Abstract
Dental pulp stromal cells (DPSCs) can be differentiated down lineages known to either express bone or dentin specific protein markers. Since the differentiation of cells can be heavily influenced by their environment, it may be possible to influence the osteogenic/odontogenic potential of DPSCs by modulating the mechanical properties of substrate on which they are grown. In this study, human DPSCs were grown with and without hydroxyapatite (HA) microparticles on a range of substrates including fibronectin-coated hydrogels and glass substrates, which represented an elastic moduli range of ~ 3 kPa – 50 GPa, over a 21 day period. Alkaline phosphatase activity, osteopontin production, and mineralization were monitored. The presence of HA microparticles increased the relative degree of mineralized matrix produced by the cells relative to those in the same substrate and media condition without the HA microparticles. In addition, cultures with cells grown on stiffer substrates had higher ALP activity and higher degree of mineralization than those grown on softer substrates. This study shows that DPSCs are affected by the mechanical properties of their underlying growth substrate and by the presence of HA microparticles. In addition, relatively stiff substrates (> 75 kPa) may be required for significant mineralization of these cultures.
Keywords: polyacrylamide hydrogels, elastic modulus, dental pulp cells, mineralization, odontogenic markers
Introduction
It is well known that bone marrow stem cells (BMSCs) differentiate into osteoblasts, adipocytes, chondrocytes, neurons, myocytes, and other mature cell types when exposed to appropriate chemical environments (growth factors and inorganic compounds)[1]. In addition, studies have reported that BMSCs are very sensitive to their mechanical environment; the elastic modulus of the underlying substrate can strongly influence BMSCs’ differentiation potential [2,3]. In more recent work, dental pulp stromal cells (DPSCs) have shown differentiation characteristics similar to BMSCs [1,4,5], and thus DPSCs may prove to be an alternative cell source for regeneration of several tissue types [6]. For instance, previous research has shown that cells isolated from human dental pulp can form a structure analogous to the dentin-pulp complex [1,4,7,8]. DPSCs have also been reported to be strongly influenced by their external chemical environment [7,9–12].
Prior studies have characterized the influence of a range of chemical cocktails on the differentiation of DPSCs. For example, an osteogenic cocktail of dexamethasone, L-ascorbic acid 2-phosphate and β-glycerophosphate (or another organic phosphate source [13]) induced DPSCs will differentiate to osteogenic/odontogenic-like lineage during extended culture times without the addition of induction factors [6,8,9,14]. Neuronal differentiation of DPSCs was achieved using the following process: first inducing epigenetic reprogramming with 5-azacytidine, followed by concurrently activating the protein kinase C (PKC) and protein kinase A (PKA) pathways, and lastly exposing the cells to a neurogenic cocktail (including neurotrophin-3, nerve growth factor, bFGF, EGF, and retinoic acid) [9,15]. The addition of β-mercaptoethanol and butylated hydroxyanisole into the cell media was also found to induce neurogenic differentiation [13]. Smooth muscle cell differentiation was been reported to occur in the presence of Transforming Growth Factor (TGF)-β [14], dexamethasone and hydrocortisone [13]. Adipocyte differentiation has been shown after intermittent exposure to dexamethasone for several weeks or by a more potent adipogenic cocktail [15,16]. These studies revealed that by carefully varying the chemical environment, DPSCs could differentiate into a range of lineages.
Although it is possible to induce both DPSCs and BMSCs to similar lineages, the magnitude of influence that an external factor or combination of factors have is not identical between these cell types. In contrast to BMSCs, prior articles indicate that it is more difficult to induce myogenic, adipogenic and chondrogenic differentiation of DPSCs compared to inducing osteogenic/odontogenic and neurogenic differentiation. This difference is possibly due to unknown naturally occuring mechanisms, since odontoblasts and neurons are readily found in teeth while adipocytes, myocytes, and chondrocytes are rarely if ever found [13]. However, it is still possible to achieve these cell types by applying differentiation factors in high enough concentrations and for an appropriate amount of time [1,6,9,13,16]. While chemical induction cues are indeed very important for cell differentiation, cells also respond to a variety of non-chemical factors including substrate mechanical properties. These non-chemical cues may prove to be crucial to the design of successful tooth and periodontal repair using tissue-engineering or regenerative medicine strategies. More work is needed to understand the combined impact of growth factors and/or inorganic compounds with the mechanical performance (stiffness) of their underlying substrate on DPSC differentiation.
Earlier reports have shown that varying a construct’s mechanical properties, specifically the elastic modulus, can altering the response of BMSCs to a tissue-engineering scaffold [2,17,18]. Using a range of elastic moduli, from less than 1 kPa up to ~ 40 kPa, Engler et al. [2] elucidated the differentiation potential of BMSCs on polyacrylamide hydrogels. They determined that a stiffness range of 0.1–1 kPa induced neurogenic differentiation, a stiffness range of 8–17 kPa resulted in myogenic differentiation, and a stiffness range of 25–40 kPa encouraged osteogenic differentiation [2].
Though research has been undertaken to explain the effects of varying substrate stiffness for the growth and differentiation of BMSCs [2], few studies have focused on DPSCs [19,20]. These studies used soft gel matrices with the mechanical properties varied over a limited range of values (less than 3.6 kPa). The gel mechanical properties were varied by altering crosslinking that can also affect the density of attachment sites for the cells. Given that the elastic modulus of dental tissues vary from about 5.5 kPa for the pulp to 70 GPa for the enamel [21,22]. Thus, the goal of this study is to elucidate the range of substrate moduli that can help to induce these dental cells towards lineages relevant for tooth repair. Here, we investigated the differentiation potential of dental pulp stromal cells to an osteoblast/odontoblast-like lineage on polyacrylamide hydrogels with a range of elastic moduli. This study includes substrates with stiffness varying between 3 kPa to 60 GPa. Since DPSCs share many properties with BMSCs, we hypothesize that a relatively stiff area of a tissue-engineering construct can induce odontoblast/osteoblast-like differentiation, while a less stiff area would induce differentiation along other lineages (e.g., neural).
Materials and Methods
Isolation and purification of dental pulp stromal cells
The DPSCs used in this study were considered to be stem/progenitor cells since they isolated from dental pulp tissue. After institutional review board approval, extracted third molar human teeth were obtained from dentists at either the Medical University of South Carolina or a local dental office. These teeth were extracted for necessary dental procedures (e.g. impaction or crowding), and thus these discarded teeth could easily be collected for research. Cells were isolated using the protocol described by Liu [23] as follows:
Extracted teeth were placed into sterile saline for initial storage, their external surfaces were subsequently cleaned and sterilized with 70% (v/v) ethanol, and then rinsed several times with sterile phosphate-buffered saline (PBS). Sterilized dental fissure burs were used to expose the pulp cavity. Next, the pulp tissue was removed and placed in αMEM using sterile forceps and minced with a sterile scalpel blade. This minced tissue was then digested in a solution of 3 mg/mL collagenase type I and 4 mg/mL dispase for 30–45 min at 37 °C. After the digestion process, additional αMEM was added to this mixture, and the cell suspension was centrifuged at 1,200 rpm for 10 min. The cell pellet was then re-suspended in mesenchymal stem cell growth medium and passed through a 70 μm cell strainer. The resultant single-cell suspension was finally cultured in mesenchymal stem cell growth medium at 37 °C and 5% CO2 [23].
To select for purified DPSCs, the cell suspension was incubated with a STRO-1 (mouse IgM) antibody (Abcam, Cambridge) for 1 hr on ice [6]. The cells were washed twice with PBS/1% (w/v) bovine serum albumin (BSA) and then incubated with either sheep anti-mouse IgG-conjugated or rat anti-mouse IgM-conjugated Dynabeads® for 1 hr, shaking at 4 °C. The bead positive cells were selected out with a Dynal® MPC-1 magnetic particle concentrator and then cultured as described below [23].
Cell culture conditions
Cells were cultured in one of the following three medium types: mesenchymal stem cell growth medium (MSCGM), odontogenic differentiation medium (ODM), or odontogenic differentiation medium with the addition of hydroxyapatite (HA) microparticles (ODM+HA). The MSCGM consisted of αMEM containing 15% fetal bovine serum, 2 mM GlutaMAX™, 100 μM L-ascorbic acid 2-phosphate, 2.5 μg/mL amphotericin B, 100 U/mL penicillin, and 100 μg/mL streptomycin. The ODM comprised of αMEM containing 15% fetal bovine serum, 2 mM GlutaMAX™, 100 μM L-ascorbic acid 2-phosphate, 1.8 mM KH2PO4, 10 nM dexamethasone, 2.5 μg/mL amphotericin B, 100 U/mL penicillin and 100 μg/mL streptomycin. The ODM+HA included spherical HA microparticles (MP Biomedicals, LLC.). These microparticles had an average particle diameter of ~ 40 μm and were porous with an average pore size of 70 nm as assessed by electron microscopy. The HA microparticles were added to deionized (DI) water at a concentration of 10 mg/mL and sterilized by steam in an autoclave. This particle suspension was briefly vortexed, and then 50 μL were added to the 3 mL ODM media for each of the ODM+HA cultures at Day 0 only.
The cells were cultured on the polyacrylamide hydrogel substrates in six well cell culture plates with approximately 3 mL of media. DPSCs were plated on each substrate at a density of ~ 2000 cells/cm2 and given 30 min to adhere before MSCGM was added. The cells were allowed one day to stabilize, and then the media was changed to the experimental conditions. In the result and discussion sections, the day when the experimental conditions were applied was considered as Day 0. Characterization of the cells were done at several time points: Days 1, 3, 5, 7 and 14 (imaging and alkaline phosphatase (ALP) assay) or at Days 7, 14, and 21 (ICC/confocal imaging of cells stained with alizarin red dye).
Substrate preparation
The polyacrylamide hydrogel used in this study has commonly been utilized within cell studies due to the relative ease with which researchers can control its elastic modulus [2,3]. In addition, other researchers have shown that a standard photoreactive crosslinker-based chemistry can be used to easily modify the surface of polyacrylamide to attach extracellular matrix (ECM) proteins and tailor cellular attachment [3]. In this study, polyacrylamide hydrogels of varying stiffness were synthesized by varying the ratio of the acrylamide monomer to the bis-acrylamide monomer in the monomer solution prior to polymerization using the protocol described earlier by Tse and Engler [24].
The polyacrylamide hydrogel substrates were deposited on glass coverslips (No. 1). First, the glass coverslips were chemically activated to allow the polyacrylamide to strongly adhere to the glass. The glass was first cleaned in a 1M NaOH solution for several min, and then removed and air dried. Next, 3-aminopropyltriethoxysilane (APTES) was dropped on top of the glass and allowed to react for five min. The glass was then rinsed with deionized (DI) water for five min while shaking to remove excess unreacted APTES. This process was repeated at least three times to ensure complete removal. Next, a solution of 0.5% (v/v) gluteraldehyde was added to the top of the glass c and allowed to react for 30 min. Again, the glass was rinsed with DI water three times while shaking. All glass coverslips were either allowed to air dry if polyacrylamide hydrogens were going to be deposited immediately or the glass coverslips stored in DI water used later within 48 hr [3].
The polyacrylamide hydrogels were polymerized on the activated glass coverslips in water. Acrylamide and bis-acrylamide monomers (in the amounts shown within Table 1) were added into a small glass beaker along with 1 M HEPES buffer and DI water while stirring gently. Next, 30 μL of ammonium persulfate (10 mg/100 μL of PBS) and 20 μL of tetramethylethylenediamine (TEMED) were added to this mixture. Immediately afterward, 25 μL of the acrylamide solution was dropped on an activated glass coverslip and then covered with an clean (un-activated) coverslip. The hydrogel on the glass coverslip was then allowed to polymerize (approximately 1–2 hr). A pair of thin forceps was used to remove the top glass coverslip and each hydrogel-glass coverslip composite was placed into an individual well (six well plate). The hydrogel-glass coverslip composite was finally rinsed with PBS twice before being stored in PBS. If needed, approximately 2 mL of 50 mM HEPES solution was added to enhance the removal of the top coverslip.
Table 1.
Acrylamide/bis-acrylamide addition table and respective elastic modulus, calculated from AFM nanoindentation measurements.
| Acrylamide (%)/Bis-Acrylamide (%) | 40% Acrylamide solution | 2% Bis-Acrylamide solution | DI Water | 1 M HEPES | Elastic Modulus (kPa) |
|---|---|---|---|---|---|
| 8/0.06 | 1000 μL | 150 μL | 3800 μL | 50 μL | 3.32 ± 0.29 |
| 8/0.1 | 1000 μL | 250 μL | 3700 μL | 50 μL | 9.78 ± 1.00 |
| 8/0.48 | 1000 μL | 1200 μL | 2750 μL | 50 μL | 45.29 ± 1.45 |
| 8/0.8 | 1000 μL | 2000 μL | 1950 μL | 50 μL | 75.07 ± 5.10 |
Since polyacrylamide does not promote cellular attachment, an ECM protein coating was necessary. Fibronectin was selected as the ECM protein and attached covalently to the hydrogel via sulfo-SANPAH (sulfosuccinimidyl 6-(4′-azido-2′-nitrophenylamino)hexanoate) chemistry. The procedure began with the PBS being removed from each well. Next, ~ 500 μL of 0.2 mg/mL sulfo-SANPAH was added to each hydrogel. The hydrogels were placed approximately 7.5 cm from an ultraviolet (UV) light source for 10 min. The sulfo-SANPAH was then removed, and the gels were rinsed twice with 50 mM HEPES to remove excess sulfo-SANPAH. Finally, 200 μL of a solution of 125 μg/mL fibronectin in 50 mM HEPES was added to each hydrogel and allowed to react with the N-hydroxysuccinimide ester in the sulfo-SANPAH overnight at 37 °C. The hydrogels were then rinsed with PBS. This method achieved ~ 5 μg/cm2 fibronectin coating density on the gel [3, 25, 26]. To sterilize the gels prior to seeding with DPSCs, the hydrogels were placed in 1 mL of sterile PBS and placed in a biohazard laminar flow hood under UV light for 30 min [3].
Atomic force microscopy (AFM) indentation was used to estimate the elastic modulus of each hydrogel. A 2.5 μm radius borosilicate spherical tip with a spring constant of 0.12 N/m was used to indent the hydrogels at a speed of 1 μm/sec to a depth of approximately 1 μm. A total of five individual locations in each hydrogel were characterized and each location was indented five times. The linear elastic Hertz model was fit to the first 500 nm of each indentation curve using an in house MATLAB (MathWorks, Natick, MA) script.
Cell imaging
Cells were imaged using both a standard inverted and confocal microscope. Images taken with an Olympus IX81 microscope (Olympus; Tokyo, Japan) were taken prior to fixing or lysing the cells. Immunocytochemistry for the protein osteopontin and standard staining of the actin filaments was performed for the confocal microscopy. For these characterization methods, the cells were initially washed with sterile PBS, then fixed in 4% (v/v) paraformaldehyde for 10 min, and finally rinsed again with PBS. The cells were permeabilized with a solution of 0.1% Triton-X and 0.01 M Glycine in PBS for 30 min. Next, they were rinsed with a 5% (w/v) BSA solution (in PBS) followed with a rinse of 5% (v/v) normal donkey serum (in a 1% BSA solution in PBS). A mouse anti-osteopontin IgG1 antibody (diluted 1:10 in 1% BSA in PBS) was added and incubated overnight at 4 °C. This antibody, developed by M. Solursh and A. Franzen, was obtained from the Developmental Studies Hybridoma Bank, created by the NIH NICHD, and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242. After incubation with the primary antibody, the cells were rinsed twice with 1% BSA (in PBS) and once with 5% (v/v) normal donkey serum (in a 1% BSA solution in PBS). Next, a 2 mg/mL Alexa Fluor® 647 donkey anti-mouse IgG (diluted 1:100 in 1% (w/v) BSA in PBS) was added for 2 hr at 37 °C. The cells were rinsed in 1% BSA (in PBS) and then in PBS. Afterwards, Alexa Fluor® 488 Phalloidin (1:100 in PBS) was added at room temperature for 1 hr while shaking. The cells were rinsed twice with PBS. Prior to mounting the hydrogels on glass slides for confocal microscopy, SlowFade® Gold antifade reagent was added. Images were taken on a Zeiss LSM 510 confocal microscope or a Nikon Ti Eclipse and C1si CLSM confocal microscope with a 40X oil lens or a 60X water lens, respectively.
Alizarin red staining
Cell cultures were analyzed for calcification with an alizarin red stain. Alizarin red is a standard histological stain used to determine calcium accumulation in tissues and cell cultures by staining them red. The cells were fixed with the same method described earlier for confocal microscopy staining. A 1% (w/v) alizarin red solution (in DI water with 10% (v/v) ammonium hydroxide) was added to the fixed cells for approximately 10 min. Excess stain was then removed and the fixed cells were washed with five or six changes of DI water. The cells were then counterstained with a 1% (w/v) light green solution (DI water with 1% (v/v) glacial acetic acid) for approximately 10 sec. Excess green stain was then removed and the cells were washed with several changes of DI water. The stained cells were stored in DI water until imaging.
Bicinchoninic Acid (BCA) and Alkaline phosphatase (ALP) specific activity
At each time point, the cells were also evaluated for alkaline phosphatase specific activity. ALP, an enzyme commonly found in osteogenic/odontogenic cultures, is an early marker of odontogenic differentiation of stem cells [11,27]. Cells reserved for this assay were rinsed once with sterile PBS and then lysed with Mammalian-Protein Extraction Reagent™ (M-PER, Thermo Fischer Scientific). 300 μL of the M-PER™ was applied to each polyacrylamide gel for 20 min. The lysates were collected in microcentrifuge tubes and spun down, and the supernatant was used for bicinchoninic acid (BCA) and ALP assays. A BCA kit (Thermo Fisher Scientific) was used to detect total protein in the supernatant by adding 200 μL of the kit working reagent to a 25 μL of the supernatant, allowing the reaction to occur for 30 min at 37 °C, and reading the color change at 562 nm. A standard curve was created by diluting a stock solution of 1 mg/mL bovine serum albumin (BSA) to various concentrations, creating a plot of concentration vs. absorbance, and determining the best-fit line. The ALP assay was also performed on a sample of the supernatant. The substrate for the ALP reaction was prepared by dissolving a pNPP tablet in 4 mL of DI water. Next, 1 mL of 5X concentrated diethanolamine buffer and 50 μL of 0.01 g/mL MgCl2 was added. A 50 μL sample of the supernatant, 50 μL of DI water, and 300 μL of the assay substrate were mixed in a microcentrifuge tube. The ALP enzyme in the sample catalyzes the dephosphorylation of the colorless pNPP (p-nitrophenol phosphate) to yellow pNP (p-nitrophenol). The concentration of pNP in solution when the reaction is stopped is an indication of the amount of ALP present in the total protein solution. The reaction was allowed to proceed for 30 min at 37 °C and then stopped with 25 μL of 2 M NaOH, and the color change was read at 405 nm. A standard curve was created by diluting a stock solution of 400 μg/mL pNP to various concentrations, adding the 2 M NaOH, creating a plot of concentration vs. absorbance, and determining the best-fit line. The ALP specific activity was determined by normalizing the amount of ALP enzyme, which is related to the amount of product formed in the reaction (pNP), to the total protein in each culture. This normalization helps to account for changes in cell numbers between cultures.
Statistical analysis of ALP specific activity
Statistical analysis was performed for the results of the BCA total protein and ALP specific activity assays using custom SAS® software (SAS Institute Inc., Cary, NC) code. A general linear model was fit with either the BCA total protein or ALP specific activity as the response, and three possible treatment effects: substrate, media, and a substrate-by-media interaction. The initial model was analyzed under the assumption of constant variance for the substrate effect, media effect, and substrate-by-media interaction effect. The variance was determined not to be constant across the time points and substrates by examining the predicted versus residual plot. Since the assumptions of constant variance were not satisfied, an appropriate mixed model was fit. In model, the variance for each substrate was estimated and applied to the corresponding substrate data. The variances were estimated for the substrates since these were causing issues with the general linear model. The mixed model tested first for overall effects due to the substrate and media and then for interactions between the substrate and media type. There were no substrate-media interactions for any of the time points. Multiple comparisons were conducted using Tukey’s adjustment for multiplicity and significance was judged using α = 0.05. Statistical analysis for the Alizarin Red quantification and elastic modulus measurements were done using a Student’s t-test assuming unequal variance and significance was judged using α = 0.05.
Results
The measured elastic moduli of the polyacrylamide hydrogel substrates spanned between 3–75 kPa (Table 1). These values were consistent with previous findings [2,3]. Results also showed that the thin fibronectin layer on top of the gels did not alter the gel’s mechanical properties of the gel; the moduli measured by AFM of the gels with and without fibronectin were not found to be statistically different using a Student’s t-test (p≫0.05).
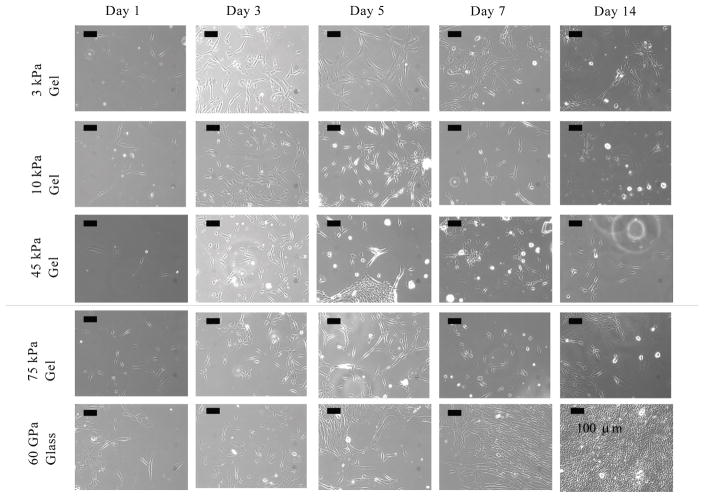
The cells initially attached and grew homogeneously on all of the polyacrylamide hydrogels until approximately Day 5. Images of the cells on all substrates in MSCGM media for Days 1, 3, 5, 7 and 14 are shown in Figure 1. These images show the initial attachment of the cells, which seemed to last until Day 5 for all culture variations. This indicated that the fibronectin coating provided a sufficient number of adhesion sites. In addition, no change in morphology was observed in any cell culture except for cells on the 3 kPa hydrogel substrate, on which the cells seemed to elongate.
Figure 1.
Phase contrast images of DPSCs at Days 1, 3, and 5 cultured on 3 kPa hydrogel (a–c respectively), 10 kPa hydrogel (d–f respectively), 45 kPa hydrogel (g–i respectively), 75 kPa hydrogel (j–l respectively) and glass in MSCGM (m–o respectively). This figure shows that the cells did not undergo a morphology change (i.e. maintained typical triangular morphology common among stromal cells) over Days 1–5 on the 10 kPa, 45 kPa, 75 kPa or glass substrates, but did undergo elongation on the relatively softer 3 kPa substrate. (All scale bars are 100 μm.)
Starting on Day 7, the cells started to detach from parts of the hydrogel and survived better on either the edges of the coverslips or in gel defects. This change in cell adhesion suggested that the fibronectin might not be effective for long-term adhesion to substrates. In these spaces, where the cells could attach to the glass directly, the cells rapidly became confluent. It was also observed that in the media with HA microparticles, the cells clustered around the particles.
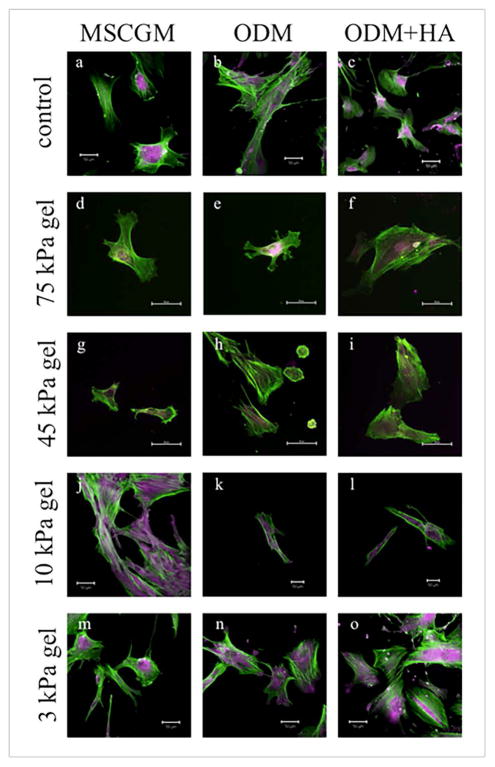
The immunocytochemistry performed on fixed cells was used as a detection method for the protein osteopontin. Osteopontin, a highly phosphorylated sialoprotein produced by odontogenic and odontogenic cells, is typically found in mineralized tissues. Thought to be a widely used protein in many biological events, osteopontin seems to be an important component in the formation, remodeling, and maintenance of bones and teeth [4,7,28]. In Figure 2, the osteopontin in the cells was stained purple and the actin was green. The presence of osteopontin protein was observed in all the cells, no matter which substrate they were on or the type of media type used (Figure 2). In addition, the osteopontin was present throughout the cytoplasm. On Day 7, it appeared that, in a few cells, vesicles containing osteopontin were pinched off (Figure 2n and o are examples). These osteopontin filled vesicles were seen only in the ODM cultures and were more prevalent in the media with the HA (ODM + HA).
Figure 2.
Confocal images of DPSCs on glass (a–c), 75 kPa hydrogel (d–f), 45 kPa hydrogel (g–i), 10 kPa hydrogel (j–l), and 3 kPa hydrogel (m–o) for each media condition at Day 7. The green color indicates actin filaments, while the purple color indicates osteopontin (OPN) within the cells. OPN was evident in all cultures by Day 7, as noted by the purple coloring seen in all images, typically in and around the center of the cell. (All scale bars are 50 μm.)
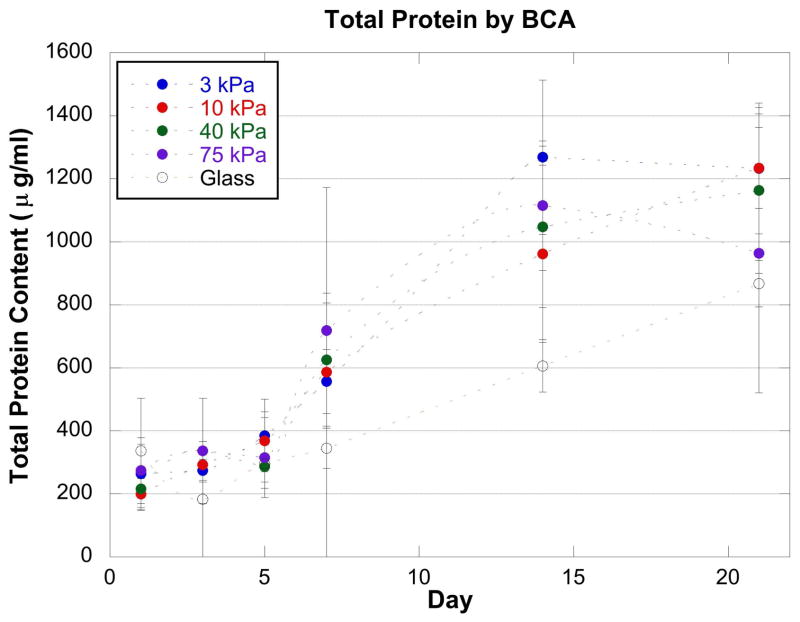
The BCA assay was used to determine the total protein in each culture (Figure 3). The total protein increased with time in all conditions. This increase is consistent with the increase in cell numbers seen with the microscopy images (Figure 1). There are no statistical differences in total protein between the different conditions on the same day. The ALP assay was used to determine the specific activity (μmol S/mg protein/min) of the enzyme alkaline phosphatase normalized to the total protein in each culture (Figure 4). Alkaline phosphatase is readily produced in osteogenic/odontogenic cells to aid with mineralization; the detection of an increase in the enzyme activity was used to indicate early osteogenic/odontogenic differentiation [4,7,29]. The confluence of the cells in the various cultures seemed to greatly affect the production of ALP. The specific activity of ALP was very low through Day 7 of the culture and then there was a sharp increase between Day 7 and Day 21. The cells that were not confluent by Day 7 of the experiment, however, became confluent by Days 14 and 21. This might explain the increase in ALP specific activity as confluency and cell crowding in culture is known to affect ALP activity in other cell types [30–32]. It was also observed that at Day 14 and 21, the experiments on the glass substrate exhibited a large variance (on the same order of magnitude as the specific activity value). A slight possible variation between the glass coverslips used for the different experiments may have caused a difference in how the cells grew on the glass or how quickly they became confluent.
Figure 3.
A BCA assay results showed that the total protein increased with time in all conditions. However, there were no statistical differences in total protein between the different conditions on the same day.
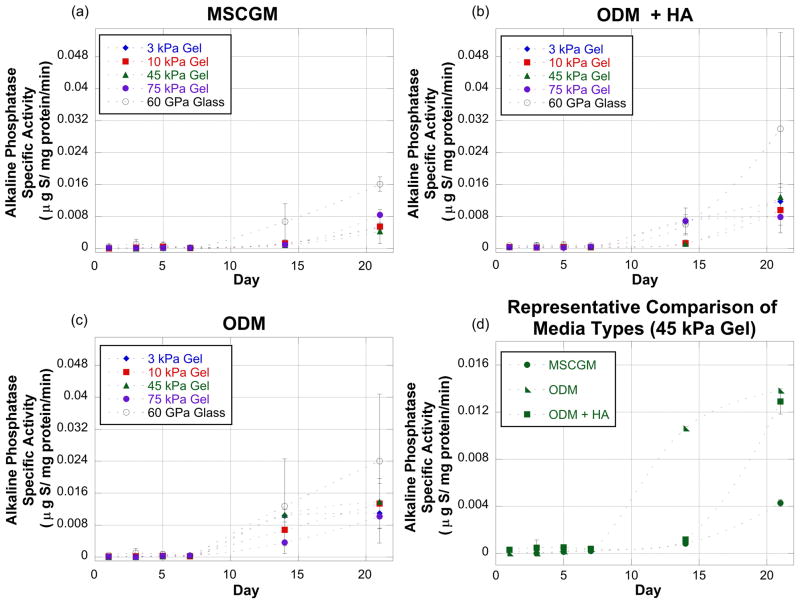
Figure 4.
Alkaline phosphatase (ALP) specific activity measured at Days 1, 3, 5, 7 and 21 for the various substrates and culture conditions. Graphs (a–c) represent ALP activity measurements for cells exposed to MSCGM (a), ODM (b), and ODM+HA (c) culture conditions on a range of substrate stiffness values. Graph (d) is a representative comparison of all three culture conditions for one substrate. The media condition graphs (a, b, and c) show that by Day 21 the stiffest substrate (glass) always induced the greatest ALP production. The representative comparison graph (d) shows that the MSCGM media condition resulted in the lowest ALP production when the substrate was glass. This trend (MSCGM resulting in the lowest ALP production) was true for all substrates.
The very low ALP specific activity for Days 1, 3, 5, and 7 in culture always was followed by a sharp increase for Days 14 and 21 in culture (Figure 4). On all substrates it was noted that by Day 7, the cells in the MSCGM generally had the lowest ALP activity over the course of the experiments. This may indicate early osteogenic/odontogenic differentiation started after Day 7 in culture. Also, it could not be determined if the addition of HA microparticles either accelerated or hindered ALP production. At some time points, the cells with HA microparticles produced less ALP, while at others they produced more ALP. Finally, though there was substantial variability in the data, the cells cultured on the glass exhibited a consistently higher ALP production throughout the experiment. This rate of production is consistent with the hypothesis that mesenchymal stem cells follow an osteogenic lineage on rigid substrates [2]. It appears, however, that the dental pulp cells were more sensitive to the hydrogel stiffness than previously reported with bone marrow stromal cells, as osteogenic differentiation could occur on substrates with an elastic modulus of 25 kPa to 40 kPa [2].
The ALP production by cells on 3 kPa gel, 10 kPa gel, and glass were analyzed. The model was fit for Day 1, 3, 5, 7, 14, and 21. Since the constant variance assumption was not satisfied for each day, a mixed model was fit with a different variance for each of the substrate types. On all days, there were no significant interaction effects between the substrate type and media type on ALP activity. In addition, there were no fixed substrate or media effects on ALP activity for Day 1, 3, 5, 14, and 21. However, the variations used in the model show an interesting trend. The variance for the glass substrate was at least one order of magnitude larger than the variances for both the 3 kPa and 10 kPa substrates, as seen in Table 2.
Table 2.
Variances for each substrate were estimated and used in the mixed model for each substrate on each day. There were no substrate-media interactions for any of the time points.
| Source: | 3 kPa gel | 10 kPa gel | Glass |
|---|---|---|---|
| Day 1 | 4.55 × 10−9 | 2.90 × 10−9 | 1.90 × 10−7 |
| Day 3 | 1.09 × 10−8 | 3.31 × 10−8 | 1.03 × 10−6 |
| Day 5 | 3.81 × 10−9 | 3.58 × 10−8 | 6.76 × 10−7 |
| Day 7 | 2.88 × 10−9 | 5.04 × 10−9 | 4.22 × 10−8 |
| Day 14 | 4.80 × 10−6 | 5.65 × 10−6 | 5.50 × 10−5 |
| Day 21 | 2.60 × 10−5 | 3.00 × 10−5 | 2.89 × 10−4 |
Though no significant effect of the substrate was observed on Day 7 (p = 0.08), there was a significant effect from the media (p = 0.04). Specifically, a significant difference was found in the average ALP specific activity between the MSCGM and ODM+HA media types (p = 0.03). As with the other days, the variance for the glass substrate was an order of magnitude larger than the variances for the other two substrates. The evidence of the media effect only on Day 7 may indicate that this was an anomaly and there are no true differences between the conditions for the whole experimental timeline. Though it appears that the glass substrates elicited the highest ALP response, the variation between samples was too great to achieve statistical significance, as is common with many biological samples.
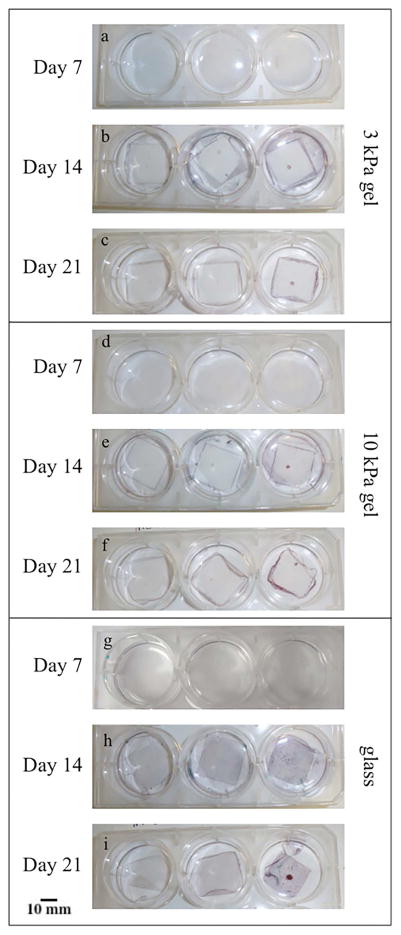
The alizarin red stain was used to visualize calcium deposits within the cell cultures, and the Light Green stain was used to visualize any collagen produced by the cells (Figure 5). It was noted that the areas where cells tended to become confluent quickly, such as in gel defects or along the edges of the gel, were the only areas with significant mineralization. This rapid confluence may indicate that the confluency of the cells in certain areas led to the mineralization and not the elasticity of the substrate. It was also noticed that there was a sheet of mineralized matrix surrounding the gel substrates, but not on the hydrogel itself. This sheet of mineralized matrix freely lifted off of the bottom of the well of the six well plate. As expected, the amount of calcium present increased with increasing culture time; the alizarin red staining was significantly higher by day 14 in all the cultures (p = 0.0017 for the 10kPa MSCGM condition, p<0.001 for all others). There was no substantial accumulation of collagen was observed in any of the cultures, as indicated by the green counterstain. In contrast, the sheet of mineralization described around the edge of the gel substrates was present across the whole glass coverslip for the control experiment.
Figure 5.
Alizarin red staining of cells on the 3 kPa, 10 kPa and glass substrates. Red staining in the images indicates Ca2+ (mineralization), while green staining indicates collagen. The areas where cells tended to become confluent quickly, such as in gel defects or along the edges of the gel, were the only areas with significant mineralization.
Image analysis was used to quantify the amount of red staining on each substrate. An image was taken of each coverslip or hydrogel individually, and was cropped down so that just the coverslip or hydrogel was visible. Next red pixels in the image were isolated and the percentage of red pixels present, with respect to the total number of pixels in the image, was calculated. The relative percentage of red stained area for each substrate (Figure 5) was calculated to quantify the differences observed in the images of the alizarin red stained cultures. The percentage of stained area increased for all substrates and media conditions while progressing through the time points, as a significant increase in staining occurred between Day 7 and Day 14. In addition, the highest percentages of staining were seen on the glass substrates and more specifically, on the glass substrate with ODM+HA.
Discussion
No neurogenic or myogenic phenotypic shift was observed through the morphology observations even on the lower modulus substrates. However, markers for those phenotypes were not specifically addressed in this study. In addition, this absence of shift was not unexpected since, unlike experiments from previously reported studies on BMSCs [2], there were no neurogenic or myogenic differentiation factors in the media in this study. DPSCs produced osteopontin on all substrates and media conditions. In contrast, Engler et al. observed osteopotin staining on BMSC in similar media conditions grown on similar polyacrylamide substrates only when the substrate stiffnesses ranged between 25–40 kPa [2]. However, DPSCs only showed significant mineralization with alizarin red staining on very stiff (> 75kPa) substrates. This is indicates that DPSCs may require stiffer substrates for full differentiation to bone/dental-lineages. The inclusion of HA microparticles (average diameter of 40 μm and pore size of 70 nm) seemed to aid the mineralization. In addition, the ODM+HA cultures exhibited more osteopontin filled vesicles at Day 7. After about Day 7, it was evident that the majority of living cells were growing on HA microparticles, in gel defects, or along the edges of the gel, where they could adhere directly to the hard materials (glass or HA microparticles). These areas were highly confluent and highly mineralized, as evidenced by the high degree of alizarin red staining in the culture. This indicates that substrate stiffness and cell confluence plays an important role in mineralization by DPSCs in culture.
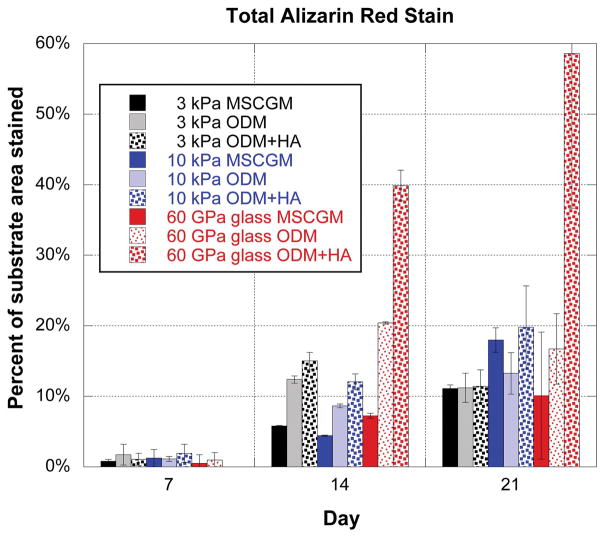
The ALP activities in the cultures increased significantly during the second week of culture. It should be noted that ALP activity is normalized by the total protein, which controls for cell number. Even with this normalization, the cultures with more ALP activity had relatively more cells than those with the lower ALP activity. The ALP activity was highest in those cultures where the cells were at or near confluency. The ALP assays also indicated that the osteogenic differentiation media (ODM condition) was successful at increasing the production of an early marker related to osteo/odontogenic differentiation. The cells in culture also produced nodules of osteopontin as seen in the immunofluorescence staining images (Figure 2). While there was statistically higher ALP activity from the cells on the glass substrates, there was no significant change in ALP as a function of substrate stiffness on the soft gels. Thus, it seems that varying the mechanical properties of the substrates in the range seen for softer musculoskeletal tissues (3–75 kPa) does not affect the ALP response of the cells. Similarly significant mineralization was only achieved in cultures with the stiffest substrates and HA particles (Figure 6).
Figure 6.
This graph shows the percentage of culture area stained during alizarin red staining process. Cell culture conditions included both a range of substrate stiffness magnitudes (3 kPa, 10 kPa and glass) and media variations (MSCGM, ODM, and ODM+HA). For each substrate type, the percentage of stained area increased from Day 7 to Day 14. The increase in staining caused by the HA microparticles is most evident for the glass substrate, as there was approximately 17% of the substrate stained on Day 21 for ODM, but almost 60% of the substrate stained in the ODM+HA media at the same time point.
In conclusion, this study shows that dental pulp stromal cells are affected by their underlying substrate stiffness and the presence of HA microparticles within the cell culture. In addition, this study indicated that relatively stiff substrates (> 75 kPa) with properties closer to dentin and enamel than pulp tissue may be required for significant mineralization of these cultures.
Acknowledgments
The completion of this work was supported by the following funding sources: SC Life/HHMI grant, National Science Foundation Graduate Research Fellowships Program, and National Institutes of Health Heart Lung and Blood Institute (NIH NHLBI) K25 grant (award number: HL092228). The authors would like to thank Ms. L. Jenkins, Dr. A. Simionescu, Ms. C. Gregory, and Dr. T. Bruce for technical assistance, and Dr. J. Sharp for assistance with statistical analysis.
References
- 1.Huang GJ, Gronthos S, Shi S. Mesenchymal stem cells derived from dental tissues vs. those from other sources: their biology and role in regenerative medicine. J Dent Res. 2009;88:792–806. doi: 10.1177/0022034509340867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Engler A, Sen S, Sweeney H, Discher D. Matrix Elasticity Directs Stem Cell Lineage Specification. Cell. 2006;126:677–689. doi: 10.1016/j.cell.2006.06.044. doi: http://dx.doi.org/10.1016/j.cell.2006.06.044. [DOI] [PubMed] [Google Scholar]
- 3.Tse J, Engler A. Preparation of hydrogel substrates with tunable mechanical properties. Curr Protoc Cell Biol. 2010:10–16. doi: 10.1002/0471143030.cb1016s47. [DOI] [PubMed] [Google Scholar]
- 4.Shi S, Bartold PM, Miura M, Seo BM, Robey PG, Gronthos S. The efficacy of mesenchymal stem cells to regenerate and repair dental structures. Orthod Craniofac Res. 2005;8:191–199. doi: 10.1111/j.1601-6343.2005.00331.x. [DOI] [PubMed] [Google Scholar]
- 5.Zhang Y, Zhi C, SONG Y, Chao L, CHEN Y. Making a tooth: growth factors, transcription factors, and stem cells. Cell Res. 2005;15:301–316. doi: 10.1038/sj.cr.7290299. [DOI] [PubMed] [Google Scholar]
- 6.Yang X, Zhang W, van den Dolder J, Walboomers XF, Bian Z, Fan M, et al. Multilineage potential of STRO-1+ rat dental pulp cells in vitro. J Tissue Eng Regen Med. 2007;1:128–135. doi: 10.1002/term.13. [DOI] [PubMed] [Google Scholar]
- 7.Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci U S A. 2000;97:13625–13630. doi: 10.1073/pnas.240309797. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC17626/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang W, Walboomers XF, Wolke JGC, Bian Z, Fan MW, Jansen JA. Differentiation ability of rat postnatal dental pulp cells in vitro. Tissue Eng. 2005;11:357–368. doi: 10.1089/ten.2005.11.357. [DOI] [PubMed] [Google Scholar]
- 9.Kadar K, Kiraly M, Porcsalmy B, Molnar B, Racz GZ, Blazsek J, et al. Differentiation potential of stem cells from human dental origin-promise for tissue engineering. J Physiol Pharmacol. 2009;60:167–175. [PubMed] [Google Scholar]
- 10.Wang W, Yi X, Ren Y, Xie Q. Effects of Adenosine Triphosphate on Proliferation and Odontoblastic Differentiation of Human Dental Pulp Cells. J Endod. 2016;42:1483–1489. doi: 10.1016/j.joen.2016.07.013. [DOI] [PubMed] [Google Scholar]
- 11.Umemura N, Ohkoshi E, Tajima M, Kikuchi H, Katayama T, Sakagami H. Hyaluronan induces odontoblastic differentiation of dental pulp stem cells via CD44. Stem Cell Res Ther. 2016;7:135. doi: 10.1186/s13287-016-0399-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alkharobi H, Alhodhodi A, Hawsawi Y, Alkafaji H, Devine D, El-Gendy R, et al. IGFBP-2 and-3 co-ordinately regulate IGF1 induced matrix mineralisation of differentiating human dental pulp cells. Stem Cell Res. 2016;17:517–522. doi: 10.1016/j.scr.2016.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang W, Walboomers X, Shi S, Fan M, Jansen J. Multilineage differentiation potential of stem cells derived from human dental pulp after cryopreservation. Tissue Eng. 2006;12:2813–2823. doi: 10.1089/ten.2006.12.2813. [DOI] [PubMed] [Google Scholar]
- 14.Laino G, D’Aquino R, Graziano A, Lanza V, Carinci F, Naro F, et al. A new population of human adult dental pulp stem cells: a useful source of living autologous fibrous bone tissue (LAB) J Bone Miner Res. 2005;20:1394–1402. doi: 10.1359/JBMR.050325. [DOI] [PubMed] [Google Scholar]
- 15.d’Aquino R, Graziano A, Sampaolesi M, Laino G, Pirozzi G, De Rosa A, et al. Human postnatal dental pulp cells co-differentiate into osteoblasts and endotheliocytes: a pivotal synergy leading to adult bone tissue formation. Cell Death Differ. 2007;14:1162–1171. doi: 10.1038/sj.cdd.4402121. http://dx.doi.org/10.1038/sj.cdd.4402121. [DOI] [PubMed] [Google Scholar]
- 16.Gronthos S, Brahim J, Li W, Fisher LW, Cherman N, Boyde A, et al. Stem cell properties of human dental pulp stem cells. J Dent Res. 2002;81:531–535. doi: 10.1177/154405910208100806. [DOI] [PubMed] [Google Scholar]
- 17.Munoz-Pinto D, Jimenez-Vergara A, Hou Y, Hayenga H, Rivas A, Grunlan M, et al. Osteogenic Potential of Poly(Ethylene Glycol)–Poly(Dimethylsiloxane) Hybrid Hydrogels. Tissue Eng Part A. 2012;18:1710–1719. doi: 10.1089/ten.tea.2011.0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.MacQueen L, Sun Y, Simmons C. Mesenchymal stem cell mechanobiology and emerging experimental platforms. J R Soc Interface. 2013;10 doi: 10.1098/rsif.2013.0179. http://rsif.royalsocietypublishing.org/content/10/84/20130179.abstract. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones TD, Kefi A, Sun S, Cho M, Alapati SB. An optimized injectable hydrogel scaffold supports human dental pulp stem cell viability and spreading. Adv Med. 2016;2016 doi: 10.1155/2016/7363579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lu Q, Pandya M, Rufaihah AJ, Rosa V, Tong HJ, Seliktar D, et al. Modulation of dental pulp stem cell odontogenesis in a tunable PEG-fibrinogen hydrogel system. Stem Cells Int. 2015;2015 doi: 10.1155/2015/525367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang Y-R, Du W, Zhou X-D, Yu H-Y. Review of research on the mechanical properties of the human tooth. Int J Oral Sci. 2014;6:61–69. doi: 10.1038/ijos.2014.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ozcan B, Bayrak E, Erisken C. Characterization of Human Dental Pulp Tissue Under Oscillatory Shear and Compression. J Biomech Eng. 2016;138:61006. doi: 10.1115/1.4033437. [DOI] [PubMed] [Google Scholar]
- 23.Liu H, Gronthos S, Shi S. Dental Pulp Stem Cells. In: IK, RLBT-M, editors. Enzymology Adult Stem Cells. Academic Press; 2006. pp. 99–113. doi: http://dx.doi.org/10.1016/S0076-6879(06)19005-9. [DOI] [PubMed] [Google Scholar]
- 24.Narayanan K, Gajjeraman S, Ramachandran A, Hao J, George A. Dentin matrix protein 1 regulates dentin sialophosphoprotein gene transcription during early odontoblast differentiation. J Biol Chem. 2006;281:19064–19071. doi: 10.1074/jbc.M600714200. [DOI] [PubMed] [Google Scholar]
- 25.Deitch S, Gao BZ, Dean D. Effect of Matrix on Cardiomyocyte Viscoelastic Properties in 2D Culture. Mol Cell Biomech. 2012;9:227–249. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3539228/ [PMC free article] [PubMed] [Google Scholar]
- 26.Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM. Force and velocity measured for single molecules of RNA polymerase. Science (80-) 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 27.Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, et al. Multilineage potential of adult human mesenchymal stem cells. Science (80-) 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 28.Sodek J, Ganss B, McKee MD. Osteopontin. Crit Rev Oral Biol Med. 2000;11:279–303. doi: 10.1177/10454411000110030101. [DOI] [PubMed] [Google Scholar]
- 29.Papagerakis P, Berdal A, Mesbah M, Peuchmaur M, Malaval L, Nydegger J, et al. Investigation of osteocalcin, osteonectin, and dentin sialophosphoprotein in developing human teeth. Bone. 2002;30:377–385. doi: 10.1016/s8756-3282(01)00683-4. [DOI] [PubMed] [Google Scholar]
- 30.Ellies LG, Aubin JE. Temporal sequence of interleukin 1α—mediated stimulation and inhibition of bone formation by isolated fetal rat calvaria cells in vitro. Cytokine. 1990;2:430–437. doi: 10.1016/1043-4666(90)90052-u. [DOI] [PubMed] [Google Scholar]
- 31.Harris SE, Bonewald LF, Harris MA, Sabatini M, Dallas S, Feng JQ, et al. Effects of transforming growth factor β on bone nodule formation and expression of bone morphogenetic protein 2, osteocalcin, osteopontin, alkaline phosphatase, and type I collagen mRNA in long-term cultures of fetal rat calvarial osteoblasts. J Bone Miner Res. 1994;9:855–863. doi: 10.1002/jbmr.5650090611. [DOI] [PubMed] [Google Scholar]
- 32.Martin JY, Schwartz Z, Hummert TW, Schraub DM, Simpson J, Lankford J, et al. Effect of titanium surface roughness on proliferation, differentiation, and protein synthesis of human osteoblast-like cells (MG63) J Biomed Mater Res. 1995;29:389–401. doi: 10.1002/jbm.820290314. [DOI] [PubMed] [Google Scholar]