Abstract
Background
Sex hormones are implicated in diabetes development. However, whether genetic variations in sex hormone pathways (SHPs) contribute to type 2 diabetes (T2D) risk remains to be delineated. We investigated the associations between genetic variations in all candidate genes in SHPs and T2D risk among a cohort of women who participated in the Women’s Health Initiative (WHI).
Methods
We comprehensively examined single nucleotide polymorphisms (SNPs) located within 30 kb upstream and 30 kb downstream of SHP genes in 8,180 African-American, 3,498 Hispanic-American, and 3,147 European-American women in the WHI. Additionally, we examined whether significant SNPs would be replicated in independent populations.
Results
After adjusting for age, region, and ancestry estimates and correcting for multiple testing, we identified seven SNPs in the PGR (progesterone receptor) gene significantly associated with the risk of T2D among Hispanic-American women, with the rs948516 SNP showing the greatest significance (odds ratio, 0.67; 95% confidence interval, 0.57–0.78; P = 8.8 × 10−7; false discovery rate, Q = 7.8 × 10−4). However, these findings were not replicated in other ethnic groups in the WHI or in sex-combined analyses in replication studies.
Conclusion
We identified significant signals implicating the PGR gene to T2D development in Hispanic-American women in the WHI, which are also consistent with GWAS findings linking the PGR gene to glucose homeostasis. Nevertheless, the PGR SNPs-T2D association was not statistically significant in other ethnic populations. Further studies, especially sex-specific analyses, are needed to confirm our findings and clarify the role of SHPs in T2D.
Keywords: diabetes risk, sex hormone pathways, single nucleotide polymorphisms
INTRODUCTION
Diabetes mellitus is a common and complex metabolic disease associated with serious complications and premature mortality. An accelerated increase in the prevalence of diabetes is expected globally and particularly in developing countries, underscoring the urgent need for curbing the burden of diabetes.1 The fundamental basis of the pathogenesis of diabetes involves the impairment of insulin action and secretion. Increasing evidence from experimental and epidemiological studies suggests that androgens, estrogens, and sex hormone-binding globulin (SHBG) play important roles in insulin resistance, impaired glucose tolerance, and the development of type 2 diabetes and gestational diabetes.2–7 Recent genetic studies have shown that single nucleotide polymorphisms (SNPs) in genes involved in sex hormone pathways (SHPs) are associated with circulating levels of sex hormones and SHBG.8–11 Thus, an investigation of the relationships between these SNPs and type 2 diabetes might provide an understanding of diabetes pathogenesis, potentially supporting the development of therapeutic or preventative strategies for diabetes. Previous genome-wide association studies (GWASs) could not identify the top signals in sex steroid pathways associated with type 2 diabetes risk.12 However, in GWAS analyses, stringent Bonferroni correction methods are used to address multiple comparisons, which may result in missing true but relatively modest associations with type 2 diabetes. An alternative approach to reduce the number of comparisons is to analyze SNP data using the candidate gene approach, which requires strong pre-existing hypotheses on the relationship between SHPs and diabetes risk.
Therefore, the present study aimed to investigate the relations between variants in SHP genes and the risk of type 2 diabetes in three large populations of African-American (AA), Hispanic-American (HA), and European-American (EA) postmenopausal women who participated in the Women’s Health Initiative (WHI)-SNP Health Association Resource (SHARe) and WHI-Genomics and Randomized Trials Network (GARNET).
METHODS
Study population
The WHI is a long-term national health study that focuses on strategies for preventing heart disease, breast and colorectal cancer, and osteoporotic fractures in postmenopausal women. Details on the design of the WHI are described elsewhere.13 The original WHI study included 161,808 postmenopausal women enrolled between 1993 and 1998. It had the following two major parts: a partial factorial randomized clinical trial (CT) and an observational study. A total of 40 clinical centers nationwide participated in both parts of the WHI study. The WHI CT enrolled 68,132 postmenopausal women (age range, 50–79 years) into trials testing three prevention strategies, while the observational study enrolled 93,676 women who did not participate in the CT. The observational study tracked the medical history and health habits of the participants. All participants provided written informed consent to participate in the study. The institutional review boards at all participating institutions approved the research protocols.
The WHI-SHARe included 8,236 AA women (6,263 controls and 1,973 diabetes cases) and 3,497 HA women (2,881 controls and 615 diabetes cases) from the WHI CT or OS, who provided consent for deoxyribonucleic acid (DNA) analysis.14–16 The WHI-GARNET included 3,147 EA women (2,127 controls and 1,020 diabetes cases) who had been enrolled in the WHI Hormone Therapy trial and who provided consent for DNA analysis. Of the approximately 27,000 women who participated in the WHI Hormone Therapy trial, incident diabetes cases and matched controls of EA women who were free of prevalent or incident diabetes, coronary heart disease, stroke, and venous thrombosis were included. The matching criteria were age (±5 years), race/ethnicity, hysterectomy status, enrollment date (±1.5 years), and length of follow-up (±48 months).
For replication, we included 6,320 diabetes cases and 9,300 controls from among AA women included in the MEta-analysis of type 2 DIabetes in African Americans (MEDIA) Consortium (sex-combined; 15 studies).17 Originally, 16 studies were included in the MEDIA Consortium, including the AA sample in the WHI-SHARe, but the WHI data were excluded from the present analysis. In addition, we used publicly available data for replication, including the trans-ethnic meta-analysis in the DIAbetes Genetics Replication and Meta-analysis (DIAGRAM) Consortium (sex-combined; n = 110,452)12,18 and the meta-analysis among Mexican populations from the Slim Initiative in Genomic Medicine for the Americas (SIGMA) Type 2 Diabetes Consortium (sex-combined; n = 8,891).18,19
Genotyping methods
DNA was extracted from the buffy coats of blood samples collected at baseline. The Affymetrix Genome-Wide Human SNP Array 6.0 (Affymetrix, Inc., Santa Clara, CA) was used to genotype all WHI-SHARe samples. During the genotyping stage of the study, laboratory technicians were blinded to the disease status. For the WHI-SHARe, we excluded samples on the basis of genotyping failure and quality control (sample call rate, <95%; n = 149), relatedness (n = 56), discordance between self-identified race and genetic ancestry assessed with the EIGENSTRAT (n = 56),12 and missing phenotypic information (n = 40). For relatedness, using concordance information to identify relatives, the relative with the highest genotyping call rate was retained, while other family members were excluded. In addition, SNPs were excluded if they had a Hardy Weinberg P-value <10−6, minor allele frequency <0.01, or call rate <90%. For the WHI-GARNET participants, genotyping was performed by using the Illumina HumanOmni1-Quad SNP platform (Illumina Inc., San Diego, CA). Sample- and SNP-level processing quality controls were performed using a standardized protocol at the GARNET Data Coordinating Center, University of Washington. Samples with a call rate <98%, related individuals, or discordance between self-identified race and genetic ancestry were excluded, and SNPs were excluded if they had a Hardy Weinberg P-value <10−4 or a call rate <98%. For the WHI-GARNET, SNPs from the 1000 Genomes Project (December 2010 release) imputed using the BEAGLE software were also analyzed, and SNPs with a minor allele frequency >5% and relatively high imputation quality (r2 > 0.5) were included in the analysis.
Selection of genes and SNPs
The genes involved in the SHPs20,21 investigated in this study included a) genes involved in sex-hormone synthesis and transport (CYP 11A1/17A1/19 [cytochrome P-450 11A1/17A1/19], HSD3B1/B2 [3β-hydroxysteroid dehydrogenase 1/2], HSD17B1/B2/B3 [17β-hydroxysteroid dehydrogenase 1/2/3], and SHBG); b) genes involved in androgen metabolism (SRD5A1/2 [steroid 5-α reductase type I/II], CYP3A4/5 [cytochrome P-450 3A4/5], UGT2B15/17 [uridine diphosphate-glucuronosyltransferase 2B15/17], HSD3B1/B2 [3β-hydroxysteroid dehydrogenase 1/2], and AR [androgen receptor]); c) genes involved in estrogen metabolism (CYP1A1 [cytochrome P-450 1A1], CYP1B1 [cytochrome P-450 1B1], COMT [catechol-O-methyltransferase], SULT1A1/1E1/2A1 [sulfotransferase 1A1/1E1/2A1], UGT1A1 [uridine diphosphate-glucuronosyltransferase 1A1], and ERα/β [estrogen receptor α/β]); and d) genes involved in progesterone metabolism (AKR1C1-4/D1 [aldo-keto reductase C1-4/D1] and PGR [progesterone receptor]).20 Using online databases (NetAffx, http://www.affymetrix.com and NCBI, http://www.ncbi.nlm.nih.gov), we searched for SNPs within 30 kb upstream and downstream of these genes and identified 1,388 genotyped SNPs covered by the Affymetrix array (AA and HA samples) and 1,198 genotyped and partly imputed SNPs derived from the Illumina array (EA samples). The numbers of SNPs identified per gene are listed in Table 2.
Table 2.
Number of SNPs covered by the genome-wide human SNP arrays
| Gene | Affy6.0 | Omni1-Quad |
|---|---|---|
| AKR1C1 | 26 | 16 |
| AKR1C2 | 31 | 19 |
| AKR1C3 | 50 | 37 |
| AKR1C4 | 112 | 48 |
| AKR1D1 | 110 | 83 |
| AR | 89 | 37 |
| COMT | 17 | 29 |
| CYP1A1 | 6 | 19 |
| CYP1B1 | 40 | 38 |
| CYP3A4 | 16 | 26 |
| CYP3A5 | 9 | 29 |
| CYP11A1 | 14 | 19 |
| CYP17A1 | 11 | 18 |
| CYP19A1 | 86 | 92 |
| ESR1 | 171 | 157 |
| ESR2 | 28 | 32 |
| HSD3B1 | 46 | 10 |
| HSD3B2 | 40 | 5 |
| HSD17B1 | 1 | 3 |
| HSD17B2 | 104 | 60 |
| HSD17B3 | 99 | 83 |
| PGR | 102 | 82 |
| SHBG | 5 | 14 |
| SRD5A1 | 37 | 34 |
| SRD5A2 | 41 | 38 |
| SULT1A1 | 2 | 4 |
| SULT1E1 | 50 | 93 |
| SULT2A1 | 13 | 27 |
| UGT1A1 | 7 | 40 |
| UGT2B15 | 24 | 5 |
| UGT2B17 | 1 | 1 |
Abbreviations: SNP = single nucleotide polymorphism; Affy6.0 = Affymetrix Genome-Wide Human SNP Array 6.0; Omni1-Quad = Illumina HumanOmni1-Quad SNP platform.
For the Omni1-Quad array, the number of genotyped and imputed SNPs is shown.
Ascertainment of type 2 diabetes
Previous validation studies confirmed that self-reported diabetes status provides a reliable measure of diagnosed diabetes status in the WHI sample.22,23 Thus, in the WHI-SHARe sample, we classified individuals as type 2 diabetes patients (1,973 AA and 615 HA women) if they self-reported having been diagnosed with diabetes when they were not pregnant or had received treatment for diabetes at baseline or during follow-up until August 14, 2009. We included prevalent cases of diabetes because the exposures (e.g., germ line mutations) are virtually guaranteed to precede the onset of diabetes. The remaining women were considered non-cases. In the WHI-GARNET sample, a total of 1,197 individuals were classified as having incident type 2 diabetes based on self-reports during any follow-up visit before July 7, 2002 in the estrogen plus progestin trial arm, or before February 29, 2004 in the estrogen-alone trial arm.
Statistical analysis
The χ2 test and Student’s t-test were used to compare dichotomous and continuous variables between case participants and non-case participants (Table 2). To assess the relation between each SNP and diabetes risk, we fitted logistic regression models to estimate odds ratios (ORs), assuming an additive genetic model. This analysis included the top three principal components defining global ancestry obtained via the principal component analysis-based method, and was performed separately for each self-identified ethnic group, with additional adjustment for age (continuous), region in the WHI-SHARe (AA and HA samples), and matching factors in the WHI-GARNET (EA) (Table 3). To correct for multiple testing, we calculated the false discovery rate (FDR) Q values for the whole set of SNPs considered in the analysis.24 Sensitivity analyses were conducted to estimate associations between SNPs and diabetes risk in dominant and recessive genetic models. FDR Q values <0.05 were considered to indicate significance.
Table 3.
Odds ratios and 95% confidence intervals for type 2 diabetes in various populations, stratified by ethnicity
| AA (WHI) | HA (WHI) | EA (WHI) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene | SNP ID | Minor allele | MAF | OR (95% CI) * | P-value | Q-value | MAF | OR (95% CI) * | P-value | Q-value | MAF | OR (95% CI) * | P-value | Q-value |
| PGR | rs948516 | G | 0.11 | 0.97 (0.86–1.10) | 0.64 | – | 0.23 | 0.67 (0.57–0.78) | 8.8 × 10−7 | 7.8 × 10−4 | 0.32 | 0.93 (0.83–1.04) | 0.32 | – |
| AA (MEDIA) | Mexican populations (SIGMA)18,19 | Trans-ethnic populations (DIAGRAM)12,18 | ||||||||||||
| Gene | SNP ID | Minor allele | MAF | OR (95% CI) * | P-value | – | MAF | OR | P-value | – | – | OR | P-value | – |
| PGR | rs948516 | G | 0.10 | 1.02 (0.93–1.12) | 0.61 | – | 0.22 | 0.964 | 0.337 | – | – | 0.971 | 0.11 | – |
Abbreviations: WHI = Women’s Health Initiative; SNP = single nucleotide polymorphism; OR = odds ratio; CIs = confidence intervals; Q-value, false discovery rate (FDR) Q-value; MAF = minor allele frequency; AA = African American; HA = Hispanic American; EA = European American; MEDIA = MEta-analysis of type 2 DIabetes in African Americans; SIGMA = Slim Initiative in Genomic Medicine for the Americas; DIAGRAM = DIAbetes Genetics Replication and Meta-analysis.
Unconditional logistic regression models were used to estimate ORs (additive genetic model) with adjustment for: age, region, and three principal components in the WHI SNP Health Association Resource (AA and HA populations); matching factors and three principal components in the WHI Genomics and Randomized Trials Network (EA population); age, sex, study sites, and principal components in the MEDIA (AA populations).
We then assessed whether SNPs found to be significant in the WHI, or their proxies, were also associated with type 2 diabetes risk in the MEDIA, SIGMA, or DIAGRAM. For the MEDIA, association analyses were conducted for each study under the additive model after adjustment for age, sex, study sites, and principal components, using logistic regression models or generalized estimating equations. The estimates were pooled using the fixed-effect model implemented in METAL.25 The statistical methods applied for the SIGMA and DIAGRAM are described elsewhere.12,19 Statistical analyses were performed using PLINK, R, MACH2DAT, SNPTEST, GWAF, SNPGWA, or Stata.
RESULTS
Our study populations included 1973 cases and 6263 non-cases among AA women with a mean age of 61–62 years, 615 cases and 2882 non-cases among HA women with a mean age of 60 years, and 1020 cases and 2127 non-cases among EA women with a mean age of 64–66 years. For all ethnicities evaluated (AA, HA, and EA), the amount and intensity of physical exercise was lower, body mass index (BMI) was greater, and family history of diabetes and use of postmenopausal hormone replacement therapy were more prevalent in case participants than in control participants (Table 1).
Table 1.
Characteristics of women with diabetes (cases) and without diabetes (controls), stratified by ethnicity
| AA | HA | EA | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Characteristic | Cases | Controls | P-value | Cases | Controls | P-value | Cases | Controls | P-value |
| n | 1,973 | 6,263 | 615 | 2,882 | 1,020 | 2,127 | |||
|
| |||||||||
| Age, years | 62.2 (6.8) | 61.4 (7.1) | <0.001 | 60.4 (6.7) | 60.2 (6.7) | 0.66 | 64.0 (6.9) | 65.6 (6.9) | <0.001 |
| BMI, kg/m2 | 33.1 (6.5) | 30.3 (6.2) | <0.001 | 31.5 (5.8) | 28.3 (5.3) | <0.001 | 32.5 (6.2) | 28.5 (5.9) | <0.001 |
| Family history of diabetes, % | 63.0 | 41.2 | <0.001 | 62.3 | 38.6 | <0.001 | 51.8 | 30.4 | <0.001 |
| Current smoker, % | 11.7 | 11.6 | 0.29 | 7.8 | 6.7 | 0.43 | 10.0 | 10.4 | 0.74 |
| Alcohol intake, servings/week | 0.7 (5.0) | 1.2 (4.2) | <0.001 | 0.6 (4.6) | 1.4 (3.6) | <0.001 | 1.5 (3.7) | 2.5 (5.2) | <0.001 |
| Physical activity* | 8.0 (11.1) | 10.2 (13.1) | <0.001 | 8.5 (12.4) | 11.2 (14.0) | <0.001 | 8.7 (11.7) | 11.2 (13.1) | <0.001 |
| Ever PMH therapy users, % | 49.0 | 55.0 | <0.001 | 52.0 | 63.2 | <0.001 | 37.5 | 36.9 | 0.79 |
Data were obtained from the Women’s Health Initiative and are presented as mean (standard deviation) or frequencies.
The chi-square test and Student’s t-test were used for comparisons of proportions and means between cases and controls.
Abbreviations: AA = African American; HA = Hispanic American; EA = European American; BMI = body mass index; MET = metabolic equivalent; PMH = postmenopausal hormone.
Total energy expenditure from recreational physical activities including walking and mild, moderate, or strenuous physical activity (MET-hours/week)
In HA women, 116 SNPs were nominally significantly associated with diabetes risk (Supplemental Table 1), and seven SNPs (rs4754732, rs948516, rs10501972, rs1942845, rs522819, rs632542, and rs485283; Supplemental Table 4) in the PGR gene showed statistical significance after FDR adjustment. Of these SNPs, the rs948516 SNP in the upstream region of the PGR gene had the most significant signal (OR = 0.67, 95% CI = 0.57–0.78, P = 8.8 × 10−7, Q= 7.8 × 10−4; Table 3). Other SNPs in high linkage disequilibrium with rs948516, including rs4754732 (complete linkage disequilibrium; D’ = 1.0; r2 = 1.0), were also significantly associated with diabetes risk in HA women (Figure 1).26,27 A conditional analysis to test which SNPs are independently associated with diabetes revealed that those SNPs originally identified as significant did not retain significant association with the risk of diabetes after conditioning on rs948516 (P-value: rs4754732, not available because of complete linkage disequilibrium with rs948516; rs10501972, 0.8842; rs1942845, 0.7256; rs522819, 0.5651; rs632542, 0.9473; rs485283, 0.9777). Among AA and EA women, 122 SNPs in AA women and 61 SNPs in EA women were nominally significantly associated with diabetes risk (Supplemental Tables 2 and 3); however, none of the SNPs showed statistical significance after FDR adjustment.
Figure 1. Regional association and linkage disequilibrium (LD) plots for the PGR gene among Hispanic-American women.
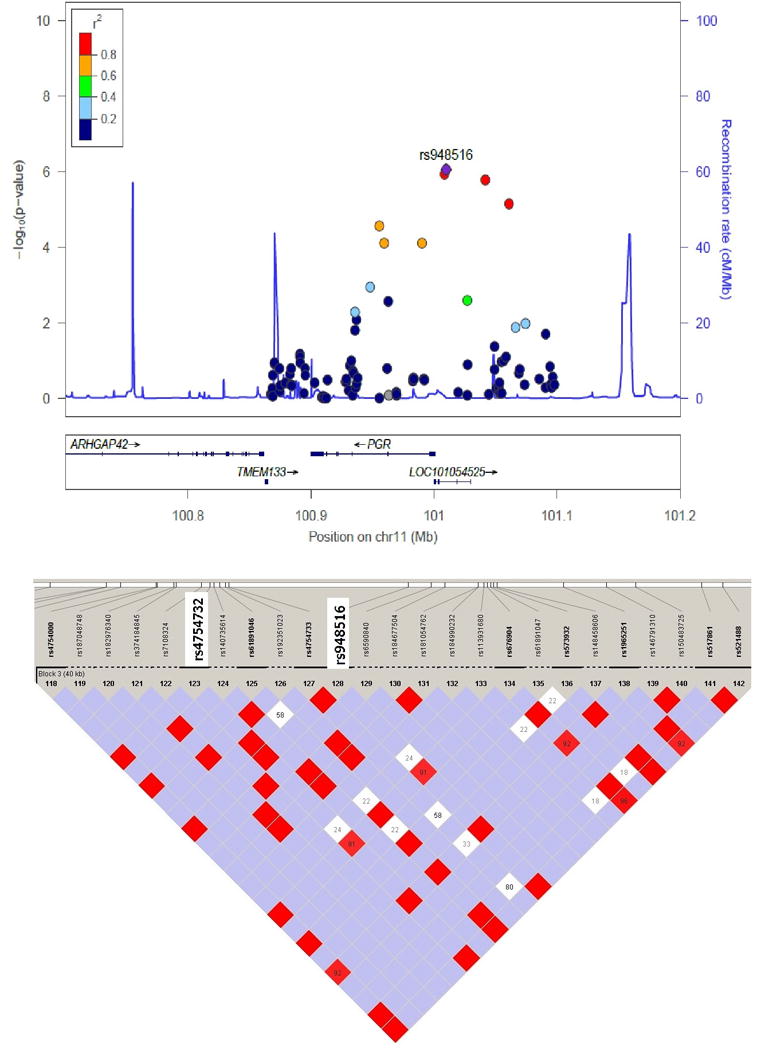
The regional association plots were created using the LocusZoom (http://locuszoom.sph.umich.edu/), and the LD plots were created using the HaploView software with phase 3 data (MXL) from the 1000 Genome Project. Blue indicates |D’| = 1 and logarithm of odds <2; bright red indicates |D’| = 1 and logarithm of odds ≥2; white indicates |D’| < 1 and logarithm of odds <2.
We further performed replication studies to examine whether the significance of the rs948516 SNP in the PGR gene among HA women could also be identified in the three independent cohorts of various ethnicities. However, we did not see any significant association among Hispanic populations (SIGMA), AA populations (MEDIA), or trans-ethnic populations (DIAGRAM) (Table 3; Supplemental Tables 5 and 6). In addition, we examined whether SNPs in high linkage disequilibrium (r2 > 0.9) with rs948516 would be associated with diabetes in the SIGMA or MEDIA populations, but found no significant association for any of the evaluated SNPs in the SIGMA or DIAGRAM samples (Supplemental Table 7). For the PGR gene, the top SNPs were rs7948940 (P-value = 2.1 × 10−3) in AA (WHI), rs516895 (P-value = 4.9 × 10−2) in EA (WHI), rs5632025 (P-value = 2.2 × 10−2) in AA (MEDIA), rs145583608 (P-value = 5.2 × 10−2) in Mexican populations (SIGMA), and rs534200 (P-value = 3.9 × 10−3) in trans-ethnic populations (DIAGRAM) (Supplementary Tables 5, 6, 8–10). For sex-stratified analyses, only the DIAGRAM investigators alone provided the results for only three SNPs on PGR (rs543215, rs481855, rs499590) and none of these showed significant association with diabetes (data not shown).
DISCUSSION
Among the three large samples of AA, HA, and EA women who participated in the WHI, we found that SNPs in the PGR gene were significantly associated with the risk of type 2 diabetes in HA women, and the significance of this association was retained after FDR adjustment. However, the significance of the rs948516 SNP was not replicated in the Hispanic population of the SIGMA Type 2 Diabetes Consortium (OR, 0.964; P = 0.337).18 Furthermore, we assessed the rs948516 SNP in sex-combined data from AA populations in the MEDIA Consortium (OR = 1.02; P = 0.61) and in the DIAGRAM Consortium (OR = 0.971; P = 0.11),18 and did not identify significant associations. Thus, the observed significant signal in the PGR gene could have been a false positive. However, the large consortiums of GWASs consist of mixed populations, which may be more subject to population stratification and considerable heterogeneity and thus may explain the inconsistencies in our results. However, several SNPs other than rs948516 on the PGR showed nominally significant associations with diabetes in other populations such as rs7948940 (P-value = 2.1 × 10−3) among AA in the WHI and rs534200 (P-value = 3.9 × 10−3) among AA in the DIAGRAM; suggesting that there is also a possibility of “gene effects” instead of “SNP effects”. Alternatively, threshold effects might explain the mostly null findings in common variants of the SHP genes, as the effects of these common variants on diabetes may be too small to reach the strict type I error level of significance. More importantly, it has been suggested that sex hormones may have a different effect on type 2 diabetes risk in men and women,2,4 and these sex-specific effects could not be addressed in our replication analyses. However, this motivates the need for future studies, especially sex-specific analyses, to confirm our findings and clarify the role of SHPs in type 2 diabetes.
Previous GWASs have identified more than 50 genetic loci associated with type 2 diabetes risk (including KCNJ11, TCF7L2, HHEX, CDKAL1, CDKN2A/B, IGF2BP2, and SLC30A8). Although these genome-wide signals were not attributable to SHPs, modest associations with variations in SHP genes may have been missed owing to the stringent criterion of genome-wide adjustment for multiple comparisons. Moreover, accumulating evidence suggests that sex hormones and SHBG play important roles in the development of type 2 diabetes.1,2 Our classic hypothesis-driven candidate gene approach can complement GWASs by decreasing the number of tests required for statistical power, while taking advantage of known biology.
We observed that SNPs located in the upstream region of the PGR gene were strongly and significantly associated with diabetes risk in HA women. In particular, two SNPs in the PGR gene (rs4754732 and rs948516) were significantly associated with type 2 diabetes and were in complete linkage disequilibrium with each other. Most likely due to heterogeneities across sexes and different populations and the strict type 1 error control, the significant effect of SNP rs948516 was not replicated in several consortia of GWAS of type 2 diabetes. Therefore, the significant signal observed in the PGR gene in relation to diabetes risk, albeit with strong biological plausibility, needs to be interpreted with caution. Of note, however, proxies of rs948516, such as rs1938953 and rs1942835, have been nominally and significantly associated with insulin sensitivity index (P = 0.001041)28 and HbA1c level (P = 0.02129),29 respectively, further supporting a potential functional role of this PGR variant in the glucose metabolism. Another study reported that the onset of gestational diabetes typically occurs during the second trimester of pregnancy, when the circulating levels of progesterone are high, suggesting a possible role of progesterone in gestational diabetes.30 Further, in a retrospective cohort study involving 2,081 pregnant women, those receiving a prophylactic intramuscular injection of 17α-hydroxyprogesterone caproate had a 2.9-fold higher risk of gestational diabetes than the risk noted in women who did not receive the injection.31 An experimental study in mice also demonstrated an active role of the PGR product in β-cell function, as female mice lacking the progesterone receptor had improved glucose tolerance resulting from enhanced insulin secretion.32 Taken together, these findings indicate a possible role of the progesterone receptor in increasing diabetes risk. Further functional studies illustrating the role of PGR in glucose metabolism are clearly warranted.
Several prospective studies have linked elevated circulating levels of estradiol to an increase in the risk of type 2 diabetes in men and women.2,40 In addition, previous case-control studies and prospective studies showed a sexually-dimorphic relationship between total testosterone levels and type 2 diabetes, with an inverse association observed in men, and a positive association observed in women.2,4 Because androgen and estrogen exert their effects through binding to androgen and estrogen receptors, respectively, polymorphisms in AR, ERα, or ERβ might be associated with the risk of type 2 diabetes. However, in our study, variants in such genes were not found to be significantly associated with diabetes risk. Further studies are required to understand the roles of these receptors in modulating diabetes risk in both men and women, and to explore potential sex-specific differences.
Previous studies reported several significant associations between variants in the SHBG gene and diabetes risk. In our study involving samples of the WHI population, the SNP in the SHBG gene with the lowest P-value was rs6258, which was nominally associated with diabetes risk in EA women (OR = 1.85, 95% CI = 1.04–3.30, P = 0.03) (Supplemental Table 3). The rs6258 SNP has a relatively low minor allele frequency, which has been shown to convert Pro156 into Leu and decrease the binding affinity of testosterone to SHBG, leading to low testosterone levels. On the other hand, the SNP with the lowest P-value was rs1641525 (OR = 0.97, 95% CI = 0.90–1.04, P = 0.35) in AA women and rs9898876 (OR = 1.17, 95% CI = 0.99–1.40, P = 0.07) in HA women in the WHI. The clinical significance of the rs6258 SNP and other functional SNPs in the SHBG gene warrant further investigation.
The main strength of the current study is the use of high-quality, well-characterized multiethnic samples of AA, HA, and EA women who were prospectively followed in a national cohort. Nevertheless, several limitations need to be kept in mind when interpreting the findings. First, the statistical power to assess less common variants, diabetes risk, and the heterogeneity of diabetes risk across ethnic groups remains marginal given that there are likely differences in allele frequencies, environmental modifiers, and genetic heterogeneity among these samples. Second, sex-stratified analyses were not available in the replication studies. To further clarify the genetic roles of SHPs, sex-specific analyses are needed. Third, considering that all individuals self-identified their ethnicity, population stratification may be a concern, although we did adjust for three principal components of global ancestry. Finally, potential misclassification due to undiagnosed diabetes, albeit most likely non-differential with respect to germline mutations, may have underestimated the magnitude of the genotype-outcome associations observed in the present study.41
In conclusion, SNPs in the PGR gene were significantly associated with diabetes risk in HA women in the WHI; however, the findings were not replicated in EA or AA women in the WHI, nor in sex-combined analyses in other studies. Although the significant signal may be false-positive, our findings suggest that further studies, including sex-specific analyses, sequencing, and fine-mapping with special emphasis on the functional loci in genes of pathways that interact with SHPs, may provide further insight into the role of sex hormones in the development of type 2 diabetes.
Supplementary Material
Key points/Highlights.
The present study investigated the role of genetic variations in sex hormone pathways and the risk of diabetes among African-American, Hispanic-American, and European-American women and found a significant signal in the PGR (progesterone receptor) gene in Hispanic-American women. However, the finding was not replicated in other populations. Our findings suggest the need for further studies, especially sex-specific analyses, to confirm our findings and clarify the role of sex hormone pathways in T2D.
Acknowledgments
This work was supported by the National Human Genome Research Institute (5U01HG005152), the Burroughs Wellcome Fund, the American Heart Association (AHA) Genome-Phenome Initiative, the WHI, the National Heart, Lung, and Blood Institute (R01DK103699), and the Brown University’s China and Brazil Initiatives. This study was also supported by a Grant-in-Aid for Young Scientists (B) from the Japan Society for the Promotion of Science (15K21389).
The WHI program is funded by the National Heart, Lung, and Blood Institute; National Institutes of Health; and U.S. Department of Health and Human Services, through contracts N01WH22110, 24152, 32100-2, 32105-6, 32108-9, 32111-13, 32115, 32118-32119, 32122, 42107-26, 42129-32, and 44221. Funding support for the WHI-GARNET was provided through the Genomics and Randomized Trials Network (GARNET) of the National Human Genome Research Institute (U01 HG005152). Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GARNET Coordinating Center (U01 HG005157). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Funding support for genotyping, which was performed at the Broad Institute of MIT and Harvard, was provided by the NIH Genes, Environment and Health Initiative (U01 HG004424).
We thank all the participants and the staff of the WHI for their dedicated and conscientious collaboration and assistance. We are grateful for the invaluable contributions of the following investigators: Nai-chieh Yuko You at the University of California, Los Angeles, Yiqing Song at Indiana University, James Y. Dai at Fred Hutchinson Cancer Research Center, Mitsuhiko Noda at Saitama Medical University, Maki Goto at JCHO Tokyo Yamate Medical Center, and Xiaochen Lin and Stephannie Shih at Brown University.
Footnotes
Conflicts of Interest
No potential conflicts of interest relevant to this article were reported.
Disclosures
None declared.
References
- 1.Guariguata L, Whiting DR, Hambleton I, Beagley J, Linnenkamp U, Shaw JE. Global estimates of diabetes prevalence for 2013 and projections for 2035. Diabetes Res Clin Pract. 2014;103:137–49. doi: 10.1016/j.diabres.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 2.Ding EL, Song Y, Malik VS, Liu S. Sex differences of endogenous sex hormones and risk of type 2 diabetes: a systematic review and meta-analysis. JAMA. 2006;295:1288–99. doi: 10.1001/jama.295.11.1288. [DOI] [PubMed] [Google Scholar]
- 3.Ding EL, Song Y, Manson JE, et al. Sex hormone-binding globulin and risk of type 2 diabetes in women and men. N Engl J Med. 2009;361:1152–63. doi: 10.1056/NEJMoa0804381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Goto A, Morita A, Goto M, et al. Associations of sex hormone-binding globulin and testosterone with diabetes among men and women (the Saku Diabetes study): a case control study. Cardiovasc Diabetol. 2012;11:130. doi: 10.1186/1475-2840-11-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heine PA, Taylor JA, Iwamoto GA, Lubahn DB, Cooke PS. Increased adipose tissue in male and female estrogen receptor-alpha knockout mice. Proc Natl Acad Sci U S A. 2000;97:12729–34. doi: 10.1073/pnas.97.23.12729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nadal A, Alonso-Magdalena P, Soriano S, Quesada I, Ropero AB. The pancreatic beta-cell as a target of estrogens and xenoestrogens: Implications for blood glucose homeostasis and diabetes. Mol Cell Endocrinol. 2009;304:63–8. doi: 10.1016/j.mce.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 7.Lin HY, Xu Q, Yeh S, Wang RS, Sparks JD, Chang C. Insulin and leptin resistance with hyperleptinemia in mice lacking androgen receptor. Diabetes. 2005;54:1717–25. doi: 10.2337/diabetes.54.6.1717. [DOI] [PubMed] [Google Scholar]
- 8.Ahn J, Schumacher FR, Berndt SI, et al. Quantitative trait loci predicting circulating sex steroid hormones in men from the NCI-Breast and Prostate Cancer Cohort Consortium (BPC3) Hum Mol Genet. 2009;18:3749–57. doi: 10.1093/hmg/ddp302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohlsson C, Wallaschofski H, Lunetta KL, et al. Genetic determinants of serum testosterone concentrations in men. PLoS Genet. 2011;7:e1002313. doi: 10.1371/journal.pgen.1002313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haiman CA, Dossus L, Setiawan VW, et al. Genetic variation at the CYP19A1 locus predicts circulating estrogen levels but not breast cancer risk in postmenopausal women. Cancer Res. 2007;67:1893–7. doi: 10.1158/0008-5472.CAN-06-4123. [DOI] [PubMed] [Google Scholar]
- 11.Dunning AM, Dowsett M, Healey CS, et al. Polymorphisms associated with circulating sex hormone levels in postmenopausal women. J Natl Cancer Inst. 2004;96:936–45. doi: 10.1093/jnci/djh167. [DOI] [PubMed] [Google Scholar]
- 12.Morris AP, Voight BF, Teslovich TM, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–90. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The Women’s Health Initiative Study Group. Design of the Women’s Health Initiative clinical trial and observational study. Control Clin Trials. 1998;19:61–109. doi: 10.1016/s0197-2456(97)00078-0. [DOI] [PubMed] [Google Scholar]
- 14.Chan KH, Huang YT, Meng Q, et al. Shared molecular pathways and gene networks for cardiovascular disease and type 2 diabetes mellitus in women across diverse ethnicities. Circ Cardiovasc Genet. 2014;7:911–9. doi: 10.1161/CIRCGENETICS.114.000676. [DOI] [PubMed] [Google Scholar]
- 15.Hutter CM, Young AM, Ochs-Balcom HM, et al. Replication of breast cancer GWAS susceptibility loci in the Women’s Health Initiative African American SHARe study. Cancer Epidemiol Biomarkers Prev. 2011;20:1950–9. doi: 10.1158/1055-9965.EPI-11-0524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crandall CJ, Manson JE, Hohensee C, et al. Association of genetic variation in the tachykinin receptor 3 locus with hot flashes and night sweats in the Women’s Health Initiative study. Menopause. 2017;24:252–61. doi: 10.1097/GME.0000000000000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ng MC, Shriner D, Chen BH, et al. Meta-analysis of genome-wide association studies in African Americans provides insights into the genetic architecture of type 2 diabetes. PLoS Genet. 2014;10:e1004517. doi: 10.1371/journal.pgen.1004517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.AMP-T2D Program; T2D-GENES Consortium, SIGMA T2D Consortium. Accessed on 2017, August 4th. Accessed at: http://www.type2diabetesgenetics.org/variantSearch/variantSearchWF.
- 19.SIGMA Type 2 Diabetes Consortium. Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population. JAMA. 2014;311:2305–14. doi: 10.1001/jama.2014.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henderson BE, Ponder BAJ, Ross RK. Hormones, Genes, and Cancer. Oxford University Press; New York: p. 2003. [Google Scholar]
- 21.Barrett KE, Brooks H, Boitano S, Barman S. Ganong’s Review of Medical Physiology. McGraw-Hill Medical; New York: 2010. [Google Scholar]
- 22.Gyamfi C, Horton AL, Momirova V, et al. The effect of 17-alpha hydroxyprogesterone caproate on the risk of gestational diabetes in singleton or twin pregnancies. Am J Obstet Gynecol. 2009;201:392.e1–5. doi: 10.1016/j.ajog.2009.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tulchinsky D, Hobel CJ, Yeager E, Marshall JR. Plasma estrone, estradiol, estriol, progesterone, and 17-hydroxyprogesterone in human pregnancy. I. Normal pregnancy. Am J Obstet Gynecol. 1972;112:1095–1100. doi: 10.1016/0002-9378(72)90185-8. [DOI] [PubMed] [Google Scholar]
- 24.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magi R, Morris AP. GWAMA: software for genome-wide association meta-analysis. BMC Bioinformatics. 2010;11:288. doi: 10.1186/1471-2105-11-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 28.Prokopenko I, Poon W, Magi R, et al. A central role for GRB10 in regulation of islet function in man. PLoS Genet. 2014;10:e1004235. doi: 10.1371/journal.pgen.1004235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soranzo N, Sanna S, Wheeler E, et al. Common variants at 10 genomic loci influence hemoglobin A1c levels via glycemic and nonglycemic pathways. Diabetes. 2010;59:3229–39. doi: 10.2337/db10-0502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Branisteanu DD, Mathieu C. Progesterone in gestational diabetes mellitus: guilty or not guilty? Trends Endocrinol Metab. 2003;14:54–6. doi: 10.1016/s1043-2760(03)00003-1. [DOI] [PubMed] [Google Scholar]
- 31.Rebarber A, Istwan NB, Russo-Stieglitz K, et al. Increased incidence of gestational diabetes in women receiving prophylactic 17alpha-hydroxyprogesterone caproate for prevention of recurrent preterm delivery. Diabetes Care. 2007;30:2277–80. doi: 10.2337/dc07-0564. [DOI] [PubMed] [Google Scholar]
- 32.Picard F, Wanatabe M, Schoonjans K, Lydon J, O’Malley BW, Auwerx J. Progesterone receptor knockout mice have an improved glucose homeostasis secondary to beta-cell proliferation. Proc Natl Acad Sci U S A. 2002;99:15644–8. doi: 10.1073/pnas.202612199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skjaerpe PA, Giwercman YL, Giwercman A, Svartberg J. Androgen receptor gene polymorphism and the metabolic syndrome in 60-80 years old Norwegian men. Int J Androl. 2010;33:500–6. doi: 10.1111/j.1365-2605.2008.00942.x. [DOI] [PubMed] [Google Scholar]
- 34.Rodriguez-Gonzalez G, Ramirez-Moreno R, Perez P, et al. The GGN and CAG repeat polymorphisms in the exon-1 of the androgen receptor gene are, respectively, associated with insulin resistance in men and with dyslipidemia in women. J Steroid Biochem Mol Biol. 2009;113:202–8. doi: 10.1016/j.jsbmb.2008.12.009. [DOI] [PubMed] [Google Scholar]
- 35.McInnes KJ, Smith LB, Hunger NI, Saunders PT, Andrew R, Walker BR. Deletion of the androgen receptor in Adipose tissue in male mice elevates retinol binding protein 4 and reveals independent effects on visceral fat mass and on glucose homeostasis. Diabetes. 2012;61:1072–81. doi: 10.2337/db11-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kanaya N, Murray PA, Damron DS. The differential effects of midazolam and diazepam on intracellular Ca2+ transients and contraction in adult rat ventricular myocytes. Anesth Analg. 2002;95:1637–44. doi: 10.1097/00000539-200212000-00030. table of contents. [DOI] [PubMed] [Google Scholar]
- 37.Margolis KL, Bonds DE, Rodabough RJ, et al. Effect of oestrogen plus progestin on the incidence of diabetes in postmenopausal women: results from the Women’s Health Initiative Hormone Trial. Diabetologia. 2004;47:1175–87. doi: 10.1007/s00125-004-1448-x. [DOI] [PubMed] [Google Scholar]
- 38.Bonds DE, Lasser N, Qi L, et al. The effect of conjugated equine oestrogen on diabetes incidence: the Women’s Health Initiative randomised trial. Diabetologia. 2006;49:459–68. doi: 10.1007/s00125-005-0096-0. [DOI] [PubMed] [Google Scholar]
- 39.Ribas V, Nguyen MT, Henstridge DC, et al. Impaired oxidative metabolism and inflammation are associated with insulin resistance in ERα deficient mice. Am J Physiol Endocrinol Metab. 2009;298:E304–19. doi: 10.1152/ajpendo.00504.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding EL, Song Y, Manson JE, Rifai N, Buring JE, Liu S. Plasma sex steroid hormones and risk of developing type 2 diabetes in women: a prospective study. Diabetologia. 2007;50:2076–84. doi: 10.1007/s00125-007-0785-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


