCR. A significant up-regulation of CD1d was observed in A549 cells (p < 0.0001; Fig. 5D) and SK-MES-1 cells (P = 0.0002; Fig. 5E) in response to SAHA treatment. This increase in CD1d expression was maintained in A549 cells but not SK-MES-1 cells after a 24 h recovery period (P = 0.0012).
We also investigated the effects of treating A549 cells with DAC and GEM on CD1d mRNA expression. We have previously shown that GEM, a chemotherapeutic agent, can function as a DNMT inhibitor with equivalent activity to DAC.46 A549 cells were treated with 200 nM and 1 μM DAC or GEM for 48 hours. For both treatments, the media and drugs were replaced at 24 hours. Following the treatment period CD1d mRNA was examined by RT-PCR. Fig. 5F shows that CD1d expression was significantly increased in A549 cells treated with DAC at 1 μM (P = 0.037), GEM at 200 nM (P = 0.0019) and GEM at 1 μM (p < 0.0001). These results show that treatment with HDAC and DNMT inhibitors can induce CD1d expression in NSCLC cell lines at the mRNA level.
Chromatin immunoprecipitation (ChIP) analysis of the CD1d promoter from A549 cells treated with TSA verified that the observed effects for HDAC inhibitors were due to increased histone hyperacetylation. Fig. 6 shows that treatment with TSA resulted in an increase in PCR product, showing enhanced histone hyperacetylation at the CD1d promoter. Histone acetylation is associated with gene expression, indicating that the CD1d promoter region was silenced in A549 cells due to aberrant histone deacetylation. Treatment with TSA prevented gene silencing by HDACs allowing for the gene to be expressed. Both lysine 9 and 14 were hyperacetylated following treatment. These are known activating marks of expression.47 In addition, a decrease in expression was observed at the histone H3 lysine 4 monomethylation repressive mark with a simultaneous increase in levels of an activating mark, H3K4me2. Methylation marks are also associated with gene silencing; a decrease in the expression of a known repressive methylation mark, H3K9me2, further indicates that gene expression has been induced. The ChIP analysis confirms that chromatin remodelling is directly involved in with the induction of CD1d gene expression.
Figure 6.
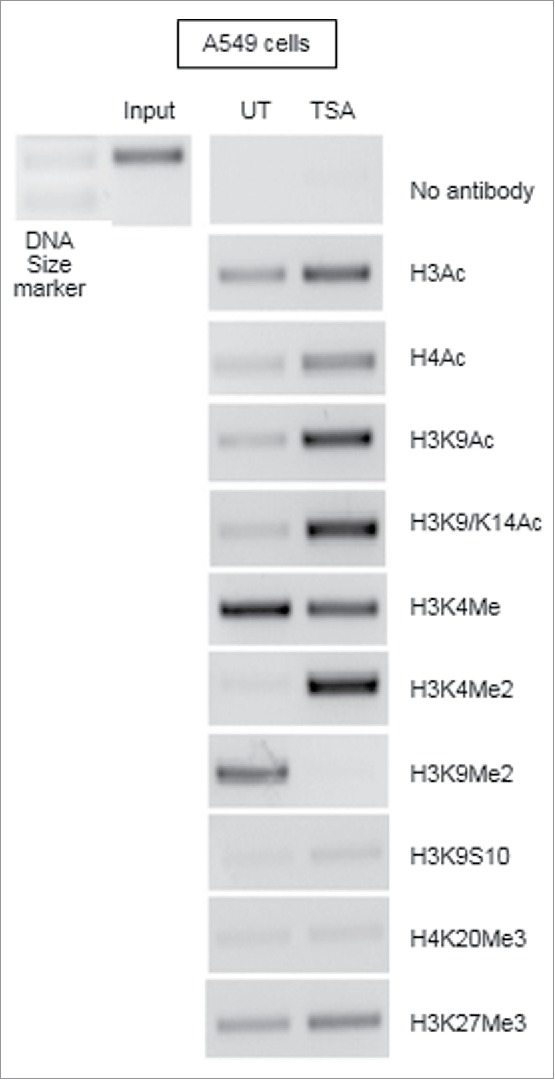
Induction of CD1d mRNA expression in A549 cells is due to increased histone hyperacetylation. A549 cells were treated with DMSO as an untreated control or with 250 ng/ml TSA for 24 hours. 1% formaldehyde was added to allow for crosslinking. Samples were sonicated thrice to shear the DNA into fragments. Cross-linked DNA fragments associated with the proteins of interest were selectively immunoprecipitated using protein-specific antibodies. Supernatants of the immunoprecipitates were subjected to PCR using primers specific for the CD1d promoter. UT samples were compared against TSA-treated samples to determine any changes to the histone marks.
Transient low-dose DAC induces CD1d protein expression by A549 cells and SK-MES-1 cells
Past trials with high doses of DAC have been plagued by extreme toxicities.48 Transient low doses of DAC have been shown to sustain long-term anti-tumor effects and are at least temporarily associated with their ability to maintain their targeting of DNMTs and alterations of gene expression.49 A549 (Fig. 7A) and SK-MES-1 cells (Fig. 7B and supplemental Fig. 1) were treated with PBS or DAC at either 50 nM, 100 nM or 1 μM over 72 hours. Media and DAC were replaced every 24 hours. Cells were assessed for expression of CD1d by flow cytometry immediately following treatment and at four days post-treatment. Fig. 7C shows a significant induction of CD1d protein expression in A549 cells at 4 days post treatment. A significant induction of CD1d protein expression in SK-MES-1 cells was observed immediately following treatment and at day 4 post treatment. In addition, the increased CD1d expression due to treatment with 1 μM DAC remained significant after 4 days of recovery (Fig. 7D). These results show that transient low dose DAC treatment is accompanied by a prolonged induction of CD1d protein expression in NSCLC cell lines.
Figure 7.
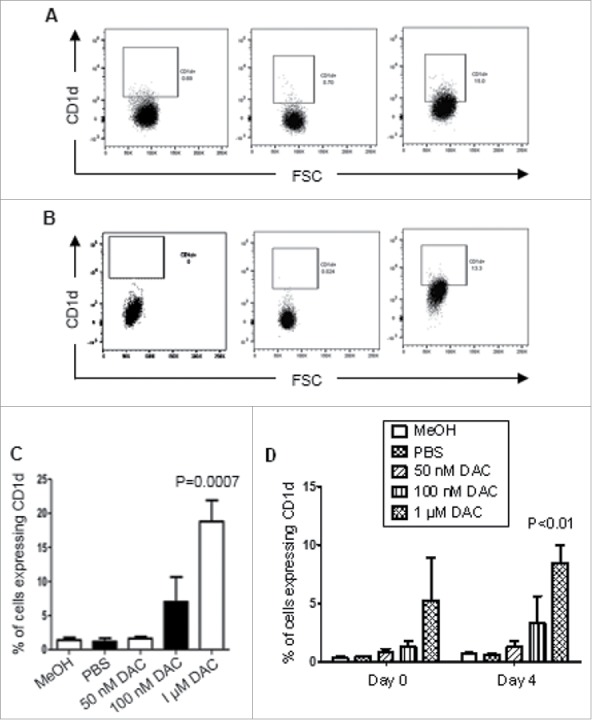
Transient low-dose DAC induces CD1d protein expression by A549 and SK-MES-1 cells. A549 and SK-MES-1 cells were treated with PBS or DAC at either 50 nM, 100 nM or 1 μM for 72 hours. Both media and treatments were replaced every 24 hours. MeOH was used as a vehicle control. After 72 hours cells were stained with an anti-CD1d mAb and analysed by flow cytometry. A and B, Flow cytometric dot plots showing CD1d expression by A549 cells (A) and SK-MES-1 cells (B), showing a fluorescence-minus-one (FMO) dot plots of DAC-treated cells (left panels), vehicle (MeOH)-treated cells (centre) and DAC-treated cells (right). C, Mean (± SEM) frequencies of A549 cells that expressed CD1d 4 days after cessation of treatment. D, Mean (± SEM) frequencies of SK-MES-1 cells that expressed CD1d immediately following cessation of treatment and 4 days after cessation of treatment. A two-way ANOVA with Bonferroni's post hoc test was used to determine significance. Results are representative of 3 experiments.
iNKT cells degranulate in response to low dose DAC-treated A549 cells
To investigate if low dose DAC treatment can increase susceptibility of A549 cells to cytoxicity by iNKT cells, A549 cells were treated with PBS or DAC at 50 nM, 100 nM and 1 μM for 72 h. The media and treatments were replaced every 24 h. Cells were then pulsed with 100 ng/ml of the glycolipids α-GalCer or 7DW8-5 or vehicle alone for 24 hours before adding iNKT cells. All iNKT cell lines used contained >98% CD3+ Vα24Jα18 TCR+ cells (Fig. 8A) and included CD4+, CD8+ and double negative CD4−CD8− subpopulations as assessed by flow cytometry (Fig. 8B). Treated and pulsed A549 cells were co-cultured with iNKT cells and a mAb specific for CD107a for 4 hours. Cell-surface CD107a expression by the iNKT cells was examined by flow cytometry (Fig. 8C). Fig. 8D shows that iNKT cells co-cultured with A549 cells treated with 50 nM – 1 μM DAC showed dose-dependent induction of CD107a when pulsed with α-GalCer (P < 0.01 – p < 0.001) or 7DW8-5 (p < 0.05 – p < 0.001). Little or no degranulation occurred in the absence of CD1d-binding glycolipids but significant degranulation occurred when DAC-untreated A549 cells were pulsed with the glycolipids. These results show that iNKT cells can kill A549 cells and that low dose DAC treatment increases their susceptibility to iNKT cell cytotoxicity. This cytolytic degranulation was reduced by up to 50% using a blocking antibody specific for CD1d (Fig. 8E), confirming at least a partial requirement for CD1d in iNKT cell mediated cytotoxicity.
Figure 8.
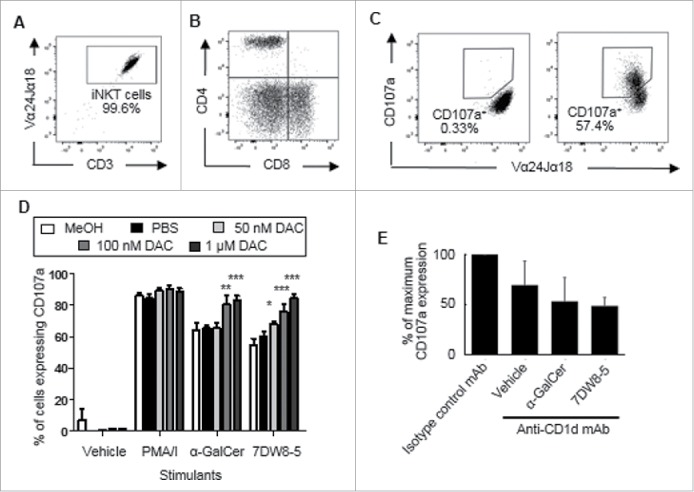
iNKT cells degranulate in response to low-dose DAC-treated A549 cells. A549 cells were treated with PBS or DAC (50 nM, 100 nM and 1 μM) at 24, 48 and 72 hours. PBS and methanol (MeOH) were used as negative and vehicle controls. After a rest period of 3 days the cells were pulsed with vehicle, α-GalCer or 7DW8-5 for 24 hours, and then co-cultured with iNKT cells and a mAb specific for CD107a. In some wells, 10 μg/ml of a blocking antibody specific for CD1d was included. PMA and ionomycin (P/I) treatment was used as a positive control. A, Representative flow cytometry dot plot showing purity of iNKT cells as lymphocytes expressing CD3 and the Vα24Jα18 TCR. B, Flow cytometric dot plot showing CD4 and CD8 expression by gated expanded iNKT cells. C, Flow cytometric dot plot showing cell-surface CD107a expression by iNKT cells after exposure to A549 cells treated with medium (left) or α-GalCer + DAC (right). D, Mean (± SEM) frequencies of iNKT cells that expressed CD107a after co-culture with DAC-treated A549 cells pulsed with glycolipids or controls. Percentages of iNKT cells that expressed CD107a in response to PBS- or DAC-treated A549 cells were compared to those exposed to A549 cells treated with vehicle alone using a two way ANOVA with Bonferroni's multiple comparison test. Results are means of 3 experiments. E, % inhibition of cytolytic degranulation by iNKT cells in response to DAC-treated A549 cells pulsed with α-GalCer or 7DW8-5 or vehicle using an anti-CD1d mAb. Results are means of 4 experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
iNKT cells degranulate in response to low dose DAC-treated SK-MES-1 cells
To assess whether lower, less toxic concentrations of DAC can increase susceptibility of SK-MES-1 cells to cytoxicity by iNKT cells, SK-MES-1 cells were treated with MeOH, PBS or DAC at 50 nM, 100 nM and 1 μM for 72 h. The media and treatments were replaced every 24 h. After three days of treatment the cells were pulsed with glycolipids and then co-cultured for 4 h with iNKT cells and a mAb specific for CD107a. CD107a expression by iNKT cells was examined by flow cytometry (Fig. 9A). Fig. 9B shows that iNKT cells exhibited significant degranulation when co-cultured with SK-MES-1 cells that had been pulsed with α-GalCer or 7DW8-5. Treatment with 50 nM DAC resulted in a statistically significant increase of CD107a when stimulated with α-GalCer (p < 0.01) and 7DW8-5 (P<0.001). SK-MES-1 cells treated with 100 nM or 1 μM DAC also showed increased susceptibility to iNKT cell degranulation when stimulated with α-GalCer (p < 0.001) and 7DW8-5 (p < 0.001). These results show that low dose DAC treatment can increase SK-MES-1 susceptibility to iNKT cell cytotoxicity. Antibody blocking experiments confirmed some involvement of CD1d in iNKT cell degranulation in response to DAC-treated SK-MES-1 cells (Fig. 9C).
Figure 9.
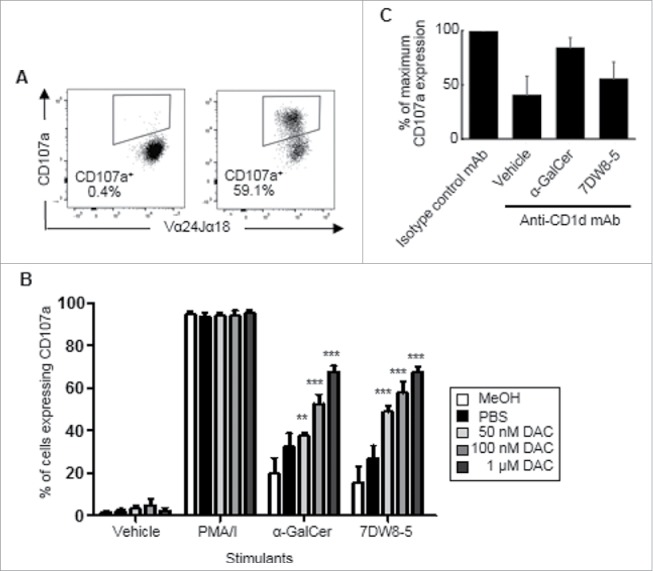
iNKT cells degranulate in response to low dose DAC-treated SK-MES-1 cells. SK-MES-1 cells were treated with PBS or DAC (50 nM, 100 nM and 1 μM) at 24, 48 and 72 hours. PBS and methanol (MeOH) were used as negative and vehicle controls. After a rest period of 3 days the cells were pulsed with vehicle, α-GalCer or 7DW8-5 for 24 hours, and then co-cultured for 4 h with iNKT cells and a mAb specific for CD107a. In some wells, 10 μg/ml of a blocking antibody specific for CD1d was included. PMA and ionomycin (P/I) treatment was used as a positive control. A, Flow cytometric dot plot showing cell-surface CD107a expression by iNKT cells after exposure to medium (left) or α-GalCer + DAC (right). B, Mean (± SEM) frequencies of iNKT cells that expressed CD107a after co-culture with control SK-MES-1 or DAC-treated SK-MES-1 cells pulsed with glycolipids. Percentages of iNKT cells that expressed CD107a in response to PBS- or DAC-treated SK-MES-1 cells were compared to those exposed to SK-MES-1 cells treated with vehicle alone using a two way ANOVA with Bonferroni's multiple comparison test. Results are representative of 3 experiments. C, % inhibition of cytolytic degranulation by iNKT cells in response to DAC-treated A549 cells pulsed with α-GalCer or 7DW8-5 or vehicle using an anti-CD1d mAb. Results are means of 4 experiments. (**p < 0.01, ***p < 0.001).
Discussion
CD1d-restricted iNKT cells promote anti-tumor immunity in humans and animals,4,5,7 but iNKT cells from cancer patients are frequently depleted and/or functionally-impaired.18-21 We enumerated iNKT cells in BAL and peripheral blood samples from NSCLC patients and compared them to those of control subjects. We found that iNKT cells were significantly depleted from both tissues in the NSCLC patients. This led us to hypothesize that iNKT cells represent an important component of anti-tumor immunity against NSCLC and that immunotherapies that boost their numbers and activities may benefit patients.
Whereas the therapeutic capacity of iNKT cells for cancer is well-documented in murine models,7,16,17 several phase I trials in humans using α-GalCer, α-GalCer-loaded DC, or ex vivo-expanded autologous iNKT cells in humans have shown little therapeutic benefit.22-24,50,51 α-GalCer caused minimal toxicity in humans but only a small subset of patients achieved disease stabilisation or partial response. The discrepancies between mouse and human trials may be the result of dramatic differences in iNKT abundancies and subset distributions between the two species,11,19 or to factors related to tumor stage and ongoing therapies. The low clinical efficacies of the iNKT cell-based immunotherapies tested in humans to date may also be due to the fact that tumor cells frequently do not express CD1d. Some tumor types such as prostate cancers, myelomonocytic leukaemia and some neurologic tumours, express CD1d and as such can be targeted for NKT-mediated cell killing.52 However, most human and mouse solid tumors are CD1d-negative. Over the last number of years increasing evidence has suggested that cancer cells are capable of downregulating MHC class I molecules in an attempt to evade immunosurveillance.53,54 Downregulation of CD1d has been documented in many tumor types including breast55 and cervical carcinoma56 and is associated with increased malignancy. We hypothesized that NSCLC cells similarly downregulate CD1d expression to evade the actions of iNKT cells. Such downregulation of CD1d expression could be the result of epigenetic modifications that take place in cancer cells.
To determine whether CD1d expression is altered in patients with NSCLC, we interrogated the Lung Cancer Explorer database and found that CD1d expression is significantly lower in the lungs of patients with lung adenocarcinoma and squamous cell carcinoma. Furthermore, by interrogating the KMPlot database, we found that low expression of CD1d was associated with significantly worse overall survival in patients with NSCLC. We then analysed NSCLC patients for variations in overall survival due to NSCLC histology, gender and smoking status. Using univariate analysis we found that high expression of CD1d was associated with significantly improved overall survival in squamous cell carcinoma patients, males and smokers. 48% of the adenocarcinoma patients and 71% of the squamous cell carcinoma patients were male, therefore the over-representation of males in the squamous cell carcinoma group, may contribute to the significance of the link between high CD1d expression and survival in this group. While low CD1d expression is associated with poor overall survival in NSCLC patients, we found that low levels of expression of a number of other markers of immune activation were also found to be associated with worse patient survival. This could either reflect a direct influence of CD1d on survival or could just be a marker for immune activation in the tumor.
Epigenetics is defined as the sum of all stable and heritable changes to gene expression that do not occur as a result of changes to the underlying DNA sequence.57 Chromatin structure defines the condition in which DNA is organised and allows for transcriptional repression or activation of genes.43 Epigenetic mechanisms such as DNA CpG methylation and histone post-translational modifications affect chromatin structure and gene expression by changing the accessibility of transcription factors to the promoter regions of genes. Aberrant acetylation patterns also contribute to cancer proliferation in various cancers, including gastric and lung,58 and over-expression of HDACs have been observed in lung cancer.59
Treatment with HDACi TSA and SAHA were shown to induce CD1d expression in A549 cells.42 We hypothesized that CD1d expression is epigenetically regulated and that treatment with DNMTi and HDACi could induce CD1d expression in NSCLC cancer cell lines. A549 and SK-MES-1 cells were treated with HDACi TSA and SAHA, and CD1d was found to be significantly increased at the mRNA level and this significant increase in expression was maintained following a recovery period in SAHA-treated A549 cell lines. HDAC inhibitors prevent the action of HDACs which silence genes by removing acetyl groups from histones. These results corroborate evidence that HDACs are over-expressed in lung cancer. To see whether a similar induction of CD1d expression was possible using DNMTi, A549 and SK-MES-1 cells were treated with DAC and GEM. Both cell lines showed an increase in CD1d expression at the mRNA following DNMTi treatment indicating that aberrant DNA methylation patterns affect the expression of CD1d in NSCLC. Treatment with DNMT inhibitors remove hypermethylation of the CD1d promoter resulting in an upregulation of CD1d expression. To assess whether the changes in CD1d expression were due to direct conformational changes at the CD1d promoter, a ChIP analysis was performed on A549 cells treated with TSA. The analysis showed that HDAC inhibition causes a conformational change in the chromatin landscape, allowing histone posttranslational modifications to occur at the promoter region of the CD1d gene.
We hypothesized that DAC could be used to induce CD1d expression at the protein level in A549 and SK-MES-1 cell lines, and that this induction would correlate with increased iNKT cell-mediated cytolytic degranulation. High dose DAC treatment is associated with extreme toxicities48 whereas transient DAC treatment have been shown to sustain long-term anti-tumor effects and to maintain its targeting of DNA methylation processes and alterations of gene expression.49 A549 and SK-MES-1 treated with a series of low dose DAC treatments were found to induce and maintain CD1d expression for up to four days. This increase in CD1d expression was associated with an increase in cytolytic degranulation by iNKT cells when the tumor cells were pulsed with the iNKT cell agonist ligands α-GalCer or 7DW8-5. No degranulation occurred in the absence of α-GalCer or 7DW8-5 but significant degranulation occurred when iNKT cells were co-cultured with DAC-untreated A549 and SK-MES-1 cells treated with these glycolipids. This suggests that the untreated tumor cell lines express low levels of CD1d and these levels are augmented by treatment with DAC. Antibody blocking of CD1d indicated that cytolytic degranulation by iNKT cells in response to DAC-untreated or DAC-treated A549 and SK-MES-1 cells is at least partially CD1d-dependent. While it is possible that stimulatory ligands other than CD1d, such as MHC or ligands for NKG2D, may contribute to iNKT cell cytotoxicity, this is unlikely because iNKT cell degranulation only occurred in the presence of the CD1d-binding glycolipids α-GalCer or 7DW8-5.
This study has shown that it is possible to increase CD1d expression in NSCLC cell lines using epigenetic targeted therapies in vitro and that this increase in expression is associated with increased cytolytic degranulation by iNKT cells. This observation is noted for both an adenocarcinoma and a squamous cell carcinoma cell line, but future studies are required to determine if the same treatment can induce CD1d expression by fresh tumor cells. Our results suggest that epigenetic modifying agents may augment the efficacies of iNKT cell based immunotherapies, which in turn may complement current chemotherapies for NSCLC. The epigenetic agents used in this study are FDA approved drugs that are indicated in the treatment of acute myeloid leukemia and cutaneous T cell lymphoma. By combining iNKT-based therapeutic strategies with epigenetic targeting agents, both which have previously been found to have therapeutic benefit in other patient populations, personalized medicines may be devised to combat the high mortality rates and poor treatment options associated with NSCLC.
Materials and methods
Subjects
Venous blood samples were obtained from 9 chemotherapy-naïve patients with NSCLC (5 females and 4 males; median age 60; range 53–75) and 13 healthy donors (8 females and 5 males; median age 59; range 50–64) after informed consent. BAL samples were obtained from 33 patients over 18 years of age who were undergoing a clinically-indicated bronchoscopy and had given written informed consent for retrieving additional bronchial washings for research.60 Of these, 7 patients had confirmed NSCLC (1 female and 5 males; median age 67.5; age range 57–81) and 26 were non-cancer subjects (15 females and 12 males; median age 62; age range 32–91) and did not have a known (or ensuing) diagnosis of malignancy or sarcoidosis. Ethical approval for this study was obtained from the Research Ethics Committee of St. James's Hospital, Dublin.
Blood and BAL preparation
PBMC were prepared from blood samples by standard density gradient centrifugation over Lymphoprep™ (Axis-Shield). BAL samples were obtained during bronchoscopy. 150 ml of saline was injected into the lungs and 50–90 ml was retrieved. Samples were plated overnight to remove macrophages by adherence purification, as previously reported.60 Cells were washed thrice with PBS and counted before analysis by flow cytometry.
Antibodies and flow cytometry
A mAb specific for CD1d was obtained from BD Biosciences (555749). MAbs specific for CD3 (300330), CD4 (344612), CD8α (300908), CD107a (328605) and the Vα24Jα18 TCR found on iNKT cells (clone 6B11; 342908) were obtained from BioLegend. Cells were stained with mAbs in PBS containing 1% bovine serum albumin and 0.02% sodium azide, and analysed using a FACSCanto II flow cytometer (BD Biosciences) and FlowJo software (Tree Star). Lymphocytes were gated on and any doublets or dead cells were excluded from the analysis. Single stained controls were used to set compensation parameters and fluorescence-minus-one controls were used to set gates. iNKT cell frequencies were expressed as percentages of lymphocytes. Absolute numbers of iNKT cells were determined from viable cell counts and expressed as cells/ml of blood or BAL.
Analysis of associations between CD1d mRNA expression and incidence and survival of NSCLC
CD1d expression levels were compared between lung adenocarcinoma, squamous cell carcinoma and healthy lung by interrogating the Lung Cancer Explorer database (https://qbrc.swmed.edu/projects/lungcancer/), which used datasets obtained by molecular profiling of patients and control subjects enrolled in the Cancer Genome Atlas Research Network.61,62 NSCLC patient groups in the KMPlot database44 were analysed to determine whether high CD1d mRNA expression is associated with improved overall survival. Patients were divided into high and low CD1d mRNA expression groups, with the best performing threshold being used as the final cut-off in a univariate Cox regression analysis. Basic univariate analysis was performed to assess overall survival in NSCLC patients of all histologies. Additional univariate analyses were performed to see whether overall survival rates differed between histological subtypes, genders and smoking status. The analysis was considered significant if p < 0.05.
NSCLC cell lines
The human adenocarcinoma alveolar basal epithelial cell line, A549, and the human lung squamous cell carcinoma cell line, SK-MES-1, were obtained from the American Tissue Culture Collection. A549 cells were cultured in F-12 (Ham) medium supplemented with 10% FBS, 2 mM L-glutamine, 50 mg/mL streptomycin and 50 U/mL penicillin. SK-MES-1 cells were maintained in Eagle's minimum essential medium supplemented with 10% FBS, 2 mM L-glutamine, 0.1 M non-essential amino acids, 50 mg/mL streptomycin and 50 U/mL penicillin (Gibco-BRL and Thermo-Scientific). Cells were maintained at 37°C in a humidified atmosphere of 5% CO2 and split when they reached 80% confluency.
Epigenetic agents
5-aza-2′deoxycytidine (DAC) was obtained from Merck (189825) and used at concentrations ranging from 50 nM to 5 µM for up to 72 hours. Gemcitabine hydrochloride (2′-deoxy-2′, 2′diflurocytidine hydrochloride; GEM) was obtained from Eli Lilly (0002-7501-01, 0002-7502-01) and used at 200 nM to 1 µM for 24 h. Trichostatin A (TSA) was obtained from Calbiochem (647926) and used at 250 ng/ml for 24 h. Suberanilohydroxamic acid (SAHA or Vorinostat) was obtained from Caymen Chemicals (10009929) and used at 5 µM for 24 h.
Analysis of CD1d mRNA expression
Total RNA was extracted using TRI reagent (Sigma-Aldrich; T9424) according to the manufacturer's instructions and was quantified using a Nanodrop ND-1000 Spectrophotometer (Labtech). cDNA was generated from 1 μg of total RNA in 1 h reactions at 42°C using RevertAid reverse transcriptase, deoxynucleotides (Thermo Scientific; EPO441 and 10297018) and oligo-dT12-18 primers (Eurofins) according to the manufacturer's instructions.
Cell lines were examined for the expression of CD1d (234 bp) and β-actin (510 bp) by RT-PCR using the following primers: forward 5′-AAGCCTGTATGGGTGAAG-3′ and reverse 5′-GACTGCCAAGGCAATCAAGC-3′ for CD1d and forward 5′-AGCACTGTGTTGGCGTACAG-3′ and reverse 5′-TGTTTGAGACCTTCAACACCC-3′ for β-actin obtained from Harvard PrimerBank.63 Cycling conditions consisted of 95°C for 5 min, followed by 35 repeat cycles of 1 min at 94°C, 1 min at the target gene annealing temperature (56°C for both CD1d and β-actin), and 1 min at 72°C with a final extension at 72°C for 10 min. Experiments conducted on cell lines were carried out in triplicate and PCR products were electrophoresed on 1% agarose gels and quantified using TINA 2.09c (Raytest) densitometry software. Target mRNA expression was standardised against β-actin controls, and expressed as a ratio of target mRNA expression:β-actin expression.
Validation of a subset of RT-PCR results was confirmed using SYBR green based quantitative real-time PCR (RT-qPCR). RT-qPCR reactions were carried out for CD1d using the same primers as above with the following primers for 18S rRNA used as the reference. Forward: 5′-GATGGGCGGCGGAAAATAG-3′ and Reverse: 5′-GCGTGGATTCTGCATAATGGT-3′.
qPCRs were carried out on a PCRmax Eco 48 using SYBR Green qPCR Master Mix (Bimake) and a 2-step qPCR program with the following cycling parameters: An initial polymerase activation of 95°C for 10 minutes followed by 40 cycles of 95°C for 15 seconds and annealing/amplification at 58°C for 60 seconds. A melting curve analysis was conducted at the end of each PCR using 95°C for 15 seconds, 55°C for 15 seconds and a final 95°C for 15 seconds. Data were analysed using the default in-built ΔΔCq analysis settings for relative quantification (Relative Expression) in the Eco Study software.
Chromatin immunoprecipitation (ChIP) assays
Chromatin immunoprecipitation (ChIP) was performed as follows: following treatments, cells were fixed with 1% formaldehyde, suspended in SDS lysis buffer (Merck Millipore; 20–163) and sonicated until DNA was fragmented into lengths of between 200 and 1000 bp. Aliquots of this sheared DNA were subsequently immunoprecipitated using the OneDayChIP Kit (Diagenode; C01010081) according to the manufacturer's instructions.
The antibodies used for immunoprecipitation were as follows: pan acetylated-histone H3 (H3Ac), pan acetylated-histone H4 (H4Ac), acetylated-histone H3 lysine 9 (H3K9Ac), acetylated-histone H3 lysine 9/14 (H3K9/K14Ac), methylated histone H3 lysine 4 (H3K4Me), di-methylated histone H3 lysine 4 (H3K4Me2), di-methylated histone H3 lysine 9 (H3K9Me2), acetylated histone H3 lysine 9 phosphoserine 10 (H3K9S10), tri-methylated histone H4 lysine 20 (H4K20Me3) and tri-methylated histone H3 lysine 27 (αH3K27Me3). All antibodies were obtained from Milipore and Sigma-Aldrich. A no-antibody control was included to test for specificity.
Primers used to study the promoter region of CD1d by ChIP (211 bp) (Forward 5′-CCACCTAGAGACATGTACTGC-3′, Reverse 5′-CGCTCACTTCAGTAGGTTTC-3′)64 were purchased from Sigma Aldrich. PCR cycling condition consisted of: 95°C for 5 min followed by 40 repeat cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 1 min with a final extension at 72°C for 10 min.
Generation and maintenance of iNKT cell lines
iNKT cells were enriched from total human PBMC by magnetic bead separation using anti-iNKT cell Microbeads (Miltenyi Biotec; 130-094-842) followed by sorting of Vα24Jα18+ CD3+ cells using a MoFlo XDP Cell Sorter (Beckman Coulter). Highly purified iNKT cells were expanded by culturing 1,000 iNKT cells in the wells of a 96-well round bottom microtitre plate, in iNKT cell medium (RPMI 1640 containing 0.05 mM L-glutamine, 10% HyClone FBS, 50 mg/mL streptomycin, 50 U/mL penicillin, 2.5 μg/ml amphotericin B – fungizone, 25 mM HEPES, 50 µM 2-mercaptoethanol, 1 mM sodium pyruvate, 1% non-essential amino acids mixture and 1% essential amino acids mixture; Gibco-BRL and Thermo-Scientific) and stimulating them with the iNKT cell glycolipid ligand α-GalCer (100 ng/ml; Funakoshi; KRN7000) and 250 U/ml IL-2 (Miltenyi Biotec; 130-097-745) in the presence of an excess (2 × 105) irradiated allogeneic PBMC prepared from two donors. Stock α-GalCer was thawed followed by heating to 80°C for 2 minutes, sonication for 10 minutes and vortexing for 1 minute before diluting to the required concentration in iNKT cell medium followed by heating to 80°C for 2 minutes, sonication for 5 minutes and vortexing for 1 minute before each further dilution. After 24 hours and again after 48 hours and subsequently every 3–4 days, medium was replaced with fresh iNKT cell medium containing 250 U/ml IL-2. Cells were expanded for a minimum of 3 weeks before being used in experiments. iNKT cells were kept at high cell densities and split 1 in 2 in fresh 96-well plates when multiple cell layers were observed using an inverted microscope. Cells were re-stimulated as above every 4–6 weeks. The purities of iNKT cells were >98% as determined by flow cytometry.
CD107a degranulation assay
Cytolytic degranulation by iNKT cells cultured with glycolipid-pulsed NSCLC cells was examined by flow cytometric analysis of CD107a expression by iNKT subsets. A549 or SK-MES-1 cells were treated with PBS or DAC (50 nM, 100 nM and 1 μM) at 24, 48 and 72 hours. Methanol was used as a vehicle control. After a rest period of 3 days the cells were pulsed with either 100 ng/ml α-GalCer or 10 ng/ml 7DW8-5 (Funakoshi) for 24 hours, and then co-cultured with iNKT cells for 4 hours at 1:1 ratios in the presence of anti-CD107a FITC mAb. In some wells, 10 μg/ml of a blocking antibody specific for CD1d (clone CD1d42; BD Biosciences) was included. Monensin (25 µM) was added after 1 hour to prevent proteolysis of the mAb conjugate upon reinternalization of CD107a. iNKT cells treated with 50 ng/ml PMA and 1 µg/ml ionomycin were used as positive controls. Cytolytic degranulation by iNKT cells was examined by flow cytometric analysis of CD107a expression. We previously have shown that CD107a expression by iNKT cells directly correlates with target cell death.11
Statistical analysis
Survival analysis of NSCLC patient groups in was carried out using Lung Cancer Explorer and KMPlot analysis.44 Statistical analysis of other data was performed using GraphPad Prism v5.0. Data were expressed as mean ± standard error of the mean and analysed using one- or two-way ANOVA with either Bonferroni or Dunnett's post-hoc test or a t test with Welch's correction. The Bonferroni Test was used for multiple comparison analysis while Dunnett's Post Test was used to compare each sample mean against a control mean. Welch's correction is used for unequal sample sizes and does not assume that the variances of the two populations are equal. The data was considered significant if p < 0.05.
Supplementary Material
Funding Statement
This research was funded by a grant from Molecular Medicine Ireland, (Clinical and Translational Research Scholars Programme PhD Project 7F76D).
Abbreviations
- α-GalCer
α-galactosylceramide
- BAL
bronchoalveolar lavage
- DAC
Decitabine (5-Aza-2′-Deoxycytidine)
- DNMT
DNA methyltransferases
- DNMTi
DNA methyltransferase inhibitor
- GEM
gemcitabine hydrochloride
- HDAC
histone deacetylase
- iNKT cell
invariant natural killer T cell
- KMPlot
Kaplan-Meier Plotter
- NSCLC
non-small cell lung cancer
- SAHA
suberoylanilide hydroxamic acid
- TSA
trichostatin A
Disclosures of potential conflicts of interest
No potential conflicts of interest are disclosed.
Acknowledgments
This research was funded by a grant from Molecular Medicine Ireland. The authors would like to thank the patients and control subjects who were enrolled in this study; the Irish Blood Transfusion Service for kindly providing buffy coat packs; Conleth Feighery, Mark Little, Andreea Petrasca, Yasmeen Ghnewa, Pádraic Dunne, Tanya Coulter, Serena Arduini, Christina Maher and Ashanty Melo Rodriguez for helpful discussions.
Author contributions
É. Dockry, S.G. Gray and D.G. Doherty designed the experiments. É. Dockry performed the experiments. S. O'Leary, L.E. Gleeson, J. Lyons and J. Keane managed the patients and obtained blood and BAL specimens. É. Dockry, S.G. Gray and D.G. Doherty wrote the manuscript.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. PMID:25651787. [DOI] [PubMed] [Google Scholar]
- 2.Currow DC, You H, Aranda S, McCaughan BC, Morrell S, Baker DF, Walton R, Roder DM. What factors are predictive of surgical resection and survival from localised non-small cell lung cancer? Med J Aust. 2014;201:475–480. doi: 10.5694/mja14.00365. PMID:25332036. [DOI] [PubMed] [Google Scholar]
- 3.Pujol JL, Barlesi F, Daurès JP. Should chemotherapy combinations for advanced non-small cell lung cancer be platinum-based? A meta-analysis of phase III randomized trials. Lung Cancer. 2006;51:335–345. doi: 10.1016/j.lungcan.2005.11.001. PMID:16478643. [DOI] [PubMed] [Google Scholar]
- 4.Bendelac A, Savage PB, Teyton L. The biology of NKT cells. Annu Rev Immunol. 2007;25:297–336. doi: 10.1146/annurev.immunol.25.022106.141711. PMID:17150027. [DOI] [PubMed] [Google Scholar]
- 5.Salio M, Silk JD, Jones EY, Cerundolo V. Biology of CD1- and MR1-restricted T cells. Annu Rev Immunol. 2014;32:323–66. doi: 10.1146/annurev-immunol-032713-120243. PMID:24499274. [DOI] [PubMed] [Google Scholar]
- 6.Kawano T, Cui J, Koezuka Y, Toura I, Kaneko Y, Motoki K, Ueno H, Nakagawa R, Sato H, Kondo E, et al.. CD1d-restricted and TCR-mediated activation of valpha14 NKT cells by glycosylceramides. Science. 1997;278:1626–1629. doi: 10.1126/science.278.5343.1626. PMID:9374463. [DOI] [PubMed] [Google Scholar]
- 7.Cui J, Shin T, Kawano T, Sato H, Kondo E, Toura I, Kaneko Y, Koseki H, Kanno M, Taniguchi M. Requirement for Vα14 NKT cells in IL-12-mediated rejection of tumors. Science. 1997;278:1623–1626. doi: 10.1126/science.278.5343.1623. PMID:9374462. [DOI] [PubMed] [Google Scholar]
- 8.Metelitsa LS, Naidenko OV, Kant A, Wu HW, Loza MJ, Perussia B, Kronenberg M, Seeger RC. Human NKT cells mediate antitumor cytotoxicity directly by recognizing target cell CD1d with bound ligand or indirectly by producing IL-2 to activate NK cells. J Immunol. 2001;167:3114–3122. doi: 10.4049/jimmunol.167.6.3114. PMID:11544296. [DOI] [PubMed] [Google Scholar]
- 9.Exley M, Garcia J, Balk SP, Porcelli S. Requirements for CD1d recognition by human invariant Valpha24+ CD4−CD8− T cells. J Exp Med 1997;186:109–120. doi: 10.1084/jem.186.1.109. PMID:9207002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gumperz JE, Miyake S, Yamamura T, Brenner MB. Functionally distinct subsets of CD1d-restricted natural killer T cells revealed by CD1d tetramer staining. J Exp Med. 200;195:625–36. doi: 10.1084/jem.20011786. PMID:11877485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O'Reilly V, Zeng SG, Bricard G, Atzberger A, Hogan AE, Jackson J, Feighery C, Porcelli SA, Doherty DG. Distinct and overlapping effector functions of expanded human CD4+, CD8α+ and CD4−CD8α− invariant natural killer T cells. PLoS One. 2011;6:e28648. doi: 10.1371/journal.pone.0028648. PMID:22174854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kitamura H, Iwakabe K, Yahata T, Nishimura S, Ohta A, Ohmi Y, Sato M, Takeda K, Okumura K, Van Kaer L, et al.. The natural killer T (NKT) cell ligand α-galactosylceramide demonstrates its immunopotentiating effect by inducing interleukin (IL)-12 production by dendritic cells and IL-12 receptor expression on NKT cells. J Exp Med 1999;189:1121–1128. doi: 10.1084/jem.189.7.1121. PMID:10190903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujii S, Shimizu K, Hemmi H, Steinman RM. Innate Vα14+ natural killer T cells mature dendritic cells, leading to strong adaptive immunity. Immunol Rev. 2007;220:183–198. doi: 10.1111/j.1600-065X.2007.00561.x. PMID:17979847. [DOI] [PubMed] [Google Scholar]
- 14.Galli G, Nuti S, Tavarini S, Galli-Stampino L, De Lalla C, Casorati G, Dellabona P, Abrignani S. CD1d-restricted help to B cells by human invariant natural killer T lymphocytes. J Exp Med. 2003;197:1051–1057. doi: 10.1084/jem.20021616. PMID:12695492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zeng SG, Ghnewa YG, O'Reilly VP, Lyons VG, Atzberger A, Hogan AE, Exley MA, Doherty DG. Human invariant NKT cell subsets differentially promote differentiation, antibody production, and T cell stimulation by B cells in vitro. J Immunol. 2013;191:1666–1676. doi: 10.4049/jimmunol.1202223. PMID:23851681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crowe NY, Coquet JM, Berzins SP, Kyparissoudis K, Keating R, Pellicci DG, Hayakawa Y, Godfrey DI, Smyth MJ. Differential antitumor immunity mediated by NKT cell subsets in vivo. J Exp Med. 2005;202:1279–1288. doi: 10.1084/jem.20050953. PMID:16275765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Macho-Fernandez E, Cruz LJ, Ghinnagow R, Fontaine J, Bialecki E, Frisch B, Trottein F, Faveeuw C. Targeted delivery of α-galactosylceramide to CD8α+ dendritic cells optimizes type I NKT cell-based antitumor responses. J Immunol. 2014;193:961–969. doi: 10.4049/jimmunol.1303029. PMID:24913977. [DOI] [PubMed] [Google Scholar]
- 18.Tahir SM, Cheng O, Shaulov A, Koezuka Y, Bubley GJ, Wilson SB, Balk SP, Exley MA. Loss of IFN-γ production by invariant NK T cells in advanced cancer. J Immunol. 2001;167:4046–4050. doi: 10.4049/jimmunol.167.7.4046. PMID:11564825. [DOI] [PubMed] [Google Scholar]
- 19.Kenna T, Golden-Mason L, Porcelli SA, Koezuka Y, Hegarty JE, O'Farrelly C, Doherty DG. NKT cells from normal and tumor-bearing human livers are phenotypically and functionally distinct from murine NKT cells. J Immunol. 2003;171:1775–1779. doi: 10.4049/jimmunol.171.4.1775. PMID:12902477. [DOI] [PubMed] [Google Scholar]
- 20.Molling JW, Kölgen W, van der Vliet HJ, Boomsma MF, Kruizenga H, Smorenburg CH, Molenkamp BG, Langendijk JA, Leemans CR, von Blomberg BM, et al.. Peripheral blood IFN-γ-secreting Vα24+Vβ11+ NKT cell numbers are decreased in cancer patients independent of tumor type or tumor load. Int J Cancer. 2005;116:87–93. doi: 10.1002/ijc.20998. PMID:15756674. [DOI] [PubMed] [Google Scholar]
- 21.Berzins SP, Smyth MJ, Baxter AG. Presumed guilty: natural killer T cell defects and human disease. Nat Rev Immunol. 201;11:131–142. doi: 10.1038/nri2904. PMID:21267014. [DOI] [PubMed] [Google Scholar]
- 22.Chang DH, Osman K, Connolly J, Kukreja A, Krasovsky J, Pack M, Hutchinson A, Geller M, Liu N, Annable R, et al.. Sustained expansion of NKT cells and antigen-specific T cells after injection of α-galactosyl-ceramide loaded mature dendritic cells in cancer patients. J Exp Med. 2005;201:1503–1517. doi: 10.1084/jem.20042592. PMID:15867097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Motohashi S, Ishikawa A, Ishikawa E, Otsuji M, Iizasa T, Hanaoka H, Shimizu N, Horiguchi S, Okamoto Y, Fujii S, et al.. A phase I study of in vitro expanded natural killer T cells in patients with advanced and recurrent non-small cell lung cancer. Clin Cancer Res. 2006;12:6079–6086. doi: 10.1158/1078-0432.CCR-06-0114. PMID:17028247. [DOI] [PubMed] [Google Scholar]
- 24.Kunii N, Horiguchi S, Motohashi S, Yamamoto H, Ueno N, Yamamoto S, Sakurai D, Taniguchi M, Nakayama T, Okamoto Y. Combination therapy of in vitro-expanded natural killer T cells and α-galactosylceramide-pulsed antigen-presenting cells in patients with recurrent head and neck carcinoma. Cancer Sci. 2009;100:1092–1098. doi: 10.1111/j.1349-7006.2009.01135.x. PMID:19302288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brock MV, Hooker CM, Ota-Machida E, Han Y, Guo M, Ames S, Glöckner S, Piantadosi S, Gabrielson E, Pridham G, et al.. DNA methylation markers and early recurrence in stage I lung cancer. N Engl J Med. 2008;358:1118–1128. doi: 10.1056/NEJMoa0706550. PMID:18337602. [DOI] [PubMed] [Google Scholar]
- 26.Sterlacci W, Tzankov A, Veits L, Zelger B, Bihl MP, Foerster A, Augustin F, Fiegl M, Savic S. A comprehensive analysis of p16 expression, gene status, and promoter hypermethylation in surgically resected non-small cell lung carcinomas. J Thorac Oncol. 2011;6:1649–57. doi: 10.1097/JTO.0b013e3182295745. PMID:21857254. [DOI] [PubMed] [Google Scholar]
- 27.Selamat SA, Chung BS, Girard L, Zhang W, Zhang Y, Campan M, Siegmund KD, Koss MN, Hagen JA, Lam WL, et al.. Genome-scale analysis of DNA methylation in lung adenocarcinoma and integration with mRNA expression. Genome Res. 2012;22:1197–211. doi: 10.1101/gr.132662.111. PMID:22613842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim H, Kwon YM, Kim JS, Han J, Shim YM, Park J, Kim DH. Elevated mRNA levels of DNA methyltransferase-1 as an independent prognostic factor in primary nonsmall cell lung cancer. Cancer. 2006;107:1042–1049. doi: 10.1002/cncr.22087. PMID:16888795. [DOI] [PubMed] [Google Scholar]
- 29.Lin RK, Hsu HS, Chang JW, Chen CY, Chen JT, Wang YC. Alteration of DNA methyltransferases contributes to 5′CpG methylation and poor prognosis in lung cancer. Lung Cancer. 2007;55:205–213. doi: 10.1016/j.lungcan.2006.10.022. PMID:17140695. [DOI] [PubMed] [Google Scholar]
- 30.Tang YA, Lin RK, Tsai YT, Hsu HS, Yang YC, Chen CY, Wang YC. MDM2 overexpression deregulates the transcriptional control of RB/E2F leading to DNA methyltransferase 3A overexpression in lung cancer. Clin Cancer Res. 2012;18:4325–4333. doi: 10.1158/1078-0432.CCR-11-2617. PMID:22733537. [DOI] [PubMed] [Google Scholar]
- 31.Sharma SV, Haber DA, Settleman J. Cell line-based platforms to evaluate the therapeutic efficacy of candidate anticancer agents. Nat Rev Cancer. 2010;10:241–253. doi: 10.1038/nrc2820. PMID:20300105. [DOI] [PubMed] [Google Scholar]
- 32.Vendetti FP, Rudin CM. Epigenetic therapy in non-small-cell lung cancer: targeting DNA methyltransferases and histone deacetylases. Expert Opin Biol Ther 2013;13:1273–1285. doi: 10.1517/14712598.2013.819337. PMID:23859704. [DOI] [PubMed] [Google Scholar]
- 33.Osada H, Tatematsu Y, Saito H, Yatabe Y, Mitsudomi T, Takahashi T. Reduced expression of class II histone deacetylase genes is associated with poor prognosis in lung cancer patients. Int J Cancer. 2004;112:26–32. doi: 10.1002/ijc.20395. PMID:15305372. [DOI] [PubMed] [Google Scholar]
- 34.Sasaki H, Moriyama S, Nakashima Y, Kobayashi Y, Kiriyama M, Fukai I, Yamakawa Y, Fujii Y. Histone deacetylase 1 mRNA expression in lung cancer. Lung Cancer. 2004;46:171–8. doi: 10.1016/j.lungcan.2004.03.021. PMID:15474665. [DOI] [PubMed] [Google Scholar]
- 35.Minamiya Y, Ono T, Saito H, Takahashi N, Ito M, Mitsui M, Motoyama S, Ogawa J. Expression of histone deacetylase 1 correlates with a poor prognosis in patients with adenocarcinoma of the lung. Lung Cancer. 2011;74:300–304. doi: 10.1016/j.lungcan.2011.02.019; PMID: 21466904. doi: 10.1016/j.lungcan.2011.02.019. PMID:21466904. [DOI] [PubMed] [Google Scholar]
- 36.Nie Y, Yang G, Song Y, Zhao X, So C, Liao J, Wang LD, Yang CS. DNA hypermethylation is a mechanism for loss of expression of the HLA class I genes in human esophageal squamous cell carcinomas. Carcinogenesis. 2001;22:1615–1623. doi: 10.1093/carcin/22.10.1615. PMID:11577000. [DOI] [PubMed] [Google Scholar]
- 37.Ye Q, Shen Y, Wang X, Yang J, Miao F, Shen C, Zhang J. Hypermethylation of HLA class I gene is associated with HLA class I down-regulation in human gastric cancer. Tissue Antigens. 2010;75:30–39. doi: 10.1111/j.1399-0039.2009.01390.x. PMID:19883394. [DOI] [PubMed] [Google Scholar]
- 38.Sigalotti L, Fratta E, Coral S, Maio M. Epigenetic drugs as immunomodulators for combination therapies in solid tumors. Pharmacol Ther. 2014;142:339–350. doi: 10.1016/j.pharmthera.2013.12.015. PMID:24384533. [DOI] [PubMed] [Google Scholar]
- 39.Armeanu S, Bitzer M, Lauer UM, Venturelli S, Pathil A, Krusch M, Kaiser S, Jobst J, Smirnow I, Wagner A, et al.. Natural killer cell-mediated lysis of hepatoma cells via specific induction of NKG2D ligands by the histone deacetylase inhibitor sodium valproate. Cancer Res. 2005;65:6321–6329. doi: 10.1158/0008-5472.CAN-04-4252. PMID:16024634. [DOI] [PubMed] [Google Scholar]
- 40.Tang KF, He CX, Zeng GL, Wu J, Song GB, Shi YS, Zhang WG, Huang AL, Steinle A, Ren H. Induction of MHC class I-related chain B (MICB) by 5-aza-2′-deoxycytidine. Biochem Biophys Res Commun. 2008;370:578–583. doi: 10.1016/j.bbrc.2008.03.131. PMID:18395517. [DOI] [PubMed] [Google Scholar]
- 41.Chávez-Blanco A, De la Cruz-Hernández E, Domínguez GI, Rodríguez-Cortez O, Alatorre B, Pérez-Cárdenas E, Chacón-Salinas R, Trejo-Becerril C, Taja-Chayeb L, Trujillo JE, et al.. Upregulation of NKG2D ligands and enhanced natural killer cell cytotoxicity by hydralazine and valproate. Int J Oncol. 2011;39:1491–1499. doi: 10.3892/ijo.2011.1144. PMID:21805029. [DOI] [PubMed] [Google Scholar]
- 42.Yang PM, Lin PJ, Chen CC. CD1d induction in solid tumor cells by histone deacetylase inhibitors through inhibition of HDAC1/2 and activation of Sp1. Epigenetics. 2012;7:390–399. doi: 10.4161/epi.19373. PMID:22419072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. PMID:19752007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Győrffy B, Surowiak P, Budczies J, Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS One. 2013;8:e82241. doi: 10.1371/journal.pone.0082241. PMID:24367507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Loza MJ, McCall CE, Li L, Isaacs WB, Xu J, Chang BL. Assembly of inflammation-related genes for pathway-focused genetic analysis. PLoS One. 2007;2:e1035. doi: 10.1371/journal.pone.0001035. PMID:17940599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gray SG, Baird AM, O'Kelly F, Nikolaidis G, Almgren M, Meunier A, Dockry E, Hollywood D, Ekström TJ, Perry AS, et al.. Gemcitabine reactivates epigenetically silenced genes and functions as a DNA methyltransferase inhibitor. Int J Mol Med. 2012;30:1505–11. doi: 10.3892/ijmm.2012.1138. PMID:23007409. [DOI] [PubMed] [Google Scholar]
- 47.Nguyen CT, Weisenberger DJ, Velicescu M, Gonzales FA, Lin JC, Liang G, Jones PA. Histone H3-lysine 9 methylation is associated with aberrant gene silencing in cancer cells and is rapidly reversed by 5-aza-2′-deoxycytidine. Cancer Res. 2002;62:6456–6461. PMID:12438235. [PubMed] [Google Scholar]
- 48.Abele R, Clavel M, Dodion P, Bruntsch U, Gundersen S, Smyth J, Renard J, van Glabbeke M, Pinedo HM. The EORTC Early Clinical Trials Cooperative Group experience with 5-aza-2′-deoxycytidine (NSC 127716) in patients with colo-rectal, head and neck, renal carcinomas and malignant melanomas. Eur J Cancer Clin Oncol 1987;23:1921–1924. doi: 10.1016/0277-5379(87)90060-5. PMID:2449354. [DOI] [PubMed] [Google Scholar]
- 49.Tsai HC, Li H, Van Neste L, Cai Y, Robert C, Rassool FV, Shin JJ, Harbom KM, Beaty R, Pappou E, et al.. Transient low doses of DNA-demethylating agents exert durable antitumor effects on hematological and epithelial tumor cells. Cancer Cell. 2012;21:430–46. doi: 10.1016/j.ccr.2011.12.029. PMID:22439938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Giaccone G, Punt CJ, Ando Y, Ruijter R, Nishi N, Peters M, von Blomberg BM, Scheper RJ, van der Vliet HJ, van den Eertwegh AJ, et al.. A phase I study of the natural killer T-cell ligand α-galactosylceramide (KRN7000) in patients with solid tumors. Clin Cancer Res. 2002;8:3702–3709. PMID:12473579. [PubMed] [Google Scholar]
- 51.Ishikawa A, Motohashi S, Ishikawa E, Fuchida H, Higashino K, Otsuji M, Iizasa T, Nakayama T, Taniguchi M, Fujisawa T. A phase I study of α-galactosylceramide (KRN7000)-pulsed dendritic cells in patients with advanced and recurrent non-small cell lung cancer. Clin Cancer Res. 2005;11:1910–1917. doi: 10.1158/1078-0432.CCR-04-1453. PMID:15756017. [DOI] [PubMed] [Google Scholar]
- 52.Metelitsa LS, Weinberg KI, Emanuel PD, Seeger RC. Expression of CD1d by myelomonocytic leukemias provides a target for cytotoxic NKT cells. Leukemia. 2003;17:1068–1077. doi: 10.1038/sj.leu.2402943. PMID:12764370. [DOI] [PubMed] [Google Scholar]
- 53.Jäger D, Jäger E, Knuth A. Immune responses to tumour antigens: implications for antigen specific immunotherapy of cancer. J Clin Pathol. 2001;54:669–674. doi: 10.1136/jcp.54.9.669. PMID:11533070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Algarra I, García-Lora A, Cabrera T, Ruiz-Cabello F, Garrido F. The selection of tumor variants with altered expression of classical and nonclassical MHC class I molecules: implications for tumor immune escape. Cancer Immunol Immunother. 2004;53:904–910. doi: 10.1007/s00262-004-0517-9. PMID:15069585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hix LM, Shi YH, Brutkiewicz RR, Stein PL, Wang CR, Zhang M. CD1d-expressing breast cancer cells modulate NKT cell-mediated antitumor immunity in a murine model of breast cancer metastasis. PLoS One. 2011;6:e20702. doi: 10.1371/journal.pone.0020702. PMID:21695190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miura S, Kawana K, Schust DJ, Fujii T, Yokoyama T, Iwasawa Y, Nagamatsu T, Adachi K, Tomio A, Tomio K, et al.. CD1d, a sentinel molecule bridging innate and adaptive immunity, is downregulated by the human papillomavirus (HPV) E5 protein: a possible mechanism for immune evasion by HPV. J Virol. 2010;84:11614–23. doi: 10.1128/JVI.01053-10. PMID:20810727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Probst AV, Dunleavy E, Almouzni G. Epigenetic inheritance during the cell cycle. Nat Rev Mol Cell Biol. 2009;10:192–206. doi: 10.1038/nrm2640. PMID:19234478. [DOI] [PubMed] [Google Scholar]
- 58.Cortez CC, Jones PA. Chromatin, cancer and drug therapies. Mutat Res. 2008;647:44–51. doi: 10.1016/j.mrfmmm.2008.07.006. PMID:18691602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bartling B, Hofmann HS, Boettger T, Hansen G, Burdach S, Silber RE, Simm A. Comparative application of antibody and gene array for expression profiling in human squamous cell lung carcinoma. Lung Cancer. 2005;49:145–154. doi: 10.1016/j.lungcan.2005.02.006. PMID:16022908. [DOI] [PubMed] [Google Scholar]
- 60.O'Leary SM, Coleman MM, Chew WM, Morrow C, McLaughlin AM, Gleeson LE, O'Sullivan MP, Keane J. Cigarette smoking impairs human pulmonary immunity to Mycobacterium tuberculosis. Am J Respir Crit Care Med. 2014;190:1430–1436. doi: 10.1164/rccm.201407-1385OC. PMID:25390734. [DOI] [PubMed] [Google Scholar]
- 61.Cancer Genome Atlas Research Network Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. PMID:25079552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–525. doi: 10.1038/nature11404. PMID:22960745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang X, Spandidos A, Wang H, Seed B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 2012;40:D1144–9. doi: 10.1093/nar/gkr1013. PMID:22086960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen QY, Zhang T, Pincus SH, Wu S, Ricks D, Liu D, Sun Z, Maclaren N, Lan MS. Human CD1D gene expression is regulated by LEF-1 through distal promoter regulatory elements. J Immunol 2010;184:5047–5054. doi: 10.4049/jimmunol.0901912. PMID:20363964. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


