Abstract
Knowledge on genetic and environmental (G × E) interaction effects on cardiometabolic risk factors (CMRFs) in children is limited. The purpose of this study was to examine the impact of G × E interaction effects on CMRFs in Mexican American (MA) children (n = 617, ages 6–17 years). The environments examined were sedentary activity (SA), assessed by recalls from “yesterday” (SAy) and “usually” (SAu) and physical fitness (PF) assessed by Harvard PF scores (HPFS). CMRF data included body mass index (BMI), waist circumference (WC), fat mass (FM), fasting insulin (FI), insulin resistance (HOMA-IR), HDL-cholesterol (HDL-C), triglycerides (TG), systolic (SBP) and diastolic (DBP) blood pressure, and metabolic syndrome risk score (MSC). We examined potential G × E interaction in the phenotypic expression of CMRFs using variance component models and likelihood-based statistical inference. Significant G × SA interactions were identified for six CMRFs: BMI, WC, FI, HOMA-IR, MSC, and HDL, and significant G × HPFS interactions were observed for four CMRFs: BMI, WC, FM, and HOMA-IR. However, after correcting for multiple hypothesis testing, only WC × SAy, FM × SAy, and FI × SAu interactions became marginally significant. After correcting for multiple testing, most of CMRFs exhibited significant G × E interactions (Red. G × E model vs. Con. model). These findings provide evidence that genetic factors interact with SA and PF to influence variation in CMRFs, and underscore the need for better understanding of these relationships to develop strategies and interventions to effectively reduce or prevent cardiometabolic risk in children.
Keywords: G × E interaction, genetic variance, genetic correlation, lifestyle modification, physical inactivity, sedentary behavior, childhood obesity
Introduction
Increasing prevalence of cardiometabolic risk factors (CMRFs) such as obesity, and metabolic syndrome (MS) in youth poses an unprecedented public health crisis with profound worldwide impact (Lloyd-Jones et al., 2009). Sedentary behaviors increase the risk of many diseases/disorders such as obesity, type 2 diabetes (T2D), and MS (Knight, 2012). Obesity-related CMRFs now appear in children and adolescents (Fowler et al., 2013; Steinberger et al., 2009), and have been shown to be strong predictors of developing T2D, MS, and CVD in later adulthood (Virdis et al., 2009). Alarmingly, between the years 1980 and 2010, obesity in US children aged 6–11 years has more than doubled, and more than tripled in adolescents aged 12–19 years (Harper, 2006; Ogden, Carroll, Kit, & Flegal, 2012). Since the rising prevalence of CMRFs is a tremendous challenge, there have been attempts to better understand the contributing factors for pediatric CMRFs. Furthermore, the association between sedentary lifestyle and CMRFs in children is controversial because some studies show that a dramatic decrease in physical activity ensues during adolescence (Kimm et al., 2000), and a decrease in physical activity levels appears to be one of the major contributors to the increase in BMI (Brownson, Boehmer, & Luke, 2005).
The rise in obesity among children and adolescents has disproportionately affected Mexican American (MA) children. Recently, our data on CMRFs in MA children aged 6–17 years in San Antonio, Texas revealed a high risk of overweight (53%), obesity (34%), MS (19%), and pre-diabetes (13%) (Fowler et al., 2013). These data stand in stark contrast to comparable National Health and Nutrition Examination Survey (NHANES) 2009–2010 data on U.S. children and adolescents (ages 2–19 years) where the prevalence of obesity was estimated to be 16.9% (Males 17.8% and Females 15.9%) with significant ethnic disparities in obesity prevalence: European Americans = 14.0%, African Americans =24.3%, and MAs = 21.2% (Ogden et al., 2012). Pediatric obesity and insulin resistance are the key factors among the CMRFs and are predictors of adult obesity, T2D, MS and CVD (Lloyd-Jones et al., 2009; Virdis et al., 2009). Particularly, abdominal fat is an important risk factor underlying the many facets of the MS and related factors, such as glucose intolerance, hyperinsulinemia, and hypertriglyceridemia (Despres et al., 2008).
CMRFs are influenced by both genetic and environmental factors (i.e., dietary intake and physical activity), and their interactions (Andreasen & Andersen, 2009; Arya et al., 2015; Jermendy et al., 2011; Santos et al., 2013). Environmental factors such as lifestyle and healthy/unhealthy behaviors and obesogenic environments (e.g., fast food restaurants, sitting or lying down and watching TV, and TV viewing time) (Bai et al., 2016; Santos et al., 2014) substantially influence variation in CMRFs. Thus, physical activity, physical fitness, and sedentary behavior are thought to be important determinants of CVD, T2D, hypertension, and MS (Blair & Haskell, 2006; DeFina et al., 2015; Hamilton, Hamilton, & Zderic, 2007; Lloyd-Jones et al., 2009; Ortega, Ruiz, Castillo, & Sjostrom, 2008). Most importantly, as (Ortega et al., 2008) stated, their role is critical during the developmental stages of childhood and adolescence since lifestyle and healthy/unhealthy behaviors are established during these early years with potential impact on behavior and health status later in life.
Sedentary lifestyle has been identified as a major risk factor for several of CMRFs including obesity, insulin resistance and heart disease. Several epidemiological and intervention studies have identified the role of sedentary activity and physical fitness for overweight and obesity in children and adolescents (Butte, Cai, Cole, & Comuzzie, 2006; Butte, Puyau, Adolph, Vohra, & Zakeri, 2007; Chung, LPY, ECK, & JWY, 2014; Esmaeilzadeh & Ebadollahzadeh, 2012; Ortega et al., 2008). On the other side of the equation, genetic factors explain 20–86% of variation in body weight, BMI, and fat mass (Choquet & Meyre, 2011; Fowler et al., 2013; Lloyd-Jones et al., 2009; Sonestedt et al., 2009). However, there is a paucity of data on how gene by environment (G × E) interaction influences CMRFs, especially in children and adolescents (Fisher, Smith, van Jaarsveld, Sawyer, & Wardle, 2015; Graff et al., 2011; T. Huang & Hu, 2015). Therefore, the purpose of this study was to examine G × E (i.e., G × PF and G × SA) interaction effects on CMRFs in MA children and adolescents (N=617), who participated in our San Antonio Family Assessment of Metabolic Risk Indicators in Youth (SAFARI) Study. Given that this study utilizes the family-based data, it has well-known advantages to assess the extent to which the clustering of CMRFs is influenced by common genetic factors and their interactions with shared common environments (Vincent P. Diego, Kent Jr, & Blangero, 2015; J. L. Hopper, 1993; J. L. Hopper, Visscher, P.M., 2005).
Subjects and Methods
The Study Subjects: San Antonio Family Assessment of Metabolic Risk Indicators in Youth (SAFARI) Study
We conducted a genetic epidemiologic study of MS, and its related cardio-metabolic risk factors (CMRFs) in Mexican American Children, called the San Antonio Family Assessment of Metabolic Risk Indicators in Youth (SAFARI) Study, involving 673 MA children (ages 6–17 years; 401 nuclear families; 3,664 relative pairs [e.g., sibling pairs = 383, and first-cousin pairs = 550]), which was described in detail previously (Fowler et al., 2013). SAFARI children were the offspring of predominantly low-income extended families previously enrolled in one of three community-based genetic epidemiologic studies of MA adults in San Antonio, Texas, as detailed in (Fowler et al., 2013). As part of the SAFARI study, we collected demographic, phenotypic, and environmental data including nutrition and physical activity information from SAFARI children using standard procedures. However, for this study, we only considered 617 SAFARI children for whom environmental (i.e., physical fitness and sedentary activity) data were available.
Phenotype Data
Family history, demographic, phenotypic, and environmental data were obtained for SAFARI participants as reported earlier by (Fowler et al., 2013). Blood samples were obtained after a 10-h overnight fast and used to measure metabolic parameters including fasting plasma glucose and insulin, high-density lipoprotein cholesterol (HDL-C), and triglycerides (TG) following standard protocols. Anthropometric measurements such as height, weight, body mass index (BMI), waist circumference (WC), and blood pressure (systolic (SBP) and diastolic blood pressure (DBP) were collected using standardized protocols, as described previously (Fowler et al., 2013). Fat mass (FM) was measured using dual-energy-X-ray absorptiometry (DXA Hologic). Using FPG and FI values, homeostasis model of assessment - insulin resistance (HOMA-IR) was derived using the University of Oxford Diabetes Trials Unit HOMA2-IR calculator (http://www.dtu.ox.ac.uk/homacalculator) (Matthews et al., 1985). The number of MS components (MSC) that we previously assessed was used as a semi-quantitative trait (range: 0–5 in our data), and the MS components include abdominal adiposity, hyperinsulinemia, glucose intolerance, hypertriglyceridemia, low HDL-C, and elevated SBP and/or DBP (Fowler et al., 2013).
Assessment of Sedentary Activity (SA)
Sedentary activities (SAs) were measured using the Girls Health Enrichment Multi-Site Studies (GEMS) Activity Questionnaire (GAQ) developed by Treuth et al. (Treuth et al., 2004). Sedentary activities performed yesterday (SAy) and usually (SAu), both having five categories of estimated time spent (none, <30 min, 30 min to 1hr, 1–3 hrs, >3 hrs), were recorded; and codes were assigned for each of the time categories as follows: none = 0; <30 min = 0.25, 30 min–1hr = 0.75, 1–3 hrs = 1.5, and >3 hrs = 2.5. The GAQ summary scores for sedentary activity (SAy and SAu) were computed as the sum of weights for all sedentary activities divided by the number of non-missing items. The numeric values assigned to answers were arbitrarily selected a priori, based on judgment to reflect monotonically increasing values (Treuth et al., 2004). Simulation studies have shown that ordinal variables with 5 categories can be safely regarded as continuous in that the type I error rate for hypothesis testing involving such traits is well controlled, power is sufficient (≥ 0.80), and associated parameter estimates are unbiased and efficient (Rhemtulla, Brosseau-Liard, & Savalei, 2012; Savalei & Rhemtulla, 2013). Thus, SAy and SAu were both treated as continuous variables in our analyses.
Assessment of Physical Fitness
We used a modified Harvard Step Test (Trevino et al., 2004) to assess physical fitness (PF). A child was asked to step up and down (both feet) from a stool (30 cm high) at the rate of thirty cycles per minute for 5 minutes or until exhaustion. Heart rates were recorded at 0, 1, and 2 minutes post exercise. The Harvard physical fitness Score (HPFS) was calculated as: total duration of exercise in seconds × 100, divided by the sum of three post-exercise heart rates.
SNP Selection and Genotyping
We selected 96 previously identified GWAS/literature reported risk SNPs for cardio-metabolic traits (Kathiresan et al., 2008; Kooner et al., 2011; Willer et al., 2008)(http://www.genome.gov/gwastudies) as described in detail in our ongoing unpublished work (Farook et al.) for this study. Genomic DNA was extracted from blood samples collected from the MA children who participated in the SAFARI study, using Qiagens’s QIAmp DNA 96 BLOOD KIT or QIAmp DNA BLOOD MINIKIT according to the manufacturer’s protocol. Genotyping of the 96 SNPs was done using the Illumina’s Goldengate Veracode Assay per the manufacturer’s protocol (Illumina, San Diego, CA; www.illumina.org).
Statistical Analysis
We used the program SOLAR (http://www.txbiomedgenetics.org/solar) to perform genetic analyses including estimation of heritabilities (h2) and G × E interaction modeling using variance components approaches (Almasy & Blangero, 1998). Each phenotype was first regressed against age, age2, sex, age-by-sex, age2-by-sex and pubertal status, and then the regression residuals thus derived for each trait were normalized using an inverse normal transformation as previously described (Blangero et al., 2013; V.P. Diego et al., 2007).
Genotype-by-Environment (G × E) Interaction Model for Continuous Environments
We examined G × E interaction effects in response to the SA and PF environments by modeling the additive genetic variance and correlation as continuous functions of the environment. We describe our methods in terms of SA for convenience but they apply to PF as well. Regarding G × SA interaction, the fundamental null hypothesis is that the expression of a polygenotype (i.e., aggregate of all genotypes related to the expression of a phenotype) is independent of SA levels. For the simplest case of a trait analyzed under two environments, the G × E interaction variance is zero if the following two conditions are simultaneously true: 1) Homogeneity in the additive genetic variance: σ2g1 = σ2g2 = σ2g, where σ2g1 and σ2g2 are the additive genetic variance in environments 1 and 2, respectively; 2) The genetic correlation (ρg) is one across environments: ρg = 1. Rejection of either hypothesis rejects the overall null hypothesis of no G × E, and is taken as evidence of G × E interaction. (Blangero, 2009; V.P. Diego, Almasy, Dyer, Soler, & Blangero, 2003). Thus, there is no G × SA interaction, for a given CMRF, if it is simultaneously true that the genetic correlation for the trait is equal to 1.0 across different levels of SA and the trait additive genetic variance is homogeneous across all levels of SA.
We modeled the variance and correlation as functions of SA levels. For the genetic variance function (and similarly for the environmental variance), we modeled the variance using an exponential function of SA levels, where the exponential function maintains positivity, which is required of a variance (Blangero, 2009; V.P. Diego et al., 2003): σ2g = exp [αg+ γg (SA)], where αg and γg are parameters to be estimated. On taking the natural logarithm of the exponential function the variance homogeneity null hypothesis holds for a slope-term equal to 0: γg = 0. The genetic correlation was modeled using the exponential decay function of the pair-wise differences in SA levels: ρg = exp [−λ|SAx − SAz|] where SAx and SAz are the values of the SA for any two individuals x and z. The null hypothesis that the genetic correlation is equal to 1 is equivalent to λ = 0 because in this event: ρg = exp [−λ|SAx − SAz|] = e0 = 1.
We carried out model evaluations and hypothesis testing in two stages as follows. In the first stage, we examined whether the overall G × E interaction model provided a better fit to the data when compared with the polygenic model by way of a likelihood ratio test (LRT). It can be shown that the polygenic model is nested within the full G × E interaction model and that relative to the polygenic model the G × E interaction model has three additional parameters (the three being γg, γe, and λ; αg and αe are reparameterized versions of the variances). The LRT statistic for this comparison is distributed as a 50:50 mixture of chi-squares with 2 and 3 df (V.P. Diego et al., 2003). In addition, we also critically evaluated the full G × E interaction model by examining the parameter values relative to their standard errors for the gamma and lambda parameters.
In the second stage, if the full G × E interaction model was found to better fit the data than the polygenic model and if the parameter MLEs were at least larger than their standard errors, then we examined the more specific G × E interaction hypotheses. Here, the full G × E model with all parameters estimated was compared with models where either gamma (γ) or lambda (λ) were constrained to 0 to respectively test the hypotheses of additive genetic variance homogeneity and a genetic correlation equal to one. The distributions of the LRT statistics are respectively a chi-square with 1 df, and a 50:50 mixture of a chi-square with a point mass at 0 and a chi-square with 1 df (V.P. Diego et al., 2003).
If parameters were judged to be unimportant (i.e. where the standard error is greater than the maximum likelihood estimate), we constrained such parameters to 0 and tested the remaining potentially important parameter(s) under a reduced version of the G × E interaction model. In most cases of the reduced G × E interaction model, we constrained two of the three additional parameters in the full G × E interaction model (the three being γg, γe, and λ) leaving one free parameter for inferring G × E interaction. The distributions of the LRTs in these cases where gamma (γ) or lambda (λ) were constrained to 0 as appropriate are respectively a chi-square with 1 df, and a 50:50 mixture of a chi-square with a point mass at 0 and a chi-square with 1 df. Power to detect an interaction effect was computed using standard methods (Blangero et al., 2013; V. P. Diego, Kent, J.W., Blangero J., 2015).
G × E Analysis with SNPs
Prior to G × E analysis with SNPs, we performed association analysis between the SNPs and three environmental variables (HPFS, SAy, and SAu) using the measured genotype approach (MGA) to select the SNPs that are associated (p < 0.05) with the environments, which were selected based on our ongoing work by our group (Farook et al.,. unpublished). We then selected the top three SNPs (one per environment) and performed the G × E interaction analysis while simultaneously modeling SNP effects.
Where necessary, multiple-hypothesis testing correction was carried out by controlling the false discovery rate at the 0.05 level (39). Further, we performed FDR control according to the hypothesis examined. Thus, FDR control was carried out for each of the two mechanisms for G × E interaction, and for the genetic association tests.
Results
The characteristics of the study subjects are presented in Table 1. The mean age of subjects was 11.7 years and 48.8% were girls. The prevalence of pre-diabetes (Impaired Fasting Glucose [IFG], Impaired Glucose Tolerance [IGT] or both), overweight, obesity, and metabolic syndrome were 13.3%, 53.8%, 33.7%, and 18.6%, respectively. The CMRF trait sample sizes varied from 599 (HOMA-IR) to 617 (MSC) reflecting missingness patterns and exclusion of trait specific outliers (i.e., values ± 4 SD from the mean were excluded from genetic analysis). The sample sizes available for environmental variables ranged from 474 (Harvard fitness scores) to 617 (sedentary activity_yesterday). Means and SDs were presented for all CMRFs and environmental variables. We found strong genetic influences for all the CMRFs. As reported in Table 1, heritabilities (h2) ranging from 0.43 [FG, p = 5.2 × 10−5] to 0.79 [BMI, p = 5.1 × 10−11] for a majority of CMRFs were strong to moderate in magnitude, and highly significant (<0.001).
Table 1.
Characteristics of Subjects Including Environmental Measures and Heritabilities for Cardiometabolic Risk Factors in 617a SAFARI Children
| Trait* | Na | Mean ± SD or % |
Definitions/Normal Thresholds |
Heritability h2± S.E |
p-value b | Variance explained by covariates (%) |
|---|---|---|---|---|---|---|
|
| ||||||
| Girls | 617 | 48.8% | - | - | - | - |
|
| ||||||
| Age | 617 | 11.7 ± 3.5 | - | - | - | - |
|
| ||||||
| Overweight | 617 | 53.8% | BMI ≥85 and <95th percentile | - | - | - |
|
| ||||||
| Obese | 617 | 33.7% | BMI ≥95 and <99th percentile | - | - | - |
|
| ||||||
| Pre-diabetes | 617 | 13.3% | FPG ≥100 and <126 mg/dl | - | - | - |
|
| ||||||
| Metabolic Syndromec | 617 | 18.6% | < 3 components of the 6 examined components | - | - | - |
|
| ||||||
| Harvard Physical Fitness Score (HPFS) | 474 | 59.1 ± 24.8 | > 96 excellent score | - | - | - |
|
| ||||||
| Sedentary Activity_Usual | 606 | 0.67 ± 0.35 | 5 categories of time spent: 0 – 3hrs | - | - | - |
|
| ||||||
| Sedentary Activity_Yesterday | 617 | 0.51 ± 0.31 | 5 categories of time spent: 0 – 3hrs | - | - | - |
|
| ||||||
| Body mass Index [BMI(kg/m2)]d | 616 | 22.9 ± 6.4 | BMI values at 6 &17 yearsf: | 0.79 ± 0.12 | 5.1 × 10−11 | 0.26 |
| Boys:16.9±0.26; 24.9± 0.70 | ||||||
| Girls:16.0±0.25; 23.9 ±0.56 | ||||||
|
| ||||||
| Waist Circumference (WC, mm)d | 615 | 773.0 ± 180.4 | WC values at 6 & 17 yearsf: | 0.67 ± 0.12 | 2.5 × 10−8 | 0.36 |
| Boys:57.4±0.74; 85.6±1.82 | ||||||
| Girls:56.5±0.60; 82.2±1.50 | ||||||
|
| ||||||
| Fat Mass (Kg)d | 615 | 14.4 ± 15.9 | Boys: 28–23% g | 0.57 ± 0.12 | 3.0 × 10−7 | 0.36 |
| Girls: 31–35% | ||||||
|
| ||||||
| Fasting Glucose (mg/dl)d | 609 | 89.6 ± 7.1 | < 100 mg/dl | 0.43 ± 0.12 | 5.2 × 10−5 | 0.02 |
|
| ||||||
| Fasting Insulin (µIU/ml)d | 600 | 13.0 ± 7.2 | ≤16.25 uIU/ml | 0.64 ± 0.12 | 1.8 × 10−8 | 0.12 |
|
| ||||||
| HOMA-IRe | 599 | 1.9 ± 1.0 | < 3 h | 0.63 ± 0.12 | 1.0 × 10−7 | 0.11 |
|
| ||||||
| HDL (mg/dl)d | 605 | 45.5 ± 10.5 | > 45 mg/dli | 0.62 ± 0.12 | 2.0 × 10−7 | 0.06 |
|
| ||||||
| TG (mg/dl)d | 602 | 73.6 ± 35.7 | <75 mg/dl (0 to 9 yrs)i | 0.75 ± 0.12 | 2.0 × 10−10 | 0.04 |
| <90 mg/dl (10–19 yrs)i | ||||||
|
| ||||||
| SBP (mmHg)d | 617 | 104.3 ± 9.7 | BP <120/80 mmHg j | 0.70 ± 0.11 | 1.0 × 10−10 | 0.20 |
|
| ||||||
| DBP (mmHg)d | 617 | 63.1 ± 7.0 | BP <120/80 mmHg j | 0.58 ± 0.11 | 1.0 × 10−7 | 0.08 |
|
| ||||||
| Metabolic Syndrome Risk Score (# of MS Components (MSC) | 617 | 1.28 ± 1.36 | risk score range: 0–5 | 0.58 ± 0.13 | 3.2 × 10−6 | 0.05 |
Sample sizes for CMRFs varied from 599 (HOMA-IR) to 617 (MSC) based on availability of data and exclusion of outliers (i.e., values ± 4 SD from the mean were excluded from genetic analysis); Sample sizes for environmental variables ranged from 474 (Harvard fitness scores) to 617 (sedentary activity_ yesterday);
From likelihood ratio tests of models with the additive genetic variance estimated and constrained to 0;
As defined in Fowler et al. 2013;
CMRF definitions and descriptions were provided in Fowler et al. 2013;
BMI and WC (cm) Reference value ranges (mean ± Standard Error of the Mean[SEM]) for boys and girls at 6 and 17 years of age (Fryar, Gu, & Ogden, 2012);
Traits were inverse-normalized for genetic analyses;
homeostasis model of assessment-insulin resistance.
(C. L. Ogden, Li, Freedman, Borrud, & Flegal, 2011);
(Keskin, Kurtoglu, Kendirci, Atabek, & Yazici, 2005);
(Hauk, 2012);
(Chow et al., 2010).
G × E Interaction Findings
The polygenic model was compared to the G × E interaction model by means of a log-likelihood ratio test (see table 2). The G × E (PF/SA) interaction model is significantly better than the polygenic model for BMI (G × SAy), FM (G × HPFS and G × SAy), FI (G × PF and G × SAy), and DBP (G × SAu), and marginally non-significant for BMI (G × PF), HIR (G × ±PF, G × SAu, and G × SAy), MSC (G × SAy), and HDL (G × SAy). For all 6 marginally non-significant test results, we found that the neither of the gamma parameters for additive genetic or environmental variance heterogeneity were important (data not shown). We therefore constrained the gamma parameters to 0 and analyzed a reduced G × E interaction model in which the alpha parameters for the additive genetic and environmental variances and the lambda parameter for the genetic correlation are free to be estimated.
Table 2.
Polygenic Model vs. Full G × E (Continuous) Interaction Models for CMRFs
| Trait | Environment | Polygenic Model Vs. Full G × E Interaction Model |
|||
|---|---|---|---|---|---|
| Model | Ln Likelihood | LR Test | p-value | ||
| BMI | HPFS | Polygenic | −246.2510 | ||
| Full G × E | −243.0798 | 6.3424 | 0.069020* | ||
| BMI | SA_y | Polygenic | −294.6440 | ||
| Full G × E | −285.2698 | 18.7484 | 0.000015* | ||
| WC | HPFS | Polygenic | −227.2178 | ||
| Full G × E | −225.2784 | 3.8788 | 0.209321* | ||
| WC | SA_u | Polygenic | −273.9042 | ||
| Full G × E | −271.671 | 4.4664 | 0.161246* | ||
| WC | SA_y | Polygenic | −281.8768 | ||
| Full G × E | −280.343 | 3.0678 | 0.298495* | ||
| FM | HPFS | Polygenic | −261.6764 | ||
| Full G × E | −254−0216 | 15.3096 | 0.001022 | ||
| FM | SA_y | Polygenic | −299.9619 | ||
| Full G × E | −295.4529 | 9.0179 | 0.020032 | ||
| FI | HPFS | Polygenic | −241.5861 | ||
| Full G × E | −237.8490 | 7.4741 | 0.041025 | ||
| FI | SA_y | Polygenic | −287.8721 | ||
| Full G × E | −283.7673 | 8.2097 | 0.029182 | ||
| HIR | HPFS | Polygenic | −235.3967 | ||
| Full G × E | −232.1142 | 6.5650 | 0.062336* | ||
| HIR | SA_u | Polygenic | −285.0120 | ||
| Full G × E | −281.6945 | 6.6350 | 0.060366* | ||
| HIR | SA_y | Polygenic | −286.9285 | ||
| Full G × E | −283.7391 | 6.3789 | 0.067879* | ||
| MSC | SA_u | Polygenic | −274.9191 | ||
| Full G × E | −271.7375 | 6.3632 | 0.068369* | ||
| HDL | SA_y | Polygenic | −270.1281 | ||
| Full G × E | −266.9251 | 6.406 | 0.06704* | ||
| DBP | SA_u | Polygenic | −296.7298 | ||
| Full G × E | −293.1876 | 7.0845 | 0.049101 | ||
Poly = polygenic; LR = likelihood ratio;
Examination of MLEs and their standard errors show that a reduced model may be important (see text)
Although the polygenic model considerably outperformed the full G × E interaction model for WC for all three environments, we decided to move forward with these trait-environment constructs because two of the three “additional” parameters had larger standard errors relative to their MLEs. For the WC × PF and WC × SAu analyses we found that only the genetic correlation parameter had the possibility of being important whereas for the WC × SAy analysis we found that only the additive genetic variance heterogeneity parameter had the potential to be important under a reduced interaction model. The above 9 situations had in common the characteristic that the full G × E interaction model perhaps did not perform well relative to the polygenic model due to being overburdened with unnecessary parameters. For the FI × SAu, FI × SAy, and DBP × SAu interaction models, however, we found that although their full G × E interaction models performed significantly better than the polygenic model at explaining the data, the significant result seemed to arise from only important parameter (the gamma parameter for the additive genetic variance for the former two and the lambda parameter for the lattermost). Thus, for these three models, we moved forward with their appropriately reduced versions of the G × E interaction model.
We then tested the specific G × E interaction null hypotheses under either the full or reduced G × E interaction model as appropriate. As shown in Table 3, BMI, WC, FM, FI, HOMA-IR, MSC, HDL, and DBP exhibited significant G × E interaction (Red. G × E model vs Con. model) after correcting for multiple testing. Specifically, significant G × HPFs interactions were detected for BMI, FM, and HOMA-IR, while G × SAy interactions were detected for BMI, FI, HOMA-IR, and HDL, and G × SAu interactions were detected for WC, MSC, HDL and DBP. Among the significant interactions, multiple testing correction was not needed for BMI_SAy, FM_HPFs, MSC_SAu, HDL_SAy, and DBP_SAu as indicated in Table 3. However, WC-SAy, FM-SAy, and FM-SAu were not significant.
Table 3.
Genotype by Environment (Continuous) Interaction Effects on CMRFs.
| Trait | Environment | Mechanism | G × E Interaction Full or Reduced Models vs. Constraint Models |
p-value* | power | ||
|---|---|---|---|---|---|---|---|
| Model | LnL | LR Test | |||||
| BMI_HPFS | HPFS | Genetic Correlation | Red. G×E Model | −243.6020 | |||
| Con_lambda | −246.2510 | 5.2981 | 0.0107a | 0.7444 | |||
| BMI_Say | SAy | Genetic Variance | Full G×E Model | −285.2698 | |||
| Con_gamma | −289.8119 | 9.0842 | 0.0025b | 0.8541 | |||
| BMI_Say | SAy | Genetic Correlation | Full G×E Model | −285.2698 | |||
| Con_lambda | −289.4178 | 8.296 | 0.0040a | 0.8917 | |||
| WC_HPFS | HPFS | Genetic Correlation | Red. G × E Model | −225.5523 | |||
| Con_lambda | −227.2178 | 3.331 | 0.0340a | 0.5718 | |||
| WC_Sau | SAu | Genetic Correlation | Red. G×E Model | −272.0554 | |||
| Con_lambda | −273.9042 | 3.6977 | 0.0272a | 0.6097 | |||
| WC_Say | SAy | Genetic Variance | Red. G × E Model | −280.984 | |||
| Con_gamma | −281.877 | 1.7849 | 0.1815 | 0.2668 | |||
| FM_HPFS | HPFS | Genetic Variance | Full G × E Model | −254.0216 | |||
| Con_gamma | −258.7501 | 9.4570 | 0.0021b | 0.8676 | |||
| FM_Say | SAy | Genetic Correlation | Full G × E Model | −295.4529 | |||
| Con_lambda | −296.4619 | 2.0179 | 0.0777 | 0.4123 | |||
| FI_Sau | SAu | Genetic Variance | Red. G × E Model | −284.343 | |||
| Con_gamma | −285.887 | 3.0895 | 0.0788 | 0.42 | |||
| FI_Say | SAy | Genetic Variance | Red. G × E Model | −284.684 | |||
| Con_gamma | −287.872 | 6.3756 | 0.0116a | 0.714 | |||
| HOMA-IR_Sau | SAu | Genetic Correlation | Red. G×E Model | −283.0792 | |||
| Con_lambda | −285.0120 | 3.8656 | 0.0246a | 0.6261 | |||
| HOMA-IR_Say | SAy | Genetic Correlation | Red. G × E Model | −285.1387 | |||
| Con_lambda | −286.9285 | 3.5796 | 0.0292a | 0.5978 | |||
| HOMA-IR_HPFS | HPFS | Genetic Correlation | Red. G × E Model | −233.6242 | |||
| Con_lambda | −23b5.3967 | 3.545 | 0.0299a | 0.5943 | |||
| MSC_Sau | SAu | Genetic Correlation | Red. G × E Model | −273.2938 | |||
| Con_lambda | −274.9191 | 3.2506 | 0.0357b | 0.5631 | |||
| HDL_Say | SAy | Genetic Correlation | Red. G × E Model | −268.7425 | |||
| Con_lambda | −270.1281 | 2.7712 | 0.048b | 0.5084 | |||
| DBP_SAu | SAu | Genetic Variance | Red. G × E Model | −294.7716 | |||
| Con_gamma | −296.7298 | 3.9164 | 0.0478b | 0.5076 | |||
from likelihood ratio (LR) tests of models with either the gamma or lambda parameters estimated and constrained (Con) to 0; (Con_gamma (γ) = genetic variance; Con_lambda (λ) = genetic correlation; Red. G × E model = reduced model;
= significant after correcting for multiple-hypothesis testing;
= significant and multiple testing is not needed because only one test was performed for this hypothesis.
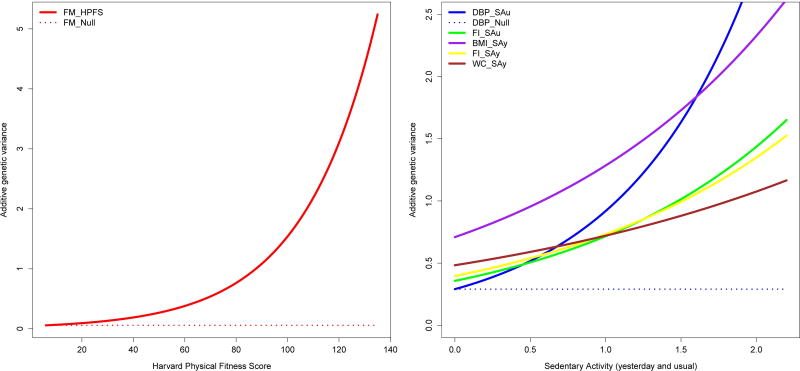
As shown in Table 3 and Figure 1, for BMI_SAy, FM_HPFs, FI_SAy and DBP_SAu, the null hypothesis of genetic variance (σ2g) homogeneity was rejected. The null hypothesis of homogeneity in the genetic variance implies a straight line graph (i.e. slope equal to 0) at the level of the natural logarithm of the heritability given that the variances are modeled as exponential functions. Thus, Figure 1 shows that the genetic variance changes as a function of the physical activity environment. In particular, the genetic variance increases with increasing HPFS for fat mass (Figure 1, left panel), and with increasing sedentary activity (yesterday or usual) or DBP, FI, BMI, and WC (Figure 1, right panel). For perspective, we plotted the expected variance lines under the null hypothesis of variance homogeneity for fat mass at the left panel and for DBP at the right panel. Taken together, these curves illustrate heterogeneity in the additive genetic variance as a function of some environmental exposure (HPFS for the left panel and sedentary activity for the right panel).
Figure 1.
Additive Genetic Variance Functions. Left panel: Fat Mass_HPFS. Right panel: DBP_SAu (blue), FI_SAu (green), BMI_SAy (purple), FI_SAy (yellow), and WC_SAy (brown).
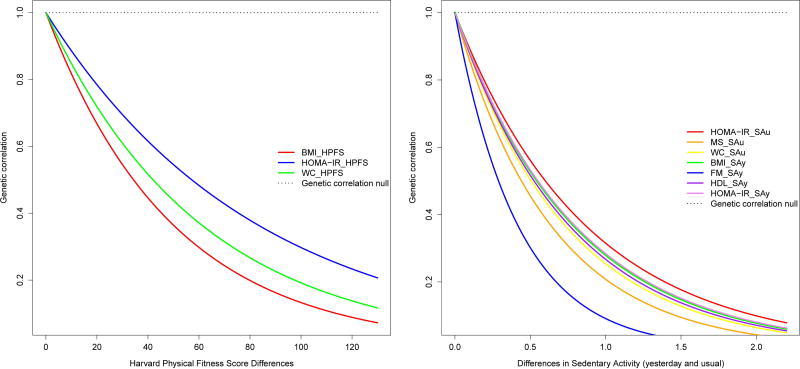
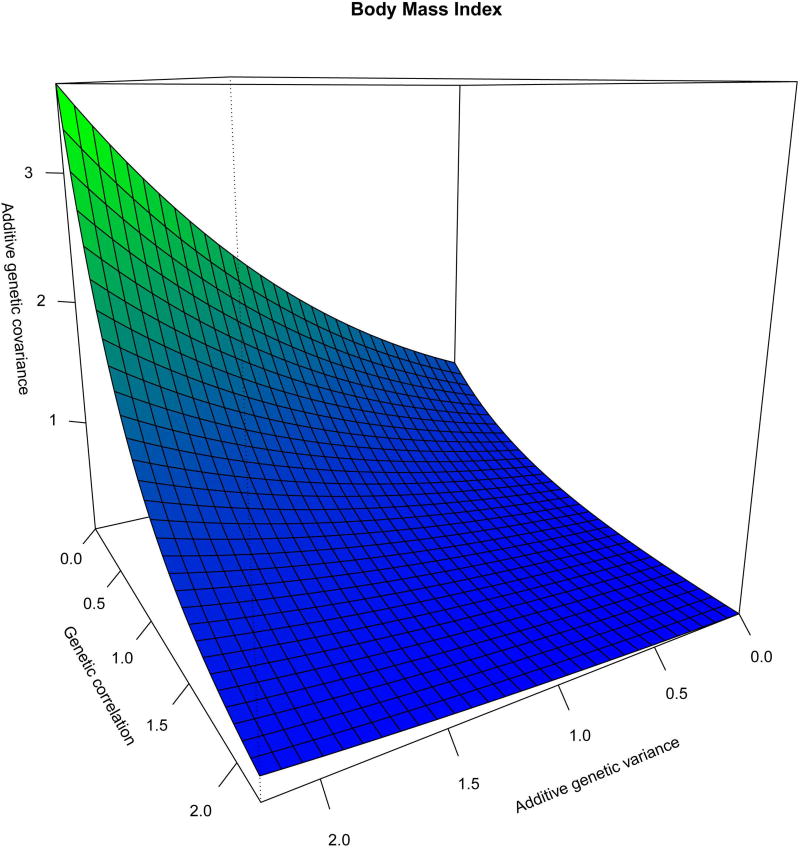
As shown in Table 3 and Figure 2, for BMI_HPFS, BMI_SAy, WC_HPFs, WC_SAu, FM_SAy, HOMA-IR_SAu/SAy/HPFS, MSC_SAu, and HDL_SAy, the null hypothesis of a genetic correlation equal to 1 was rejected. The null hypothesis of a genetic correlation equal to 1 as a function of pairwise differences in the environmental exposure is illustrated by the horizontal dotted line at a value of 1 for the genetic correlation. When this hypothesis is rejected, the lambda parameter is significant and we then observe genetic correlation functions that decay away from 1 with increasing differences in environmental exposure. In Figure 2, G × SAy interactions, both significant (BMI) and suggestive (HOMA-IR) were observed (solid lines). Due to the small sample size of our study, most trait-environment test cases in Table 3 were underpowered for G × E detection with only BMI_SAy (for the genetic variance and correlation) and FM_HPFS having power greater than 0.8 for G × E detection. Suggestive G × SAu interaction was observed for WC, HOMA-IR and MSC (dotted lines). Similarly, G × PF (i.e., Harvard fitness score) interactions, both significant (BMI) and suggestive (WC), due to the genetic correlation were observed (Figure 2). It can be seen that the genetic correlation decreases as the pair-wise differences for an environmental measure increases. BMI was the only trait for which the null hypotheses for the genetic correlation and the genetic variance were rejected, in relation to SAy. Figure 3 depicts the variance and correlation functions for BMI simultaneously as a covariance function, demonstrating that G × SAy interaction for BMI is a joint function of genetic variance heterogeneity and a genetic correlation different than one.
Figure 2.
Genetic Correlation Functions. Left panel: BMI_HPFS (red), HOMA-IR_HPFS (blue), and WC_HPFS (green). Right panel: HOMA_IR_SAu (red), MSC_SAu (orange), WC_SAu (yellow), BMI_SAy (green), FM_SAy (blue), HDL_SAy (purple), and HOMA-IR_SAy (violet).
Figure 3.
Additive Genetic Covariance Function for BMI. The covariance function is here expressed as a joint function of the additive genetic variance and genetic correlation functions.
As reported in Table 4, our G × E models were successful in detecting genetic association with the 3 SNPs in 34 out of 45 trait-environment test scenarios (10 for G × HPFS, 11 for G × SAu, and 13 for G × SAy) after multiple testing correction (FDR = 0.05).
Table 4.
mg-G × E (Continuous) Interaction Models for CMRFs
| Trait | Environment/SNP | G × E Model vs. G × E Model with SNP | |||
|---|---|---|---|---|---|
| Model | Ln Likelihood | LR Test | p-value* | ||
| BMI | HPFs | G × E | −243.601971 | ||
| rs1333049 | mg-G × E | −234.296911 | 18.61012 | 1.6 × 10−05 | |
| rs16986921 | mg-G × E | −236.050597 | 15.102748 | 1.0 × 10−04 | |
| rs12695382 | mg-G × E | −234.814916 | 17.57411 | 2.8 × 10−05 | |
| SA_y | G × E | −285.269841 | |||
| rs1333049 | mg-G × E | −278.646201 | 13.24728 | 2.7 × 10−04 | |
| rs16986921 | mg-G × E | −281.860341 | 6.819 | 9.0 × 10−03 | |
| rs12695382 | mg-G × E | −280.859309 | 8.821064 | 3.0 × 10−03 | |
| WC | HPFs | G × E | −225.552263 | ||
| rs1333049 | mg-G × E | −215.260173 | 20.58418 | 5.7 × 10−06 | |
| rs16986921 | mg-G × E | −217.893680 | 15.317166 | 9.9 × 10−05 | |
| rs12695382 | mg.G × E | −216.345399 | 9.206864 | 2.4 × 10−03 | |
| SA_u | G × E | −272.055371 | |||
| rs1333049 | mg-G × E | −264.180104 | 15.750534 | 7.2 × 10−05 | |
| rs16986921 | mg-G × E | −268.201910 | 7.706922 | 5.5 × 10−03 | |
| rs12695382 | mg-G × E | −266.696222 | 10.718298 | 1.1 × 10−03 | |
| SA_y | G × E | −280.98437 | |||
| rs1333049 | mg-G × E | −273.575208 | 14.818324 | 1.2 × 10−04 | |
| rs16986921 | mg-G × E | −276.766953 | 8.434834 | 3.7 × 10−03 | |
| rs12695382 | mg-G × E | −275.082909 | 11.802922 | 5.9 × 10−04 | |
| FM | HPFS | G × E | −254.258788 | ||
| rs1333049 | mg-G × E | −245.938561 | 16.640454 | 4.5 × 10−05 | |
| rs16986921 | mg-G × E | −247.401557 | 13.714462 | 2.1 × 10−04 | |
| rs12695382 | mg-G × E | −247.328433 | 13.86071 | 2.0 × 10−04 | |
| SA_y | G × E | −295.452938 | |||
| rs1333049 | mg-G × E | −290.640463 | 9.624950 | 1.9 × 10−03 | |
| rs16986921 | mg-G × E | −292.985805 | 4.934266 | 2.6 × 10−02 | |
| rs12695382 | mg-G × E | −291.236897 | 8.432082 | 3.7 × 10−03 | |
| FI | SA_u | G × E | −284.342451 | ||
| rs1333049 | mg-G × E | −280.948679 | 6.787544 | 9.2 × 10−03 | |
| rs16986921 | mg-G × E | −283.886050 | 0.912802 | 0.3393721a | |
| rs12695382 | mg-G × E | −281.444604 | 5.795694 | 1.6 × 10−02 | |
| SA_y | G × E | −284.684313 | |||
| rs1333049 | mg-G × E | −281.619929 | 6.128768 | 1.3 × 10−02 | |
| rs16986921 | mg-G × E | −284.359511 | 0.649604 | 0.4202543a | |
| rs12695382 | mg-G × E | −281.660447 | 6.047732 | 1.4 × 10−02 | |
| HOMA-IR | HPFs | G × E | −233.624161 | ||
| rs1333049 | mg-G × E | −232.027806 | 3.19271 | 0.0739673a | |
| rs16986921 | mg-G × E | −233.469271 | 0.30978 | 0.5778152a | |
| rs12695382 | mg-G × E | −231.407711 | 4.43290 | 3.5 × 10−02 | |
| SA_u | G × E | −283.079218 | |||
| rs1333049 | mg-G × E | −280.748401 | 4.661634 | 3.1 × 10−02 | |
| rs16986921 | mg-G × E | −282.714627 | 0.729182 | 0.3931485a | |
| rs12695382 | mg-G × E | −280.560791 | 5.036854 | 2.5 × 10−02 | |
| SA_y | G × E | −285.138691 | |||
| rs1333049 | mg-G × E | −282.592845 | 5.091692 | 2.4 × 10−02 | |
| rs16986921 | mg-G × E | −284.678495 | 0.920392 | 0.3373721a | |
| rs12695382 | mg-G × E | −282.243916 | 5.789550 | 1.6 × 10−02 | |
| MSC | SA_u | G × E | −273.293816 | ||
| rs1333049 | mg-G × E | −271.051664 | 4.484304 | 3.4 × 10−02 | |
| rs16986921 | mg-G × E | −271.921924 | 2.743784 | 0.0976333a | |
| rs12695382 | mg-G × E | −272.280778 | 2.026076 | 0.1546194a | |
| HDL | SA_y | G × E | −268.742515 | ||
| rs1333049 | mg-G × E | −267.394514 | 2.696002 | 0.1006002a | |
| rs16986921 | mg-G × E | −267.246377 | 2.992276 | 0.0836625a | |
| rs12695382 | mg-G × E | −268.714120 | 0.056790 | 0.7723941a | |
| DBP | SA_u | G × E | −294.771576 | ||
| rs1333049 | mg-G × E | −287.273606 | 14.995940 | 1.1 × 10−04 | |
| rs16986921 | mg-G × E | −289.493242 | 10.556668 | 1.2 × 10−03 | |
| rs12695382 | mg-G × E | −291.119736 | 7.30368 | 6.9 × 10−03 | |
LR = likelihood ratio;
Significant after multiple testing correction (FDR = 0.05) unless indicated by a
As shown in Table 4, we used our G × E models to study any potential genetic association with three SNPs that showed nominal associations (p < 0.05) with three environments: rs1333049 to be associated with HPFS, rs1698692 to be associated with SAu, and rs12695382 to be associated with SAy. We refer to this model as a measured genotype (mg)-G × E model because it jointly accounts for a measured genotype effect at a SNP while simultaneously accounting for potentially important G × E interaction effects.
As can be seen in Table 4, the mg-G × E model is quite successful at establishing proof of principle that we are able to simultaneously account for a measured genotype effect at a SNP and for a G × E interaction effect. Out of 45 test cases, 34 were significant after multiple testing correction (FDR = 0.05). All the three SNPs that were included in our G × E models found to be associated with cardio-metabolic traits in our data. e.g. rs1333049 (LOC729983) with triglycerides (TG) [P = 3.9 × 10−3], MS [P = 0.01], MSC-N [P = 9.2 × 10−3]; rs16986921 (CTNNBL1) with DBP (p = 0.03); rs12695382 (B4GALT4) with Fasting Insulin (FI, p = 0.03), and HOMA-IR [p = 0.03].
Discussion
We investigated the impact of G × E interaction on CMRF variation in MA children, who were previously found to be at higher risk for childhood obesity and its clinical correlates (Fowler et al., 2013). We determined moderate to high heritabilities for key CMRFs prior to performing G × E interaction analyses. It is important to note that sedentary behavior measures were not correlated with the physical fitness measure in our study, which is consistent with a previous report (Kerner, Kurrant, & Kalinski, 2004). Also, as reported earlier, the correlation between obesity measures and PF was negative, whereas it was positive with SA measures (Must & Tybor, 2005). Significant G × E (SA or PF] interactions were detected for the following CMRFs: BMI, FM, WC (obesity/abdominal obesity), HOMA-IR (insulin resistance), and MSC. The inference of G × E (SA or PF] interactions was drawn from observations of heteroscedasticity in the additive genetic
As shown in Figure 1, the genetic variance increases with increasing SA or PF. Likewise, as can be seen from Figure 3, the proportion of phenotypic variance in BMI that is attributable to additive genetic variance is larger in children with low sedentary activity compared to children with high levels of sedentary behavior. These findings suggest that there may be different genetic mechanisms underlying the physiology of sedentary behavior versus physical fitness. This observation warrants further studies to differentiate between the unique molecular and physiologic effects underlying sedentary behavior compared with physical fitness (Hamilton et al., 2007).
It is evident from these analyses that the phenotypic expression of a given trait may be influenced by the interactions between genotype (i.e., polygenotype) and environmental exposure. Thus, if a child is already at significantly increased risk for a given health condition due to genetic susceptibility, the environmental exposures assessed in this study (SA or PF) appear to significantly alter or modulate the child’s already heightened metabolic risk. Taken together, the interaction findings of this study specifically correspond to the interrelated metabolic conditions of obesity/abdominal obesity and insulin resistance with direct relevance to prediabetes, T2D, and MS. Our interaction observations can be interpreted to support the possibility that physical inactivity/sedentary lifestyle may either up-regulate genes with normal homeostatic functions for both weight maintenance and insulin sensitivity, or increase expression of susceptibility genes for obesity and insulin resistance – or do both simultaneously, across different sets of genes (Bey et al., 2003; Hamilton et al., 2007). For example, it was found that inactivity upregulated the expression of a number of genes in skeletal muscle in a rat model (Bey et al., 2003; Hamilton et al., 2007). Hojbjerre et al. (Hojbjerre et al., 2011) reported in human subjects before and after bed rest that physical inactivity was associated with higher levels of tumor necrosis factor α, a potent mediator of inflammation-related gene expression (Hotamisligil, 2006).
Sedentary activity has been associated with increased risk of obesity, T2D, CVD, and premature mortality. Bai and colleagues (Bai et al., 2016) demonstrated that screen time is a stronger predictor of weight status than physical activity in children and adolescents, and that physical activity is strongly correlated with cardiorespiratory fitness only in adolescents. Graf et al. (Graff et al., 2011) found significant interactions between screen time and genetic factors during adolescence that influence body mass changes between adolescence and young adulthood. Consistent with these studies, our findings revealed remarkable G × SA interaction influences on CMRFs (i.e., BMI, WC, HOMA-IR, and MSC) in MA children.
A number of studies have highlighted the detrimental effects of physical inactivity and the burden of inactivity-induced chronic diseases (Katzmarzyk, 2010; Tremblay, Colley, Saunders, Healy, & Owen, 2010). While exercise or physical activity is prescribed as the best treatment and/or prevention option for many chronic metabolic diseases, one of the contributing factors to the development of many metabolic diseases is considered to be the sedentary activity or physical inactivity. For example, cessation of exercise led to significant accumulation of intra-abdominal fat within 21 days in an animal model (Laye, Thyfault, Stump, & Booth, 2007). Slentz et al. (Slentz, Houmard, & Kraus, 2009) examined the effects of exercise on CMRFs. They found that there were cumulative detrimental effects in the inactive control group over only six months, and observed significant increase in weight and visceral fat. Within another six months of sedentary activity, they also noticed further deterioration in health.
The deleterious effects of abdominal obesity and its consistent association with other key CMRFs such as T2D, MS, and CVD in children and adolescents have been highlighted in several studies (Despres et al., 2008; Fowler et al., 2013). Sedentary activity is positively associated with abdominal obesity, while physical activity shows inverse association (Butte et al., 2007; Y. Kim & Lee, 2009). Studies based on directly measured PA suggest that high levels of PA or increased time spent in vigorous PA are associated with lower abdominal obesity (Y. Kim & Lee, 2009; Saelens, Seeley, van Schaick, Donnelly, & O'Brien, 2007). Thus physical inactivity or sedentary activity is considered to be an important causal factor of abdominal obesity. Given these complex relationships between abdominal obesity and physical in activity/sedentary behavior, it is noteworthy that our study revealed that waist circumference, a surrogate measure of abdominal obesity, is not only under substantial additive genetic influences, but also is influenced by G × SA or PF interaction effects in MA children.
With recent advances in susceptibility gene discoveries using genome-wide association scans, there has been an increased interest in identifying genes that may influence susceptibility to adiposity and its clinical correlates through gene-environmental interactions (Andreasen & Andersen, 2009; Bai et al., 2016; T. Huang & Hu, 2015; Jermendy et al., 2011; J. Y. Kim et al., 2016; Marti, Martinez-Gonzalez, & Martinez, 2008). There is strong evidence that genetic susceptibility to obesity can be altered through physical activity (Choquet & Meyre, 2011; Marti et al., 2008). It was shown that carriers of the Trp64Arg mutation in the ADRB3 gene with low levels of recreational physical activity have a higher risk of developing obesity (Marti et al., 2008). Several studies showed a strong interaction between the FTO genotype and physical activity on obesity risk in adults and adolescents (Andreasen & Andersen, 2009; Andreasen et al., 2008; Sonestedt et al., 2009). Li et al. (Li et al., 2010) used a genetic predisposition score based on the information from 12 obesity associated SNPs and found that high level of physical activity accounted for 40% reduction in the genetic predisposition to common obesity. In another study, Ochoa et al. (Ochoa, Moreno-Aliaga, Martinez-Gonzalez, Martinez, & Marti, 2006) assessed the relationship between the ADRB2 Gln27Glu polymorphism and TV viewing in Spanish children and adolescents and found interaction between this polymorphism and the number of hours spent watching TV with a significant effect on obesity risk (Ochoa et al., 2006). In European populations, gene-environment studies have reported that the association between the FTO gene and BMI is attenuated by physical activity levels (Andreasen et al., 2008; Rampersaud et al., 2008; Vimaleswaran et al., 2009). In the Old Order Amish population, physical activity was found to be inversely associated with lower BMI (Rampersaud et al., 2008). Although such association was only observed in individuals homozygous for a FTO risk allele, it was not observed in individuals with the protective allele (Rampersaud et al., 2008). Thus gene-environment studies indicate an association between variance due to specific genotypes and lifestyle as demonstrated by the FTO gene and physical activity.
Our findings from an exploratory G × E interaction analysis with SNPs further demonstrate their potential contribution to the observed interaction effects in CMRFs. Interestingly, all the 3 SNPs examined in our study were associated with several CMRFs. For example, rs1333049 is associated with coronary artery disease (Samani et al., 2007); rs16986921 is associated with BMI and fat mass (Liu et al., 2008); and rs12695382 is associated with LDL (Willer et al., 2008). Several of these SNP associations in our study had the same direction of effect reported previously in the European GWAS and the same risk alleles. For example, these included the risk alleles for: rs1333049 (C; MAF = 0.48, reported 0.47 – 0.49). These results are in agreement with our earlier findings. As a group, we have previously demonstrated that G × E modeling can increase power to detect quantitative trait loci (QTLs) using short tandem repeat marker data (V.P. Diego et al., 2003; Puppala et al., 2007; Voruganti et al., 2011) and expression QTLs using gene expression data (Kent et al., 2012).
In addition to environmental stimuli, sedentary behavior can be caused by neurological disorders and their underlying genetic factors. Given that there is a neurobehavioral basis for increased sedentary behavior or low physical activity, it follows that sedentary behavior may in fact be substantially underlain by genetic factors. Indeed, a number of neurological and psychiatric disorders, known to have a strong genetic component, manifest lethargy, or deficits in motor functioning. Such disorders include depression (Kendler, Kuhn, Vittum, Prescott, & Riley, 2005), cerebral palsy (Peterson, Gordon, & Hurvitz, 2013), movement disorders (Y. Huang, Yu, Wu, & Tang, 2014), and Parkinson Disease and Multiple Sclerosis (Ellis & Motl, 2013). Thus, it is quite possible that our observed gene-sedentary behavior interactions may be mediated by the genetic effects underlying sedentary behavior (which imply a kind of gene-gene interaction) as opposed to its truly environmental component. Future investigations are very much needed to pursue or develop approaches to clearly delineate the genetic and environmental components of sedentary behavior, and its role in the phenotypic determination of cardiometabolic risk factors. We plan to explore this issue in our future studies.
Our study has some limitations that warrant discussion. First, the sample size of our family-based data was modest. Second, assessment of sedentary activity through a self-administered questionnaire is sometimes poor in children due to cognitive ability. Third, common household effects were not accounted for in the estimation of heritabilities, which may be slightly inflated. Lastly, the environments used in the study are shown to have a genetic component; therefore, there is a possibility for our interaction models to reflect gene × gene interaction.
Conclusions
We found strong and highly significant genetic influences for all the examined CMRFs in this study involving Mexican American children and adolescents who participated in our SAFARI study. We observed significant G × SA and G × PF interaction effects on CMRFs, specifically in measures of obesity/abdominal obesity and insulin resistance. We also identified the measured genotype effects of specific genetic variants and G × E interaction effects simultaneously that influence susceptibility to cardiometabolic risk factors. These findings provide evidence that sedentary lifestyle and physical fitness patterns interact with additive genetic factors to influence the phenotypic expression of CMRFs, perhaps with distinct molecular and physiological repertoires. The ability to identify children with elevated susceptibility to obesity-related traits would be critical to initiating effective early prevention strategies or to treat children in these at-risk groups.
Acknowledgments
We thank Dr. Nancy F. Butte and Ms. Anne L. Adolph for their help with physical activity questionnaires. We thank the participants of the SAFARI study and their parents for their cooperation and participation.
Footnotes
Conflict of Interest: The authors declare no conflict of interest.
Author Contributions: Conceived and designed the G × E Interaction study: VPD, RA, RD; Physical Activity Questionnaire design and data collection/tabulation: SPF, SP, VSF, GC, RGR, RD; Clinical protocol development: DEH, JLL, RAD, RD; Contributed reagents/materials for lab measurements: VSF, JEC, DML, CPJ; Statistical Analyses and models: RA, VPD, JB; Manuscript preparation: RA; Manuscript review: SM, JV, LA, JEC, AGC, CPJ, JLL, JB, RD, VPD.
References
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 1998;62(5):1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasen CH, Andersen G. Gene-environment interactions and obesity--further aspects of genomewide association studies. Nutrition. 2009;25(10):998–1003. doi: 10.1016/j.nut.2009.06.001. [DOI] [PubMed] [Google Scholar]
- Andreasen CH, Stender-Petersen KL, Mogensen MS, Torekov SS, Wegner L, Andersen G, Hansen T. Low physical activity accentuates the effect of the FTO rs9939609 polymorphism on body fat accumulation. Diabetes. 2008;57(1):95–101. doi: 10.2337/db07-0910. [DOI] [PubMed] [Google Scholar]
- Arya R, Puppala S, Farook V, Chittoor G, Jenkinson C, Blangero J, Almasy L. Mapping of Susceptibility Genes for Obesity, Type 2 Diabetes, and the Metabolic Syndrome in Human Populations. In: Duggirala R, Almasy L, Williams-Blangero S, Paul S, Kole C, editors. Genome Mapping and Genomics in Human and Non-Human Primates. Heidelberg New York: Springer -Verlag; 2015. pp. 181–245. (Reprinted from: Not in File) [Google Scholar]
- Bai Y, Chen S, Laurson KR, Kim Y, Saint-Maurice PF, Welk GJ. The Associations of Youth Physical Activity and Screen Time with Fatness and Fitness: The 2012 NHANES National Youth Fitness Survey. PLoS. One. 2016;11(1):e0148038. doi: 10.1371/journal.pone.0148038. [doi];PONE-D-15-34989 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bey L, Akunuri N, Zhao P, Hoffman EP, Hamilton DG, Hamilton MT. Patterns of global gene expression in rat skeletal muscle during unloading and low-intensity ambulatory activity. Physiol Genomics. 2003;13(2):157–167. doi: 10.1152/physiolgenomics.00001.2002. [DOI] [PubMed] [Google Scholar]
- Blair SN, Haskell WL. Objectively measured physical activity and mortality in older adults. JAMA. 2006;296(2):216–218. doi: 10.1001/jama.296.2.216. doi:296/2/216 [pii];10.1001/jama.296.2.216 [doi] [DOI] [PubMed] [Google Scholar]
- Blangero J. Statistical genetic approaches to human adaptability. 1993. Hum. Biol. 2009;81(5–6):523–546. doi: 10.3378/027.081.0603. [DOI] [PubMed] [Google Scholar]
- Blangero J, Diego VP, Dyer TD, Almeida M, Peralta J, Kent JW, Jr, Goring HH. A kernel of truth: statistical advances in polygenic variance component models for complex human pedigrees. Adv. Genet. 2013;81:1–31. doi: 10.1016/B978-0-12-407677-8.00001-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownson RC, Boehmer TK, Luke DA. Declining rates of physical activity in the United States: what are the contributors? Annu Rev Public Health. 2005;26:421–443. doi: 10.1146/annurev.publhealth.26.021304.144437. [DOI] [PubMed] [Google Scholar]
- Butte NF, Cai G, Cole SA, Comuzzie AG. Viva la Familia Study: genetic and environmental contributions to childhood obesity and its comorbidities in the Hispanic population. Am. J. Clin. Nutr. 2006;84(3):646–654. doi: 10.1093/ajcn/84.3.646. [DOI] [PubMed] [Google Scholar]
- Butte NF, Puyau MR, Adolph AL, Vohra FA, Zakeri I. Physical activity in nonoverweight and overweight Hispanic children and adolescents. Med. Sci. Sports Exerc. 2007;39(8):1257–1266. doi: 10.1249/mss.0b013e3180621fb6. [DOI] [PubMed] [Google Scholar]
- Choquet H, Meyre D. Molecular basis of obesity: current status and future prospects. Curr. Genomics. 2011;12(3):154–168. doi: 10.2174/138920211795677921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung L, LPY C, ECK T, JWY C. Can a cardiorespiratory field parameter assess both cardiovascular and respiratory fitness in schoolchildren. Health. 2014;6(1):33–43. [Google Scholar]
- DeFina LF, Haskell WL, Willis BL, Barlow CE, Finley CE, Levine BD, Cooper KH. Physical activity versus cardiorespiratory fitness: two (partly) distinct components of cardiovascular health? Prog. Cardiovasc. Dis. 2015;57(4):324–329. doi: 10.1016/j.pcad.2014.09.008. doi:S0033-0620(14)00140-6 [pii];10.1016/j.pcad.2014.09.008 [doi] [DOI] [PubMed] [Google Scholar]
- Despres JP, Lemieux I, Bergeron J, Pibarot P, Mathieu P, Larose E, Poirier P. Abdominal obesity and the metabolic syndrome: contribution to global cardiometabolic risk. Arterioscler. Thromb. Vasc. Biol. 2008;28(6):1039–1049. doi: 10.1161/ATVBAHA.107.159228. [DOI] [PubMed] [Google Scholar]
- Diego VP, Almasy L, Dyer TD, Soler JM, Blangero J. Strategy and model building in the fourth dimension: a null model for genotype × age interaction as a Gaussian stationary stochastic process. BMC Genet. 2003;4(Suppl 1):S34. doi: 10.1186/1471-2156-4-S1-S34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diego VP, Kent JW, Jr, Blangero J. International Encyclopedia of the Social & Behavioral Sciences (Second Edition) Oxford: Elsevier; 2015. Familial Studies: Genetic Inferences A2 - Wright, James D; pp. 715–724. [Google Scholar]
- Diego VP, Kent JW, Blangero J. Familial studies: Genetic inferences, International Encyclopedia of Social and Behavioral Sciences. Second. Elsevier; 2015. pp. 715–724. [Google Scholar]
- Diego VP, Rainwater DL, Wang XL, Cole SA, Curran JE, Johnson MP, Blangero J. Genotype × adiposity interaction linkage analyses reveal a locus on chromosome 1 for lipoprotein-associated phospholipase A2, a marker of inflammation and oxidative stress. Am J Hum. Genet. 2007;80(1):168–177. doi: 10.1086/510497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis T, Motl RW. Physical activity behavior change in persons with neurologic disorders: overview and examples from Parkinson disease and multiple sclerosis. J Neurol Phys Ther. 2013;37(2):85–90. doi: 10.1097/NPT.0b013e31829157c0. [DOI] [PubMed] [Google Scholar]
- Esmaeilzadeh S, Ebadollahzadeh K. Physical fitness, physical activity and sedentary activities of 7 to 11 year old boys with different body mass indexes. Asian J. Sports Med. 2012;3(2):105–112. doi: 10.5812/asjsm.34709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher A, Smith L, van Jaarsveld CH, Sawyer A, Wardle J. Are children's activity levels determined by their genes or environment? A systematic review of twin studies. Prev. Med. Rep. 2015;2:548–553. doi: 10.1016/j.pmedr.2015.06.011. [doi];S2211-3355(15)00083-2 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler SP, Puppala S, Arya R, Chittoor G, Farook VS, Schneider J, Duggirala R. Genetic epidemiology of cardiometabolic risk factors and their clustering patterns in Mexican American children and adolescents: the SAFARI Study. Hum. Genet. 2013;132(9):1059–1071. doi: 10.1007/s00439-013-1315-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graff M, North KE, Monda KL, Lange EM, Lange LA, Guo G, Gordon-Larsen P. The combined influence of genetic factors and sedentary activity on body mass changes from adolescence to young adulthood: the National Longitudinal Adolescent Health Study. Diabetes Metab Res. Rev. 2011;27(1):63–69. doi: 10.1002/dmrr.1147. [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton MT, Hamilton DG, Zderic TW. Role of low energy expenditure and sitting in obesity, metabolic syndrome, type 2 diabetes, and cardiovascular disease. Diabetes. 2007;56(11):2655–2667. doi: 10.2337/db07-0882. [DOI] [PubMed] [Google Scholar]
- Harper MG. Childhood obesity: strategies for prevention. Fam. Community Health. 2006;29(4):288–298. doi: 10.1097/00003727-200610000-00007. [DOI] [PubMed] [Google Scholar]
- Hojbjerre L, Sonne MP, Alibegovic AC, Nielsen NB, Dela F, Vaag A, Stallknecht B. Impact of physical inactivity on adipose tissue low-grade inflammation in first-degree relatives of type 2 diabetic patients. Diabetes Care. 2011;34(10):2265–2272. doi: 10.2337/dc11-0631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper JL. Review papers : Variance components for statistical genetics: applications in medical research to characteristics related to human diseases and health. Statistical Methods in Medical Research. 1993;2(3):199–223. doi: 10.1177/096228029300200302. [DOI] [PubMed] [Google Scholar]
- Hopper JL, Visscher PM. Variance Component Analysis. In: Armitage P, Colton T, editors. Encyclopedia of Biostatistics. 2. Vol. 8. West Sussex, England: Wiley; 2005. [Google Scholar]
- Hotamisligil GS. Inflammation and metabolic disorders. Nature. 2006;444(7121):860–867. doi: 10.1038/nature05485. [DOI] [PubMed] [Google Scholar]
- Huang T, Hu FB. Gene-environment interactions and obesity: recent developments and future directions. BMC. Med. Genomics. 2015;8(Suppl 1):S2. doi: 10.1186/1755-8794-8-S1-S2. doi:1755-8794-8-S1-S2 [pii];10.1186/1755-8794-8-S1-S2 [doi] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Yu S, Wu Z, Tang B. Genetics of hereditary neurological disorders in children. Transl Pediatr. 2014;3(2):108–119. doi: 10.3978/j.issn.2224-4336.2014.03.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jermendy G, Horvath T, Littvay L, Steinbach R, Jermendy AL, Tarnoki AD, Osztovits J. Effect of genetic and environmental influences on cardiometabolic risk factors: a twin study. Cardiovasc. Diabetol. 2011;10:96. doi: 10.1186/1475-2840-10-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Orho-Melander M. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 2008;40(2):189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzmarzyk PT. Physical activity, sedentary behavior, and health: paradigm paralysis or paradigm shift? Diabetes. 2010;59(11):2717–2725. doi: 10.2337/db10-0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Kuhn JW, Vittum J, Prescott CA, Riley B. The interaction of stressful life events and a serotonin transporter polymorphism in the prediction of episodes of major depression: a replication. Arch Gen Psychiatry. 2005;62(5):529–535. doi: 10.1001/archpsyc.62.5.529. [DOI] [PubMed] [Google Scholar]
- Kent JW, Jr, Goring HH, Charlesworth JC, Drigalenko E, Diego VP, Curran JE, Williams-Blangero S. Genotypexage interaction in human transcriptional ageing. Mech Ageing Dev. 2012;133(9–10):581–590. doi: 10.1016/j.mad.2012.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerner M, Kurrant A, Kalinski M. Leisure-Time Physical Activity, Sedentary Behavior, and Fitness of High School Girls. European Journal of Sport Science. 2004;4(2):1–19. [Google Scholar]
- Kim JY, DeMenna JT, Puppala S, Chittoor G, Schneider J, Duggirala R, Coletta DK. Physical activity and FTO genotype by physical activity interactive influences on obesity. BMC. Genet. 2016;17(1):47. doi: 10.1186/s12863-016-0357-6. [doi];10.1186/s12863-016-0357-6 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Lee S. Physical activity and abdominal obesity in youth. Appl Physiol Nutr. Metab. 2009;34(4):571–581. doi: 10.1139/H09-066. [DOI] [PubMed] [Google Scholar]
- Kimm SYS, Glynn NW, Kriska AM, Fitzgerald SL, Aaron DJ, Similo SL, Barton BA. Longitudinal changes in physical activity in a biracial cohort during adolescence. Medicine and Science in Sports and Exercise. 2000;32(8):1445–1454. doi: 10.1097/00005768-200008000-00013. doi: [DOI] [PubMed] [Google Scholar]
- Knight JA. Physical inactivity: associated diseases and disorders. Ann. Clin. Lab Sci. 2012;42(3):320–337. doi:42/3/320 [pii] [PubMed] [Google Scholar]
- Kooner JS, Saleheen D, Sim X, Sehmi J, Zhang W, Frossard P, Chambers JC. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat. Genet. 2011;43(10):984–989. doi: 10.1038/ng.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laye MJ, Thyfault JP, Stump CS, Booth FW. Inactivity induces increases in abdominal fat. J Appl Physiol (1985.) 2007;102(4):1341–1347. doi: 10.1152/japplphysiol.01018.2006. [DOI] [PubMed] [Google Scholar]
- Li S, Zhao JH, Luan J, Ekelund U, Luben RN, Khaw KT, Loos RJ. Physical activity attenuates the genetic predisposition to obesity in 20,000 men and women from EPIC-Norfolk prospective population study. PLoS. Med. 2010;7(8) doi: 10.1371/journal.pmed.1000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YJ, Liu XG, Wang L, Dina C, Yan H, Liu JF, Deng HW. Genome-wide association scans identified CTNNBL1 as a novel gene for obesity. Hum. Mol. Genet. 2008;17(12):1803–1813. doi: 10.1093/hmg/ddn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd-Jones D, Adams R, Carnethon M, De Simone G, Ferguson TB, Flegal K, Hong Y. Heart disease and stroke statistics-2009 update: a report from the American Heart Association Statistics Committee and Stroke Statistics Subcommittee. Circulation. 2009;119(3):e21–181. doi: 10.1161/CIRCULATIONAHA.108.191261. [DOI] [PubMed] [Google Scholar]
- Marti A, Martinez-Gonzalez MA, Martinez JA. Interaction between genes and lifestyle factors on obesity. Proc. Nutr. Soc. 2008;67(1):1–8. doi: 10.1017/S002966510800596X. [DOI] [PubMed] [Google Scholar]
- Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28(7):412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- Must A, Tybor DJ. Physical activity and sedentary behavior: a review of longitudinal studies of weight and adiposity in youth. Int. J. Obes. (Lond) 2005;29(Suppl 2):S84–S96. doi: 10.1038/sj.ijo.0803064. [DOI] [PubMed] [Google Scholar]
- Ochoa MC, Moreno-Aliaga MJ, Martinez-Gonzalez MA, Martinez JA, Marti A. TV watching modifies obesity risk linked to the 27Glu polymorphism of the ADRB2 gene in girls. Int. J Pediatr. Obes. 2006;1(2):83–88. doi: 10.1080/17477160600650386. [DOI] [PubMed] [Google Scholar]
- Ogden CL, Carroll MD, Kit BK, Flegal KM. Prevalence of obesity and trends in body mass index among US children and adolescents, 1999–2010. JAMA. 2012;307(5):483–490. doi: 10.1001/jama.2012.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega FB, Ruiz JR, Castillo MJ, Sjostrom M. Physical fitness in childhood and adolescence: a powerful marker of health. Int. J. Obes. (Lond) 2008;32(1):1–11. doi: 10.1038/sj.ijo.0803774. doi:0803774 [pii];10.1038/sj.ijo.0803774 [doi] [DOI] [PubMed] [Google Scholar]
- Peterson MD, Gordon PM, Hurvitz EA. Chronic disease risk among adults with cerebral palsy: the role of premature sarcopoenia, obesity and sedentary behaviour. Obes Rev. 2013;14(2):171–182. doi: 10.1111/j.1467-789X.2012.01052.x. [DOI] [PubMed] [Google Scholar]
- Puppala S, Arya R, Thameem F, Arar NH, Bhandari K, Lehman DM, Abboud HE. Genotype by diabetes interaction effects on the detection of linkage of glomerular filtration rate to a region on chromosome 2q in Mexican Americans. Diabetes. 2007;56(11):2818–2828. doi: 10.2337/db06-0984. [DOI] [PubMed] [Google Scholar]
- Rampersaud E, Mitchell BD, Pollin TI, Fu M, Shen H, O'Connell JR, Snitker S. Physical activity and the association of common FTO gene variants with body mass index and obesity. Arch. Intern. Med. 2008;168(16):1791–1797. doi: 10.1001/archinte.168.16.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhemtulla M, Brosseau-Liard PE, Savalei V. When can categorical variables be treated as continuous? A comparison of robust continuous and categorical SEM estimation methods under suboptimal conditions. Psychol. Methods. 2012;17(3):354–373. doi: 10.1037/a0029315. [DOI] [PubMed] [Google Scholar]
- Saelens BE, Seeley RJ, van Schaick K, Donnelly LF, O'Brien KJ. Visceral abdominal fat is correlated with whole-body fat and physical activity among 8-y-old children at risk of obesity. Am J Clin. Nutr. 2007;85(1):46–53. doi: 10.1093/ajcn/85.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Schunkert H. Genomewide association analysis of coronary artery disease. N. Engl. J Med. 2007;357(5):443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos DM, Katzmarzyk PT, Diego VP, Souza MC, Chaves RN, Blangero J, Maia JA. Genotype by energy expenditure interaction with metabolic syndrome traits: the Portuguese healthy family study. PLoS. One. 2013;8(11):e80417. doi: 10.1371/journal.pone.0080417. [doi];PONE-D-13-19869 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savalei V, Rhemtulla M. The performance of robust test statistics with categorical data. Br. J Math. Stat Psychol. 2013;66(2):201–223. doi: 10.1111/j.2044-8317.2012.02049.x. [DOI] [PubMed] [Google Scholar]
- Slentz CA, Houmard JA, Kraus WE. Exercise, abdominal obesity, skeletal muscle, and metabolic risk: evidence for a dose response. Obesity (Silver. Spring) 2009;17(Suppl 3):S27–S33. doi: 10.1038/oby.2009.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonestedt E, Roos C, Gullberg B, Ericson U, Wirfalt E, Orho-Melander M. Fat and carbohydrate intake modify the association between genetic variation in the FTO genotype and obesity. Am. J. Clin. Nutr. 2009;90(5):1418–1425. doi: 10.3945/ajcn.2009.27958. [DOI] [PubMed] [Google Scholar]
- Steinberger J, Daniels SR, Eckel RH, Hayman L, Lustig RH, McCrindle B, Mietus-Snyder ML. Progress and challenges in metabolic syndrome in children and adolescents: a scientific statement from the American Heart Association Atherosclerosis, Hypertension, and Obesity in the Young Committee of the Council on Cardiovascular Disease in the Young; Council on Cardiovascular Nursing; and Council on Nutrition, Physical Activity, and Metabolism. Circulation. 2009;119(4):628–647. doi: 10.1161/CIRCULATIONAHA.108.191394. [DOI] [PubMed] [Google Scholar]
- Tremblay MS, Colley RC, Saunders TJ, Healy GN, Owen N. Physiological and health implications of a sedentary lifestyle. Appl Physiol Nutr. Metab. 2010;35(6):725–740. doi: 10.1139/H10-079. [DOI] [PubMed] [Google Scholar]
- Treuth MS, Sherwood NE, Baranowski T, Butte NF, Jacobs DR, Jr, McClanahan B, Obarzanek E. Physical activity self-report and accelerometry measures from the Girls health Enrichment Multi-site Studies. Prev. Med. 2004;38(Suppl):S43–S49. doi: 10.1016/j.ypmed.2003.01.001. [DOI] [PubMed] [Google Scholar]
- Trevino RP, Yin Z, Hernandez A, Hale DE, Garcia OA, Mobley C. Impact of the Bienestar school-based diabetes mellitus prevention program on fasting capillary glucose levels: a randomized controlled trial. Arch. Pediatr. Adolesc. Med. 2004;158(9):911–917. doi: 10.1001/archpedi.158.9.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vimaleswaran KS, Li S, Zhao JH, Luan J, Bingham SA, Khaw KT, Loos RJ. Physical activity attenuates the body mass index-increasing influence of genetic variation in the FTO gene. Am J Clin. Nutr. 2009;90(2):425–428. doi: 10.3945/ajcn.2009.27652. [DOI] [PubMed] [Google Scholar]
- Virdis A, Ghiadoni L, Masi S, Versari D, Daghini E, Giannarelli C, Taddei S. Obesity in the childhood: a link to adult hypertension. Curr. Pharm. Des. 2009;15(10):1063–1071. doi: 10.2174/138161209787846900. [DOI] [PubMed] [Google Scholar]
- Voruganti VS, Diego VP, Haack K, Cole SA, Blangero J, Goring HH, Comuzzie AG. A QTL for genotype by sex interaction for anthropometric measurements in Alaskan Eskimos (GOCADAN Study) on chromosome 19q12-13. Obesity (Silver Spring) 2011;19(9):1840–1846. doi: 10.1038/oby.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Abecasis GR. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 2008;40(2):161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]