Abstract
Slits ligands and their Robo receptors are involved in quite disparate cell signaling pathways that include axon guidance, cell proliferation, cell motility and angiogenesis. Neural crest cells emerge by delamination from neural cells in the dorsal neural tube, and give rise to various components of the peripheral nervous system in vertebrates. It is well established that these cells change from a non-migratory to a highly migratory state allowing them to reach distant regions before they differentiate. However, but the mechanism controlling this delamination and subsequent migration are still not fully understood. The repulsive Slit ligand family members, have been classified also as true tumor suppressor molecules. The present study explored in further detail what possible Slit/Robo signals are at play in the trunk neural cells and neural crest cells by carrying out a microarray after Slit2 gain of function in trunk neural tubes. We found that in addition to molecules known to be downstream of Slit/Robo signaling, there were a large set of molecules known to be important in maintaining cells in non-motile, epithelia phenotype. Furthermore, we found new molecules previously not associated with Slit/Robo signaling: cell proliferation markers, Ankyrins and RAB intracellular transporters. Our findings suggest that neural crest cells use and array of different Slit/Robo pathways during their transformation from non-motile to highly motile cells.
Keywords: Neural tube cells, Neural crest, Cell migration, Cell motility, Slit2 and Robo signaling
1. Introduction
Slits ligands and their Robo receptors are large proteins involved in quite disparate cell signaling pathways that include axon guidance, cell proliferation, cell motility and angiogenesis (Blockus and Chedotal, 2016; Dickinson and Duncan, 2010). Initially discovered as axonal guidance molecules, Slits, and especially Slit2, inhibit neural crest migration and are true tumor suppressor genes (Dallol et al., 2003; Dickinson et al., 2004). They attenuate cancer metastasis progression by regulating beta-catenin expression as well as regulate intermediate progenitor cell transition in brain cortex and target Ezh2 repression (Chatterjee et al., 2001; Yu et al., 2010). However, though we know relevant key downstream signals from Slit molecules in these processes in different cell lineages (Blockus and Chedotal, 2016), we do not have an overall understanding of the downstream signals for Slit2 inside neural crest cells.
Neural crest cells (NCC) start as a non-migratory population from the dorsal portion of the neural tube in the developing embryo. These cells undergo an epithelial-to-mesenchymal transition (EMT) process that changes these cells from differentiated epithelial cells into motile, mesenchymal cells (Guarino et al., 2007). Understanding EMT has farther repercussions that go beyond understanding this process as one that plays during development when NCC are migrating throughout the embryo because EMT changes are what transform cancer cells into metastatic aggressive cancers (Kitzing et al., 2007; Olson and Sahai, 2008; Thiery et al., 2009 #21915). Neural crest cells (NCC) are characterized by their persistent motility in reaching their targets and differentiating into a vast number of cells types (Kulesa and Gammill, 2010; Rogers et al., 2012; Selleck et al., 1993). However, EMT is not the most crucial step in NCC development, but their special migratory pattern is what sets them apart from other mesenchymal cells. Neural crest cells move at probably the fastest speed in an embryo, travel along specific pathways (ventromedial and dorsolateral), in continuous waves and finally stop at their destination to give rise to very different types of cells (Serbedzija et al., 1989 #11287; Bronner-Fraser and Fraser, 1988 #1494; Baggiolini et al., 2015 #14623; Gammill and Roffers-Agarwal, 2010 #16598). Central to their successful migration is the timely coordination of a complex regulatory network of signals that regulate: cytoskeleton re-arrangements, transcription factors and cell adhesion molecules distribution (Sauka-Spengler and Bronner-Fraser, 2008; Taneyhill, 2008).
Our lab has been interested in the role that Slit molecules play during neural crest development (Giovannone et al., 2012; Zuhdi et al., 2015). Of especial relevance is the fact that pre-migratory neural crest cells (NCC) express Slits and Robo receptor simultaneously (Giovannone et al., 2012). While we know that trunk neural crest cells are repelled by Slit molecules (Giovannone et al., 2012; Vaughen and Igaki, 2016; Zuhdi et al., 2015), and that neural crest cells over-expressing Robo1-Δ-67 do not migrate along their ventromedial pathway (Jia et al., 2005), we still do not have a clear picture among the many possible downstream pathways of Robo, which ones are involved in these responses (Blockus and Chedotal, 2016). Are Slit molecules modulating the proliferation of neural crest cells? What are the cytoskeletal changes that Robo signaling causes in pre-migratory neural crest cells before delamination? Do Slit molecules alter their cytoskeleton organization? In this study, we aimed to answer these questions by doing a Slit microarray after constitutively expressing Slit2 in trunk neural tube cells. Our results showed many new gene candidate interactions via Slit/Robo signaling, especially for trunk neural crest cells.
2. Results
2.1. Results from a chicken neural tube microarray after Slit2 overexpression
Past studies from our lab showed evidence that Slit family ligands have a role in the timely migration of trunk neural crest cells (Giovannone et al., 2012). In order to find the players involved in Slit/Robo signaling during the initiation of migration of trunk neural crest cells we performed a neural tube/pre-migratory neural crest microarray after Slit2 electroporation in chicken HH13–14 embryo neural tubes (Fig. 1A).
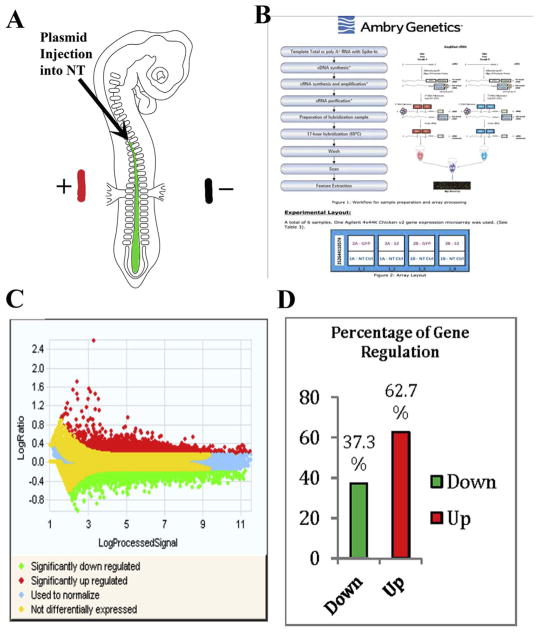
Fig. 1.
Microarray of Slit2 produced log ratios, expression and classification of genes.
(A) Cartoon of a chicken embryo electroporation. In green we show the putative plasmid solution with fast green being injected inside the neural tube. And the ± shows the position of the electrodes when giving them the electric pulses necessary for the plasmid to enter cells in the NT. (B) Workflow and sample preparation and array processing. For the microarray we processed 6 Total RNA samples (1 Control, 2 experimental in duplicates) for gene expression. RNA quality and concentration was determined on the Agilent Bioanalyzer and NanoDrop spectrophotometer. One-color labeling reactions were prepared using the Agilent Two-Color Protocol (Version 6.6) with 400 ng total RNA input. (C) Log ratio values collected after microarray show significantly upregulated genes (red), significantly down regulated genes (green), and not differentially expressed (yellow). Control samples were labeled with Cy5 and Experimental sample labeled with Cy3. RNA Spike A mix was used for Cy3 (experimental) samples and RNA spike B mix was used for all Cy5 (Control) samples and scanned at 3 μM resolution on an Agilent G2565CA High Resolution Scanner. (D) Percentage of gene expression upon Slit2 gain of function in microarray. The bar graphed represents 37.3% down-regulation genes and 62.7% of incidents generated up-regulated genes of NCCs Slit2 GOF vs control.
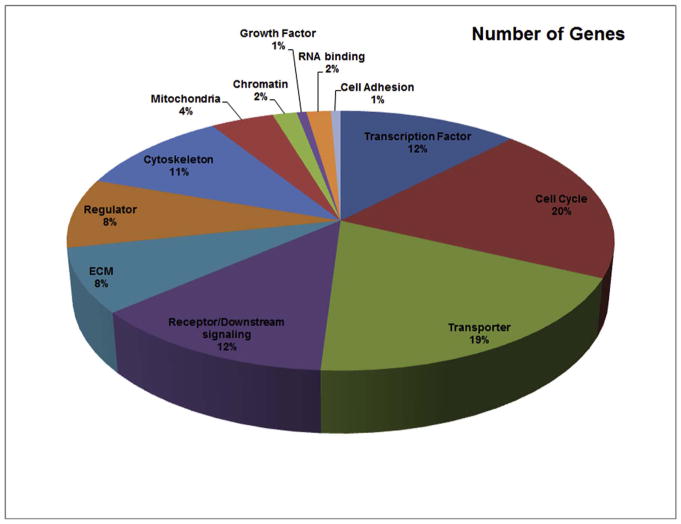
This screen allowed us to collect old and new information on genes that are regulated during Slit2 gain of function (GOF) in trunk neural tubes. A heat-map analysis for all detected gene expression measurements allowed us to observe the overall differential gene expression, where yellow corresponds to no difference, green for down-regulated and red for up-regulated genes (Fig. 1B). The genes with fold change values of ≤2, and ≥2 were plotted and the generated normalization is indicated by the blue color. Overall, we found that in 37.3% of cases, Slit2 GOF down-regulated genes, compared with 62.7% of up-regulated genes (Fig. 1C). Further analysis with IPA software provided a pie chart with biological distribution of genes based on their functions and percentage of the gene expression microarray data (Fig. 2). From this distribution of genes we found that among the many genes with increased or decreased expression after Slit2 constitutive expression, around 40% are involved in cytoskeleton, ECM, Rho pathways (included under heading “Receptor/Downstream signaling”) and 20% shared responses with cancer genes (Supp. Fig. 1). Interestingly, cell cycle molecules (i.e. cdks, STATs, etc.) and transporting machinery (i.e. RABs) amounted each to 19% of total affected genes.
Fig. 2.
GO analysis of biological function and the percentage of the genes in each category.
(A) Heat map representation of microarray gene signaling. A two-color heat-map, with the brightest green, yellow, and brightest red colors of the color scale used for values to generate up and down regulation of microarray samples. (B).
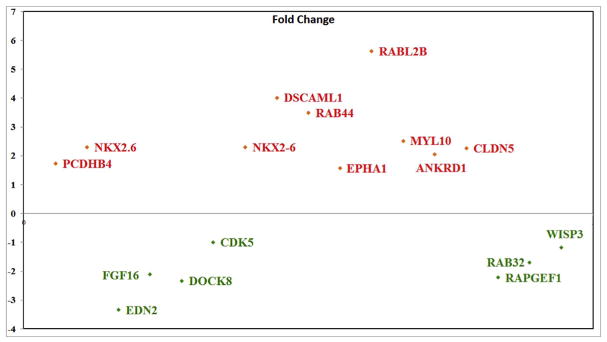
Among the large number of genes showing substantial increase between control un-transfected neural tube and neural tube over-expressing Slit2, the most notables because they have been shown to be involved in mesenchymal cell transformation were Nkx2.6, Fibronectin-1 (FN1), Claudin5, EphA1 and Cadherin-17 (Cdh17), while endothelin-2 (Edn2), Dock8, cdk5, Pax7 and FGF16 were reduced (see Fig. 3 and Table 1). We grouped changed genes based on their relation to cell shape, motility, etc. In Table 1: A) Those known to be involved in neural crest epithelial-to-mesenchymal transition (EMT): cadherins, catenin, Ezh1, Notch1 TGFbeta, ADAMs, matrix metallopeptidases (MMPs), Dock5 and Claudins among others (Powell et al., 2013; Rabadan et al., 2013) (see also Supp. Table I). B) Those known to be important in regulating cytoskeleton: Rabl2B, AnkyrinD1 and DSCAML1, WASP, contactin. C) Those also known to be downstream of classic Slit-Robo signaling or integrins (Supp. Fig. 1 and 2). D) Finally, those molecules previously not associated with Robo signaling like Claudins, Shh, HoxA, ezrin, merlin, Delta, GDF5, Bcl2-A1, DSCAML1, Nkx2.6, Otx2, NCAM2, and glial genes (S100, FABP, gcm2), (see also Supp. Table I).
Fig. 3.
Genes with marked expression.
Genes with substantial expression difference of −2<x>+2 between control and Slit2 treatment were graphed.
Table 1.
List shows the most important and/or interesting genes changed in our Slit2 GOF microarray. The fold change corresponds to the change (increase or decrease) compared with control neural tubes.
| Gene Symbol | Gene Name | |
|---|---|---|
| Rho proteins | ARHGEF4 | Rho guanine nucleotide exchange factor (GEF) 4 |
| LOC417537 | merlin-like | |
| EZR | ezrin | |
| ARHGEF12 | Rho guanine nucleotide exchange factor (GEF) 12 | |
| RAC2 | ras-related C3 botulinum toxin substrate 2 (rho family) | |
| KALRN | kalirin, RhoGEF kinase | |
| PAK6 | p21 protein (Cdc42/Rac)-activated kinase 6 | |
| RHOH | ras homolog gene family, member H | |
| RAB44 | ras-related protein Rab-44-like | |
| RAB11B | RAB11B, member RAS oncogene family | |
| ARHGAP15 | Rho GTPase activating protein 15 | |
| Cell Division | CDK5 | cyclin-dependent kinase 5 |
| NOTCH1 | notch 1 | |
| MYCL1 | v-myc myelocytomatosis viral oncogene homolog 1 | |
| CDK2 | cyclin-dependent kinase 2 | |
| OLIG3 | oligodendrocyte transcription factor 3 | |
| DLK2 | delta-like 2 homolog (Drosophila) | |
| EZH1 | enhancer of zeste homolog 1 (Drosophila) | |
| SPRY1 | sprouty homolog 1,antagonist of FGF signaling (Drosophila) | |
| PCGF5 | polycomb group ring finger 5 | |
| G0S2 | G0/G1switch 2 | |
| JAG2 | jagged 2 | |
| STAT4 | signal transducer and activator of transcription 4 | |
| NEDD1 | neural precursor cell expressed, developmentally down-regulated 1 | |
| GAS2 | growth arrest-specific 2 | |
| CDKAL1 | CDK5 regulatory subunit associated protein 1-like 1 | |
| DNA Modifiers | HDAC9 | Histone deacetylase 9 |
| EZH1 | Enhancer of zeste homolog 1 | |
| JMJD6 | Jumonji domain containg 6 | |
| MBD2 | Methyl-CpG binding domain protein 2 | |
| HIST2HAC | Histone cluster 2, H2ac | |
| Cytoskeleton | ANKRD33B | ankyrin repeat domain 33B |
| ANKRD10 | ankyrin repeat domain 10 | |
| TUBB | tubulin, beta class I | |
| ACTB | actin, beta | |
| COL2A1 | collagen, type II, alpha 1 | |
| CNTN4 | contactin 4 | |
| ACTN1 | actinin, alpha 1 | |
| VIM | vimentin | |
| ACTBL2 | actin, beta-like 2 | |
| ANK3 | ankyrin 3, node of Ranvier (ankyrin G) | |
| TUBA8 | tubulin, alpha 8 | |
| ACTBL2 | actin, beta-like 2 | |
| ANKS4B | ankyrin repeat and sterile alpha motif domain containing 4B | |
| ACTN2 | actinin, alpha 2 | |
| LOC422323 | wiskott-Aldrich syndrome protein family member 3-like | |
| GFAP | glial fibrillary acidic protein | |
| Receptors | FGFR1 | Firoblast growth factor receptor 1 |
| EFNB1 | Ephrin-B1 | |
| EPHA6 | EPH receptor A6 | |
| RET | Ret proto-oncogene | |
| FZD5 | Frizzled family receptor 5 | |
| CXCR2 | Chemokine (C-X-C motif) receptor 2 | |
| FGFR3 | Fibroblast growth factor receptor 3 | |
| TLR5 | Toll-like receptor 5 | |
| CXCR4 | Chemokine (C-X-C motif) receptor 4 | |
| SEMA3E | semaphorin 3E | |
| EDN1 | Endothelin 1 | |
| MET | Met proto-oncogene (hepatocyte growth factor receptor) | |
| ERBB4 | v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) | |
| TLR2-2 | Toll-like receptor 2 | |
| CELSR3 | Cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog) | |
| CSF2RA | Colony stimulating factor 2 receptor, alpha, low-affinity | |
| EPHA1 | EPH receptor A1 | |
| ERBB4 | v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) | |
| TLR2-2 | Toll-like receptor 2 | |
| GFRA3 | GDNF family receptor alpha 3 | |
| Transcription Factors | MYCL1 | v-myc myelocytomatosis viral oncogene homolog 1 |
| MYCT1 | myc target 1 | |
| FOXO1 | forkhead box O1 | |
| GATA5 | GATA binding protein 5 | |
| OLIG3 | oligodendrocyte transcription factor 3 | |
| HOXA2 | homeobox A2 | |
| KLF7 | Kruppel-like factor 7 (ubiquitous) | |
| PHOX2B | paired-like homeobox 2b | |
| LHX2 | LIM homeobox 2 | |
| FOXO1 | forkhead box O1 | |
| STAT4 | signal transducer and activator of transcription 4 | |
| SIX6 | SIX homeobox 6 | |
| GFI1 | growth factor independent 1 transcription repressor | |
| FBXO32 | F-box protein 32 | |
| NKX2-6 | NK2 homeobox 6 |
In order to detect genes that are shared by different functions we arranged the data according to Venn diagrams. We specifically explored “cancer” genes since this group encompasses markers for migratory mesenchymal cells, a key characteristic of NCC. 1) Venn diagram of EMT, cell adhesion and cancer grouped genes provided only CDH17 for all three (Fig. 4A). 2) Venn diagram between cytoskeleton, cancer and DNA modifiers did not show a common one, although EZH1 was present between cancer and DNA modifiers (Fig. 4B). 3) Venn diagram between oncogenes, EMT and cell proliferation did not show any common molecule. While EMT and cell proliferation combination showed Notch1 and MycL1 (Fig. 4C). Cell proliferation, cytoskeleton and cell motility did not show any common genes despite the large number of genes and cytoskeletal molecules affected by Slit2. Only highlighted the Ankyrin family members from the microarray (see Table 1) between cytoskeleton and cell proliferation (Fig. 4D). When looking at cell proliferation and cell motility, ELMO2 protein was the only common one. ELMO2 works as a GAP in conjunction with ARL2 (Bowzard et al., 2007), and ELMO1 is known to work with Dock1 in cytoskeletal re-arrangements and cell migration (Gumienny et al., 2001).
Fig. 4.
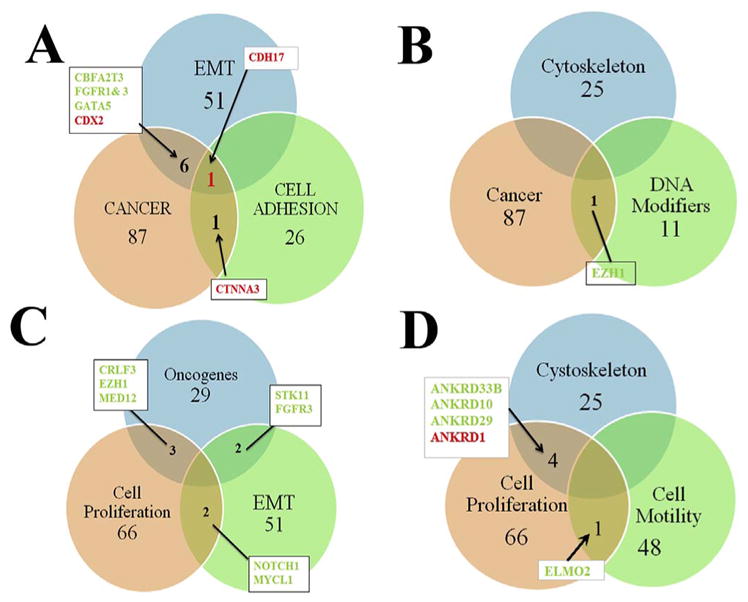
Slit2 microarray results in Venn diagrams arrangements.
We generated Venn diagrams from our microarray as a way of representing group of genes affected by Slit2. A shows diagram for EMT, cancer and cell adhesion molecules. CDH17 was the only common gene among these three groups. B shows cytoskeleton, cancer and DNA modifiers, we only found EZH1 gene from cancer and DNA modifiers combined. C shows oncogenes, EMT and cell cycle gene arrangement. We did not have a triple positive among the three but NOTCH1 and MYCL1 between EMT and cell cycle. D shows cytoskeleton, cell cycle and cell motility combination. Here Ankyrins were common among cell cycle and cytoskeleton, while between cell cycle and motility only ELMO was the common gene. Names in red indicates upregulated genes, in green, downregulated.
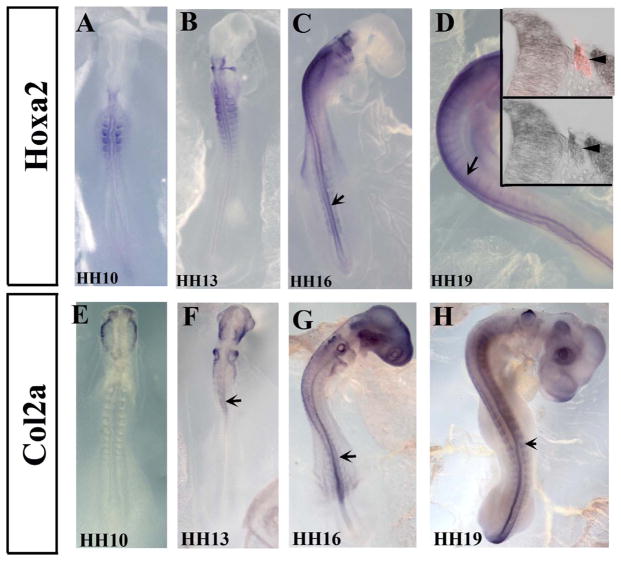
Among the many genes that showed upregulation or downregulation, some are well known to be expressed and are required for neural crest cell (NCC) development or migration (i.e. Notch, Spry1, Merlin, Endothelin, ErbB receptor, Sema proteins, EphA1, etc). But we also observed many that are new for NCC development: HoxA2, EZH1, Collagen 2, Ankyrins, Rabs, etc. (see Table 1). We carried out in situ hybridization experiment for the downregulated HoxA and Col2a to determine their expression in trunk chicken embryos. HoxA2, known to be expressed in neural tubes (Paxton et al., 2010), showed a noticeable, prominent pattern which has no expression in the most anterior portion of the head and strong expression in the hindbrain rhombomeres NCC streams, showed also to be strongly expressed in the trunk. In the HH10 and HH13 its expression is in the trunk somite. But by HH16 and HH19 it is expressed in the neural tube and migrating NCC (Fig. 5A–D). Sections through HH19 embryos at the level of mid-trunk showed that HoxA2 clearly overlapped with migrating NCCs that are HNK1 positive (Fig. 5D insets). Collagen Type II Alpha 1 (Col2A1) expression pattern has been explored in limb development before (Bendall et al., 2003). Here we observed that it is expressed by migrating cranial (Fig. 4E and F) and trunk (Fig. 5G–H) NCC by HH13–19, as well as in the neural tube.
Fig. 5.
HoxA2 and Col2a are expressed by pre-migratory and migrating neural crest cells.
Wholemount in situ hybridization images of HH10–19 chicken embryos with HoxA2 (A–D) and Collagen2a (E–H) anti-sense probes. Chicken embryos showed expression in neural tubes (arrows in A–H) and migrating neural crest cells (arrowheads in A–H). Sections through mid-trunk showed HoxA2 expression in delaminated, migrating HNK1-positive NCC (insert D′ showing in situ and HNK1 in red, and D″ just in situ stain).
2.2. Validation studies
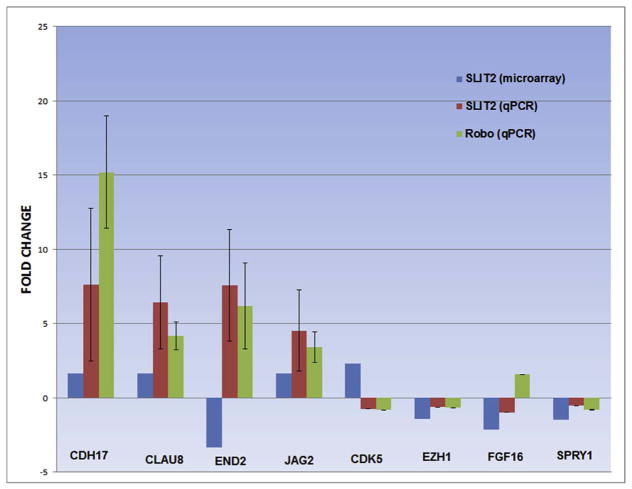
Next we carried out RT-PCR and QPCR of genes selected showing significant fold change expression difference in order to validate microarray findings. We observed that qPCR for CDH-17, Claudin-8, Endothelin-2 (Edn2) and Jagged-2 (Jag2) showed changes in these genes levels in line with our microarray findings (Fig. 6). In addition to repeating it after Slit2 GOF we also included Robo GOF electroporation of neural tubes to determine if overexpression of the ligand and its receptor provide similar results. Indeed, CDH-17 showed a +8 fold change difference for Slit2 treatment in comparison with our control treatment (p < 0.01), and a +16 fold change difference for Robo GOF treatment in comparison with our control treatment (p < 0.01). Claudin-8 showed +4–8 fold change difference for Slit2 and Robo GOF treatments (p < 0.01). Endothelin-2 showed a +~7 fold change for both Slit2 and Robo GOF (p < 0.05). Jagged-2 had a +4 fold change induction for both Slit2 and Robo GOF treatment (p < 0.05). Of this set of molecules, only End-2 gave opposite results compared to our microarray findings.
Fig. 6.
Slit2 microarray validation by qPCR.
Quantification of fold change difference of CDH17, CLAUD8, END2, JAG2 CDK5, EZH1, FGF16 and SPRY1 in chicken neural tubes transfected with control Slit2 or Robo in comparison with microarray results. The validation followed microarray results except for EDN2, CDK5 and FGF16 which showed downregulation in the Slit2 microarray. Each experiment was repeated at least twice and always in triplicate. Asterisks denote significance using t-test: *P < 0.05; **P < 0.01.
In order to further validate our observations, we also tested by qPCR a set of key cell regulators: cdk5, Ezh1, FGF16 and Spry1 after Slit and Robo GOF (Fig. 6). We found that for Slit GOF cdk5, Ezh1, FGF16 and Spry1 all showed down regulation (Fig. 6) as mostly observed in the microarray with the exception of FGF16. When we repeated the same experiment under Robo GOF to further validate Slit GOF findings, we found down regulation of Cdk5, Ezh1, and Spry1; and up regulation of FGF16.
We further looked to corroborate our microarray findings by electroporating neural tubes under Slit2 GOF or Robo GOF (Supp. Fig. 2). We found that Dock8 showed down regulation by Slit2 and Robo. EphA1 and FGF16 exhibited up regulation after Robo GOF, while Slit2 GOF effects were not different from untreated NCC. However, we did not observe changes in Nkx2.6 after Slit2 or Robo GOF.
2.3. The role Robo signaling on neural crest cell cytoskeleton
One of the best known roles of Robo signaling is its effect on the cells cytoskeleton (Blockus and Chedotal, 2016; McConnell et al., 2016; Vaughen and Igaki, 2016). Results from our microarray further supported the hypothesis that Slit/Robo signaling has a wider effect on neural crest cells cytoskeleton than previously thought. Several molecules known to be responsible for cell motility and/or cell shape came from the screen: cdc42, Rac, several Rho guanine nucleotide exchange factors (ARHGEFs) and Rho GTPase activating proteins (ARHGAPs), vimentin, ezrin, merlin, WASP and ankyrin among the best known (Table 1 and Supplementary Table I).
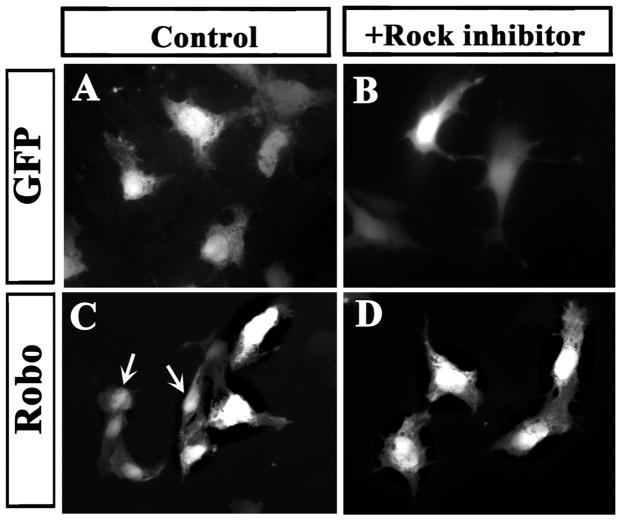
We had previously reported on the effect of Slit2 GOF/LOF altered actin and tubulin cytoskeletal organization in neural crest cells (Giovannone et al., 2012). In order to determine if these changes after GOF were mediated by canonical Rho pathway, especially given the changes observed in our microarray along the Robo-Rho pathway (Supp. Fig. 3), we electroporated Robo (GOF) and explanted neural tubes in the presence or absence of the Rock inhibitor, Y-27632. We found that inhibiting ROCK signaling reversed NCC rounder shape after Robo GOF while cells expressing control GFP did not change their shape (Fig. 7). This is the first time that we have been able to reverse the rounder shape of NCC after Slit2/Robo GOF {Giovannone et al., 2012 #16733}. Our results show that: A) the inhibitor rescued the Robo phenotype of reduced migration in trunk NCC neural crest cells (data not shown). B) Rock inhibitor partially rescued the epithelized phenotype in neural crest cells after Robo GOF. Robo GOF cells (rounder in Fig. 7C) became more mesenchymal after ROCK inhibitor (Fig. 7D).
Fig. 7.
ROCK inhibitions reverses Robo GOF and LOF phenotype.
Chicken neural tubes (HH14–15) were electroporated with control-GFP or Robo plasmids, isolated and cultured for 24 h. A, C shows GFP staining of cells under control conditions and B, D cells treated with ROCK inhibitor. Control GFP cells showed classic mesenchymal, more extended shape while many Robo cells did not exhibit mesenchymal shape but a more rounder one (arrows in C). After ROCK inhibitor, GFP control cell remained looking mesenchymal while Robo GOF cell regained a mesenchymal/migratory shape (D).
2.4. Robo loss of function enhances cell division
Among the genes regulated by Slit2 GOF in our microarray, there was a wide range of molecules involved in cell division: Ezh1, NOTCH1, cdk5, NEDD1, GAS proteins and STAT4 among them (Table 1 and Supplementary Table). Recently Marin and co-workers showed that loss of Robo signaling enhances the proliferation of cortical precursors (Borrell et al., 2012). Importantly for Robo signaling, Sasai and coworkers showed that application of a Rho-associated kinase (ROCK) inhibitor to hES cells significantly diminished apoptosis (Watanabe et al., 2007).
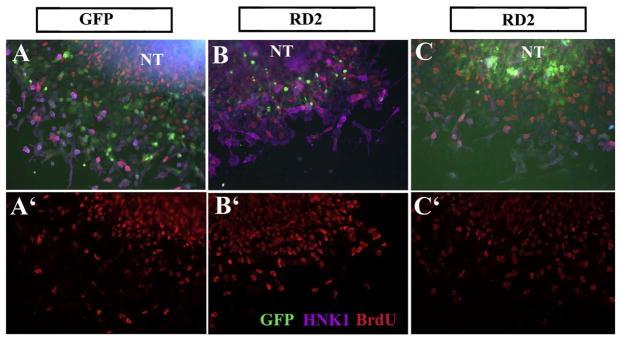
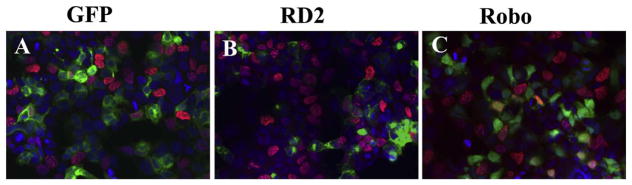
Following on these findings we replicated Borrell’s approach by electroporating a dominant negative form of Robo (RD2) and looking at BrdU incorporation in trunk neural crest cells (Fig. 8). This Robo receptor has a truncated cytoplasmic portion that while it can bind and form heterodimers with other Robos, it blocks any downstream signaling (Stein and Tessier-Lavigne, 2001). Thus, we will refer to this experiments as Robo loss-of-function (Robo LOF). These Robo LOF experiments showed similar findings to Marin’s, Robo LOF almost doubled neural crest cell proliferation (total BrdU incorporation was 42% higher in RD2 cultures compared with GFP control, p < 0.002, N=5 experiments with minimum 20 cells counted at least per treatment). These findings were specific for trunk neural crest cells since repeating RD2 transfections on HEK293 cells and carrying out BrdU labeling did not show any changes in the proliferation rate of these cells, while using a neural crest cell line (SpL201) showed increased BrdU incorporation (Table 2 and Fig. 9).
Fig. 8.
Robo LOF increases BrdU incorporation in trunk NCC.
Chicken neural tubes (HH14–15) were electroporated with control-GFP or RoboD2-GFP (RD2) plasmids, isolated and cultured for 24 h. A–C shows GFP (green), HNK1 (fuchsia) and BrdU (red nuclei) staining of cells under control conditions (A) or RD2 (B–C). Panels A′, B′, C′ shows the BrdU channel only.
Table 2.
BrdU incorporation in cells after RD2 transfection.
| GFP | RD2 | Robo2 | |
|---|---|---|---|
| NCC | 24.31 + 8.11 | 34.56 + 5.29 | ND |
| 142% p < 0.0023 | |||
| SpL201 | 0.17 + 0.07 | 0.22 + 0.05 | 0.18 + 0.06 |
| 133% p < 0.04 | 110% p < Not sig | ||
| HEK293 | 9.22 + 0.08 | 0.20 + 0.02 | 0.19 + 0.04 |
| 87% p < Not sig | 84% p < Not sig |
NCC= Chicken neural tubes (HH14–15) were electroporated with control-GFP, Robo2-GFP (Robo2) or RoboD2-GFP (RD2) plasmids, isolated and cultured for 24 h SpL201=Cell line that is neural crest-like. HEK293=ATCC cell line used as control in several of our experiments. For BrdU treatment, neural crest or cell cultures were treated with 10mM of BrdU for 30min and cultures were fixed and stained with anti-BrdU. Labeling with anti-BrdU cultures were washed, and stained with Alexa594, mounted with Permafluor and visualized using a conventional fluorescence microscope to count cells that are DAPI, GFP and BrdU positive. P < T-test.
Fig. 9.
Robo LOF increases BrdU incorporation in SpL201 cells.
SpL201 cells were transfected with control-GFP or RoboD2-GFP (RD2) plasmids and cultured for 24 h. Next day they were incubated for 30min with BrdU and fixed. A, B, C shows GFP (green), BrdU (red nuclei) and DAPI (blue) staining of cells under control conditions (A) or RD2 (B) or Robo (C).
3. Discussion
Robo signaling is known for its involvement in many cellular processes, especially cell guidance (Ypsilanti et al., 2010). While Slit molecules have been shown to repel neural crest cells (De Bellard et al., 2003; Jia et al., 2005; Shiau and Bronner-Fraser, 2009) and affec their migratory properties (Giovannone et al., 2012; Kirby and Hutson, 2010), we still do not know what are the specific intracellular events in cells responding to Slits. Here we aimed to begin dissecting out which molecules are downstream of Slit/Robo signaling during trunk neural crest cells development by overexpressing Slit2 in trunk neural tubes. This is the first time that a combined screen for neural cells and a specific ligand (Slit2) are explored in combination. Our findings confirmed many of the known signaling pathways for Robo as well as brought new players in neural crest migration.
3.1. Slit/Robo signaling
Overall our screen brought up a set of well-known molecules involved in neural and more pertinent, NCC development. For example, we observed that Slit2 GOF increased Cadherins and catenin expression, molecules known for their role in regulating epithelial morphology in cells (Coles et al., 2007; Lien et al., 2006; Rogers et al., 2013; Takeichi et al., 2000) and their cross talk with Slits (Shiau and Bronner-Fraser, 2009; Tseng et al., 2010). In addition to these two classic major players, we also observed increased levels in a cohort of other important epithelial markers with a counterpart decrease in mesenchymal markers (See Supplementary Table I). These observations in combination with our GO analysis, suggest that Slit/Robo signaling is involved in the process that NCC undergo when becoming mesenchymal/migratory. Recent findings by Vaughen and Igaki in Drosophila further supports this hypothesis. They present data that demonstrates a role for Slit/Robo via E-cadherin (E-cad) deregulation in extruding tumorigenic cells from epithelia (Vaughen and Igaki, 2016). Their findings help to explain how Slit/Robo signaling acts both as tumor suppressor and/or promoter in variety of cancers, and how this can be applied to the process of NCC becoming migratory upon delamination.
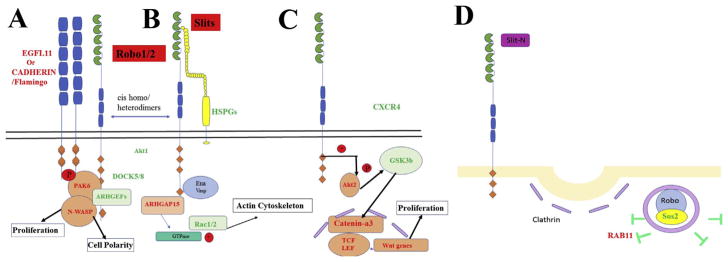
Results from the Venn diagrams suggested a hypothetical Slit/Robo signaling that we want to refer as a “Rho non-canonical pathway”. This hypothesis, that Robo is more than a “repulsive” molecule, has been recently highlighted by Chedotal in a recent review (Blockus and Chedotal, 2016). In our microarray the group with the largest set of shared genes were cancer and mesenchymal markers (Fig. 3A), with CDH17 shared among all 3 with cell adhesion genes. This is a new finding, because while cadherins have been well known for their role in neural crest emigration (Nakagawa and Takeichi, 1995) and migration (Clay and Halloran, 2011) Cadherin17 has not. CDH17 encodes a cadherin-like protein, of which a similar one in Xenopus has been shown to impair migration of cranial neural crest cells (Bartolome et al., 2014; Borchers et al., 2001). What is relevant for our findings is that CDH17 can regulate α2β1 integrin signaling in cell adhesion and proliferation in colon cancer cells (Bartolome et al., 2014), and in our microarray, we found that ITGA4 and ITGB1 were upregulated (α4β1) by Slit2 (Supp. Fig. 4).
Recently, it has been hypothesized that there might be two distinct phases of neural crest EMT: detachment and mesenchymalization, with the latter involving a novel requirement for Sip1 in regulation of cadherin expression during completion of neural crest EMT (Rogers et al., 2013). From the overall findings in here, combined with past research, we would like to propose following Rogers hypothesis, that Slit/Robo signaling is involved in the mesenchymalization of the NCC as they migrate out of the neural tube. Among the known NCC migration pathway receptors there are several that stood out, validating our hypothesis that Slit molecules play relevant roles in regulating the timeliness and migration of NCC. Thus, molecules that correlate with enhanced NCC migration (EDN2, FGFR1, Ret, CXCR4, MET, EphA6, FZD5, NRG2, LPAR6 and NOTCH1) were down regulated by over-expressing Slit2, while molecules that correlate with non-migratory phenotypes were up-regulated (ErbB4, MUC-1, Nidogen, PAK6, ANKRD1, CSF2RA and EphA1). In summary, we find an overall correlation between Slit2 GOF and crucial genes required for proper NCC migration/mesenchymalization.
A surprise finding from the microarray was the upregulation of Toll-like receptor-2 (TLR2) which has not been shown yet to play a role in neural crest migration. These receptors are well-known for their role in innate immunity, but they can also induce pro-metastatic inflammatory responses that could lead to tumor growth and metastasis (Fabbri et al., 2012). The only correlation with NCC has been from a 2008 transcriptome study with isolated human neural crest cells whose profile was compared with murine NCC (Thomas et al., 2008). Analogously to our microarray, they found upregulation of TLRs in NCC from both species. Toll-Like receptors have emerged as important participants in giving shape to the microenvironment of tumors by their involvement in tumorigenic pathways (Ridnour et al., 2013), to the extent that TLR5 agonists are under study as candidates for organ-specific immunotherapy to prevent metastases (Brackett et al., 2016). These findings regarding Toll receptors fit along a role for Slits in impairing proper NCC migration in our GOF experiments and open the door for future research (Giovannone et al., 2012).
The other significant finding was that Slit GOF upregulated two set of family molecules: RABs and Serpins. RABs, (RAB11B and RAB44) which are part of the largest family of small Ras-like GTPases, were among the genes with the highest upregulation in our microarray, while interestingly, we did not observe reduction among any of its members. RAB11B is known for its involvement in Golgi to plasma membrane cycling while RAB44 is unknown (Hutagalung and Novick, 2011). Serpins were initially discovered as serine proteases; the best known are the ones belonging to clade A (antitrypsin type), but clade B (oval-bumin), are known too for regulating other proteases through cleavage (Law et al., 2006; Silverman et al., 2001). More importantly, besides playing relevant roles in some neurological disorders, Massague’s lab recently showed that they are part of a cascade in brain cancer metastasis (Valiente et al., 2014). These two findings combined give a new facet to Slit/Robo signaling: 1) possible role in intra-cellular trafficking and 2) upregulating proteases. Both of these functions are required for normal cell migration (Fortenberry, 2015; Laflamme et al., 2012; Stefansson and Lawrence, 1996; Tzeng and Wang, 2016).
3.2. Slit/Robo signaling in neural crest cytoskeleton
Cellular morphology and cell shape are dependent on cytoskeletal components such as actin, tubulin and intermediate filaments. Data presented here further supports our past studies and a hypothetical role of Slit/Robo signaling for proper cytoskeletal distribution/reorganization and regulation in the initiation and maintenance of migration of neural crest cells.
Another set of important molecules from our microarray are those involved in cytoskeletal motility. Among the most prominent ones we found are: myosin, tropomyosin, titin, tektin and troponin. These findings fit well with our observations that cells during Slit GOF develop impaired motility and their cytoskeletal assembly becomes disrupted (Giovannone et al., 2012). Halloran’s group has shown the need for both myosin and ROCK signaling in neural crest migration. They found that loss of cell adhesion and membrane blebbing appears before filopodia extension and onset of migration, showing an interplay of myosin II and ROCK during NCC mesenchymal transition (Berndt et al., 2008). Our past Slit2 GOF research showed increased membrane blebbing (De Bellard et al., 2003; Giovannone et al., 2012), and our current result that ROCK inhibitors rescue the cell shape phenotype after Slit GOF supports a role for Robo signaling via ROCK kinase in regulating these cell changes in migrating NCC. Thus our current findings closes the loop from these two observations.
Another significant observation from our microarray was the presence of cytoskeletal associated molecules necessary for its reorganization and cell motility. Our past research showed that under Slit LOF conditions there were much fewer actin stress fibers in migrating neural crest cells (Giovannone et al., 2012). Stress fibers are associated with Myosin (Tojkander et al., 2012), in our microarray we found many myosins highly down-regulated, especially the heavy chains (MYH1 and MYH6) or up-regulated, especially their binding proteins (MYH3, MYBPC and MYL10). Another important set of molecules (tropomyosin-1 and cofilin or ADF), have been shown by Gammill and co-workers to be expressed in pre-migratory and migratory neural crest cells (Vermillion et al., 2014). They found cofilin to be necessary for EMT but not for NCC migration. We did not observe cofilin in our microarray, but tropomyosin was downregulated, suggesting that Slit/Robo signaling regulates its expression during NCC migration.
3.3. Robo loss of function affects neural crest proliferation
Findings from this microarray and other labs highlight the complexity of Slit-Robo interactions on the neural crest or other neural progenitors, especially one as unusual as the increase in the number of cell proliferation. Marin and co-workers demonstrated that deleting or silencing Robo increases the number of cortical interneurons (Andrews et al., 2008; Borrell et al., 2012). Our current findings that: 1) Slit GOF affected a set of cell cycle molecules (CDK5, NOTCH1, MYCL1, Spry1, STAT4, GAS2 and NEDD1), and 2) Robo LOF significantly increased proliferation of migrating NCC; leads us to hypothesize that Slit/Robo downstream signaling could also interacts with cell cycle signals. This is not the first time that Slit is shown to have a role in cell proliferation (Borrell et al., 2012; Gu et al., 2016), although we did not observe cell proliferation reduction after Slit2 GOF (Giovannone et al., 2012).
A possible mechanism for the cell proliferation increase in NCC after Robo LOF is via the TGF pathway. In our microarray, TGFB1 was reduced while SMAD7B (TGF-beta signal pathway antagonist) was increased. It has been shown that mammary gland morphogenesis is impaired in Slit and Robo mutants and that the signaling underlying this phenotype involves TGFβRII signaling when Slit-Robo signaling is active, leading to inhibition of pro-proliferative genes (Blockus and Chedotal, 2016; Macias et al., 2011). Another mechanism could be via Notch receptor, which we found reduced in Slit2 GOF. Recently, it has been found that the defects in cardiac cushion derivatives observed in Slit and Robo knockout mice seem to be due to the regulation of Notch pathway by Slit-Robo signaling (Mommersteeg et al., 2015). Thus, it seems that Slit/Robo signaling might be involved in the proliferation of migrating NCC.
3.4. Conclusions
In summary, our overall results suggest that Slit/Robo signaling in neural crest cells participates in a variety of cascade/cross talking signaling pathways among different genes, such as cell division, adhesion, cytoskeletal and known guidance molecules. Fig. 10 shows a modified cartoon that highlights known and new pathways in Slit/Robo signaling tailored for neural crest cells based on current observations (Fig. 10).
Fig. 10.
Results from the Slit2 microarray along Slit2-Robo signaling pathway.
We generated a set of Slit-Robo signaling pathways following Chedotal’s review but followed pathways marking genes that changed in our microarray. The molecules are shown in green or red if reduced/increased respectively.
4. Materials and Methods
4.1. Electroporation, RNA extraction, and Slit2 microarray
Chicken embryos at HH13–14 were electroporated on both sides of the neural tube with pMES-Slit2 plasmid with electrodes on either side of the neural tube 3mm apart. 5 pulses at 20 V, 100 ms on, 50 ms off. Embryos were incubated at 38 °C for 3hpe, removed and rinsed in ice cold ringers before placed in ice cold L15 media for dissection of neural tubes. RNA was isolated from neural tubes (n=5–7) using the GeneJet RNA purification (Fermentas) recommended protocol. For the microarray we processed 6 Total RNA samples (1 Control, 2 experimental in duplicates) for gene expression. RNA quality and concentration was determined on the Agilent Bioanalyzer and NanoDrop spectrophotometer (see supplied Bioanalyzer pdfs). One-color labeling reactions were prepared using the Agilent Two-Color Protocol (Version 6.6) with 400 ng total RNA input. Data was processed through Agilent’s Feature Extraction software version 11.0.1.1 using the protocol GE2_1100_Jul11 and the 44K grid file 026441_D_F_2013012. Quality metric data were produced for each sample and can be analyzed on the supplied Feature Extraction pdf files. The protocol consists of two parts: Labeling of the RNA and hybridization. First, 500–1200 ng of total RNA with the appropriate concentration of RNA spike-in controls is converted into double stranded cDNA using an oligo (dT) primer linked to the T7 promoter sequence and MMLV-RT. The double stranded cDNA is then in vitro transcribed by T7 RNA polymerase, which simultaneously amplifies and incorporates cyanine 3-labeled CTP and cyanine 5-labeled CTP into the resulting cRNA. Control samples were labeled with Cy5 and Experimental sample labeled with Cy3. RNA SpikeA mix was used for Cy3 (experimental) samples and RNA spike B mix was used for all Cy5 (Control) samples. Labeled cRNA is then purified using Qiagen RNeasy columns and the labeling efficiency determined by the NanoDrop spectrophotometer (see Table 2). Next, the labeled cRNA is fragmented and placed on the chicken 4×44K Gene Expression microarray and hybridized at 65 °C for ~17 h. Finally, the arrays are washed and scanned at 3 μM resolution on an Agilent G2565CA High Resolution Scanner.
To convert RNA into cDNA we used SuperScript Vilo cDNA synthesis kit (Invitrogen), which uses Reverse Transcriptase to synthesized DNA from RNA. PCR was carried out in a 20 μL reaction mixture containing 10 nM of the respective primers, APEX 2.0x Taq Master Mix (Genesee Scientific). Analyses of PCR Products: The PCR products were electrophoresed through 2% agarose (BP 160–500 Fisher Scientific) gel, stained with EtBr, run at 35V for 3Hrs with Nanodrop DNA concentrations at ~2 μg/μL and visualized under a UV (DyNA Light) lamp. After, we used Fluorchem analyzer to analyze these bands.
4.2. RT-PCR and qPCR analysis
1 mg of total RNA was used in RT-PCR and qPCR experiments using specific primers (see Table 3). Amplification was done over 35 cycles (95 °C 5 min, 94 °C 30 s, 58 °C 30 s, 72 °C 1 min) for RT-PCR (Eppendorf master cycler gradient). For qPCR (Bio-Rad CFX-96) the following parameters were used: Initial temperature at 95 °C for 3 min, denature at 95 °C for 10 s, annealing at 59 °C for 30 s (+plate read), a melt curve was introduce at 65 °C–95 °C, increment of 0.5 °C for 5 s (+plate read), the program was set for 39 cycles. The following control primers were used: Actin-1 5′-TGGGAC GAT ATG GAI AAI ATC TGG CA -3′, Actin-3R 5′-TCG GGC AAT TCI TAG GAC TTT TC -3′ was used for RT-PCR, and Gapdh forward 5′-GGA CAC TTC AAG GGC ACT GT -3′. Gapdh reverse 5′-TCT CCA TGG TGG TGG AGA CA -3′. For qPCR we used a Bio-rad CFX96 and iTaq Universal SYBR Green Supermix (Bio-Rad) with ROX and fluorescein reference dyes for dye-based quantitative PCR (qPCR). QPCR analysis: ΔΔCT, normalized expression, the expression of the target relative to reference gene(s) normalized to a control sample. ΔΔCT = ΔCT sample – ΔCT control.
Table 3.
List of primers used.
| Name | Sequence | Regulation | Specie |
|---|---|---|---|
| ELM02 | GCC CCT GGC ATC CAT TAT CA (Forward) | Down | Homo Sapiens |
| GGA GTG CTC TTT CAG TCG GA (Reverse) | |||
| D0CK2 | CTT CCA GAC CAG CTC TGA ACT T (Forward) | Up | Homo Sapiens |
| CTC TCA GGA AGA CCC TGT TTT GA (Reverse) | |||
| CDH17 | GGA CAG AGA AGC CGG AAG TC (Forward) | Down | Homo Sapiens |
| GAA CAA GCC CGT GTA GTC CTT (Reverse) | |||
| CLAU8 | GGG ACT TCT ACA ACC CCG TG (Forward) | Up | Gailus Gallus |
| CTG GTT TTC TCA CAG CAG CG (Reverse) | |||
| END2 | ACA GAG TGC TGA TGT GAA GCC (Forward) | Down | Gallus Gallus |
| CTT CCA AGG CAA AAC TGC TGG (Reverse) | |||
| JAG2 | Up | ||
| FMNL1 | GAC CAT CAA GCT GAC CCC A (Forward) | Up | Homo Sapiens |
| ATG ACA AGG CTG AAG CCA GA (Reverse) | |||
| SHH | GGC TGA TGA CTC AGA GGT GTA A (Forward) | Down | Homo Sapiens |
| CTC TGA GTG GTG GCC ATC TT (Reverse) | |||
| EZH1 | GCT TCC TTC ACC CTT TTC ATG CCA CCC (Forward) | Down | Homo Sapiens |
| CGA CGA CCA GAG CAC TTG GAG (Reverse) | |||
| EZH2 | CCC TGA CCT CTG TCT TAC TTG TGG A (Forward) | Homo Sapiens | |
| ACG TCA GAT GGT GCC AGC AAT A (Reverse) | |||
| PCGF5 | CAA GGT GTG GCA ACC AAG TT (Forward) | Up | Homo Sapiens |
| TTC ACG CTC AAG TTC TTG TTC TC (Reverse) | |||
| CXCR4 | CAG GTA GCA GTG ACC CTC TGA (Forward) | Down | Rattus Norvegicus |
| GAA GCA GGG TTC CTT GTT GGA (Reverse) | |||
| CXCR2 | TCC ACT CCC AGC ATC GTA GA (Forward) | Down | Rattus Norvegicus |
| CAG AGT AAA GGG CGG GTC AG (Reverse) | |||
| CXCL12 | CCT CAA CAC TCC AAA CTG TGC (Forward) | Down | Rattus Norvegicus |
| TTG GGC TGT TGT GCT TAC TTG (Reverse) | |||
| TRPC6 | CAG CAG CCA CCT TAT GGC TA (Forward) | Down | Rattus Norvegicus |
| TCA CGA AGA ACT GTC TGC CG (Reverse) | |||
| LAMA1 | GAG ACC TCG ACC CGA TTG TC (Forward) | Down | Rattus Norvegicus |
| CTT CTT CAT CCC AGG GGC AG (Reverse) | |||
| D0CK8 | GCA TGC TTG AAG ATC GCA GC (Forward) | Down | Gallus Gallus |
| GCG GAT TCC TCC AGC ACA TT (Reverse) | |||
| EPHA1 | AGA CAC AAG CAC TGC ACA GG (Forward) | Up | Gallus Gallus |
| GCC AGT GAT CAG TGT CAC CC (Reverse) | |||
| FGF16 | CAC CTC AAG GGC ATC CTG C (Forward) | Up | Gallus Gallus |
| CTG CCA GGC TGA TGA ACT CC (Reverse) | |||
| NKX2-6 | GGT GCT GGT CAG AAA TGG CA (Forward) | Up | Gallus Gallus |
| TCA CCC CGT AGG CAC TGT AG (Reverse) | |||
| CDK5 | GAA GAG GAT CTT CCG GCT GC (Forward) | Down | Gallus Gallus |
| TTG GGC ACC ACA TTG ACG AG (Reverse) |
4.3. In vitro neural crest culture
Electroporated chicken neural tubes (HH14–16) were isolated 2 h post-electroporation (HPE) by incubating trunks in 1.5 mg/ml of Dispase for 30–60 min, followed by dissecting neural tubes into small pieces and culturing on Fibronectin coated (0.1 mg/ml) Nunc-glass chamber slides for 18 h In Dulbecco’s Modified Eagle Medium (DMEM), 10% fetal bovine serum (FBS) and 100 mg/ml and 100U of penicillin and streptomycin respectively. Cultures were then fixed in 4% PFA. For immunostaining, cultures were blocked for 30min with PBS, 1% Triton-X100, 10% goat serum. Primary antibodies were directed against HNK1 for visualizing neural crest cells. Tyrosinated Tubulin antibody was from purchased from Sigma. Alexa-594 phalloidin was purchased from Invitrogen. Secondary antibodies were anti-mouse or anti-rabbit-Alexa 488/594 (Invitrogen). DAPI was used to visualize cell nuclei.
For BrdU treatment, neural crest or cell cultures were treated with 10mM of BrdU (Sigma) was added for 30min and cultures were fixed and stained with anti-BrdU dissolved in double distilled H2O. After 2N HCl for 20 min, washed in Borate buffer, then PBS and incubated at 4 °C overnight with blocking (PBS with 1% triton x-100 and 10% goat serum), next day cultures were incubated with 1:150 mouse IgG anti-BrdU (Beckton Dickinson catalog #347580). The cultures were washed in PBS extensively, treated at room temperature for 3 h with 1:500 Alexa594-labeled goat-anti-mouse IgG before being washed in PBS, mounted with Permafluor and visualized using a conventional fluorescence microscope.
4.4. Electroporation with Slit or Robo
A ~2 mg/ml solution of DNA (pMES-GFP: control, pMES-Slit2: Slit GOF, pCax-Robo1: Robo GOF, pCab-Robo-Delta2: Robo LOF) was injected into the chicken embryos neural tubes using a mouth pipette and immediately electroporated with two 50 ms pulses of 25 mV each. Embryos were sealed with tape and re-incubated for 1–2 in the instance of culturing the neural tube or for 24 h, until fixation for wholemount embryo analysis.
4.5. In situ hybridization
Chicken embryos were fixed in 4% paraformaldehyde (PFA) overnight before being stored in 0.1M Phosphate Buffered Saline (PBS). Patterns of gene expression were determined by whole-mount in situ hybridization using DIG-labeled RNA antisense probes as previously described (Henrique et al., 1995).
4.6. Cell lines transfection
Cultures of SpL201 cells were transfected using Lipofectamine 2000 (Gibco) following the company’s protocol. Cells were treated with Lipofectamine mixtures and the addition of plasmids (pMES-GFP, pCAB-Robo and pCAB-ROBO-Delta2) and incubated for approximately 24 h (or until they reach>80% confluency). All the experiments were done 3–5 biological replicates.
Supplementary Material
Acknowledgments
We thank Chris Walheim and Jennie Soniega-Sherwood for technical assistance; Jane Wu for Slit2 plasmids and Sarah Guthrie for Robo-D2 (Dominant Negative-Robo2) plasmids. This work was supported by an NIH/NINDS AREA grant 1R15-NS060099-03 to MEdB.
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.gep.2018.01.002.
References
- Andrews W, Barber M, Hernadez-Miranda LR, Xian J, Rakic S, Sundaresan V, Rabbitts TH, Pannell R, Rabbitts P, Thompson H, Erskine L, Murakami F, Parnavelas JG. The role of Slit-Robo signaling in the generation, migration and morphological differentiation of cortical interneurons. Dev Biol. 2008;313:648–658. doi: 10.1016/j.ydbio.2007.10.052. [DOI] [PubMed] [Google Scholar]
- Baggiolini A, Varum S, Mateos JM, Bettosini D, John N, Bonalli M, Ziegler U, Dimou L, Clevers H, Furrer R, Sommer L. Premigratory and migratory neural crest cells are multipotent in vivo. Cell Stem Cell. 2015;16(3):314–322. doi: 10.1016/j.stem.2015.02.017. [DOI] [PubMed] [Google Scholar]
- Bartolome RA, Barderas R, Torres S, Fernandez-Acenero MJ, Mendes M, Garcia-Foncillas J, Lopez-Lucendo M, Casal JI. Cadherin-17 interacts with alpha2beta1 integrin to regulate cell proliferation and adhesion in colorectal cancer cells causing liver metastasis. Oncogene. 2014;33:1658–1669. doi: 10.1038/onc.2013.117. [DOI] [PubMed] [Google Scholar]
- Bendall AJ, Hu G, Levi G, Abate-Shen C. Dlx5 regulates chondrocyte differentiation at multiple stages. Int J Dev Biol. 2003;47:335–344. [PubMed] [Google Scholar]
- Berndt JD, Clay MR, Langenberg T, Halloran MC. Rho-kinase and myosin II affect dynamic neural crest cell behaviors during epithelial to mesenchymal transition in vivo. Dev Biol. 2008 Dec 15;324(2):236–244. doi: 10.1016/j.ydbio.2008.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blockus H, Chedotal A. Slit-Robo signaling. Development. 2016;143:3037–3044. doi: 10.1242/dev.132829. [DOI] [PubMed] [Google Scholar]
- Borchers A, David R, Wedlich D. Xenopus cadherin-11 restrains cranial neural crest migration and influences neural crest specification. Development. 2001;128:3049–3060. doi: 10.1242/dev.128.16.3049. [DOI] [PubMed] [Google Scholar]
- Borrell V, Cardenas A, Ciceri G, Galceran J, Flames N, Pla R, Nobrega-Pereira S, Garcia-Frigola C, Peregrin S, Zhao Z, Ma L, Tessier-Lavigne M, Marin O. Slit/Robo signaling modulates the proliferation of central nervous system progenitors. Neuron. 2012;76:338–352. doi: 10.1016/j.neuron.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowzard JB, Cheng D, Peng J, Kahn RA. ELMOD2 is an Arl2 GTPase-activating protein that also acts on Arfs. J Biol Chem. 2007;282:17568–17580. doi: 10.1074/jbc.M701347200. [DOI] [PubMed] [Google Scholar]
- Brackett CM, Kojouharov B, Veith J, Greene KF, Burdelya LG, Gollnick SO, Abrams SI, Gudkov AV. Toll-like receptor-5 agonist, entolimod, suppresses metastasis and induces immunity by stimulating an NK-dendritic-CD8+ T-cell axis. Proc Natl Acad Sci Unit States Am. 2016;113:E874–E883. doi: 10.1073/pnas.1521359113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner-Fraser M, Fraser SE. Cell lineage analysis reveals multipotency of some avian neural crest cells. Nature. 1988;335:161–164. doi: 10.1038/335161a0. [DOI] [PubMed] [Google Scholar]
- Chatterjee B, Li YX, Zdanowicz M, Sonntag JM, Chin AJ, Kozlowski DJ, Valdimarsson G, Kirby ML, Lo CW. Analysis of Cx43alpha1 promoter function in the developing zebrafish embryo. Cell Commun Adhes. 2001;8:289–292. doi: 10.3109/15419060109080739. [DOI] [PubMed] [Google Scholar]
- Clay MR, Halloran MC. Regulation of cell adhesions and motility during initiation of neural crest migration. Curr Opin Neurobiol. 2011;21:17–22. doi: 10.1016/j.conb.2010.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coles EG, Taneyhill LA, Bronner-Fraser M. A critical role for Cadherin6B in regulating avian neural crest emigration. Dev Biol. 2007;312:533–544. doi: 10.1016/j.ydbio.2007.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallol A, Morton D, Maher ER, Latif F. SLIT2 axon guidance molecule is frequently inactivated in colorectal cancer and suppresses growth of colorectal carcinoma cells. Canc Res. 2003;63:1054–1058. [PubMed] [Google Scholar]
- De Bellard ME, Rao Y, Bronner-Fraser M. Dual function of Slit2 in repulsion and enhanced migration of trunk, but not vagal, neural crest cells. J Cell Biol. 2003;162:269–279. doi: 10.1083/jcb.200301041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson RE, Dallol A, Bieche I, Krex D, Morton D, Maher ER, Latif F. Epigenetic inactivation of SLIT3 and SLIT1 genes in human cancers. Br J Canc. 2004;91:2071–2078. doi: 10.1038/sj.bjc.6602222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson RE, Duncan WC. The SLIT-ROBO pathway: a regulator of cell function with implications for the reproductive system. Reproduction. 2010;139:697–704. doi: 10.1530/REP-10-0017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, Lovat F, Fadda P, Mao C, Nuovo GJ, Zanesi N, Crawford M, Ozer GH, Wernicke D, Alder H, Caligiuri MA, Nana-Sinkam P, Perrotti D, Croce CM. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci Unit States Am. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortenberry Y. The role of serpins in tumor cell migration. Biol Chem. 2015;396:205–213. doi: 10.1515/hsz-2014-0254. [DOI] [PubMed] [Google Scholar]
- Gammill LS, Roffers-Agarwal J. Division of labor during trunk neural crest development. Dev Biol. 2010;344:555–565. doi: 10.1016/j.ydbio.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannone D, Reyes M, Reyes R, Correa L, Martinez D, Ra H, Gomez G, Kaiser J, Ma L, Stein MP, de Bellard ME. Slits affect the timely migration of neural crest cells via Robo receptor. Dev Dynam. 2012;241:1274–1288. doi: 10.1002/dvdy.23817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu JJ, Gao GZ, Zhang SM. MiR-218 inhibits the tumorgenesis and proliferation of glioma cells by targeting Robo1. Canc Biomarkers. 2016;16:309–317. doi: 10.3233/CBM-160568. [DOI] [PubMed] [Google Scholar]
- Guarino M, Rubino B, Ballabio G. The role of epithelial-mesenchymal transition in cancer pathology. Pathology. 2007;39:305–318. doi: 10.1080/00313020701329914. [DOI] [PubMed] [Google Scholar]
- Gumienny TL, Brugnera E, Tosello-Trampont AC, Kinchen JM, Haney LB, Nishiwaki K, Walk SF, Nemergut ME, Macara IG, Francis R, Schedl T, Qin Y, Van Aelst L, Hengartner MO, Ravichandran KS. CED-12/ELMO, a novel member of the CrkII/Dock180/Rac pathway, is required for phagocytosis and cell migration. Cell. 2001;107:27–41. doi: 10.1016/s0092-8674(01)00520-7. [DOI] [PubMed] [Google Scholar]
- Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D. Expression of a Delta homologue in prospective neurons in the chick. Nature. 1995;375:787–790. doi: 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]
- Hutagalung AH, Novick PJ. Role of rab GTPases in membrane traffic and cell physiology. Physiol Rev. 2011;91:119–149. doi: 10.1152/physrev.00059.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Cheng L, Raper J. Slit/Robo signaling is necessary to confine early neural crest cells to the ventral migratory pathway in the trunk. Dev Biol. 2005;282:411–421. doi: 10.1016/j.ydbio.2005.03.021. [DOI] [PubMed] [Google Scholar]
- Kirby ML, Hutson MR. Factors controlling cardiac neural crest cell migration. Cell Adhes Migrat. 2010;4:609–621. doi: 10.4161/cam.4.4.13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitzing TM, Sahadevan AS, Brandt DT, Knieling H, Hannemann S, Fackler OT, Grosshans J, Grosse R. Positive feedback between Dia1, LARG, and RhoA regulates cell morphology and invasion. Genes Dev. 2007;21:1478–1483. doi: 10.1101/gad.424807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulesa PM, Gammill LS. Neural crest migration: patterns, phases and signals. Dev Biol. 2010;344:566–568. doi: 10.1016/j.ydbio.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laflamme C, Assaker G, Ramel D, Dorn JF, She D, Maddox PS, Emery G. Evi5 promotes collective cell migration through its Rab-GAP activity. J Cell Biol. 2012;198:57–67. doi: 10.1083/jcb.201112114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law RH, Zhang Q, McGowan S, Buckle AM, Silverman GA, Wong W, Rosado CJ, Langendorf CG, Pike RN, Bird PI, Whisstock JC. An overview of the serpin superfamily. Genome Biol. 2006;7:216. doi: 10.1186/gb-2006-7-5-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien WH, Klezovitch O, Vasioukhin V. Cadherin-catenin proteins in vertebrate development. Curr Opin Cell Biol. 2006;18:499–506. doi: 10.1016/j.ceb.2006.07.001. [DOI] [PubMed] [Google Scholar]
- Macias H, Moran A, Samara Y, Moreno M, Compton JE, Harburg G, Strickland P, Hinck L. SLIT/ROBO1 signaling suppresses mammary branching morphogenesis by limiting basal cell number. Dev Cell. 2011;20:827–840. doi: 10.1016/j.devcel.2011.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell RE, Edward van Veen J, Vidaki M, Kwiatkowski AV, Meyer AS, Gertler FB. A requirement for filopodia extension toward Slit during Robo-mediated axon repulsion. J Cell Biol. 2016 doi: 10.1083/jcb.201509062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mommersteeg MT, Yeh ML, Parnavelas JG, Andrews WD. Disrupted Slit-Robo signalling results in membranous ventricular septum defects and bicuspid aortic valves. Cardiovasc Res. 2015;106:55–66. doi: 10.1093/cvr/cvv040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Takeichi M. Neural crest cell-cell adhesion controlled by sequential and subpopulation-specific expression of novel cadherins. Development. 1995;121:1321–1332. doi: 10.1242/dev.121.5.1321. [DOI] [PubMed] [Google Scholar]
- Olson MF, Sahai E. The actin cytoskeleton in cancer cell motility. Clin Exp Metastasis. 2008;26:273. doi: 10.1007/s10585-008-9174-2. [DOI] [PubMed] [Google Scholar]
- Paxton CN, Bleyl SB, Chapman SC, Schoenwolf GC. Identification of differentially expressed genes in early inner ear development. Gene Expr Patterns. 2010;10:31–43. doi: 10.1016/j.gep.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell DR, Blasky AJ, Britt SG, Artinger KB. Riding the crest of the wave: parallels between the neural crest and cancer in epithelial-to-mesenchymal transition and migration. Wiley interdisciplinary reviews. Syst biol med. 2013;5:511–522. doi: 10.1002/wsbm.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabadan MA, Usieto S, Lavarino C, Marti E. Identification of a putative transcriptome signature common to neuroblastoma and neural crest cells. Developmental neurobiology. 2013;73:815–827. doi: 10.1002/dneu.22099. [DOI] [PubMed] [Google Scholar]
- Ridnour LA, Cheng RYS, Switzer CH, Heinecke JL, Ambs S, Glynn S, Young HA, Trinchieri G, Wink DA. Molecular pathways: toll-like receptors in the tumor microenvironment—poor prognosis or new therapeutic opportunity. Clin Canc Res. 2013;19:1340–1346. doi: 10.1158/1078-0432.CCR-12-0408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers CD, Jayasena CS, Nie S, Bronner ME. Neural crest specification: tissues, signals, and transcription factors. Wiley interdisciplinary reviews. Dev Biol. 2012;1:52–68. doi: 10.1002/wdev.8. [DOI] [PubMed] [Google Scholar]
- Rogers CD, Saxena A, Bronner ME. Sip1 mediates an E-cadherin-to-N-cadherin switch during cranial neural crest EMT. J Cell Biol. 2013;203:835–847. doi: 10.1083/jcb.201305050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauka-Spengler T, Bronner-Fraser M. A gene regulatory network orchestrates neural crest formation. Nat Rev Mol Cell Biol. 2008;9:557–568. doi: 10.1038/nrm2428. [DOI] [PubMed] [Google Scholar]
- Selleck MA, Scherson TY, Bronner-Fraser M. Origins of neural crest cell diversity. Dev Biol. 1993;159:1–11. doi: 10.1006/dbio.1993.1217. [DOI] [PubMed] [Google Scholar]
- Serbedzija GN, Bronner-Fraser M, Fraser SE. A vital dye analysis of the timing and pathways of avian trunk neural crest cell migration. Development. 1989;106:809–816. doi: 10.1242/dev.106.4.809. [DOI] [PubMed] [Google Scholar]
- Shiau CE, Bronner-Fraser M. N-cadherin acts in concert with Slit1-Robo2 signaling in regulating aggregation of placode-derived cranial sensory neurons. Development. 2009;136:4155–4164. doi: 10.1242/dev.034355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PG, Irving JA, Lomas DA, Luke CJ, Moyer RW, Pemberton PA, Remold-O’Donnell E, Salvesen GS, Travis J, Whisstock JC. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J Biol Chem. 2001;276:33293–33296. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- Stefansson S, Lawrence DA. The serpin PAI-1 inhibits cell migration by blocking integrin [alpha]v[beta]3 binding to vitronectin. Nature. 1996;383:441–443. doi: 10.1038/383441a0. [DOI] [PubMed] [Google Scholar]
- Stein E, Tessier-Lavigne M. Hierarchical organization of guidance receptors: silencing of netrin attraction by slit through a Robo/DCC receptor complex. Science. 2001;291:1928–1938. doi: 10.1126/science.1058445. [DOI] [PubMed] [Google Scholar]
- Takeichi M, Nakagawa S, Aono S, Usui T, Uemura T. Patterning of cell assemblies regulated by adhesion receptors of the cadherin superfamily. Philos Trans R Soc Lond B Biol Sci. 2000;355:885–890. doi: 10.1098/rstb.2000.0624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneyhill LA. To adhere, or not to adhere: the role of Cadherins in neural crest development. Cell Adhes Migrat. 2008;2:1–8. doi: 10.4161/cam.2.4.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RYJ, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Thomas S, Thomas M, Wincker P, Babarit C, Xu P, Speer MC, Munnich A, Lyonnet S, Vekemans M, Etchevers HC. Human neural crest cells display molecular and phenotypic hallmarks of stem cells. Hum Mol Genet. 2008;17:3411–3425. doi: 10.1093/hmg/ddn235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tojkander S, Gateva G, Lappalainen P. Actin stress fibers – assembly, dynamics and biological roles. J Cell Sci. 2012;125:1855–1864. doi: 10.1242/jcs.098087. [DOI] [PubMed] [Google Scholar]
- Tseng RC, Lee SH, Hsu HS, Chen BH, Tsai WC, Tzao C, Wang YC. SLIT2 attenuation during lung cancer progression deregulates beta-catenin and E-cadherin and associates with poor prognosis. Canc Res. 2010;70:543–551. doi: 10.1158/0008-5472.CAN-09-2084. [DOI] [PubMed] [Google Scholar]
- Tzeng HT, Wang YC. Rab-mediated vesicle trafficking in cancer. J Biomed Sci. 2016;23:70. doi: 10.1186/s12929-016-0287-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valiente M, Obenauf Anna C, Jin X, Chen Q, Zhang Xiang HF, Lee Derek J, Chaft Jamie E, Kris Mark G, Huse Jason T, Brogi E, Massagué J. Serpins promote cancer cell survival and vascular Co-Option in brain metastasis. Cell. 2014;156:1002–1016. doi: 10.1016/j.cell.2014.01.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughen J, Igaki T. Slit-robo repulsive signaling extrudes tumorigenic cells from epithelia. Dev Cell. 2016;39:683–695. doi: 10.1016/j.devcel.2016.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermillion KL, Lidberg KA, Gammill LS. Expression of actin-binding proteins and requirement for actin-depolymerizing factor in chick neural crest cells. Dev Dynam. 2014;243:730–738. doi: 10.1002/dvdy.24105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K, Ueno M, Kamiya D, Nishiyama A, Matsumura M, Wataya T, Takahashi JB, Nishikawa S, Nishikawa S-i, Muguruma K, Sasai Y. A ROCK Inhibitor Permits Survival of Dissociated Human Embryonic Stem Cells. 2007;25:681. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- Ypsilanti AR, Zagar Y, Chedotal A. Moving away from the midline: new developments for Slit and Robo. Development. 2010;137:1939–1952. doi: 10.1242/dev.044511. [DOI] [PubMed] [Google Scholar]
- Yu J, Cao Q, Wu L, Dallol A, Li J, Chen G, Grasso C, Cao X, Lonigro RJ, Varambally S, Mehra R, Palanisamy N, Wu JY, Latif F, Chinnaiyan AM. The neuronal repellent SLIT2 is a target for repression by EZH2 in prostate cancer. Oncogene. 2010 doi: 10.1038/onc.2010.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuhdi N, Ortega B, Giovannone D, Ra H, Reyes M, Asencion V, McNicoll I, Ma L, de Bellard ME. Slit molecules prevent entrance of trunk neural crest cells in developing gut. Int J Dev Neurosci. 2015;41:8–16. doi: 10.1016/j.ijdevneu.2014.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.