The completion of the human genome project1 has enabled the identification of many genes, including variants that cause disease.2–4 An important discovery, although less celebrated, is that more than half the human genome is derived from mobile pieces of DNA called transposable elements (colloquially known as “jumping genes”). Transposable elements were discovered in corn by Barbara McClintock more than 60 years ago,5 but few people would have guessed that the aggregate length of these sequences exceeds that of protein-coding exons by a factor greater than 40 (Fig. 1).1 Although the bulk of transposable element–derived DNAs are remnants of their former selves and cannot transpose, some retain the ability to mobilize.11,12 The insertion of mobile elements into the DNA of gametes or the early embryo can disrupt genes, leading to sporadic cases of disease,13 and their insertion into the DNA of somatic cells may contribute to cancers and neuropsychiatric disease.9,14 Clearly, mobile DNA has been instrumental in shaping the structure, function, and evolution of the human genome. Here we discuss the biology of mobile DNA, emphasizing key discoveries that illustrate how it contributes to human disease.
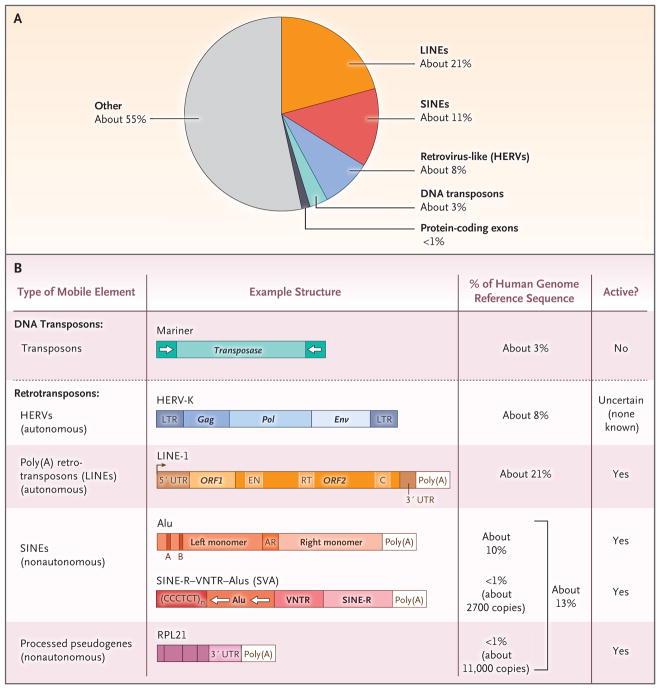
Figure 1. Transposable Elements in the Human Genome.
Panel A shows protein-coding exons and classes of human transposable elements as a percentage of the genome.1 As shown in Panel B, DNA transposons encode transposase, an enzyme that binds at or near inverted repeats flanking the element (white arrows flanking transposase) to promote mobility by means of a cut-and-paste mechanism.6 Retrotransposons mobilize by means of a copy-and-paste mechanism through an RNA intermediate.7 Human endogenous retroviruses (HERVs) contain sequences analogous to the gag, pol, and env genes of retroviruses, but most HERVs, if not all of them, are not active in humans.8 Elements are considered to be autonomous when they encode the protein activities necessary for retrotransposition (e.g., an endonuclease [EN] and a reverse transcriptase [RT]). The arrow at the 5′ end of poly(A) retrotransposons denotes the transcription start site from their internal promoter. Nonautonomous elements do not encode proteins; their retrotransposition depends on the proteins encoded by autonomous elements.9 Nonautonomous elements include Alu elements and SVAs. Alu elements have a size of approximately 280 bp and consist of two monomers separated by an adenosine-rich (AR) linker; the left Alu monomer has an internal RNA polymerase III promoter (bars labeled A and B). SVAs are composite elements with a portion of a HERV (SINE-R [short interspersed element of HERV origin]), a variable number of tandem repeats (VNTRs), and a backward Alu, as well as a CCCTCT multimer. Processed pseudogenes are copies of cellular messenger RNAs that have been reverse-transcribed into DNA and lack introns. C denotes cysteine-rich domain (encoded by ORF2), HERV-K a HERV of the K type, LINE long interspersed element, LTR long terminal repeat, ORF1 open reading frame 1, ORF2 open reading frame 2, and UTR untranslated region. The diagram in Panel B is adapted from Beck et al.10
MOBILE DNA IN HUMANS
DNA TRANSPOSONS
DNA transposons are a major class of mobile elements (Fig. 1). They are active in many lower organisms, including bacteria,6 but have been inactivated by mutations and can no longer transpose in humans.1 However, transposon-derived sequences have been repurposed over evolutionary time and affect human biology. For example, the recombination-activating genes RAG1 and RAG2 were probably derived from an ancient DNA transposon and are critical for adaptive immunity because they encode enzymes that generate diverse immunoglobulin proteins.15,16 Moreover, DNA sequences derived from ancient DNA transposons have served to rewire endometrial-cell gene expression in eutherian mammals and may have influenced the evolution of pregnancy.17
RETROTRANSPOSONS
The second major class of mobile DNA is retrotransposons (see the Glossary).10 The DNA of a retrotransposon is copied into RNA, which is then copied back into DNA (the “retro” step) by a reverse-transcriptase enzyme encoded by the retrotransposon. The reverse-transcribed DNA is then integrated into the genome.7
Retrotransposons fall into two classes: the human endogenous retroviruses (HERVs; also known as long-terminal-repeat [LTR] retrotransposons, owing to the long repeat at each end) and the poly(A) retrotransposons. Much like DNA transposons, almost all HERVs are mutated and cannot retrotranspose in humans.10,18 However, sequences within HERVs influence host gene expression in the early embryo,19 and HERV-derived proteins are important for placental development.20 Moreover, stretches of DNA derived from endogenous retroviruses have shaped, over evolutionary time, a transcriptional network involved in the interferon response of innate immunity.21 HERV expression (i.e., the level of HERV messenger RNA [mRNA]) is elevated in the affected tissues of persons with rheumatoid arthritis, multiple sclerosis, or amyotrophic lateral sclerosis, and the expression of HERV-encoded accessory proteins may be involved in the development of certain cancers.8,22 However, these are only associations; it is not known whether HERV expression is pathogenic.
The retrotransposition of LINE-1 (long interspersed element 1) poly(A) retrotransposons (which can autonomously replicate),23 as well as the non-autonomous retrotransposon RNAs (e.g., Alu), is mediated by the LINE-1–encoded proteins, as are reverse-transcribed mRNAs (i.e., processed pseudogenes).9 LINE-1, Alu, SINE-R (short interspersed element of HERV origin)–VNTR (variable-number tandem repeat)–Alu (SVA), and processed pseudogenes account for a remarkable one third, or 1 billion bp, of human DNA.1,9 There are more than 500,000 LINE-1 poly(A) retrotransposon sequences in each human genome, about 100 of which are active,24,25 but only a small number (termed “hot LINE-1” sequences) cause most cases of LINE-1–mediated disease.25
RETROTRANSPOSONS AS MUTAGENS
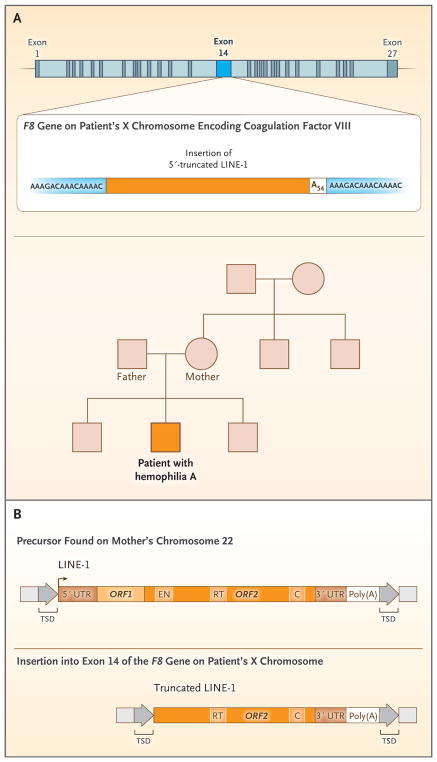
In the late 1980s, Kazazian and colleagues discovered that LINE-1 elements can cause disease. Of 240 boys with hemophilia A, an X-linked disorder, 2 had mutagenic LINE-1 insertions; each insertion disrupted an exon of F8 (on the X chromosome), which is the gene encoding coagulation factor VIII.26 These LINE-1 insertions were not full length and therefore could not undergo further retrotransposition. The hypothesis was that both insertions were derived from full-length LINE-1 elements, and support for this hypothesis was obtained by showing that one full-length LINE-1 was present on chromosome 22 in the mother of one of the affected boys (Fig. 2). A portion of this full-length LINE-1 was identical to her son’s truncated LINE-1 insertion.11 Pathogenic retrotransposition events mediated by LINE-1 have been reported in 130 persons with various diseases, including Duchenne’s muscular dystrophy, β-thalassemia trait, factor IX hemophilia, and cancers (both inherited and sporadic [i.e., occurring without a family history]).13,27 LINE-1–mediated retrotransposition events account for approximately 1 in every 250 pathogenic human mutations28; they occur in exons, introns, or regulatory regions and adversely affect gene expression.10 Moreover, the overexpression of Alu RNA in the cells of the retinal pigment epithelium has been linked to geographic atrophy, a form of age-related macular degeneration.29
Figure 2. LINE-1 Retrotransposition in a Boy with Hemophilia A.
Panel A shows the family pedigree. The solid orange square denotes the affected boy, and the other symbols indicate unaffected family members. The boy’s mother is not a carrier of the insertion in exon 14 of F8 (the gene encoding factor VIII). In Panel B, the precursor LINE-1 element isolated from the mother’s DNA is shown above the insertion. The precursor was a full-length LINE-1 in chromosome 22. The smaller pathogenic insertion was identical in sequence to a portion of the precursor LINE-1. The brown arrow at the 5′ end of the precursor element denotes the transcription start site from the LINE-1 internal promoter. TSD denotes target-site duplication, and the light-gray box next to TSD indicates genomic DNA. The diagram in Panel A is adapted from Kazazian et al.,26 and the diagram in Panel B is adapted from Dombroski et al.11
MECHANISM OF LINE-1 RETROTRANSPOSITION
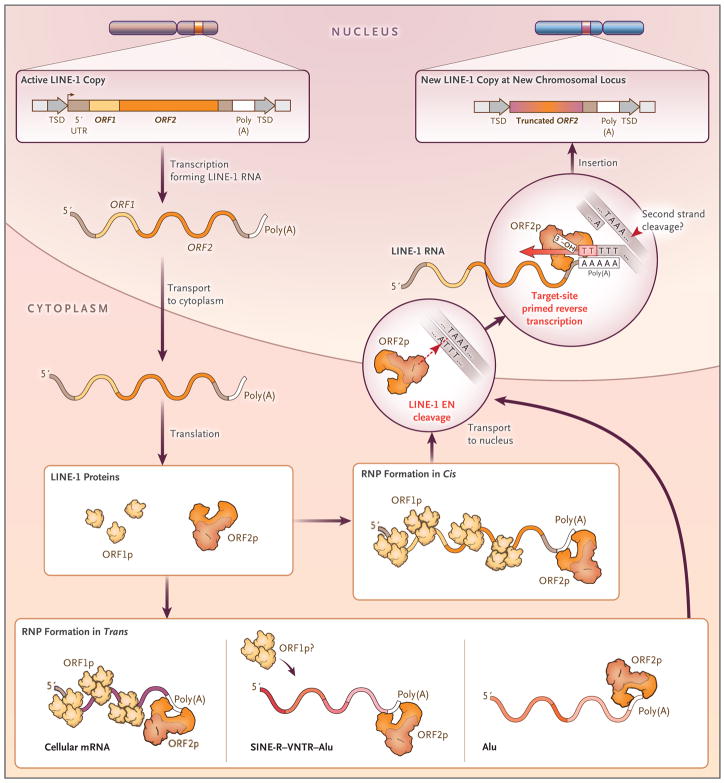
Retroviruses and HERVs “jump” by means of a rather complicated mechanism. First, their viral RNA is reverse-transcribed into DNA within a virus like particle in the cytoplasm of the cell. The full-length viral DNA is then transported into the nucleus and integrated into the genome.8,30 In contrast, LINE-1 retrotransposition (also known as target-site–primed reverse transcription) occurs through a somewhat simpler process (Fig. 3).31,32 Active LINE-1 elements contain two open reading frames, ORF1 and ORF2; an endonuclease encoded by ORF2 nicks one strand of DNA at the site of the new integration.39 The reverse-transcriptase enzyme (also encoded by LINE-1 ORF2) then copies LINE-1 RNA, beginning at the chromosomal integration site. It is not yet clear how the second strand of LINE-1 DNA is made, although it probably involves the LINE-1 reverse transcriptase.40
Figure 3. A Model of LINE-1 Retrotransposition.
LINE-1 RNA is transcribed from a promoter located within its 5′ UTR. The RNA is exported into the cytoplasm, where it undergoes translation. The LINE-1–encoded proteins ORF1p and ORF2p bind to the LINE-1–encoding RNA by a process known as cis-preference,33,34 leading to the formation of a cytoplasmic complex.35–38 Components of this complex (at least ORF2p and LINE-1 RNA) gain nuclear access, at which point the ORF2p endonuclease (EN) cleaves a single strand of chromosomal DNA at a consensus sequence (i.e., 5′-TTTT/A-3′), liberating a 3′ hydroxyl group that is used by ORF2p reverse transcriptase to copy LINE-1 RNA and integrate the resultant LINE-1 DNA into this new chromosomal location.32,39 It is not known how the second DNA strand at the insertion site is cleaved and how second-strand LINE-1 DNA is synthesized, but the ORF2 protein probably mediates these processes. The ORF2 protein is also required for the retrotransposition of nonautonomous RNAs, such as cellular mRNAs (creating processed pseudogenes), SVA RNAs, and Alu RNAs.9 The ORF1 protein may aid in SVA and Alu retrotransposition.9 RNP denotes ribonucleoprotein particle. The diagram is adapted from Richardson et al.9
Knowledge of how LINE-1 elements retrotranspose has led to insight into how other retrotransposons use LINE-1–encoded proteins to move to new genomic locations. For example, the LINE-1 endonuclease and reverse-transcriptase activities of LINE-1 ORF2 are critical for the mobilization of Alu RNAs.41 In contrast, the proteins encoded by both LINE-1 ORF1 and LINE-1 ORF2 appear to be critical for the mobilization of SVA elements,42,43 other noncoding RNAs,44,45 and processed pseudogenes (Fig. 3).33,34
ROLE OF LINE-1 AND ALU IN GENOMIC REARRANGEMENTS AND DISEASE
LINE-1–mediated retrotransposition not only creates insertional mutations but also results in other structural rearrangements in both LINE-1 itself and genomic DNA.10,46–48 Such rearrangements can obstruct subsequent retrotransposition.49,50 Indeed, only 7 of the 31 known disease-producing LINE-1 insertions are preserved, full-length LINE-1s.13 Examples involving genomic disruption of the sequence flanking the insertion site are a large deletion involving PDHX, leading to pyruvate dehydrogenase deficiency,51 and deletions affecting NF1, causing neurofibromatosis.52
LINE-1–mediated retrotransposition generally requires the endonuclease activity encoded by its ORF2. However, on rare occasions, LINE-1 elements can act as chromosomal bandages by integrating at preexisting DNA lesions, such as dysfunctional telomeres, through a process known as endonuclease-independent (ENi) retrotransposition.53,54 Indeed, an ENi-retrotransposition–mediated insertion of LINE-1 into EYA1, accompanied by a genomic DNA deletion, was found in a person with the branchiootorenal syndrome.55
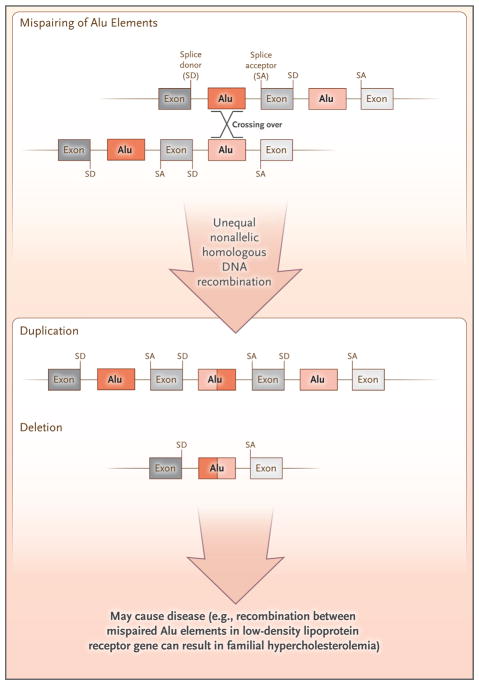
The abundance of retrotransposon-derived DNA provides ample templates for disease-producing DNA recombination events (Fig. 4). For example, unequal crossing-over events mediated by mispaired Alu elements in the low-density lipoprotein receptor gene have caused familial hypercholesterolemia and more than 70 cases of other diseases.57,58 In addition to causing monogenic disease by disrupting single genes, Alu-mediated recombination events can generate copy-number variations in the human genome.59 Nearly 500 Alu-mediated deletions are known to have occurred since the divergence of chimpanzees and humans.60 Mispairing and unequal crossing over of LINE-1s, although less common than Alu-mediated recombination events, has resulted in sporadic cases of genetic disease.10 Increased use of whole-genome sequencing to diagnose disease will probably uncover more retrotransposon-mediated pathogenic events.
Figure 4. DNA-Level Recombination Events and Genome Instability.
Mispairing of Alu elements or LINE-1 elements, followed by crossing over and unequal nonallelic homologous recombination, leads to the deletion or duplication of the genomic sequence. Exons and introns (black lines) of the hypothetical gene are shown. The diagram is adapted from Hulme et al.56
LINE-1–MEDIATED RETROTRANSPOSITION EVENTS
Retrotransposition events mediated by LINE-1 are estimated to occur, at a minimum, in 1 of 20 meioses for Alu, 1 of 20 to 200 meioses for LINE-1, and 1 of 900 meioses for SVA.59 These events are the cause of a great deal of interindividual variation in the population. For example, the genomes of any two unrelated persons are likely to differ at about 300 LINE-1 insertion sites.61 Similarly, any two unrelated persons are likely to have many differences in Alu and SVA insertion sites; some of these differences probably affect gene expression.
Some disease-producing LINE-1–mediated insertions occurred many generations ago. For example, an SVA insertion into an intron of FCMD (the gene that, when mutated, causes Fukuyama-type congenital muscular dystrophy) causes mis-splicing and the production of a mutant form of the fukutin protein.62 This SVA insertion is specific to the Japanese population, causes most cases of Fukuyama-type congenital muscular dystrophy, and has been segregating within the population for many generations.
LINE-1–mediated retrotransposition events occur in somatic cells of the early human embryo63,64 and can occur in human embryonic stem cells.65 Similarly, studies in transgenic mice and rats revealed that engineered human LINE-1 retrotransposition was more prevalent in the blastocyst and morula stages of early development than during spermatogenesis.66
What other somatic cells are susceptible to LINE-1–mediated insertions? Human LINE-1 can retrotranspose in various regions of the brain in transgenic mice and in neural progenitor cells differentiated from embryonic stem cells in humans.67,68 Studies of DNA derived from either bulk brain tissues or single neurons have confirmed that LINE-1 can retrotranspose in the brain.69–71 It remains unclear whether brain-cell types differ in their capacity for LINE-1 retrotransposition and whether somatic insertions influence behavior and susceptibility to psychiatric disorders.14,72 There have been no detailed studies of somatic retrotransposition in human tissues other than the brain and the gastrointestinal tract.
LINE-1 retrotransposition occurs in various cancers.73–83 Studies have shown that somatic LINE-1 insertions primarily occur in epithelial cancers (e.g., those in the gastrointestinal tract); that the number of LINE-1 insertions varies among epithelial tumors (with some having >50 somatic insertions and others having none); that certain clonal somatic insertions in esophageal and gastric tumors are present at low frequencies in normal tissue, suggesting that a normal cell harboring a somatic LINE-1 insertion may be clonally expanded in the cancer78,79,81; and that some types of cancers (e.g., certain hematopoietic and brain cancers) lack somatic LINE-1 insertions.
Whether LINE-1 insertions represent “driver” or “passenger” mutations in cancers is unclear. Most somatic LINE-1 insertions found in epithelial cancers probably represent passenger mutations.84 However, some somatic LINE-1 insertions, such as those reported to inactivate the tumor-suppressor genes APC27,85 and PTEN,77 probably promote tumorigenesis.
DEFENSES AGAINST MOBILE ELEMENTS
The ability to replicate is critical for the evolutionary survival of retrotransposons. Because the resultant insertions can act as mutagens, evidence supporting the evolution of mechanisms to combat retrotransposition comes as no surprise.86
Methylation of DNA at CpG dinucleotides (sites at which a cytosine precedes a guanine in the DNA sequence) restricts retrotransposon expression in both the germline and soma. Gene-knockout experiments in mice have shown that disruption of a gene encoding an enzyme that aids in the methylation of CpG motifs87 leads to a loss of CpG methylation, derepression of LINE-1s and certain endogenous retroviruses in the male germline, and meiotic arrest, resulting in male infertility. Whether the derepression of retrotransposon expression causes meiotic catastrophe or leads to increased retrotransposition requires further study. Small noncoding RNAs are also known to affect CpG methylation and transcriptional silencing of retrotransposons in the male germline.86,88
Certain zinc-finger proteins can recruit protein complexes to sequences that reside within retrotransposons, leading to the deposition of repressive histone modifications and transcriptional silencing of retrotransposons.89–92 For example, zinc-finger protein 91 (ZNF91) was found to direct the deposition of repressive histone modifications on human SVA elements, and zinc-finger protein 93 (ZNF93) has a similar effect on older LINE-1 subfamily members.93 An active LINE-1 has a deletion that allows it to escape ZNF93-mediated repression.93
Various cytoplasmic pathways influence the stability of retrotransposon RNAs, their translation, or both, and certain proteins inhibit retroviral replication and retrotransposon mobility.9,94,95 Some of these proteins colocalize with LINE-1 RNAs in cytoplasmic stress granules, suggesting that sequestration to stress granules may play a role in the degradation or translational repression of LINE-1 RNAs.96,97 Finally, since full-length LINE-1 RNAs contain functional splice sites and polyadenylation signal sequences, proteins involved in RNA processing may inhibit LINE-1 retrotransposition.98,99
CONCLUSIONS
Since the discovery nearly 30 years ago that LINE-1 insertions can cause human disease, research has yielded insights into how LINE-1 mobilizes, disrupts genes, and causes disease. Many questions remain to be addressed. Does somatic retrotransposition play an important role in the development of certain cancers? Can somatic retrotransposition in the brain affect human behavior? How often are sequences within retrotransposons coopted by the host for functional purposes? These are just a few areas for future research. Although we have focused on the harmful effects of retrotransposons in this review, it is clear that transposable elements are not just weeds in the garden. They provide the fertile soil that is the fodder for the evolution of genomes.100
Acknowledgments
We thank members of our laboratories for critical discussions and Dr. Barbara Migeon for constructive comments on the manuscript.
Glossary
- Accessory protein
A protein that is not required for viral or retrotransposon replication but that plays an indirect regulatory role in the function of a virus or retrotransposon.
- Alu
A short (approximately 280 bp) element of which 1.1 million copies are present in the human genome. Some Alu elements move to new genomic locations by means of the protein encoded by the second open reading frame (ORF2) of LINE-1.
- Autonomous transposable elements
Elements that encode proteins that are required for the mobility of the transposable elements throughout the genome.
- Copy-number variation
Interindividual genetic variation caused by either the increase or decrease of a block of DNA sequence within the genome.
- CpG dinucleotides
Sites at which a cytosine precedes a guanine in the DNA sequence and DNA methylation occurs at the cytosine in the human genome.
- DNA transposon
A DNA mobile element containing transposase, an enzyme that allows the transposon to cut itself out of one genomic site and insert itself into another site.
- Exon
The protein-coding and untranslated-region (UTR) sequences of a messenger RNA (mRNA). Exons remain in the mRNA after splicing.
- Human endogenous retrovirus (HERV)
A retrovirus-like sequence accounting for approximately 8% of the human genome. These DNAs are immobile retrotransposons and usually contain defective envelope genes. Over evolutionary time, sequences within HERVs have been “exapted,” or co-opted, for functional use in humans (e.g., placental syncytins).
- Histone
An octameric protein complex that packages DNA into structures called nucleosomes.
- Intron
A sequence that resides between exons in the precursor of mRNA and is spliced out of the RNA.
- Long interspersed element 1 (LINE-1 or L1)
Repeated DNA elements that are present at approximately 500,000 copies in the human genome and account for approximately 17% of the human genome. Some autonomous LINE-1 elements can retrotranspose. The LINE-1–encoded proteins can also retrotranspose nonautonomous elements, such as Alu and SVA. LINE-1 retrotransposons are unable to move from cell to cell.
- LINE-1 endonuclease
An enzyme encoded by LINE-1 that can cut one strand of a DNA double helix at a consensus sequence at the insertion site for a retrotransposition event.
- Mispairing and unequal crossing over
Mispairing of chromosomal homologues when two homologous sequences are in proximity on the chromosome. For example, Alu1 is close to Alu2. Mispairing leads Alu1 on one chromosome 1 to pair with Alu2 on the other chromosome 1. Recombination of Alu1 and Alu2 results in unequal crossing over, leading to deletion on one chromosomal homologue and duplication on the other.
- Nonautonomous retrotransposon
A transposable element that does not encode proteins but instead relies on the proteins encoded by an autonomous transposable element (LINE-1) to move to a new genomic location.
- Polyadenylation signal sequence
A specific signal sequence (5′-AAUAAA-3′) near the back end (3′ end) of an RNA that signals cleavage of the RNA roughly 20 nucleotides downstream of the signal. After cleavage of the RNA, a poly(A) tail is added at the cleavage site.
- Processed pseudogene
A reverse-transcribed copy of an mRNA that is inserted back into the genome with the use of LINE-1 proteins. Processed pseudogene sequences lack the introns present in precursor mRNAs.
- Retrotransposon
A piece of DNA that can be transcribed into RNA and then reverse-transcribed into complementary DNA (cDNA), with the cDNA copy then reinserted into the genome at a new location by a copy-and-paste process. Human retrotransposons are LINE-1, Alu, and SVA.
- Retrovirus
A virus that encodes gag, pol, and env proteins. The gag protein forms a capsule around the viral RNA. The pol protein contains reverse-transcriptase and integrase activity. The env protein aids the virus in entering and exiting cells. Retroviruses and HERVs have similar sequences, but the HERV env protein is usually defective, precluding its movement from one cell to another.
- Reverse transcriptase
An enzyme that copies RNA back into DNA. Some LINE-1 elements and HERVs encode reverse-transcriptase activity.
- SINE-R–VNTR–Alu (SVA)
A primate-specific retrotransposon present in approximately 2700 copies in the human genome that can retrotranspose with the help of LINE-1–derived proteins. SINE-R denotes short interspersed element of HERV origin, and VNTR variable-number tandem repeat.
- Target-site–primed reverse transcription
The mechanism by which LINE and SINE retrotransposons, as well as processed pseudogenes, move to new genomic locations.
- Transposable elements
Segments of DNA that can move from one location in DNA to another.
- Untranslated region (UTR)
A sequence in mRNA or LINE-1 RNA that is not translated into protein.
Footnotes
Disclosure forms provided by the authors are available with the full text of this article at NEJM.org.
References
- 1.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9:356–69. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 3.Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–8. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.National Human Genome Research Institute. A catalog of published genome-wide association studies. 2015 https://www.genome.gov/26525384/catalog-of-published-genomewide-association-studies/
- 5.McClintock B. The origin and behavior of mutable loci in maize. Proc Natl Acad Sci U S A. 1950;36:344–55. doi: 10.1073/pnas.36.6.344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Craig NL, Chandler M, Gellert M, Lambowitz AM, Rice PA, Sandmeyer SB. Mobile DNA III. Washington, DC: ASM Press; 2015. [Google Scholar]
- 7.Boeke JD, Garfinkel DJ, Styles CA, Fink GR. Ty elements transpose through an RNA intermediate. Cell. 1985;40:491–500. doi: 10.1016/0092-8674(85)90197-7. [DOI] [PubMed] [Google Scholar]
- 8.Mager DL, Stoye JP. Mammalian endogenous retroviruses. Microbiol Spectr. 2015;3:MDNA3-0009. doi: 10.1128/microbiolspec.MDNA3-0009-2014. [DOI] [PubMed] [Google Scholar]
- 9.Richardson SR, Doucet AJ, Kopera HC, Moldovan JB, Garcia-Perez JL, Moran JV. The influence of LINE-1 and SINE retrotransposons on mammalian genomes. Microbiol Spectr. 2015;3:MDNA3-0061. doi: 10.1128/microbiolspec.MDNA3-0061-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beck CR, Garcia-Perez JL, Badge RM, Moran JV. LINE-1 elements in structural variation and disease. Annu Rev Genomics Hum Genet. 2011;12:187–215. doi: 10.1146/annurev-genom-082509-141802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dombroski BA, Mathias SL, Nanthakumar E, Scott AF, Kazazian HH., Jr Isolation of an active human transposable element. Science. 1991;254:1805–8. doi: 10.1126/science.1662412. [DOI] [PubMed] [Google Scholar]
- 12.Kazazian HH, Jr, Moran JV. The impact of L1 retrotransposons on the human genome. Nat Genet. 1998;19:19–24. doi: 10.1038/ng0598-19. [DOI] [PubMed] [Google Scholar]
- 13.Hancks DC, Kazazian HH., Jr Roles for retrotransposon insertions in human disease. Mob DNA. 2016;7:9. doi: 10.1186/s13100-016-0065-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richardson SR, Morell S, Faulkner GJ. L1 retrotransposons and somatic mosaicism in the brain. Annu Rev Genet. 2014;48:1–27. doi: 10.1146/annurev-genet-120213-092412. [DOI] [PubMed] [Google Scholar]
- 15.Kapitonov VV, Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3(6):e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang S, Tao X, Yuan S, et al. Discovery of an active RAG transposon illuminates the origins of V(D)J recombination. Cell. 2016;166:102–14. doi: 10.1016/j.cell.2016.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lynch VJ, Leclerc RD, May G, Wagner GP. Transposon-mediated rewiring of gene regulatory networks contributed to the evolution of pregnancy in mammals. Nat Genet. 2011;43:1154–9. doi: 10.1038/ng.917. [DOI] [PubMed] [Google Scholar]
- 18.Weiss RA. Human endogenous retroviruses: friend or foe? APMIS. 2016;124:4–10. doi: 10.1111/apm.12476. [DOI] [PubMed] [Google Scholar]
- 19.Grow EJ, Flynn RA, Chavez SL, et al. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522:221–5. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dupressoir A, Lavialle C, Heidmann T. From ancestral infectious retroviruses to bona fide cellular genes: role of the captured syncytins in placentation. Placenta. 2012;33:663–71. doi: 10.1016/j.placenta.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 21.Chuong EB, Elde NC, Feschotte C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science. 2016;351:1083–7. doi: 10.1126/science.aad5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, Lee MH, Henderson L, et al. Human endogenous retrovirus-K contributes to motor neuron disease. Sci Transl Med. 2015;7:307ra153. doi: 10.1126/scitranslmed.aac8201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH., Jr High frequency retrotransposition in cultured mammalian cells. Cell. 1996;87:917–27. doi: 10.1016/s0092-8674(00)81998-4. [DOI] [PubMed] [Google Scholar]
- 24.Sassaman DM, Dombroski BA, Moran JV, et al. Many human L1 elements are capable of retrotransposition. Nat Genet. 1997;16:37–43. doi: 10.1038/ng0597-37. [DOI] [PubMed] [Google Scholar]
- 25.Brouha B, Schustak J, Badge RM, et al. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci U S A. 2003;100:5280–5. doi: 10.1073/pnas.0831042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kazazian HH, Jr, Wong C, Youssoufian H, Scott AF, Phillips DG, Antonarakis SE. Haemophilia A resulting from de novo insertion of L1 sequences represents a novel mechanism for mutation in man. Nature. 1988;332:164–6. doi: 10.1038/332164a0. [DOI] [PubMed] [Google Scholar]
- 27.Scott EC, Gardner EJ, Masood A, Chuang NT, Vertino PM, Devine SE. A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome Res. 2016;26:745–55. doi: 10.1101/gr.201814.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wimmer K, Callens T, Wernstedt A, Messiaen L. The NF1 gene contains hotspots for L1 endonuclease-dependent de novo insertion. PLoS Genet. 2011;7(11):e1002371. doi: 10.1371/journal.pgen.1002371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaneko H, Dridi S, Tarallo V, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–30. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu WS, Hughes SH. HIV-1 reverse transcription. Cold Spring Harb Perspect Med. 2012;2(10):a006882. doi: 10.1101/cshperspect.a006882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. doi: 10.1016/0092-8674(93)90078-5. [DOI] [PubMed] [Google Scholar]
- 32.Cost GJ, Feng Q, Jacquier A, Boeke JD. Human L1 element target-primed reverse transcription in vitro. EMBO J. 2002;21:5899–910. doi: 10.1093/emboj/cdf592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–7. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 34.Wei W, Gilbert N, Ooi SL, et al. Human L1 retrotransposition: cis preference versus trans complementation. Mol Cell Biol. 2001;21:1429–39. doi: 10.1128/MCB.21.4.1429-1439.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hohjoh H, Singer MF. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 1996;15:630–9. [PMC free article] [PubMed] [Google Scholar]
- 36.Kulpa DA, Moran JV. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum Mol Genet. 2005;14:3237–48. doi: 10.1093/hmg/ddi354. [DOI] [PubMed] [Google Scholar]
- 37.Kulpa DA, Moran JV. Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles. Nat Struct Mol Biol. 2006;13:655–60. doi: 10.1038/nsmb1107. [DOI] [PubMed] [Google Scholar]
- 38.Doucet AJ, Hulme AE, Sahinovic E, et al. Characterization of LINE-1 ribonucleoprotein particles. PLoS Genet. 2010;6(10):6. doi: 10.1371/journal.pgen.1001150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng Q, Moran JV, Kazazian HH, Jr, Boeke JD. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell. 1996;87:905–16. doi: 10.1016/s0092-8674(00)81997-2. [DOI] [PubMed] [Google Scholar]
- 40.Christensen SM, Eickbush TH. R2 target-primed reverse transcription: ordered cleavage and polymerization steps by protein subunits asymmetrically bound to the target DNA. Mol Cell Biol. 2005;25:6617–28. doi: 10.1128/MCB.25.15.6617-6628.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dewannieux M, Esnault C, Heidmann T. LINE-mediated retrotransposition of marked Alu sequences. Nat Genet. 2003;35:41–8. doi: 10.1038/ng1223. [DOI] [PubMed] [Google Scholar]
- 42.Raiz J, Damert A, Chira S, et al. The non-autonomous retrotransposon SVA is transmobilized by the human LINE-1 protein machinery. Nucleic Acids Res. 2012;40:1666–83. doi: 10.1093/nar/gkr863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hancks DC, Goodier JL, Mandal PK, Cheung LE, Kazazian HH., Jr Retrotransposition of marked SVA elements by human L1s in cultured cells. Hum Mol Genet. 2011;20:3386–400. doi: 10.1093/hmg/ddr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Buzdin A, Ustyugova S, Gogvadze E, Vinogradova T, Lebedev Y, Sverdlov E. A new family of chimeric retrotranscripts formed by a full copy of U6 small nuclear RNA fused to the 3′ terminus of l1. Genomics. 2002;80:402–6. doi: 10.1006/geno.2002.6843. [DOI] [PubMed] [Google Scholar]
- 45.Garcia-Perez JL, Doucet AJ, Bucheton A, Moran JV, Gilbert N. Distinct mechanisms for transmediated mobilization of cellular RNAs by the LINE-1 reverse transcriptase. Genome Res. 2007;17:602–11. doi: 10.1101/gr.5870107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilbert N, Lutz S, Morrish TA, Moran JV. Multiple fates of L1 retrotransposition intermediates in cultured human cells. Mol Cell Biol. 2005;25:7780–95. doi: 10.1128/MCB.25.17.7780-7795.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gilbert N, Lutz-Prigge S, Moran JV. Genomic deletions created upon LINE-1 retrotransposition. Cell. 2002;110:315–25. doi: 10.1016/s0092-8674(02)00828-0. [DOI] [PubMed] [Google Scholar]
- 48.Symer DE, Connelly C, Szak ST, et al. Human l1 retrotransposition is associated with genetic instability in vivo. Cell. 2002;110:327–38. doi: 10.1016/s0092-8674(02)00839-5. [DOI] [PubMed] [Google Scholar]
- 49.Grimaldi G, Skowronski J, Singer MF. Defining the beginning and end of KpnI family segments. EMBO J. 1984;3:1753–9. doi: 10.1002/j.1460-2075.1984.tb02042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ostertag EM, Kazazian HH., Jr Twin priming: a proposed mechanism for the creation of inversions in L1 retrotransposition. Genome Res. 2001;11:2059–65. doi: 10.1101/gr.205701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miné M, Chen JM, Brivet M, et al. A large genomic deletion in the PDHX gene caused by the retrotranspositional insertion of a full-length LINE-1 element. Hum Mutat. 2007;28:137–42. doi: 10.1002/humu.20449. [DOI] [PubMed] [Google Scholar]
- 52.Vogt J, Bengesser K, Claes KB, et al. SVA retrotransposon insertion-associated deletion represents a novel mutational mechanism underlying large genomic copy number changes with non-recurrent breakpoints. Genome Biol. 2014;15:R80. doi: 10.1186/gb-2014-15-6-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Morrish TA, Garcia-Perez JL, Stamato TD, Taccioli GE, Sekiguchi J, Moran JV. Endonuclease-independent LINE-1 retrotransposition at mammalian telomeres. Nature. 2007;446:208–12. doi: 10.1038/nature05560. [DOI] [PubMed] [Google Scholar]
- 54.Morrish TA, Gilbert N, Myers JS, et al. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet. 2002;31:159–65. doi: 10.1038/ng898. [DOI] [PubMed] [Google Scholar]
- 55.Morisada N, Rendtorff ND, Nozu K, et al. Branchiootorenal syndrome caused by partial EYA1 deletion due to LINE-1 insertion. Pediatr Nephrol. 2010;25:1343–8. doi: 10.1007/s00467-010-1445-x. [DOI] [PubMed] [Google Scholar]
- 56.Hulme AE, Kulpa DA, Garcia-Perez JL, Moran JV. The impact of LINE-1 retrotransposition on the human genome. In: Lupski JR, Stankiewicz P, editors. Genomic disorders: the genomic basis of disease. Totowa, NJ: Humana Press; 2006. pp. 35–72. [Google Scholar]
- 57.Lehrman MA, Schneider WJ, Südhof TC, Brown MS, Goldstein JL, Russell DW. Mutation in LDL receptor: Alu-Alu recombination deletes exons encoding transmembrane and cytoplasmic domains. Science. 1985;227:140–6. doi: 10.1126/science.3155573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Konkel MK, Batzer MA. A mobile threat to genome stability: the impact of non-LTR retrotransposons upon the human genome. Semin Cancer Biol. 2010;20:211–21. doi: 10.1016/j.semcancer.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cordaux R, Batzer MA. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sen SK, Han K, Wang J, et al. Human genomic deletions mediated by recombination between Alu elements. Am J Hum Genet. 2006;79:41–53. doi: 10.1086/504600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ewing AD, Kazazian HH., Jr High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res. 2010;20:1262–70. doi: 10.1101/gr.106419.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Taniguchi-Ikeda M, Kobayashi K, Kanagawa M, et al. Pathogenic exon-trapping by SVA retrotransposon and rescue in Fukuyama muscular dystrophy. Nature. 2011;478:127–31. doi: 10.1038/nature10456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van den Hurk JA, Meij IC, Seleme MC, et al. L1 retrotransposition can occur early in human embryonic development. Hum Mol Genet. 2007;16:1587–92. doi: 10.1093/hmg/ddm108. [DOI] [PubMed] [Google Scholar]
- 64.de Boer M, van Leeuwen K, Geissler J, et al. Primary immunodeficiency caused by an exonized retroposed gene copy inserted in the CYBB gene. Hum Mutat. 2014;35:486–96. doi: 10.1002/humu.22519. [DOI] [PubMed] [Google Scholar]
- 65.Garcia-Perez JL, Marchetto MC, Muotri AR, et al. LINE-1 retrotransposition in human embryonic stem cells. Hum Mol Genet. 2007;16:1569–77. doi: 10.1093/hmg/ddm105. [DOI] [PubMed] [Google Scholar]
- 66.Kano H, Godoy I, Courtney C, et al. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–12. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muotri AR, Chu VT, Marchetto MC, Deng W, Moran JV, Gage FH. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–10. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- 68.Coufal NG, Garcia-Perez JL, Peng GE, et al. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–31. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Upton KR, Gerhardt DJ, Jesuadian JS, et al. Ubiquitous L1 mosaicism in hippocampal neurons. Cell. 2015;161:228–39. doi: 10.1016/j.cell.2015.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baillie JK, Barnett MW, Upton KR, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–7. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Evrony GD, Cai X, Lee E, et al. Single-neuron sequencing analysis of L1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–96. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bundo M, Toyoshima M, Okada Y, et al. Increased l1 retrotransposition in the neuronal genome in schizophrenia. Neuron. 2014;81:306–13. doi: 10.1016/j.neuron.2013.10.053. [DOI] [PubMed] [Google Scholar]
- 73.Iskow RC, McCabe MT, Mills RE, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–61. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee E, Iskow R, Yang L, et al. Landscape of somatic retrotransposition in human cancers. Science. 2012;337:967–71. doi: 10.1126/science.1222077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Shukla R, Upton KR, Muñoz-Lopez M, et al. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell. 2013;153:101–11. doi: 10.1016/j.cell.2013.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tubio JM, Li Y, Ju YS, et al. Mobile DNA in cancer: extensive transduction of nonrepetitive DNA mediated by L1 retrotransposition in cancer genomes. Science. 2014;345:1251343. doi: 10.1126/science.1251343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Helman E, Lawrence MS, Stewart C, Sougnez C, Getz G, Meyerson M. Somatic retrotransposition in human cancer revealed by whole-genome and exome sequencing. Genome Res. 2014;24:1053–63. doi: 10.1101/gr.163659.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Doucet-O’Hare TT, Rodić N, Sharma R, et al. LINE-1 expression and retrotransposition in Barrett’s esophagus and esophageal carcinoma. Proc Natl Acad Sci U S A. 2015;112:E4894–E4900. doi: 10.1073/pnas.1502474112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Doucet-O’Hare TT, Sharma R, Rodić N, Anders RA, Burns KH, Kazazian HH., Jr Somatically acquired LINE-1 insertions in normal esophagus undergo clonal expansion in esophageal squamous cell carcinoma. Hum Mutat. 2016;37:942–54. doi: 10.1002/humu.23027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rodić N, Steranka JP, Makohon-Moore A, et al. Retrotransposon insertions in the clonal evolution of pancreatic ductal adenocarcinoma. Nat Med. 2015;21:1060–4. doi: 10.1038/nm.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ewing AD, Gacita A, Wood LD, et al. Widespread somatic L1 retrotransposition occurs early during gastrointestinal cancer evolution. Genome Res. 2015;25:1536–45. doi: 10.1101/gr.196238.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Solyom S, Ewing AD, Rahrmann EP, et al. Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res. 2012;22:2328–38. doi: 10.1101/gr.145235.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pitkänen E, Cajuso T, Katainen R, et al. Frequent L1 retrotranspositions originating from TTC28 in colorectal cancer. Oncotarget. 2014;5:853–9. doi: 10.18632/oncotarget.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bozic I, Antal T, Ohtsuki H, et al. Accumulation of driver and passenger mutations during tumor progression. Proc Natl Acad Sci U S A. 2010;107:18545–50. doi: 10.1073/pnas.1010978107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Miki Y, Nishisho I, Horii A, et al. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res. 1992;52:643–5. [PubMed] [Google Scholar]
- 86.Levin HL, Moran JV. Dynamic interactions between transposable elements and their hosts. Nat Rev Genet. 2011;12:615–27. doi: 10.1038/nrg3030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bourc’his D, Bestor TH. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature. 2004;431:96–9. doi: 10.1038/nature02886. [DOI] [PubMed] [Google Scholar]
- 88.Siomi MC, Sato K, Pezic D, Aravin AA. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 2011;12:246–58. doi: 10.1038/nrm3089. [DOI] [PubMed] [Google Scholar]
- 89.Trono D. Transposable elements, polydactyl proteins, and the genesis of human-specific transcription networks. Cold Spring Harb Symp Quant Biol. 2015;80:281–8. doi: 10.1101/sqb.2015.80.027573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wolf D, Goff SP. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature. 2009;458:1201–4. doi: 10.1038/nature07844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wolf G, Greenberg D, Macfarlan TS. Spotting the enemy within: targeted silencing of foreign DNA in mammalian genomes by the Krüppel-associated box zinc finger protein family. Mob DNA. 2015;6:17. doi: 10.1186/s13100-015-0050-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wolf G, Yang P, Füchtbauer AC, et al. The KRAB zinc finger protein ZFP809 is required to initiate epigenetic silencing of endogenous retroviruses. Genes Dev. 2015;29:538–54. doi: 10.1101/gad.252767.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jacobs FM, Greenberg D, Nguyen N, et al. An evolutionary arms race between KRAB zinc-finger genes ZNF91/93 and SVA/L1 retrotransposons. Nature. 2014;516:242–5. doi: 10.1038/nature13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Moldovan JB, Moran JV. The zinc-finger antiviral protein ZAP inhibits LINE and Alu retrotransposition. PLoS Genet. 2015;11(5):e1005121. doi: 10.1371/journal.pgen.1005121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Goodier JL, Pereira GC, Cheung LE, Rose RJ, Kazazian HH., Jr The broad-spectrum antiviral protein ZAP restricts human retrotransposition. PLoS Genet. 2015;11(5):e1005252. doi: 10.1371/journal.pgen.1005252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Goodier JL, Zhang L, Vetter MR, Kazazian HH., Jr LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol Cell Biol. 2007;27:6469–83. doi: 10.1128/MCB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hu S, Li J, Xu F, et al. SAMHD1 inhibits LINE-1 retrotransposition by promoting stress granule formation. PLoS Genet. 2015;11(7):e1005367. doi: 10.1371/journal.pgen.1005367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Perepelitsa-Belancio V, Deininger P. RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat Genet. 2003;35:363–6. doi: 10.1038/ng1269. [DOI] [PubMed] [Google Scholar]
- 99.Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–74. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 100.Chuong EB, Elde NC, Feschotte C. Regulatory activities of transposable elements: from conflicts to benefits. Nat Rev Genet. 2017;18:71–86. doi: 10.1038/nrg.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]