Abstract
Genetic mutations in genes encoding proteins involved in epigenetic machinery have been reported in individuals with autism spectrum disorder, intellectual disability, congenital heart disease, and other disorders. H1 histone linker protein, the basic component in nucleosome packaging and chromatin organization, has not been implicated in human disease until recently. We report a de novo deleterious mutation of histone cluster 1 H1 family member e (HIST1H1E; c.435dupC; p.Thr146Hisfs*50), encoding H1 histone linker protein H1.4, in a 10-year-old boy with autism and intellectual disability diagnosed through clinical whole exome sequencing. The c.435dupC at the 3' end of the mRNA leads to a frameshift and truncation of the positive charge in the carboxy-terminus of the protein. An expression study demonstrates the mutation leads to reduced protein expression, supporting haploinsufficiency of HIST1H1E protein and loss of function as an underlying mechanism of dysfunction in the brain. Taken together with other recent cases with mutations of HIST1H1E in intellectual disability, the evidence supporting the link to causality in disease is strong. Our finding implicates the deficiency of H1 linker histone protein in autism. The systematic review of candidate genes implicated in ASD revealed that 42 of 215 (19.5%) genes are directly involved in epigenetic regulations and the majority of these genes belong to histone writers, readers, and erasers. While the mechanism of how haploinsufficiency of HIST1H1E causes autism is entirely unknown, our report underscores the importance of further study of the function of this protein and other histone-linker proteins in brain development.
Keywords: whole exome sequencing, epigenetic machinery, neurodevelopment, behavior characterization
Introduction
Eukaryotic DNA is packaged and maintained within highly regulated chromatin structures through association with various histone proteins. The fundamental unit of chromatin, the nucleosome, consists of ~146 bp units of DNA wrapped around an octamer of core histone proteins, including H2A, H2B, H3, and H4 [Zlatanova et al., 1999]. H1 linker, the fifth histone protein, exists near the base of the nucleosome at DNA entry and exit sites, and is involved in the folding and stabilization of the 30 nm chromatin fiber [Brown, 2003; Bustin et al., 2005]. The H1 histone family is the most divergent class of histones, with at least 11 different types present in humans. They have a tripartite structure consisting of a short N-terminus enriched in basic amino acids, a central conserved globular H15 domain important for DNA binding, and a long C-terminal tail enriched in lysine, serine, and proline. H1 linker histone proteins are believed to contribute to the organization and stabilization of chromosomal DNA, in addition to the folding of nucleosome filaments into higher-order structures and modulation of gene transcription [Harvey and Downs, 2004; Happel and Doenecke, 2009; Trollope et al., 2010; Xiao et al., 2012; Harshman et al., 2013]. Mouse models knocking out expression of various linker histones, including specific H1 subtypes, confirm these proteins are involved in regulating specific genes and specific processes [Sancho et al., 2008]. Sequentially inactivating multiple H1 family genes in mutant mice demonstrates the total amount of H1 present is critical for proper embryonic development. Different H1 variants have different temporal- and spatial- expression profiles [Happel and Doenecke, 2009], suggesting a distinct role for individual histone linkers during development. However, this has not been investigated.
HIST1H1E (histone cluster 1 H1 family member e) is one of 11 H1 linker genes, and encodes histone H1.4 or H1E. It belongs to the subgroup of replication dependent histones mainly expressed in the S phase of the cell cycle, believed to be involved in heterochromatin formation through their interaction with HP1 [Vaquero et al., 2004]. Depletion of H1.4 leads to arrest of cell proliferation [Sancho et al., 2008]. The C-terminus of H1.4 typically has an abundance of lysine residues, and is almost completely devoid of acidic and highly hydrophobic amino acids [Kasinsky et al., 2001], resulting in 30–50 net positive charges [Subirana 1990]. This region is important for stable folding of nucleosome arrays in chromatin fibers [Allan et al., 1986] and is required for high-affinity binding to chromatin in vivo [Hendzel et al., 2004].
Here, we report a de novo frameshift mutation in the HIST1H1E gene in a patient with major features of autism and intellectual disability. Our finding supports a role for linker histones in brain development and expands a spectrum of genes encoding epigenetic machinery in autism etiology. This underscores the importance of understanding the function of H1 linker histones in brain development.
Materials and Methods
This study was approved by the Duke institutional review board, and written consent for the case report and pictures was obtained from the individual’s parent.
Whole blood sample preparation
Sample was collected in K2-EDTA-coated blood collection tubes. After collection, blood was prepared as described in Lin et al., 2012. Briefly, samples were lysed using an equal volume of lysis buffer [20mM Tris (pH 7.5), 150mM NaCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 2.5 mM sodium pyrophosphate (Na4P2O4), 1 mM β-glycerophosphate, 1mM sodium orthovanadate (Na3VO4), and 1 µg/mL leupeptin], vortexed for 2 minutes, placed on ice for 5 minutes, and vortexed for an additional 2 minutes. Samples were placed at −80°C for at least 30 minutes, then thawed in a 37°C water bath for 3 minutes. Samples were vortexed 2 minutes and centrifuged at 14,000×g for 10 minutes at 4°C. Supernatant was transferred to a new tube and immediately used.
Quantitative immunoblot
Protein from the affected patient and an unaffected patient were separated by SDS-PAGE. Proteins were transferred to PVDF membranes (Bio-Rad, Hercules, CA). Membranes were blocked with 5% non-fat milk in TRIS-buffered saline (pH 7.4, TBS) for 1 hour at room temperature. Blots were then incubated with primary antibodies overnight at 4°C. Blots were washed in TBS containing 0.1% Tween-20 (TBST) three times for 10 minutes each, with shaking, before being incubated with their respective HRP-conjugated secondary antibodies for 180 minutes at room temperature. Following 3 TBST washes (10 minutes each with shaking), blots wer incubated with ECL reagent (GE Healthcare Life Sciences, Piscataway, NJ) and exposed using a Bio-Rad ChemiDoc system. For quantification, the gray values of H1.4 proteins were analyzed by ImageJ software (NIH, Bethesda, MD; http://rsb.info.nih.gov/ij/) and normalized to the corresponding value of the GAPDH internal control.
Antibodies
Primary – Anti-H1.4 (Abcam, ab10552, rabbit, 1:500); Anti-GAPDH (Abcam, ab9483, goat, 1:500)
Secondary – Donkey, anti-goat (Abcam, ab97110, 1:3000); Goat, anti-rabbit (SantaCruz, sc-2004, 1:3000)
Whole exome sequencing procedure
Genomic DNA was extracted from whole blood from the affected child and their parents. Exome sequencing at GeneDx was performed on exon targets isolated by capture with the Agilent SureSelect Human All Exon V4 (50 Mb) Kit (Agilent Technologies). One microgram of DNA from blood specimen was sheared into 350- to 400-bp fragments then repaired, ligated to adaptors, and purified for subsequent PCR amplification. Amplified products were captured by biotinylated RNA library baits in solution according to the manufacturer’s instructions. Bound DNA was isolated with streptavidin-coated beads and re-amplified. The final isolated products were sequenced with the Illumina HiSeq 2000 sequencing system with 100-bp paired-end reads. At the time of sequencing, DNA sequence was mapped to the reference human genome sequence (UCSC Genome Browser hg19) with the latest internally validated version of the Burrows-Wheeler Aligner (BWA), progressing from BWA v.0.5.8 to BWA-Mem v.0.7.8 [Li 2012; Li and Durbin, 2009]. Targeted coding exons and splice junctions of known protein-coding RefSeq genes were assessed for average depth of coverage, and a minimum depth of 10× was required for inclusion in downstream analysis. Local realignment around insertion-deletion sites was performed with the Genome Analysis Toolkit v.1.6 [DePristo et al., 2011].Variant calls were generated simultaneously on all sequenced family members with SAMtools v.0.1.18 [Li et al., 2009]. All coding exons and surrounding intron-exon boundaries were analyzed. Automated filtering removed common sequence changes (defined as >10% frequency present in 1000 Genomes). The targeted coding exons and splice junctions of the known protein-coding RefSeq genes were assessed for the average depth of coverage and data-quality threshold values. WES data for all sequenced family members were analyzed with GeneDx’s XomeAnalyzer (a variant annotation, filtering, and viewing interface for WES data), which includes nucleotide and amino acid annotations, population frequencies (from the NHLBI Exome Sequencing Project Exome Variant Server and 1000 Genomes), in silico prediction tools, amino acid conservation scores, and mutation references. Variants were filtered on the basis of inheritance patterns, lists of genes of interest, and phenotype and population frequencies, as appropriate. Resources including the Human Gene Mutation Database, 1000 Genomes, the NHLBI Exome Variant Server, OMIM, PubMed, and ClinVar were used for evaluating genes and detecting sequence changes of interest. Additional searches were performed with specific lists of genes related to ID. Identified sequence changes of interest were confirmed in all members of the trio by classic Sanger sequencing on an ABI3730 (Life Technologies) according to standard protocols with an additional DNA preparation. The general assertion criteria for variant classification are publicly available on the GeneDx ClinVar submission page.
GeneDx ClinVar submission page: http://www.ncbi.nlm.nih.gov/clinvar/submitters/26957
ExAc: Exome Aggregation Consortium, http://exac.broadinstitute.org
dbSNP: Database of Single Nucleotide Polymorphisms, http://www.ncbi.nlm.nih.gov/SNP
1000G: 1000 Genomes, http://www.1000genomes.org
ESP: Exome Variant Server, http://evs.gs.washington.edu/EVS
Results
General Description of Patient
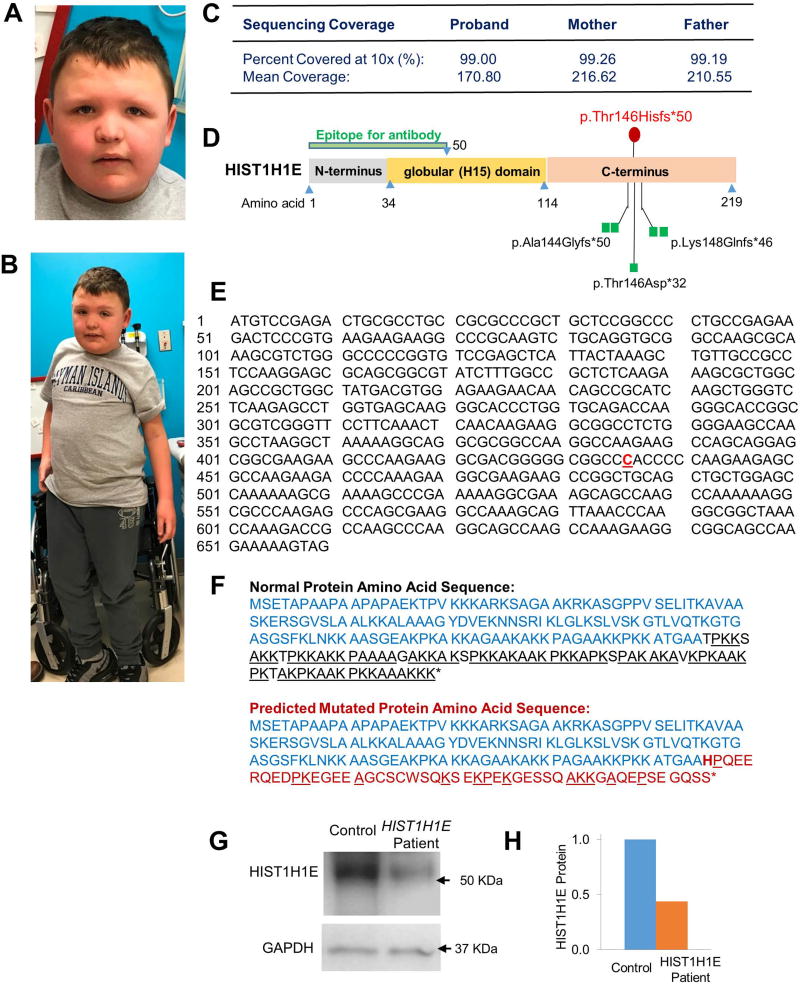
The patient is currently a 10-year-old male with autism, intellectual disability, and other behavioral problems. At the age of 10 years, his weight is 54.5kg (98th percentile), height is 144.8cm (70th percentile), and head circumference is 53cm (45th percentile). He has minor dysmorphic facial features, including downward-slanting palpebral fissures, hypertelorism, light eyebrows, micro and retrognathia, and wide philtrum (Figure 1A–B). In addition, he has bilateral fifth finger clinodactyly, inverted nipples, pes planus, exotropia, and strabismus of the right eye.
Figure 1. Dysmorphic facial and physical features of the patient.
A. Facial features. B. Full-body photograph displays relative overweight. C. Sequencing depth and coverage captured through Whole Exome Sequencing. D. Schematic illustrating the domains of the HIST1H1E protein and mutations. Locations of previously-reported mutations are illustrated with green squares, while the patient presented in this case study is represented with a red circle. The epitope used for figures G and H is against the N-terminus of the protein, and targets amino acids 1 – 50. E. Nucleotide sequence for human HIST1H1E gene. The bold, red, underlined nucleotide illustrates the patient’s c.435dupC mutation. F. The normal amino acid sequence for HIST1H1E (top) has a C-terminus (black) largely composed of K, A, and P (underlined) residues. The predicted mutated amino acid sequence (bottom) is predicted to change at the insertion point (bold), drastically reducing the content of K, A, and P residues (underlined) found in the C-terminus (red). G. Western blot using whole-cell lysate from patient and control blood samples shows a reduction in HIST1H1E protein expression. H. Quantification of western blot examining HIST1H1E protein content.
Birth History
He was born at 38 weeks gestation to a G1P0 21-year-old mother via emergency C-section due to fetal distress precipitated by a maternal motor vehicle accident. His birth weight was 3.195kg (25th percentile) and length was 49.5cm (~25th percentile). He was noted to have micrognathia and increased muscle tone at birth. He stayed in the NICU for 2 weeks due to jaundice, apnea, and poor feeding requiring orogastric feeds.
Developmental Milestones
He first sat unsupported at 9 months, and walked at 2 years. He presented with increased tone at birth but subsequently demonstrated low tone later in childhood. He began receiving developmental services, including home-based developmental therapy, then center-based speech, occupational, and physical therapy, when he was 6 months of age. At 4 year old, he had no clear spoken words but produced unintelligible, open-mouthed sounds. He does not make good eye contact and his mother feels that “he is in his own world”. He has restricted, repetitive behaviors and interests, and is obsessed with hair. He has unusual fears (noted to fear biscuits). He is still not toilet trained at current age.
Family History
Both maternal and paternal sides of families are Caucasian and deny any consanguinity. There is a paternal family history of developmental delay and neuropsychiatric disease, with bipolar disorder in the father, a great uncle with developmental delay, and a first cousin with paranoid schizophrenia. There is a diagnosis of depression in the mother and maternal grandmother.
Neurobehavioral Characterization (Table 1)
Table 1. Summary of developmental assessment performance.
Tests administered are arranged according to date, and compare the patient’s raw scores to average scores.
| Date | Test | Average Standard Scores | Patient Standard Score |
|---|---|---|---|
| 2014 | Peabody Picture Vocabulary Test – 4th Edition (PPVT) | 90–109 | <45 |
|
| |||
| 2014 | Beery-Buktenica Developmental Test of Visual-Motor Integration (Beery VMi) | 90–109 | <45 |
|
| |||
| 2014 | Vineland Adaptive Behavior Scales | 90–109 | Communication: 56 |
| 90–109 | Daily Living Skills: 66 | ||
| 90–109 | Socialization: 62 | ||
| 90–109 | Adaptive Behavior Composite: 61 | ||
|
| |||
| 2017 | Autism Diagnostic Interview-Revised (ADI-R) | Score of ‘Autism’ | |
Expressive Language
The patient communicates through some single words (vocabulary size <10 words) and some sign language, with his overall expressive language level approximating that of an 18 month child.
Receptive Language
At 7 years of age he was administered the Peabody Picture Vocabulary Test - Fourth Edition. His performance reached a standard score of <45 (raw score of 2 with a mean score of 100, and standard deviation of 15) which falls in the Extremely Low range.
Visual Motor Integration
Beery-Buktenica Developmental Test of Visual-Motor Integration (Beery VMI) was administered at 7 years of age to assess the individual’s ability to integrate his visual and motor abilities. He attained a standard score of <45 (Raw score <2) with a mean score of 100, and standard deviation of 15.
Adaptive Development
The Vineland Adaptive Behavior Scales, Second Edition–Survey Interview was administered at 7 years of age to assess the patient’s Adaptive Development. His scores for each category are as follows: Communication 56; Daily Living Skills 66; Socialization 62; Adaptive Behavior Composite 61. The mean score for each category is 100 and standard deviation is 15.
Intellectual Disability
He has moderate intellectual disability determined through the clinical assessment given at age 10 years.
Autism Diagnostic Evaluation
The patient received the diagnosis of autism at age 4 using criteria set forth by the DSM-IV. The Autism Diagnostic Interview – Revised (ADI-R) was administered at the age of 10 years, with his score reaching a diagnosis of Autism.
Communication Skills
The patient expresses hunger by getting items of food himself (his mother stresses about his independence with such tasks). He does follow one step commands, intermittently follows two step commands, and does not follow three step commands. His speech intelligibility is less than 25%. He can wave “goodbye”, blow a kiss, nod “yes” and shake his head “no”, and he is able to point. His vocabulary size is 5 to 10 words, including “ook” for look, “whoa”, “don't”, and “bye”. He jargons frequently. He signs “sorry”, “thank you”, and “more”.
Social Skills
Patient does not share his interests with others and does not engage in solitary or interactive pretend play. He will interact with adults, but not children. He often shows unusual social overtures and lacks empathy toward others. He does not comfort others in distress. He does not exhibit descriptive gestures. His eye contact is consistently cast downward. He plays ball interactively with adults only. He is “very lovable” and affectionate with family members. His temperament is calm and easy-going, with some periods of hyperactivity and unmanageable behavior.
Restricted and Repetitive Behaviors or Interests (RRB)
The patient displays restricted interests and possesses highly obsessive behaviors. He has a fixated interest in playing with water and others' hair, and preferentially interested in watching YouTube videos. He likes to line up cars in a particular order.
Other Comorbidities
At six months of age, he underwent outpatient surgery for correction of hydrocele and hernia. At 2 years old, a brain MRI revealed an arachnoid cyst and mild hydrocephalus, while a metabolic bone survey showed mild scoliosis. Both gross and fine motor skills are delayed. He is able to use a spoon and fork, and requires help when brushing his teeth. He can zip up a zipper but is unable to button large or small buttons. He is able to pedal, and can ascend and descend stairs using alternating feet while holding on to a railing. Hearing evaluations have been normal.
At 7 years of age, the patient experienced a single grand mal seizure. However, all EEGs have been normal and he is not currently on seizure medication. Sleep disturbances are particularly challenging, as he frequently wakes up during the night.
Genetic evaluation and basic genetic and metabolic testing
The patient was initially evaluated at the Medical Genetics clinic at Duke Children’s Hospital at 5 years of age. He received basic genetic and biochemical evaluations including tests for FMR1 triplet repeat expansion, SNP chromosomal microarray, plasma amino acids, an acylcarnitine profile, urine and plasma creatine/guanidinoacetate and urine organic acid analysis, SHANK3 gene sequencing (Athena Diagnostics), plasma lactate, and a biochemical screen for congenital disorder of glycosylation (Emory University Medical Genetics Lab). The results from these tests were unremarkable.
Whole Exome Sequencing
Trio whole exome sequencing (WES) was performed as previously described [Tanaka et al, 2015]. The sequencing depth and coverage are shown in Figure 1C. The initial WES analysis did not reveal any significant clinical variant, however upon re-analysis, WES revealed a de novo and frameshift mutation in HIST1H1E gene (c.435dupC; p. Thr146Hisfs*50, NM_005321.2). The c.435dupC variant in HIST1H1E was validated by Sanger sequencing and this variant was not found in ExAc, dbSNP, 1000G, or ESP. In addition, the re-analysis also revealed an incidental findings of a compound heterozygous mutation in the GALT gene, known to cause Duarte galactosemia (data not shown), and a heterozygous c.871C>T; p.R291* mutation (NM_004328.4) in the BCS1L gene. Duarte galactosemia is a mild and specific variant of galactosemia, usually not having any clinical consequences. His parents report his newborn screen for this was negative, and he did not have clinical issues with galactose metabolism. Homozygous mutations in BCS1L gene are the cause for several allelic disorders (OMIM:262000, 603358, 256000, 124000) with recessive inheritance. Because only one mutation was found and his clinical presentation, this is not consistent with BCS1L being disease causing. From this, we conclude he is likely a carrier for this BCS1L-related disorder. Further bioinformatics filtering did not identify any other homozygous, compound heterozygous, X-linked, or de novo variants consistent with the proband’s phenotype.
Mutation Annotation and Expression Analysis
HIST1H1E is a single exon and intron-less gene mapped to chromosome 6p22.1. It encodes a protein with 219 amino acids and molecular weight of ~22kD. The HIST1H1E protein contains an N-terminus region enriched in basic amino acids, a central conserved globular H15 domain involved in DNA binding, and a long C-terminal tail enriched in lysine, serine, and proline (Figure 1D). The c.435dupC results in a frameshift at Thr146His and introduces a premature stop codon resulting in a truncated protein, 25 amino acids shorter than the normal protein (Figure 1E–F).
To better understand the impact of the c.435dupC mutation on HIST1H1E protein, we performed quantitative immunoblot staining using protein isolated from whole-blood lymphocytes. As shown in Figure 1G–H, the patient has ~50% reduction of HIST1H1E protein compared to a similarly-aged control. Because the epitope of the antibody against HIST1H1E targets the N-terminus (Figure 1D), reduced HIST1H1E protein expression suggests the mutation results in haploinsufficiency due to either mutation-induced RNA decay, or an unstable truncated protein.
Catalog of mutations in genes encoding epigenetic regulators in ASD
The role of epigenetics in autism spectrum disorders (ASDs) has been recognized for more than two decades. Historically, MECP2, a gene encoding the X-linked methyl-CpG-binding protein 2, is known to cause Rett Syndrome, a prototype for syndromic autism in females [Amir et al., 1999]. MECP2 specifically interacts with methylated DNA to mediate the transcriptional regulation through both its interaction with methylated DNA and histone methylation [Ho et al., 2008]. Similarly, the full triplet repeat expansion mutation in FMR1, a gene responsible for fragile X syndrome and the common syndromic ASD in males, is abnormally methylated at the 5' promoter. It represses transcription of the FMR1 gene [Li et al., 2001] and leads to excess translation of epigenetic regulators [Korb et al., 2017], causing fragile X syndrome. With the rapid advance of next generation sequencing techniques in genetic studies, a growing list of genes encoding proteins acting as epigenetic modifiers at different levels has been identified in individuals with ASD (Table 2).
Table 2. Summary of epigenetic regulator genes currently implicated in autism.
Genes are arranged by their gene score, which are based on confidence levels for currently available evidence implicating these genes in autism.
| Evidence | Gene | Protein function | Function | Co-morbidity | Reference |
|---|---|---|---|---|---|
| High | ARID1B‡ | SNF/SWI chromatin remodeling complex | C | CSS, ID | 1–15 |
| ASH1L | H3K36 methyltransferase | W | ID, DD | 3,4,14–18 | |
| ASXL3‡ | Likely acts via methylation of histones | W | BRS, ID, DD | 3,4,19 | |
| CHD8 | nucleosome binding/remodeling | C | SCZ, DD/NDD, ID | 3,4,6,7,9,11,14–16,20–25 | |
| KMT5B | H4K20 methyltransferase | W | 3,4,15,26,27 | ||
| NAA15 | acetyltransferase | W | 3,4,15 | ||
|
| |||||
| Strong | ANKRD11‡ | chromatin regulator controlling histone acetylation | W | KBGS, EPS, ID | 9,13,14,16,28–30 |
| CHD2 | nucleosome binding/remodeling | R | DD/NDD, EPS, ID, EP | 3,4,14–16,31–35 | |
| CHD7‡ | nucleosome binding/remodeling | R | CHARGE; DD, EPS, ID | 4,7,14,51,75 | |
| HDAC4‡ | HDAC | E | BMRS; ID; SCZ | 76 | |
| KAT2B | HAT for K | W | 4,14,16,34,36 | ||
| KDM5B | H3K4 demethylase | E | DD/NDD | 3,10,16 | |
| KMT2A‡ | H3K4 methyltransferase | W | WSS, EP, EPS, DD, ID | 3,9,16,37 | |
| KMT2C | H3K4 methyltransferase | W | DD | 3,4,6,10,14,16,38 | |
| MECP2‡ | methylation-dependent transcriptional repressor | R | Rett, EPS, DD, ID, EP | 12,14,39–50 | |
| WAC | regulates H2BK120ub1 | R | . | 4,14,16,18 | |
|
| |||||
| Suggestive | C11orf30 | Interacts with chromatin remodeling complex | C | 3,4,76 | |
| CTCF | binds HATs and HDACs to regulate transcription | R | DD | 10,16,35 | |
| CUX1 | homeodomain family of DNA-binding proteins and repressor of H2B | C | 13,80,81 | ||
| DNMT3A | DNA methyltransferase | M | 3,16,36,51 | ||
| EHMT1‡ | H3K9 methyltransferase | W | KS, DD, ID | 12,21,51,53–56 | |
| ELP4 | HAT component; acetylates H3 and likely H4 | W | 13,35,57,58 | ||
| EP400 | component of NuA4 histone acetyltransferase complex (H4 and H2A) | W | 3,4,27,59 | ||
| HDAC3 | HDAC | E | 16 | ||
| HERC2‡ | E3 ubiquitin-protein ligase regulating damaged chromosomes | C | DD, EPS | 4,16 | |
| HMGN1 | binds nucleosomal DNA altering interaction between DNA and histone | C | 60,79 | ||
| JMJD1C | Histone demethylase | E | 4,32,77,78 | ||
| KAT6A | H3K9 HAT | W | EPS, DD, EP | 4,16,61,62 | |
| KDM5C | H3K4 histone demethylase | E | 12,63,64 | ||
| KDM6B | H3K27 histone demethylase | E | 3,4,16,27 | ||
| KMT2E | H3K4me1 and H3K4me2 methyltransferase | W | 14,16,65 | ||
| MBD5 | member of methyl-CpG -binding domain family | C | EP, EPS, DD, ID | 4–6,21,31,47,66–70 | |
| PHF2 | Lysine demethylase | E | 4,14,16,27 | ||
| SETD2 | H3K36 methyltransferase | W | 6,7,11,12,31,71,72 | ||
| SETDB1 | H3K9 methyltransferase | W | 47 | ||
| SETDB2 | Histone methyltransferase | W | EPS | 47 | |
| SIN3A | MECP2 and histone interacting protein | R | EPS | 3,16,52 | |
| SMARCA2 | SWI/SNP chromatin remodeling complex | C | CSS, NS,DD,EP,EPS,ID | 13 | |
| SMARCC2 | Regulates transcription via SNF/SWI chromatin remodeling complex | C | 4,14,16,32 | ||
| SRCAP | incorporates H2A.Z into nucleosomes | C | 3,15,16 | ||
| ZMYND11 | transcription corepressor by modulating RNA pol II and H3K36M3 | R | 4,27,73,74 | ||
M: DNA modifying; W: histone writer; R: histone reader; E: histone eraser; C: chromatin remodeling complex; CSS: Coffin-Siris syndrome; ID: intellectual disability; DD/NDD: developmentally delayed/non developmentally delayed; BBS: Bainbridge-Ropers syndrome; SCZ: schizophrenia; KBGS: KBG syndrome; EPS: epilepsy; EP: encephalopathy; CHARGE: CHARGE syndrome; BMRS: Byachydactyly mental retardation syndrome; WSS: Wiedemann-Steiner Syndrome; Rett: Rett syndrome; KS: Kleefstra syndrome; NBS: Nicolaides-Baraitser syndrome.
indicates genes involved in syndromic disorders
To catalog the epigenetic-related genes in ASD, we systematically reviewed genes implicated in ASD in literature or other online autism-specific genetic databases. Two autism genetics database: SFARI gene (https://gene.sfari.org/) [Abrahams et al., 2013] and autDB (http://autism.mindspec.org/autdb/Welcome.do) [Basu et al., 2009] are included in our review. There are a total of 845 autism candidate genes in autDB and 668 genes in SFARI gene. Because evidence supporting the causality for ASD varies among them, our review focused on a total of 215 genes, scored in the categories of high (23 genes), strong (42 genes), and suggestive evidence (150), using a scoring framework described in SFARI gene and the additional annotation for review the function data in literature by our own team (Table 2). Our review determined 42 out of 215 genes (19.5%) are directly implicated in epigenetic machinery. Among them, 6 out 23 (26.1%) fall in the category of high confidence, 10 out 42 (23.8%) in strong confidence, and 26 out 150 (17.3%) in a category of suggestive evidence.
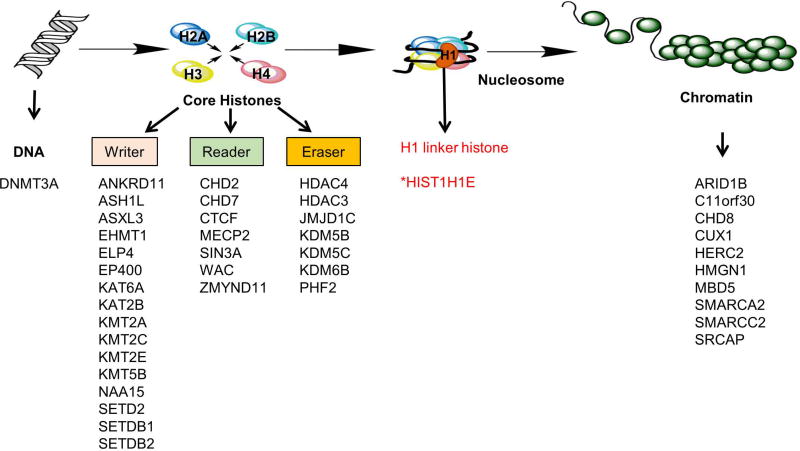
The recognition of the specific modifications to histones and DNA is necessary to properly regulate the state of chromatin packaging within a cell, which in turn regulates gene expression. The proteins responsible for maintaining epigenetic regulation of gene expression include those which modify DNA, add histone modifications to chromatin (“writers”), recognize and bind chromatin through modifications (“readers”), remove histone modifications from chromatin (“erasers"), and pack and remodel chromatin [Allis and Jenuwein, 2016; Janzen et al., 2010]. Understanding how these proteins interact with chromatin is necessary to uncover the role for epigenetic modifications in human disease. The molecular pathway for epigenetic modifications can be divided into 4 levels: DNA modifications at the nucleotide level, histone modifications, nucleosome packing, and chromatin remodeling (Kizaki et al., 2014) (Figure 2). Genes encoding proteins involved in histone modification, including histone writers, readers, and erasers, are predominantly represented in the ASD candidate gene list (Figure 2). Genes encoding proteins involved in chromatin remodeling are also frequently implicated but the exact function of some of these proteins on chromatin remains to be further elucidated. The HISTH1E gene is the first linker histone involved in nucleosome packing to be implicated in ASD. However, it should be noted the actual number of epigenetic regulator genes implicated in ASD is likely more than 42, as we did not include genes with indirect involvement in epigenetic regulation, or genes whose functions are currently unknown.
Figure 2. Schematic illustrating the distribution of epigenetic regulator genes implicated in ASD, and the predicted mechanism of disruption within epigenetic regulation pathways.
HIST1H1E is the first gene reported to be implicated in autism through interference with the basic machinery of epigenetic regulation.
Discussion
The etiological role of epigenetic dysregulation in ASD has long been recognized from studies of syndromic ASD, such as Rett and fragile X syndromes [Zoghbi and Beaudet, 2016]. Only recent discoveries of mutations in genes encoding proteins specifically involved in epigenetic regulation provide direct evidence to support the causal role of epigenetic machinery in ASD. Our finding implicates the family of H1 linker histones in the etiology of ASD, and expands the spectrum to involve the epigenetic machinery in autism etiology. Due to the molecular nature of the frameshift and de novo mutation of the c.435dupC variant in HIST1H1E, genetic evidence to support the pathogenicity of this variant is strong. Expression analysis in the patient carrying the mutation also supports a mechanism of loss of function and haploinsufficiency underling the pathophysiology. Causally linking this mutation to the clinical presentation is also strengthened by the recent report of 3 mutations of HIST1H1E (c.430dupC, c.441dupC, and c.430-458del23) in 5 individuals presenting with clinical features of overgrowth and intellectual disability [Tatton-Brown et al., 2017] (Figure 1D). Interestingly, the c.430dupC and c.441dupC are recurrent mutations reported in 2 out of 5 cases. Together with the mutation of c.435dupC in our patient, these findings suggest the 3' end of the HIST1H1E gene as a mutation “hot-spot”. The C-terminus of linker histones typically has a net positive charge [Subirana 1990] necessary for high-affinity binding to chromatin in vivo [Hendzel et al., 2004]. The reduced HIST1H1E protein will likely affect the chromatin structure and function.
The clinical features including overgrowth and intellectual disability reported in 5 cases with HIST1H1E mutations in literature are variable, even in cases with the same or similar mutations. It was also noted the documentation of clinical features in some of these cases in the report is probably not complete [Tatton-Brown et al., 2017]. For example, the feature of large head sizes at different ages is described in 3 out of 5 cases. However, the increase in height and weight is much less consistent among cases, and varies at different ages. The birth weight in 4 cases is within the normal range, but higher in one case. Abnormally tall height and high weight was only reported during infancy in one case, but these measures normalized during the patient’s teenage years. Similarly, intellectual disability varies from mild to severe in two cases with the same mutation of c.430dupC. A diagnosis of autism or autistic-like behaviors is not mentioned in any of these 5 cases. It is possible that autism or autistic behaviors are present in these cases but have to not been fully assessed. In our case with the c.435dupC mutation, we note some overlapping clinical features. These include similar dysmorphic facial features such as high hairline, full checks, and micrognathia. Additionally, we report a relative high weight at age 10, but normal height. Our patient has a history of mild ventriculomegaly in infancy, but his current FOC is in the 50th percentile. His feature of newborn hypertonia progressing to hypotonia is similar to the case reported to have a c.436-458del23 mutation in literature [Tatton-Brown et al., 2017]. However, we have provided findings from extensive behavioral assessments for our case. Most importantly, we have confirmed the diagnosis of autism based on the criteria outlined in the DSM-IV at the age of 4 years and the ADI-R at the age of 10 years. Our patient displays significant impairment in social interaction behavior. Both his expressive and receptive languages are significantly affected. Repetitive behaviors and restricted interests are evident, as is significant intellectual disability. Other clinical behavioral issues we report include anxiety, poor visual and spatial coordination, sleep disruptions, and maladaptive behaviors. While the behavioral profiling in our case is significantly different from other cases in literature, it remains to be seen whether these features, particularly the diagnosis of ASD, are unique to the c.435dupC variant. Of interesting note, neuropsychiatric disorders, including bipolar disorder in the patient’s father and depression in the patient’s mother, are prominent in this family. Because of the de novo nature of the c.435dupC mutation in the proband, other genetic variants yet to be identified may be responsible for his parents’ psychiatric presentations. The strong genetic background of neuropsychiatric disease may modify the clinical presentation caused by the c.435dupC mutation in HIST1H1E, making the patient’s presentation unique or distinct. Nevertheless, our report does suggest additional behavioral assessments, including autism evaluations, are valuable to assess the genotype and phenotype correlation of HIST1H1E mutations.
While the genetic evidence strongly supports causality in autism, little is known about how deficiency in HIST1H1E, a basic component of epigenetic machinery, causes brain dysfunction manifesting primarily as autism and intellectual disability. Triple knockout of H1 linker histones (H1TKO), including HIST1H1E, result in early embryonic lethality and ~50% reduction of total H1 histones in mice [Fan et al., 2003]. Analysis in H1TKO embryonic stem cells (ESCs) revealed altered expression of select genes via modulation of DNA and histone methylation. The study of neural differentiation in H1TKO ESCs revealed selective impairments in specification and maturation of glutamatergic and dopaminergic neurons, and their association with deregulation of expression in forebrain-derived neural stem cells [Alami et al., 2003]. Mutant mice lacking Hist1h1e only are viable, and do not have apparent developmental defects [Fan et al., 2001; Fan et al., 2003]. Unfortunately, behavioral and other studies of brain function in Hist1h1e mutant mice have not been reported. An interesting hypothesis in Hist1h1e mutant mice would be to test whether haploinsufficiency of the gene selectively reduces H1 linker histone in certain neuronal cell lineages, and alters subsequent expression of genes important for brain development and function.
In summary, we report the first case of a HIST1H1E mutation associated with autism, expanding the spectrum of epigenetic-related genes in the etiology of autism. This study underscores the importance of future studies to evaluate autism in other patients carrying HIST1H1E mutations and to understand the role of H1 linker histones in brain development and function.
Acknowledgments
We wish to thank the family for their participation. LJD is supported by the Duke University Postdoctoral Program in Fundamental & Translational Neuroscience grant 5T32NS051156-10. YHJ is supported by grants from National Institute of Health: MH098114, HD077197, and MH104318 and HD088007
References
- Abrahams BS, Arking DE, Campbell DB, Mefford HC, Morrow EM, Weiss LA, Menashe I, Wadkins T, Banerjee-Basu S, Packer A. SFARI Gene 2.0: a community-driven knowledgebase for the autism spectrum disorders (ASDs) Mol Autism. 2013;4(1):36. doi: 10.1186/2040-2392-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alami R, Fan Y, Pack S, Sonbuchner TM, Besse A, Lin Q, Greally JM, Skoultchi AI, Bouhassira EE. Mammalian linker-histone subtypes differentially affect gene expression in vivo. Proc Natl Acad Sci USA. 2003;100(10):5920–5925. doi: 10.1073/pnas.0736105100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allan J, Mitchell T, Harborne N, Bohm L, Crane-Robinson C. Roles of H1 domains in determining higher order chromatin structure and H1 location. J Mol Biol. 1986;187(4):591–601. doi: 10.1016/0022-2836(86)90337-2. [DOI] [PubMed] [Google Scholar]
- Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17(8):487–500. doi: 10.1038/nrg.2016.59. [DOI] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23(2):185–188. doi: 10.1038/13810. [DOI] [PubMed] [Google Scholar]
- Basu SN, Kollu R, Banerjee-Basu S. AutDB: a gene reference resource for autism research. Nucleic Acids Res. 2009;37:D832–836. doi: 10.1093/nar/gkn835. (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DT. Histone H1 and the dynamic regulation of chromatin function. Biochem Cell Biol. 2003;81(3):221–227. doi: 10.1139/o03-049. [DOI] [PubMed] [Google Scholar]
- Bustin M, Catex F, Lim JH. The dynamics of histone H1 function in chromatin. Mol Cell. 2005;17(5):617–620. doi: 10.1016/j.molcel.2005.02.019. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Nikitina T, Morin-Kensicki EM, Zhao J, Magnuson TR, Woodcock CL, Skoultchi AI. H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol Cell Biol. 2003;23(13):4559–4572. doi: 10.1128/MCB.23.13.4559-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Sirotkin A, Russell RG, Ayala J, Skoultchi AI. Individual somatic H1 subtypes are dispensable for mouse development even in mice lacking the H1(0) replacement subtype. Mol Cell Biol. 2001;21(23):7933–7943. doi: 10.1128/MCB.21.23.7933-7943.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Happel N, Doenecke D. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene. 2009;431(102):1–12. doi: 10.1016/j.gene.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Harshman SW, Chen MM, Branson OE, Jacob NK, Johnson AJ, Byrd JC, Freitas MA. Isolation and analysis of linker histones across cellular compartments. J Proteomics. 2013;91:595–604. doi: 10.1016/j.jprot.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey AC, Downs JA. What function do linker histones provide? Mol Microbiol. 2004;53(3):771–775. doi: 10.1111/j.1365-2958.2004.04195.x. [DOI] [PubMed] [Google Scholar]
- Hendzel MJ, Lever MA, Crawford E, Th’ng JP. The C-terminal domain is the primary determinant of histone H1 binding to chromatin in vivo. J Biol Chem. 2004;279(19):20028–20034. doi: 10.1074/jbc.M400070200. [DOI] [PubMed] [Google Scholar]
- Ho KL, McNae IW, Schmiedeberg L, Klose RJ, Bird AP, Walkinshaw MD. MeCP2 binding to DNA depends upon hydration at methyl-CpG. Mol Cell. 2008;29(4):525–531. doi: 10.1016/j.molcel.2007.12.028. [DOI] [PubMed] [Google Scholar]
- Janzen WP, Wigle TJ, Jin J, Frye SV. Epigenetics: tools and technologies. Drug Discov Today Technol. 2010;7(1):e59–e65. doi: 10.1016/j.ddtec.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasinsky HE, Lewis JD, Dacks JB, Ausió J. Origin of H1 linker histones. FASEB J. 2001;15(1):34–42. doi: 10.1096/fj.00-0237rev. [DOI] [PubMed] [Google Scholar]
- Kizaki S, Suzuki Y, Takenaka T, Endo M, Sugiyama H. AFM analysis of changes in nucleosome wrapping induced by DNA epigenetic modification. Biomater Sci. 2014;2:1399–1403. doi: 10.1039/c4bm00113c. [DOI] [PubMed] [Google Scholar]
- Korb E, Herre M, Zucker-Scharff I, Gresack J, Allis CD, Darnell RB. Excess translation of epigenetic regulators contributes to Fragile X Syndrome and is alleviated by Brd4 inhibition. Cell. 2017;170(6):1209–1223. doi: 10.1016/j.cell.2017.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. Exploring single-sample SNP and INDEL calling with whole-genome de novo assembly. Bioinformatics. 2012;28(14):1838–1844. doi: 10.1093/bioinformatics/bts280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;15(25):278–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang Y, Ku L, Wilkinson KD, Warren ST, Feng Y. The fragile X mental retardation protein inhibits translation via interaction with mRNA. Nucleic Acids Res. 2001;29(11):2276–2283. doi: 10.1093/nar/29.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin X, Cook TJ, Zabetian CP, Leverenz JB, Peskind ER, Hu S-C, Cain KC, Pan C, Edgar JS, Goodlett DR, Racette BA, Checkoway H, Montine TJ, Shi M, Zhang J. DJ-1 isoforms in whole blood as potential biomarkers of Parkinson disease. Scientific Reports. 2012;2:954. doi: 10.1038/srep00954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho M, Diani E, Beato M, Jordan A. Depletion of human histone H1 variants uncovers specific roles in gene expression and cell growth. PLoS Genet. 2008;4(10):e1000227. doi: 10.1371/journal.pgen.1000227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subirana JA. Analysis of the charge distribution in the C-terminal region of histone H1 as related to its interaction with DNA. Biopolymers. 1990;29(10–11):1351–1357. doi: 10.1002/bip.360291003. [DOI] [PubMed] [Google Scholar]
- Tanaka AJ, Cho MT, Millan F, Juusola J, Retterer K, Joshi C, Niyazov D, Garnica A, Gratz E, Deardorff M, Wilkins A, Oritz-Gonzalez X, Mathews K, Panzer K, Brilstra E, van Gassen KL, Volker-Touw CM, van Binsbergen E, Sobreira N, Hamosh A, McKnight D, Monaghan KG, Chung WK. Mutations in SPAT5 are associated with microcephaly, intellectual disability, seizures, and hearing loss. Am J Hum Genet. 2015;97(3):457–464. doi: 10.1016/j.ajhg.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatton-Brown K, Loveday C, Yost S, Clarke M, Ramsay E, Zachariou A, Elliott A, Wylie H, Ardissone A, Rittinger O, Stewart F, Temple IK, Cole T, Childhood Overgrowth Collaboration. Mahamdallie S, Seal S, Ruark E, Rahman N. Mutation in epigenetic regulation genes are a major cause of overgrowth with intellectual disability. Am J Hum Genet. 2017;100(5):725–736. doi: 10.1016/j.ajhg.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trollope AF, Sapojnikova N, Thorne AW, Crane-Robinson C, Myers FA. Linker histone subtypes are not generalized gene repressors. Biochim Biophys Acta. 2010;1799(9):642–652. doi: 10.1016/j.bbagrm.2010.08.007. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16(1):93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Xiao B, Freedman BS, Miller KE, Heald R, Marko JF. Histone H1 compacts DNA under force during chromatin assembly. Mol Biol Cell. 2012;23(24):4864–4871. doi: 10.1091/mbc.E12-07-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlatanova J, Leub SH, van Holde K. Chromatin structure revisited. Crit Rev Eukaryot Gene Expr. 1999;9(3–4):245–255. doi: 10.1615/critreveukargeneexpr.v9.i3-4.90. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY, Beaudet AL. Epigenetics and human disease. Cold Spring Harb Prespect Biol. 2016;8(2):a019497. doi: 10.1101/cshperspect.a019497. [DOI] [PMC free article] [PubMed] [Google Scholar]