Abstract
Light controls the translation of several mRNAs in fully developed chloroplasts via at least two regulatory pathways. In the first, the light signal is transduced as a thiol-mediated signal that modulates translation in parallel to light intensity. The second light-controlled pathway, termed priming, is a prerequisite to the thiol-mediated regulatory pathway. Light regulation is rapid and requires intrachloroplast photoreceptor(s). To delineate the signaling pathways controlling each of these regulatory events, we assayed the effect of photosynthetic inhibitors and electron donors on the translation of chloroplastic psbA mRNA. We show that the thiol-mediated signal is generated by photosystem I and transduced by vicinal dithiol-containing proteins. We also found that the priming signal probably initiates on reduction of plastoquinone. These findings suggest that translation of chloroplast psbA mRNA is controlled by both linear photosynthetic electron transport, exerted by the reduction of the ferredoxin–thioredoxin system, and the relative activities of photosystems I and II, signaled by the redox state of the plastoquinone pool. These data underscore the function of the light-capturing reactions of photosynthesis as chloroplast photoreceptors.
Light dramatically stimulates the translation of several chloroplast mRNAs in plant and algae cells (1–6). The protein showing the highest induction by light, 50- to 100-fold, is D1 [a core protein of photosystem (PS) II encoded by psbA] (2, 7, 8). However, the translation of psbD (encoding the D2 protein), rbcL (encoding the large subunit of ribulose bisphosphate carboxylase/oxygenase), and psaA-psaB (encoding the 65- and 70-kDa chlorophyll a apoproteins of PS I) has been shown to be light-regulated as well (4, 5, 9). Both initiation and elongation steps of translation are probably controlled by light (1, 5, 10–16).
The light signal(s) controlling translation of chloroplast mRNAs may originate from an extrachloroplastic or an intrachloroplastic source. The preservation of light-induced translation in isolated intact chloroplasts (10, 17, 18) suggests that the factors mediating light-signal perception and transduction are localized within this organelle. To date, only the light-capturing reactions of photosynthesis have been shown to act as photoreceptors within the chloroplast. Light signals emanating from photosynthetic electron transport have been implicated in the initiation of several regulatory pathways controlling both nuclear and chloroplastic gene expression by transcriptional and posttranscriptional events (19–23). Further analyses have implicated the electron acceptor on the reducing side of each of the two PS, plastoquinone (PQ) of PS II and ferredoxin (Fd) of PS I, as the redox-active signaling molecules (19, 20, 22, 23). In both cases, photosynthetic light perception results in the accumulation of the reduced forms of PQ and Fd, which are then proposed to initiate signal transduction. PQ has been implicated as a regulator of the reversible State I–II transition responsible for adjusting the relative light absorption of PS I and II (24, 25) and the transcription of several chloroplast genes (22). Furthermore, PQ has been implicated in signaling, which controls nuclear gene expression such as CAB and APX1 and APX2 (19, 20). Transduction of signals originating in PQ is probably mediated by at least two protein kinases showing distinct patterns of induction and substrate specificities (26).
Fd has been suggested to regulate the activities of key enzymes of carbon fixation and ATP synthesis in response to light (27–29). The signal-transduction pathway originating in Fd, termed the Fd–thioredoxin system, is comprised of a series of electron-transfer reactions involving Fd–thioredoxin reductase and thioredoxin. An important characteristic shared by proteins regulated by the Fd–thioredoxin system is their preferential reduction by the dithiol reductant thioredoxin (27). The chemical dithiol reductant DTT mimics the dithiol reduction by thioredoxin.
Light has been proposed to modulate psbA translation via the Fd–thioredoxin system. A reductive signal transduced by thioredoxin was suggested to activate a protein complex (5′ protein complex) showing high affinity to the 5′ untranslated region of psbA mRNA (23). Activation of the 5′ protein complex was suggested to occur by reduction of a regulatory disulfide of the complex. The regulatory redox-responsive site was later identified as a vicinal dithiol site (VDS) carried by RB60, a component of the 5′ protein complex (18). Reduction of the regulatory VDS of RB60 was predicted to consequently activate translation of psbA mRNA (18). Recently, two additional components of light-signal transduction controlling psbA mRNA translation have been proposed. First, a light-activated signaling pathway, termed priming, is required to allow the thiol-mediated regulatory pathway. In addition, the thiol-modulated translation of psbA mRNA has been shown to include an oxidative component acting in a counterbalancing fashion to the reductive signal (18). After priming (by a yet unidentified signal), a counteraction of reducing (stimulatory) and oxidizing (inhibitory) activities may modulate psbA mRNA translation via regulatory thiol-containing proteins in parallel with fluctuating light intensities. These observations suggested that the main function of the thiol-mediated pathway is to modulate psbA mRNA translation in proportion with light intensity and not as a dark-to-light switch (18).
To delineate the signaling pathways controlling each of these regulatory events, we assayed the effect of photosynthetic inhibitors and electron donors on the translation of psbA mRNA. Here we show that the priming signal initiates on reduction of PQ by PS II and that the reductive signal is generated by PS I and transduced via VDS-containing proteins. These findings suggest that synthesis of D1 is coordinated with both linear photosynthetic electron transport (signaled by reduction of Fd) and the relative activities of PS I and II (signaled by the PQ redox state).
Materials and Methods
Algae Growth Conditions and Chloroplast Isolation.
Chlamydomonas reinhardtii cw15 cells were grown in TAP medium (30) under a 12-h-light/12-h-dark regime at 25°C, to a density of ≈1 × 107 cells/ml. Intact chloroplasts were isolated as described in ref. 31. Cells were washed with 50 mM Tricine-NaOH buffer, pH 8, containing 330 mM sorbitol, 1 mM MgCl2, 10 mM NaCl, 2 mM EDTA, 1 mM MnCl2, 0.5 mM KH2PO4, and 5 mM Na-ascorbate, followed by a second wash in 50 mM Hepes-KOH buffer, pH 7.6, containing 330 mM sorbitol, 1 mM MgCl2, 1 mM MnCl2, 2 mM NaNO3, 20 mM NaCl, 2 mM EDTA, and 5 mM Na-ascorbate. Cells were broken in an ice-cold Yeda pressure cell (32). Broken cells were fractionated on a discontinuous Percoll gradient, and intact chloroplasts were collected from a 45%/70% interface (18, 31, 32). Chlorophyll concentration was determined spectrophotometrically (33). Isolated chloroplasts were kept in the dark for at least 30 min before subsequent treatments.
Translation in Intact Chloroplasts.
Protein synthesis in intact chloroplasts (200 μg chlorophyll/ml) was performed according to Trebitsh et al. (18), with slight modifications. All translation reactions included 10 mM MgATP. Dark-adapted chloroplasts were treated in the dark with or without 15 μM of the translation initiation inhibitor lincomycin or the vicinal dithiol inhibitor phenyl arsine oxide (PAO), the photosynthetic electron transport inhibitors 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone (DBMIB) and 3-(3′,4′-dichlorophenyl)-1,1′-dimethyl urea (DCMU), or the PS I electron donor 2,6-dichlorophenol-indo-phenol (DCPIP) and 20 mM ascorbate, and the chloroplasts were transferred to the dark or light (150 μmol m−2⋅sec−1) for a 5-min preincubation. To minimize the potential direct effect of DTT on DBMIB (34), 5 mM DTT was added, where indicated, after the preincubation, and protein pulse-labeling was limited to 5 min. Chloroplasts were pulse-labeled with 10 μCi [35S]-methionine (1,000 Ci/mmol specific activity), then allowed to complete nascent protein synthesis in the presence of 5 mM unlabeled methionine for an additional 5 min. The reaction was stopped by placing tubes in liquid N2. For analysis, chloroplast proteins (corresponding to ≈1 μg of chlorophyll) were incubated in SDS sample buffer (3% SDS/2.25% β-mercaptoethanol/6.7% glycerol/0.133 M Tris⋅HCl, pH 6.8), fractionated by SDS/PAGE (35), and transferred onto nitrocellulose membranes. Radiolabeled proteins were detected by autoradiography. The location of the proteins D1, D2, and coupling factor 1 was determined by immunoblot assays by using the corresponding antisera.
Measurements of Photosynthetic Activity.
The PS II activity of C. reinhardtii cw15 cells was measured as steady-state oxygen evolution with a Clark-type oxygen electrode. Similar to the protein pulse-labeling experiments, oxygen evolution was measured for 5 min in the dark or light (150 μmol m−2⋅sec−1) in the presence or absence of DBMIB or DCMU. Where indicated, DTT was added to a final concentration of 5 mM, and oxygen evolution was measured for an additional 5 min.
Results
Translation of psbA mRNA Requires a Reductive Signal Generated by Photosynthetic Electron Transport.
Translation of psbA mRNA has been proposed to be activated by a reductive signal originating from PS I, transduced by the Fd–thioredoxin system, and perceived by a regulatory disulfide-containing translational factor(s) (23). To test this model, we first determined whether translation of psbA mRNA depends on photosynthetic electron transport. Protein pulse-labeling reactions were performed with intact chloroplasts isolated from C. reinhardtii cells showing induction of psbA mRNA translation by light (18). To focus on photoregulatory events that are independent of the energy status of the chloroplast, we included 10 mM ATP in all translation reactions. Under these conditions, a comparison of proteins synthesized in the dark (Fig. 1A, lane 1) and in the light (Fig. 1A, lane 5) showed D1 synthesis to be highly stimulated by light. Incubating illuminated chloroplasts with the photosynthetic electron transport inhibitor DBMIB clearly diminished the photoinduced synthesis of D1 (Fig. 1A, compare lanes 5 to 7). These data show that the light-regulated synthesis of D1 is controlled, at least in part, by photosynthetic electron transport.
Figure 1.
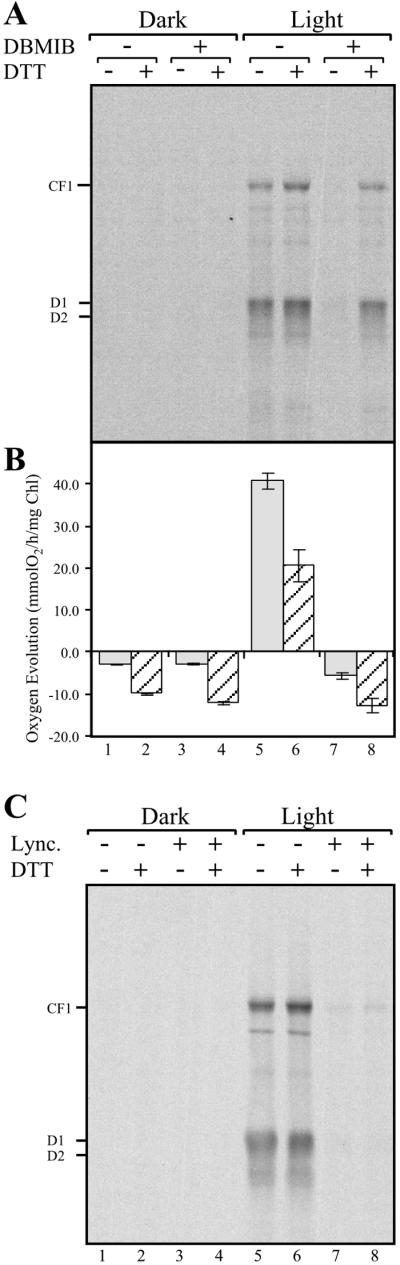
DTT restores DBMIB-inhibited light-regulated translation of chloroplast mRNAs but not activity of PS II. (A) Dark-adapted intact chloroplasts were treated in the dark with or without 2.5 μM DBMIB, then an aliquot was transferred for 5 min to the light (150 μmol m−2⋅sec−1). The translational activities of the dark- and light-incubated chloroplasts were then assayed by labeling newly synthesized proteins for 5 min with [35S]methionine, in the presence or absence of 5 mM DTT. Translation reactions were chased by incubating for an additional 5 min in the presence of 5 mM nonradioactive methionine and stopped by placing tubes in liquid N2. Chloroplasts were lysed, and extracted proteins were fractionated by SDS/PAGE, blotted onto nitrocellulose membranes, and visualized by autoradiography. (B) Net oxygen exchange was measured in C. reinhardtii cw15 cells with an oxygen electrode. Measurements were performed for 5 min in the dark or light (150 μmol m−2⋅sec−1) in the presence or absence of 2.5 μM DBMIB, and then for an additional 5 min with or without 5 mM DTT. (C) Initiation of translation is required for light-regulated translation of chloroplast mRNAs. Dark-adapted intact chloroplasts were treated with or without 15 μM lincomycin (Lync.), then an aliquot was transferred for 5 min to the light (150 μmol m−2⋅sec−1), and treatment followed as in A.
What is the nature of the photosynthesis-derived signal regulating D1 synthesis? Previously, it was shown that light-induced translation of psbA mRNA is stimulated by a reductive signal, which could be mimicked by the thiol reductant DTT (18). Therefore, we set out to determine whether the signal derived from photosynthetic electron transport is reductive. We reasoned that if inhibition of photosynthesis by DBMIB diminishes translation by turning off the stimulatory reductive signal, then treating the DBMIB-inhibited chloroplasts with DTT would bypass the lack of photosynthetic-derived reductive signal and restore translation. The inhibition of psbA mRNA translation by DBMIB was alleviated by the addition of the thiol reductant DTT (Fig. 1A, compare lanes 7 to 8), illustrating that the translational stimulatory signal derived from photosynthetic electron transport is reductive.
To ensure that DTT did not affect DBMIB directly, the inhibition of photosynthetic electron transport by DBMIB was assayed by monitoring oxygen evolution (Fig. 1B). The addition of DTT resulted in similar rates of oxygen evolution in DBMIB-treated cells in the dark or light (Fig. 1B, compare lanes 4 to 8), indicating that DTT did not alleviate DBMIB blockage of photosynthetic electron transport. DTT had a similar negative effect on net oxygen evolution under all treatments in the dark or light (Fig. 1B), possibly by increasing oxygen consumption by respiration. This potential stimulatory effect on mitochondrial activity may reflect a previously identified redox-regulated step in this organelle (36). To assay whether light stimulates initiation of translation, we treated dark-adapted chloroplasts with lincomycin, an inhibitor of translation initiation, before transfer to light. We found that lincomycin diminished translation in illuminated chloroplasts, and that this effect was not reversed by DTT (Fig. 1C). We concluded that, under these experimental conditions, translation initiation is required for light-activated translation. These findings, however, do not exclude additional light regulated steps, such as elongation.
The Reductive Signal Stimulating Translation of psbA mRNA Emanates from PS I.
PS I has been shown to produce a regulatory reductive signal transduced by the Fd–thioredoxin system (27). An important characteristic shared by proteins regulated by this system is their preferential reduction by thioredoxins (mimicked by the dithiol reductant, DTT) (27). Both translation of psbA mRNA and the RNA-binding activity of a protein complex implicated as translational regulator psbA mRNA are redox-regulated and display these characteristics (18, 23). These observations suggested that the source of the reductive signal controlling translation of psbA mRNA is PS I. The demonstration that psbA mRNA translation is regulated by a reductive signal produced by photosynthetic electron transport (Fig. 1) further promotes this hypothesis. To verify this hypothesis, we tested whether reinstating electron flow through PS I in DBMIB-inhibited chloroplasts could restore translation of psbA mRNA. In this protocol, photosynthetic electron transport from PS II to I is inhibited by DBMIB, but electron flow through PS I is restored chemically by DCPIP, an ascorbate-dependent electron donor to PS I. Fig. 2 shows that D1 synthesis in DBMIB-inhibited chloroplasts recovered progressively in response to increasing concentrations of DCPIP (Fig. 2, compare lanes 3 to 4–6). This finding demonstrates that electron flow through PS I produces a reductive signal responsible for modulating translation of psbA mRNA in response to light.
Figure 2.
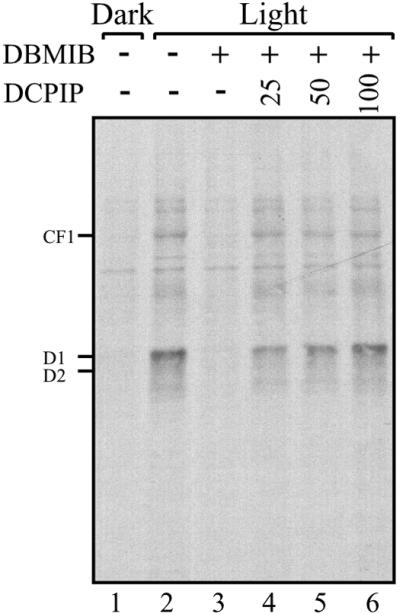
Reversal of DBMIB inhibition of chloroplast proteins synthesis by PS I electron transport. Translation reactions of intact chloroplasts were performed in the dark or light, in the presence or absence of 5 μM DBMIB. The inhibition of PS I electron transport by the photosynthetic inhibitor DBMIB was bypassed by incubating with increasing micromolar concentrations of the PS I electron donor DCPIP, as indicated above each lane. Intact chloroplasts translation reactions were performed as in Fig. 1A.
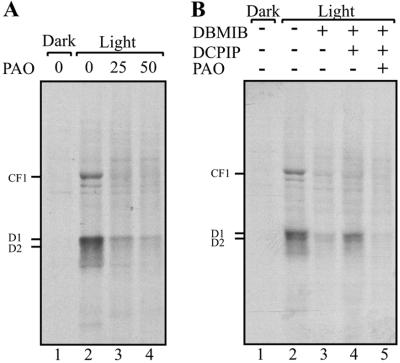
The Stimulatory Reductive Signal of PS I Is Mediated by Regulatory Vicinal Dithiol-Containing Protein(s).
The light signal produced by PS I and transduced by the Fd–thioredoxin system is typically perceived by proteins containing a regulatory disulfide that is preferentially responsive to thioredoxin (27). Such a regulatory site has been identified in the RB60 protein, a component of a protein complex implicated as translational regulator of psbA mRNA (18, 23, 37). The binding of PAO to RB60 in vitro has characterized the regulatory site as being comprised of vicinal dithiol (18). Therefore, to assay whether regulatory VDS-containing proteins participate in the control of D1 synthesis, we challenged illuminated chloroplasts with PAO. Inclusion of PAO in the translation reactions arrested the light-induced translation of psbA mRNA (Fig. 3A, compare lanes 2 to 3, 4). Next, we tested whether the transduction pathway of the reductive signal emanating from PS I is inhibited by PAO. Incubating illuminated and DBMIB-inhibited chloroplasts, in which electron transport through PS I was chemically restored by DCPIP, with PAO also diminished D1 synthesis (Fig. 3B, compare lanes 4 to 5). Together, these findings imply that the PS I-generated reductive signal, responsible for stimulating translation of psbA mRNA, initiates a transduction pathway that requires regulatory VDS-containing proteins, further implicating RB60 or an as-yet-unknown protein.
Figure 3.
Vicinal dithiol inhibitor blocks the stimulatory effect of PS I electron transport on chloroplast mRNA translation. Translation reactions of intact chloroplasts were performed as in Fig. 1A, in the dark or light, (A) in the presence or absence of micromolar concentrations of PAO, as indicated above each lane, and (B) in the presence or absence of 5 μM DBMIB, 100 μM DCPIP, or 25 μM PAO.
Reduced PQ Is Implicated in Signaling the Priming of Thiol-Regulated Translation.
The regulatory steps of light-induced psbA mRNA translation have been proposed to comprise at least two light signals. The first signal induces a priming event, which triggers thiol-mediated modulation of psbA mRNA translation on transfer to light (18). The thiol-mediated pathway includes the reductive signal emanating from PS I in response to light (Figs. 1 and 2). Next, we wished to study the origin of the light signal, which primes the thiol-mediated modulation of psbA mRNA translation.
To date, only the light-capturing reactions of photosynthesis have been shown to act as photoreceptors within the chloroplast. Namely, each of the electron acceptors of the reducing side of the two PS, PQ of PS II and Fd of PS I, has been implicated as a redox-active signaling molecule controlling gene expression (19, 20, 22, 23). That illuminated and DBMIB-treated chloroplasts translate psbA mRNA on addition of DTT (Fig. 1A) or DCPIP (Fig. 2) indicated that thiol-mediated modulation of translation had already been primed by light. These indications imply that PS I is not a likely candidate for generating the signal controlling the priming event and suggests two main alternatives as the source of the priming signal: (i) a novel chloroplastic photoreceptor or (ii) a reduced PQ pool.
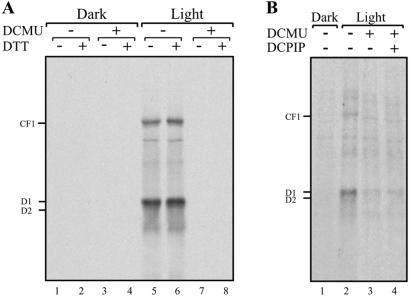
To differentiate between these two main alternatives, we used a second photosynthetic electron transport inhibitor, DCMU. In contrast to DBMIB treatment, which results in net reduction of the PQ pool, inhibition of electron transport by DCMU leads to net oxidation of the PQ pool (25). This protocol has been previously used to implicate the redox state of PQ as a regulator of gene expression (19, 20, 22). We reasoned that if the priming signal originates in a novel chloroplastic photoreceptor, then DCMU treatment would be similar to that of DBMIB and would be reversed by either reduction with DTT or DCPIP treatment. Alternatively, if net reduction of the PQ pool induces the priming signal, then neither DTT nor DCPIP would induce psbA mRNA translation after incubation with DCMU.
The results in Fig. 4 show that, similar to DBMIB (Fig. 1A), the inclusion of DCMU in the translation reactions inhibited the light-induced translation of psbA mRNA (Fig. 4A, compare lanes 5 to 7). However, the addition of DTT (Fig. 4A, compare lanes 6 to 8), or DCPIP (Fig. 4B, compare lanes 2 to 3, 4) did not restore D1 synthesis. These data show that, unlike DBMIB, which blocked only the thiol-modulated signal transduction pathway, the inhibition of photosynthetic electron transport by DCMU blocked, in addition, the priming component of light-induced translation. These findings suggest that reduction of the PQ pool on illumination triggers the priming signal-transduction pathway, which in turn enables thiol-mediated modulation of psbA mRNA translation.
Figure 4.
DCMU inhibition of chloroplast proteins synthesis is not reversed by PS I electron transport. Translation reactions of intact chloroplasts were performed, as in Fig. 1A, in the dark or light, (A) in the presence or absence of 5 mM DTT and 5 μM DCMU, and (B) in the presence or absence of 4 μM DCMU or 100 μM of the PS I electron donor DCPIP.
Discussion
Translation of psbA mRNA is regulated in response to light. Here we show that the light-regulated translation is controlled by at least two main signaling pathways: one that is initiated via reduction of the PQ pool (Fig. 4) and a second, by PS I electron transport (Fig. 2). The finding that the two light signals are both perceived by the light-capturing reactions of photosynthesis underscores the function of the membranal photosynthetic reactions as the main photoreceptor of the chloroplast.
PS I is the primary reducer of the Fd–thioredoxin system. This system regulates the activities of several key enzymes in the Benson–Calvin cycle, as well as coupling factor 1, by reduction of a regulatory VDS in the thioredoxin-regulated enzymes (27). The characteristics of redox-regulated psbA mRNA translation were shown to be reminiscent of the thioredoxin-regulated enzymes. Thus, it was proposed that the Fd–thioredoxin system transduces a reductive signal from PS I, controlling a regulatory VDS in the 5′ protein complex of psbA mRNA. Reduction of the regulatory VDS by thioredoxin activates the 5′ protein complex, thereby stimulating the translation of psbA mRNA (23). The stimulation of psbA mRNA translation, under conditions that restrict electron transport to PS I (Fig. 2), further implicates the Fd–thioredoxin system as transducer of the light signal. In addition, the finding that reduction by DTT bypasses the translational arrest imposed by the inhibition of photosynthetic electron transport (Fig. 1A) corroborates the reductive nature of the light signal emanating from PS I. Finally, that the VDS inhibitor PAO blocks the stimulatory effect of PS I-restricted electron transport (Fig. 3B) suggests that the reductive signal is transduced by regulatory VDS-containing protein(s). Together, these findings indicate that a light-generated reductive signal originates from PS I and is probably transduced by regulatory VDS-containing proteins to control psbA mRNA translation (Fig. 5).
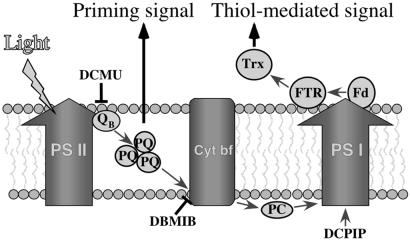
Figure 5.
Model of light-signaling control of translation. Translation of several chloroplast mRNAs is regulated in response to light via at least two major pathways. In the first, the reduced PQ pool activates a signal-transduction pathway that leads to the priming of the translational factors required for thiol-mediated modulation of translation. In the second, a thiol-mediated signal, proportional to the reducing potential generated by PS I, is transduced by Fd, Fd–thioredoxin reductase (FTR), and thioredoxin (Trx) to stimulate translation. DCMU binds to the QB site of PS II, resulting in oxidation of the PQ pool. DBMIB inhibits cytochrome b6f (Cyt bf), leading to increased reduction of the PQ pool. DCPIP is an artificial electron donor of PS I that can be used to allow electron transport through PS I under conditions of inhibited PS II to I electron transport.
Signaling from Fd is mediated by a cascade of redox-active proteins, transferring reducing equivalents generated by PS I. In contrast, the redox state of the PQ pool initiates a signaling cascade that has so far been shown to be mediated by associated kinase(s) (24). The reduced PQ pool has been implicated in the regulation of phosphorylation of the antenna protein LHC II in a phenomenon termed State I–II transition (25). A more detailed analysis suggested that the binding of reduced PQ to Cyt b6f may trigger signaling (24). A reduced PQ pool has also been implicated in the regulation of phosphorylation of PS II proteins (38–40) and regulation of transcription of nuclear photosynthetic genes by activating protein kinase/phosphatase activities (20). In contrast to the reductive signal of PS I, the priming signal, shown here to originate by net reduction of the PQ pool, could not be replaced by protein–thiol reduction (Fig. 4 and ref. 18). Hence, it is tempting to speculate that the priming signal is mediated by activation of a PQ-regulated kinase, which may render the VDS-containing translational regulators competent to redox modulation by the Fd–thioredoxin system.
Why is there a need for multiple signaling pathways? One possible explanation may derive from the apparent increase in the complexity of regulation of chloroplast gene expression during evolution. In the cyanobacteria, thought to be closely related to the progenitor of the chloroplast, the regulation of gene expression in response to light is primarily transcriptional (41). In the chloroplast, on the other hand, the regulation is mostly posttranscriptional (reviewed in refs. 12, 42–45). A concomitant increase in the complexity of redox regulation during chloroplast evolution is evidenced by the appearance of the chloroplast-unique thioredoxin f, which probably originated in the eukaryotic donor genome (46), and the increase in PQ-regulated kinases (26).
Such increased complexity of regulatory pathways may reflect better adaptation to environmental conditions. Coupling factor 1 activity is regulated by a proton gradient and by superimposed redox regulation via the Fd–thioredoxin system (47). Moreover, phosphorylation of chlorophyll a/b-binding proteins of PS II has been recently shown to be regulated by both the PQ and cytochrome b6f complex and via the redox state of the chloroplast (48). By analogy, Fd–thioredoxin-mediated redox regulation of psbA mRNA translation may be superimposed over the priming pathway controlled by the reduced PQ pool. An integrated regulation by light was also established for activity of key enzymes in the Benson–Calvin cycle. In addition to regulation via the Fd–thioredoxin system, light modulates enzyme activity by stromal pH and Mg2+ concentration changes (27). The multiplicity of signals regulating coupling factor 1, chlorophyll a/b-binding proteins of PS II, enzymes in the Benson–Calvin cycle, and psbA mRNA translation implies that a unique combination of specific signals is tailored for regulation that is optimal for each system. For example, carbon fixation is a major sink of reducing equivalents produced by PS I. Therefore, it is possible that carbon fixation reactions are rapidly activated by low light intensities (49) to minimize production of reactive oxygen species by photosynthesis (50). On the other hand, D1 is a membrane protein that plays a key role in charge separation reactions of PS II, leading to its photodegradation and de novo synthesis (2, 7, 8). The identification of a priming pathway that regulates psbA mRNA translation may reflect the unique requirements of synthesis of photosynthetic membrane proteins. It is possible that the Fd–thioredoxin system and PQ are designed to sense different environmental changes. PQ may better sense the relative activities of PS I and II, and Fd may be better adapted to sensing the overall production of photosynthetic reducing equivalents. Both may be affected by the consumption rate of the photosynthetic reducing equivalents by the carbon-fixation cycle. Integration of these two types of signals may be required for the efficient adjustment of photosynthetic membrane protein synthesis to fluctuating light intensity and/or quality.
Acknowledgments
We thank I. Ohad for helpful discussions and U. Pick for assistance with the oxygen-evolution experiments. A.D. holds the Judith and Martin Freedman Career Developmental Chair. This work was supported by Grant 651/00 from the Israel Science Foundation and by a grant from the Minerva Foundation.
Abbreviations
- DBMIB
2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone
- DCMU
3-(3′,4′-dichlorophenyl)-1,1′-dimethyl urea
- DCPIP
2,6-dichlorophenol-indo-phenol
- Fd
ferredoxin
- PAO
phenyl arsine oxide
- PQ
plastoquinone
- PS
photosystem
- VDS
vicinal dithiol site
References
- 1.Klein R R, Mullet J E. J Biol Chem. 1987;262:4341–4348. [PubMed] [Google Scholar]
- 2.Fromm H, Devic M, Fluhr R, Edelman M. EMBO J. 1985;4:291–295. doi: 10.1002/j.1460-2075.1985.tb03628.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Krupinska K, Apel K. Mol Gen Genet. 1989;219:467–473. [Google Scholar]
- 4.Malnoë P, Mayfield S P, Rochaix J-D. J Cell Biol. 1988;106:609–616. doi: 10.1083/jcb.106.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berry J O, Breiding D E, Klessig D F. Plant Cell. 1990;2:795–803. doi: 10.1105/tpc.2.8.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng X-W, Gruissem W. EMBO J. 1988;7:3301–3308. doi: 10.1002/j.1460-2075.1988.tb03200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mattoo A K, Pick U, Hoffman-Falk H, Edelman M. Proc Natl Acad Sci USA. 1981;78:1572–1576. doi: 10.1073/pnas.78.3.1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wettern A K, Ohad I. Isr J Bot. 1984;33:253–263. [Google Scholar]
- 9.Klein R R, Mason H S, Mullet J E. J Cell Biol. 1988;106:289–301. doi: 10.1083/jcb.106.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Edhofer I, Muhlbauer S K, Eichacker L A. Eur J Biochem. 1998;257:78–84. doi: 10.1046/j.1432-1327.1998.2570078.x. [DOI] [PubMed] [Google Scholar]
- 11.Kim J, Mullet J E. Plant Mol Biol. 1994;25:437–448. doi: 10.1007/BF00043872. [DOI] [PubMed] [Google Scholar]
- 12.Mayfield S P, Yohn C B, Cohen A, Danon A. Annu Rev Plant Physiol Plant Mol Biol. 1995;46:147–166. [Google Scholar]
- 13.Minami E-I, Shinohara K, Kawakami N, Watanabe A. Plant Cell Physiol. 1988;29:1303–1309. [Google Scholar]
- 14.Muhlbauer S K, Eichacker L A. J Biol Chem. 1998;273:20935–20940. doi: 10.1074/jbc.273.33.20935. [DOI] [PubMed] [Google Scholar]
- 15.Zhang L X, Paakkarinen V, van Wijk K J, Aro E M. Plant Cell. 2000;12:1769–1781. doi: 10.1105/tpc.12.9.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kuroda H, Kobashi K, Kaseyama H, Satoh K. Plant Cell Physiol. 1996;37:754–761. [Google Scholar]
- 17.Kettunen R, Pursiheimo S, Rintamaki E, Van Wijk K J, Aro E M. Eur J Biochem. 1997;247:441–448. doi: 10.1111/j.1432-1033.1997.00441.x. [DOI] [PubMed] [Google Scholar]
- 18.Trebitsh T, Levitan A, Sofer A, Danon A. Mol Cell Biol. 2000;20:1116–1123. doi: 10.1128/mcb.20.4.1116-1123.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karpinski S, Escobar C, Karpinska B, Creissen G, Mullineaux P M. Plant Cell. 1997;9:627–640. doi: 10.1105/tpc.9.4.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Escoubas J M, Lomas M, LaRoche J, Falkowski P G. Proc Natl Acad Sci USA. 1995;92:10237–10241. doi: 10.1073/pnas.92.22.10237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petracek M E, Dickey L F, Huber S C, Thompson W F. Plant Cell. 1997;9:2291–2300. doi: 10.1105/tpc.9.12.2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pfannschmidt T, Nilsson A, Allen J. Nature (London) 1999;397:625–628. [Google Scholar]
- 23.Danon A, Mayfield S P. Science. 1994;266:1717–1719. doi: 10.1126/science.7992056. [DOI] [PubMed] [Google Scholar]
- 24.Vener A V, Ohad I, Andersson B. Curr Opin Plant Biol. 1998;1:217–223. doi: 10.1016/s1369-5266(98)80107-6. [DOI] [PubMed] [Google Scholar]
- 25.Allen J F, Bennett J, Steinback K E, Arntzen C J. Nature (London) 1981;291:25–29. [Google Scholar]
- 26.Pursiheimo S, Rintamaki E, Baena-Gonzalez E, Aro E M. FEBS Lett. 1998;423:178–182. doi: 10.1016/S0014-5793(98)00088-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buchanan B B. Arch Biochem Biophys. 1991;288:1–9. doi: 10.1016/0003-9861(91)90157-e. [DOI] [PubMed] [Google Scholar]
- 28.Ruelland E, Miginiac-Maslow M. Trends Plant Sci. 1999;4:136–141. doi: 10.1016/s1360-1385(99)01391-6. [DOI] [PubMed] [Google Scholar]
- 29.Schürmann P. Methods Enzymol. 1995;252:274–283. doi: 10.1016/0076-6879(95)52030-9. [DOI] [PubMed] [Google Scholar]
- 30.Gorman D S, Levine R P. Proc Natl Acad Sci USA. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Goldschmidt-Clermont M, Malnoe P, Rochaix J-D. Plant Physiol. 1989;89:15–18. doi: 10.1104/pp.89.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belknap W R. Plant Physiol. 1983;72:1130–1132. doi: 10.1104/pp.72.4.1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arnon D. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trebst A. Methods Enzymol. 1980;69:675–715. [Google Scholar]
- 35.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 36.Vanlerberghe G C, McIntosh L, Yip J Y. Plant Cell. 1998;10:1551–1560. doi: 10.1105/tpc.10.9.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim J, Mayfield S P. Science. 1997;278:1954–1957. doi: 10.1126/science.278.5345.1954. [DOI] [PubMed] [Google Scholar]
- 38.Gal A, Zer H, Ohad I. Physiol Plant. 1997;100:869–885. [Google Scholar]
- 39.Carlberg I, Andersson B. Photosynth Res. 1996;47:145–156. doi: 10.1007/BF00016177. [DOI] [PubMed] [Google Scholar]
- 40.Carlberg I, Rintamaki E, Aro E M, Andersson B. Biochemistry. 1999;38:3197–3204. doi: 10.1021/bi982506o. [DOI] [PubMed] [Google Scholar]
- 41.Bustos S A, Schaefer M R, Golden S S. J Bacteriol. 1990;172:1998–2004. doi: 10.1128/jb.172.4.1998-2004.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stern D B, Higgs D C, Yang J J. Trends Plant Sci. 1997;2:308–315. [Google Scholar]
- 43.Rochaix J-D. Plant Mol Biol. 1996;32:327–341. doi: 10.1007/BF00039389. [DOI] [PubMed] [Google Scholar]
- 44.Mullet J E. Plant Physiol. 1993;103:309–313. doi: 10.1104/pp.103.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Danon A. Plant Physiol. 1997;115:1293–1298. doi: 10.1104/pp.115.4.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hartman H, Syvanen M, Buchanan B B. Mol Biol Evol. 1990;7:247–254. doi: 10.1093/oxfordjournals.molbev.a040602. [DOI] [PubMed] [Google Scholar]
- 47.Schwarz O, Schurmann P, Strotmann H. J Biol Chem. 1997;272:16924–16927. doi: 10.1074/jbc.272.27.16924. [DOI] [PubMed] [Google Scholar]
- 48.Rintamaki E, Martinsuo P, Pursiheimo S, Aro E M. Proc Natl Acad Sci USA. 2000;97:11644–11649. doi: 10.1073/pnas.180054297. . (First Published September 26, 2000; 10.1073/pnas.180054297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kramer D M, Wise R R, Frederick J R, Alm D M, Hesketh J D, Ort D R, Crofts A R. Photosynth Res. 1990;26:213–222. doi: 10.1007/BF00033134. [DOI] [PubMed] [Google Scholar]
- 50.Asada K. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:601–639. doi: 10.1146/annurev.arplant.50.1.601. [DOI] [PubMed] [Google Scholar]