Abstract
The androgen receptor (AR) plays a critical role in the development of the normal prostate as well as prostate cancer. Using an integrative transcriptomic analysis of prostate cancer cell lines and tissues, we identified ARLNC1 (AR-regulated long non-coding RNA 1) as an important long non-coding RNA that is strongly associated with AR signaling in prostate cancer progression. Not only was ARLNC1 induced by AR protein, ARLNC1 stabilized the AR transcript via RNA-RNA interaction. ARLNC1 knockdown suppressed AR expression, global AR signaling, and prostate cancer growth in vitro and in vivo. Taken together, these data support a role for ARLNC1 in maintaining a positive feedback loop that potentiates AR signaling during prostate cancer progression, and identifies ARLNC1 as a novel therapeutic target.
Keywords: androgen receptor, prostate cancer, long non-coding RNA, ARLNC1
Introduction
Long non-coding RNAs (lncRNAs) are a class of transcripts with diverse and largely uncharacterized biological functions1–3. Through cross-talk with chromatin, DNA, RNA species, and proteins, lncRNAs function via chromatin remodeling, transcriptional and post-transcriptional regulation4–9. High-throughput RNA sequencing (RNA-Seq) has enabled the identification of lncRNAs with suggested oncogenic and tumor suppressive roles, including involvement in the pathogenesis of prostate cancer (PCa)7,10–12. Primary PCa is often hormone-dependent and relies on signaling through the androgen receptor (AR); therefore, the majority of patients are responsive to front-line treatment with androgen deprivation therapy (ADT)13–15. However, approximately 20% of cases progress to an incurable stage of the disease known as castration-resistant prostate cancer (CRPC), which still critically relies on AR signaling16,17, as evidenced by the clinical benefit afforded through the use of enzalutamide18–21 or abiraterone22–24. While substantial efforts have been undertaken to identify mechanisms of sustained AR signaling in CRPC (i.e., AR mutations, AR splice variants, and alternative activation pathways)25–31, few studies have investigated the role of AR-regulated lncRNAs. Therefore, we initiated a comprehensive RNA-Seq profiling investigation of AR-regulated, cancer-associated lncRNAs from prostate cancer cell lines and patient tissue samples.
Results
Analysis of androgen receptor-regulated transcriptome in prostate cancer
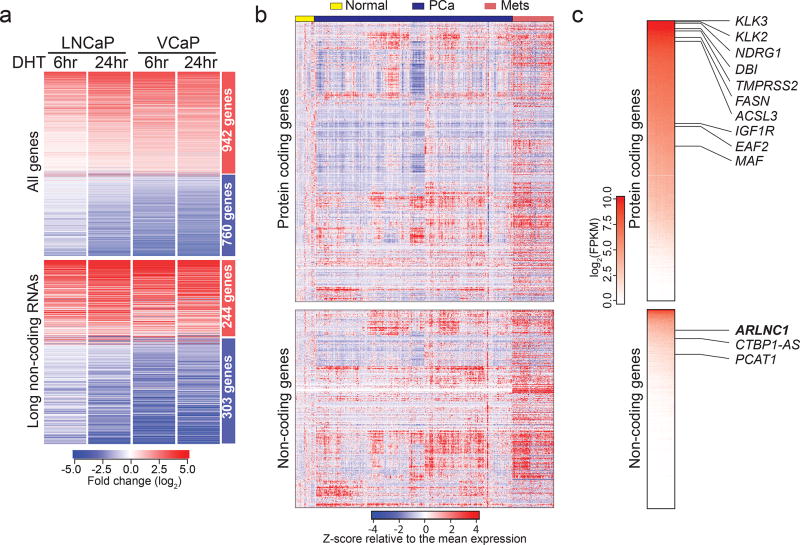
To nominate AR-regulated genes (ARGs), RNA-Seq was performed on AR-dependent VCaP and LNCaP prostate cancer cell lines that were stimulated with an AR ligand, dihydrotestosterone (DHT), for 6 and 24 hours (Supplementary Fig. 1a). 1702 genes were identified to be concordantly induced or repressed in VCaP and LNCaP at both time points (Fig. 1a, Supplementary Fig. 1b–c, Supplementary Table 1), including over 500 lncRNAs (Fig. 1a, Supplementary Fig. 1d); these data indicate that a large portion of the AR transcriptome remains uncharacterized.
Fig. 1. Identification of AR regulated genes in prostate cancer.
a, The androgen-regulated transcriptome of prostate cancer cells. A heatmap representation of the 1702 genes (including 547 lncRNAs) differentially regulated in LNCaP and VCaP cells, following 6 and 24 hours of DHT treatment. b, The landscape of transcriptomic alterations of prostate cancer progression. A heatmap depicting 1155 protein-coding genes and 547 lncRNAs across benign prostate (normal, n = 52 samples), localized (PCa, n = 500 samples), and metastatic prostate cancer (Mets, n = 100 samples) in the TCGA prostate and SU2C–PCF RNA-Seq data, with rows representing genes and columns representing patients. Patients were grouped by clinical stages and genes were subject to hierarchical clustering. Expression variability is quantified for each gene as a Z-score relative to the mean expression in normal prostate samples. c, A heatmap representation of ranked gene expression levels in prostate tissues. Canonical prostate-lineage and prostate cancer markers are listed. (Upper panel: protein-coding genes. Lower panel: non-coding genes.)
To differentiate between direct and indirect ARGs, previously published AR chromatin immunoprecipitation (ChIP)-seq data from LNCaP and VCaP cells were analyzed32. As expected for direct AR targets, increased levels of AR binding at transcription start sites (TSS) in both LNCaP and VCaP cells were observed (Supplementary Fig. 1e). The binding levels decreased following treatment with an AR antagonist (enzalutamide) (Supplementary Fig. 1f–g), and the binding sites revealed a de novo motif identical to the canonical AR response element33 (Supplementary Fig. 1h). A total of 987 genes were categorized as direct ARGs, including 341 lncRNAs (lncARG) (Supplementary Table 1). Within these genes, we observed an enrichment of chromatin marks associated with “open” chromatin (H3K27ac, H3K4me1), active promoters (H3K4me3), and transcription (H3K36me3), which, together with Pol-II occupancy, are recognized as manifestations of active gene expression (Supplementary Fig. 1i). Bromodomain and extra-terminal (BET) family proteins, such as BRD4, recognize acetylated histones and have been shown to promote AR transcriptional activity32. Consistently, we observed the co-localization of BRD4 and AR protein at promoters of direct AR responsive genes and the loss of AR following treatment with a bromodomain inhibitor (JQ1) (Supplementary Fig. 1f, i). We further sought to determine whether ARGs identified from cell lines were also targeted by AR in normal prostate tissues and primary tumors. We leveraged the dataset from Pomerantz et. al and queried for the presence of AR peaks within ARG promoters34. Remarkably, the majority of ARG promoters were TSS-proximally bound by AR in both tissues and cell lines (Supplementary Fig. 1j–k); conversely, AR-independent genes were distal to AR binding sites (Supplementary Fig. 1l).
Finally, we confirmed that the AR-regulated genes were also expressed in human prostate tissues. We interrogated RNA-Seq data from normal prostate, clinically-localized PCa (The Cancer Genome Atlas, TCGA)35, and metastatic CRPC (Stand Up to Cancer-Prostate Cancer Foundation, SU2C–PCF)30 (Fig. 1b). This revealed remarkable heterogeneity in the expression of ARGs during prostate cancer progression to metastatic disease. As expected, compared to protein-coding genes, non-coding ARGs were detected at lower overall levels (Fig. 1c), although ~10% of them showed robust expression of over 10 FPKM on average across prostate cancer samples.
ARLNC1 is a prostate lineage-specific lncRNA with elevated expression in cancer
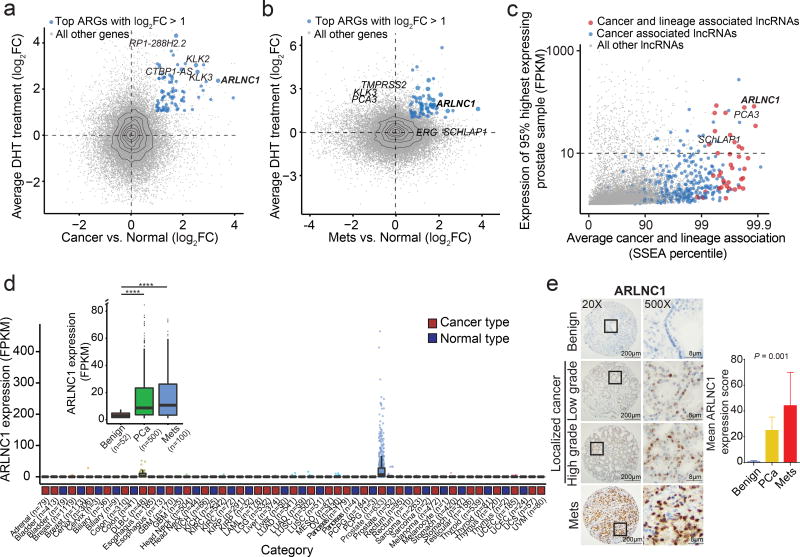
We hypothesized that lncRNAs associated with PCa progression and castration-resistance should be either upregulated if they enhance AR signaling or, conversely, downregulated if they attenuate AR signaling. Their expression is also expected to be AR-dependent and lineage-restricted if they are part of bona fide physiological feedback loops. Accordingly, a top-down strategy was developed in order to establish and prioritize clinically-relevant, prostate cancer- and lineage-specific lncARGs. First, we identified genes that were both directly regulated by AR in VCaP/LNCaP cell lines and upregulated in primary (Fig. 2a) or metastatic PCas (Fig. 2b) compared to normal prostate tissues. As expected, canonical AR targets, including KLK3, KLK2, and TMPRSS2, were among the most differentially expressed protein coding genes. Importantly, this approach highlighted several novel lncARGs, including ARLNC1 (ENSG00000260896, PRCAT4710), and validated previously identified lncARGs, such as CTBP1-AS36 (Fig. 2a–b). Interestingly, ARLNC1 was found to be one of the most differentially expressed AR-regulated genes in both localized and metastatic PCa (Fig. 2a–b, Supplementary Fig. 2a–b).
Fig. 2. Nomination and in situ characterization of ARLNC1 in prostate cancer.
a–b, Identification of androgen-regulated transcripts elevated in prostate cancer progression. Scatterplots showing AR-regulation and cancer-association of ARGs identified in Fig. 1a. Y-axis depicts log2-fold change of gene expression upon DHT stimulation, and x-axis indicates log2-gene expression level difference between benign (n = 52 samples) and localized prostate cancer (n = 500 samples) (a), or expression level differences between benign (n = 52 samples) and metastatic prostate cancer (n = 100 samples) (b). Significant genes with log2 fold-change > 1 were ranked according to the combined P values (limma moderated t-test). c, Nomination of prostate cancer- and lineage- associated lncRNAs based on expression levels. Scatterplot shows the expression level, prostate tissue specificity, and prostate cancer association of lncRNAs. Expression level is the FPKM value at the 95th percentile across TCGA prostate samples (total n = 7,256 samples). Average cancer and lineage associations are represented by the percentile rank for each gene in SSEA analysis. d, Relative expression (FPKM) of ARLNC1 across different cancer types in the TCGA cohort. Inset: relative expression (FPKM) of ARLNC1 across benign (n = 52 samples), localized (n = 500 samples), and metastatic (n = 100 samples) prostate cancer. PCa vs. Normal: ****P < 2.2e-16; Mets vs. Normal: ****P = 2.6e-7 (two-sided t-test). Box-plot definition: center - median, box limits 1st and 3rd quartile, whiskers follow the 1.5 rule. eIn situ hybridization of ARLNC1 in human prostate cancer tissue microarray. Representative ARLNC1 staining is shown for benign prostate, localized, and metastatic prostate cancer tissue. Bar plot represents mean ARLNC1 expression scores across benign (n = 11), localized (n = 85), and metastatic (n = 37) tissues, with vertical bars indicating bootstrapped 95% CI of the means. Significance was calculated by a Kruskal-Wallis rank sum test.
Next, we sought to establish the prostate lineage and cancer specificity of prostate cancer-associated lncRNAs. We leveraged the MiTranscriptome assembly10, an online resource, to interrogate lncRNA expression across a multitude of tissue and tumor types, and we calculated Sample Set Enrichment Analysis (SSEA) scores, which indicate the strength of cancer and lineage association10. After applying an expression level filter (10 FPKM at the 95th percentile), we identified 12 of the most prostate lineage- and prostate cancer-specific lncRNAs (Fig. 2c, Supplementary Fig. 2c–d); five of these lncRNAs were regulated by AR. Across these analyses, ARLNC1 was the top prioritized transcript and thus warranted further investigation.
Expression of ARLNC1 was interrogated across cancer and normal tissue RNA-Seq samples from TCGA and the Genotype-Tissue Expression (GTEx) project37,38, respectively. In the TCGA cohort, ARLNC1 exhibited a highly prostate cancer-specific expression pattern, with little to no expression in other tumor types (Fig. 2d). Similarly, in the GTEx normal tissue cohort, its expression was limited to the prostate (Supplementary Fig. 2e). Among prostate samples, ARLNC1 expression was significantly higher in localized and metastatic prostate cancers compared to benign tissues, as assessed by RNA-Seq (Fig. 2d inset) and in situ hybridization (Fig. 2e). In an extensive differential expression analysis using MiTranscriptome, ARLNC1 was found to be among the top 1% of transcripts most upregulated in prostate cancer and specific to the prostate lineage, with no significant associations in other tissues (Supplementary Fig. 2f). Additionally, the protein-coding genes that were most correlated with ARLNC1 were found to be associated with prostate cancer progression in ONCOMINE concept analyses performed on multiple clinical datasets39 (Supplementary Fig. 2g). Together, these results confirm that ARLNC1 expression is restricted to prostate lineage, elevated in prostate cancer, and associated with AR signaling throughout prostate cancer progression.
To functionally characterize ARLNC1, we first identified appropriate prostate cancer cell lines with moderate to high levels of ARLNC1 expression using in-house RNA-Seq data (Supplementary Fig. 3a). Supporting the association of AR with ARLNC1, ARLNC1 expression was highly enriched in AR-positive cell lines, with the highest expression in MDA-PCa-2b and LNCaP cells. In addition, qPCR analysis for the ARLNC1 transcript also demonstrated that this gene was expressed highest in MDA-PCa-2b and LNCaP cell lines (Supplementary Fig. 3b). As existing annotations of ARLNC1 (located on chromosome 16) predicts the presence of several transcript isoforms that differ in exon and TSS usage, we determined the exact structure in MDA-PCa-2b and LNCaP cells, by Random amplification of cDNA ends (RACE). A common TSS for ARLNC1 was found in both cell lines, and the ~2.8 kb transcript isoform was further confirmed by northern blot analysis (Supplementary Fig. 3c). Single molecule fluorescent in situ hybridization (smFISH) revealed that approximately 100 molecules of ARLNC1 transcripts existed per MDA-PCa-2b cell (Supplementary Fig. 3d–e). Using smFISH and qPCR, we also found that ARLNC1 molecules were distributed equally between the nuclear and cytoplasmic cellular compartments (Supplementary Fig. 3f–g).
ARLNC1 transcription is directly regulated by AR
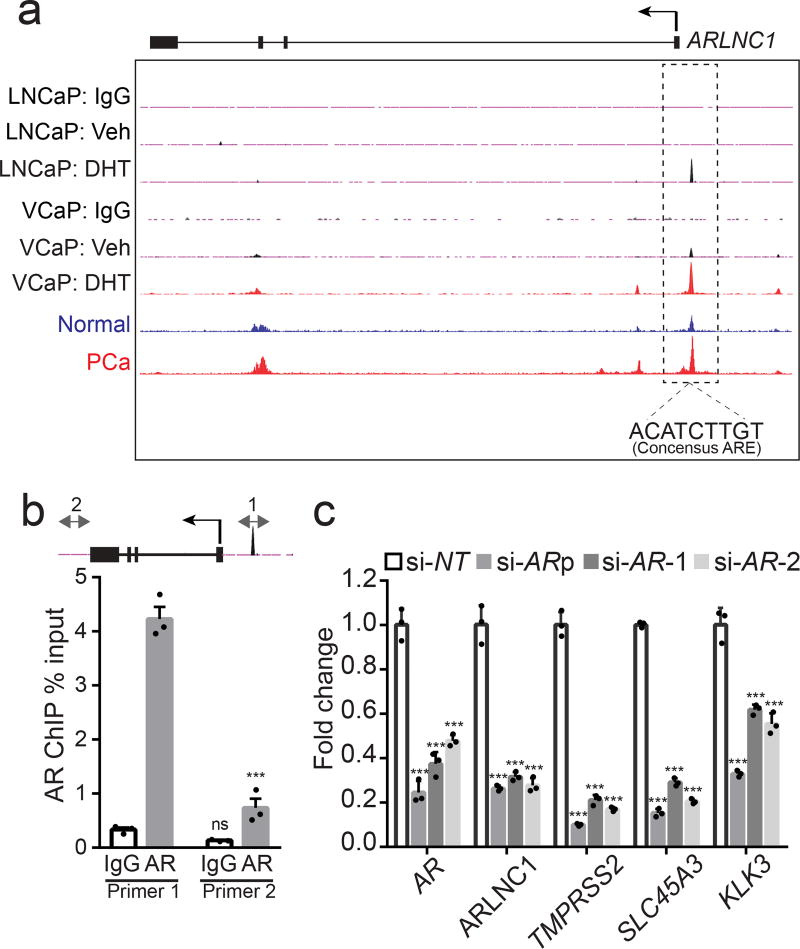
Since ARLNC1 was identified as an AR-regulated lncRNA, we inspected ARLNC1 promoter region for AR occupancy and identified an androgen-induced AR peak in AR ChIP-Seq data from both DHT-stimulated VCaP and LNCaP cells (Fig. 3a). Importantly, this AR binding site was also observed in prostate tissue samples and contained a canonical androgen response element (ARE)33 (Fig. 3a). These observations were corroborated by ChIP-qPCR in MDA-PCa-2b cells, which showed the highest level of ARLNC1 expression (Fig. 3b). Considering the observation that ARLNC1 expression is prostate tissue-specific, while AR expression is not as much, we searched for additional regulators (transcription factors and epigenetic modifiers) of this gene (Supplementary Fig. 4a). Motif analysis of the ARLNC1 promoter region identified several transcription factor binding sites, including a FOXA1-response element. To further validate ARLNC1 gene regulation by AR and FOXA1, we evaluated ARLNC1 transcript levels following AR or FOXA1 knockdown. AR or FOXA1 loss resulted in decreased expression of ARLNC1, along with other canonical AR target genes that served as positive controls (Fig. 3c, Supplementary Fig. 4b). ChIP-seq and ChIP-PCR analysis additionally confirmed the putative FOXA1 binding motif on the ARLNC1 promoter (Supplementary Fig. 4c). Together, these observations suggest that ARLNC1 is directly regulated by AR and modestly regulated by FOXA1, which, partially explains the tissue-specific expression pattern of ARLNC1, as expression of these two factors overlaps nearly exclusively in prostate tissue37,38 (Supplementary Fig. 4d).
Fig. 3. ARLNC1 is directly regulated by AR.
a, AR ChIP-Seq in prostate cancer cell lines and tissues. Normalized ChIP-Seq enrichment. Top, AR or control (IgG) ChIP-Seq results across the ARLNC1 locus in LNCaP and VCaP cells with vehicle (ethanol) treatment or DHT treatment. Bottom, AR ChIP-Seq in benign prostate and clinically-localized prostate cancer tissue. b, ChIP-qPCR in MDA-PCa-2b cells showing AR or IgG enrichment (ChIP/input) over ARLNC1 promoter region (Primer 1) or control region (Primer 2). Error bars represent mean ± s.e.m. (n = 3 biologically independent samples). ***Adjusted P < 0.0001, ns: P = 0.5746, compared to control Primer 2, by ANOVA analysis with Sidak correction for multiple comparisons. Top: schematic of amplicon locations for ChIP-qPCR validation. cAR and AR target gene (ARLNC1, TMPRSS2, SLC45A3, and KLK3) expression in MDA-PCa-2b cells transfected with control siRNA (si-NT) or siRNAs against AR (si-AR-pool, si-AR-1, si-AR-2). Mean ± s.e.m. are shown, n = 3 biologically independent samples. ***P = 0.0001 determined by ANOVA with Dunnett’s multiple comparisons test.
ARLNC1 regulates AR signaling
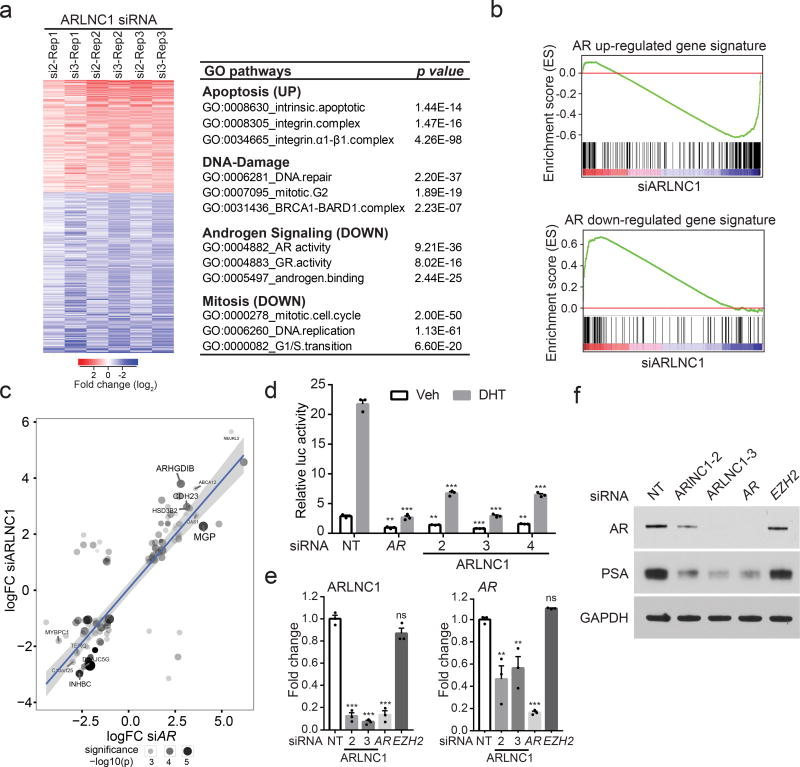
To elucidate the function of ARLNC1 in prostate cancer, we performed gene expression profiling of wildtype and ARLNC1-knockdown MDA-PCa-2b cells (Fig. 4a). Gene Ontology (GO) pathway enrichment analysis of the differentially expressed genes revealed deregulation of four main biological activities: apoptosis, cell proliferation, DNA damage response, and androgen signaling (Fig. 4a). A significant decrease in AR target gene expression is particularly interesting given the fact that ARLNC1 is regulated by AR, suggesting a positive feedback loop between ARLNC1 and AR signaling. To confirm this observation, we generated an AR target gene signature from MDA-PCa-2b cells stimulated with DHT (Supplementary Fig. 5a, Supplementary Table 2) and performed Gene Set Enrichment Analysis (GSEA) using this gene signature (Fig. 4b). Knockdown of ARLNC1 led to suppression of genes positively regulated by AR and upregulation of genes negatively regulated by AR (Fig. 4b–c, Supplementary Fig. 5b). This was further confirmed by AR reporter activity assay (Fig. 4d, Supplementary Fig. 5c), as well as qPCR analysis of AR target genes (Supplementary Fig. 5d). Interestingly, ARLNC1 knockdown also had a significant effect on the mRNA and protein levels of AR (Fig. 4e–f), suggesting direct regulation of AR by ARLNC1. We, however, found that ARLNC1 overexpression did not affect AR and its signaling cascade (Supplementary Fig. 5e).
Fig. 4. ARLNC1 loss attenuates AR signaling.
a, Gene expression profiling for ARLNC1 knockdown in MDA-PCa-2b cells (n = 3 biologically independent cell cultures for each siRNA). The chart presents top enriched pathways upon ARLNC1 knockdown, identified using Gene Ontology (GO) enrichment analysis (RandomSet test). b, Gene Set Enrichment Analysis (GSEA) showing significant enrichment of ARLNC1- regulated gene set with respect to the AR target gene sets (n= 3 independent gene expression profiles). Shown are the enrichment plots for gene sets consisting of genes positively regulated by AR (upper panel), and genes negatively regulated by AR (lower panel). c, Comparison of ARLNC1-regulated and AR target genes based on RNA-seq following knockdown of AR and ARLNC1. Significant genes and their log-fold changes in either of the conditions are shown (n= 2 independent cell cultures per-condition). Combined significance levels, determined by limma moderated t-test (across both knockdowns) are indicated by circle size. d, siRNA knockdown of ARLNC1 in MDA-PCa-2b cells impairs AR signaling by AR reporter gene assay. siRNA against AR serves as a positive control for inhibition of AR signaling. Mean ± s.e.m. are shown, n = 3 biologically independent cell cultures. **P < 0.01, ***P = 0.0001 determined by ANOVA with Dunnett correction. e, qRT–PCR analysis of ARLNC1 and AR in MDA-PCa-2b cells transfected with siRNAs against ARLNC1, AR, EZH2, or non-specific control (NT). siRNA against AR serves as a positive control for inhibited AR signaling, while siRNA against EZH2 serves as a negative control. Mean ± s.e.m. are shown, n = 3. **P < 0.01, ***P = 0.0001 determined by ANOVA with Dunnett correction. f, Immunoblot of AR, PSA, and GAPDH in MDA-PCa-2b cells transfected with siRNAs against ARLNC1, AR, EZH2, or non-specific control (NT). Experiments were repeated 3 times independently with similar results. Uncropped images are shown in Supplementary Fig. 9.
In situ co-localization of ARLNC1 and AR transcripts
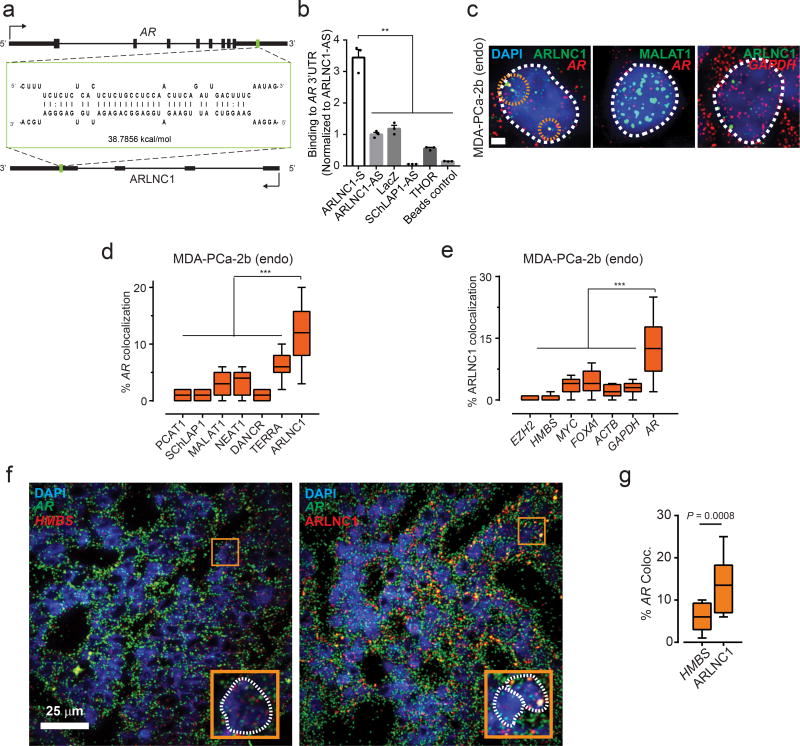
Non-coding RNAs have been shown to target mRNAs via direct or indirect RNA-RNA interaction9,40–42. To identify target mRNAs that could interact with ARLNC1, we performed an unbiased prediction of RNA-RNA interactions using IntraRNA43,44. Interestingly, the 3’ UTR of the AR transcript was identified as a target of ARLNC1 (Fig. 5a, Supplementary Fig. 6a). An in vitro RNA-RNA interaction assay between the 3’UTR of AR and full-length ARLNC1 confirmed this in silico prediction (Fig. 5b). To evaluate this interaction in the context of the cellular environment, multiplexed smFISH for AR and ARLNC1 transcripts was performed in MDA-PCa-2b cells. Upon co-staining MDA-PCa-2b cells with either AR and a panel of lncRNAs, or ARLNC1 and a panel of mRNAs, we observed specific colocalization between AR and ARLNC1 transcripts in the nucleus within foci that were typically larger than individual molecules (Fig. 5c–e). The extent of colocalization was much higher than that expected from co-incidental colocalization with an abundant transcript, such as MALAT1 or GAPDH (Fig. 5c–e). More specifically, colocalization typically occurred at a stoichiometry of 2:1 ARLNC1:AR, which accounted for ~10–20% of all AR and ARLNC1 transcripts in the cell (Supplementary fig. 6b). Furthermore, AR-ARLNC1 colocalization was observed in ARLNC1-positive prostate cancer tissues (Fig. 5f–g).
Fig. 5. In situ co-localization between AR mRNA and ARLNC1 in prostate cancer cells.
a, Schematic of predicted RNA-RNA interaction between ARLNC1 and 3’UTR of ARb, ARLNC1 interacts with AR 3’UTR in an in vitro RNA-RNA interaction assay. Compared to a panel of control RNAs (ARLNC1 antisense, LacZ, SCHLAP1-AS, THOR), ARLNC1 binds to AR 3’UTR-1-980 with high affinity. Binding affinity was quantified by qPCR analysis of AR 3’UTR. Data were normalized to ARLNC1-AS control. Mean ± s.e.m. are shown, n = 3. **P < 0.001 by two-tailed Student’s t-test. c–e, smFISH depiction of AR-ARLNC1 colocalization in situ. Representative pseudocolored images of MDA-PCa-2b cell nuclei (c) stained for the appropriate endogenous (endo) transcripts (green, red) and DAPI (nucleus, blue). Scale bar, 5 µm. Quantification of the percentage of AR or ARLNC1 molecules co-localizing with a panel of lncRNAs (d) or mRNAs (e) respectively. Orange circles represent regions of colocalization. Center line and whiskers depict the median and range respectively and box extends from 25th to 75th percentiles (n= 50 cells for each sample aggregated from 3 independent experiments). ***P < 0.0001 by two-tailed Student’s t-test. f–g, Representative pseudo-colored images of ARLNC1 positive prostate cancer tissues (f) stained with DAPI (nucleus, blue) and AR (green), HMBS (red), or ARLNC1 (red) transcripts (smFISH). Scale bar, 25 µm. Inset, 5.5×5.5 µm2 zoomed-in view of box within large panel. Quantification of the percentage of AR molecules (g) colocalizing with HMBS or ARLNC1 is also depicted in box plot. Center line and whiskers depict the median and range respectively and box extends from 25th to 75th percentiles (n = 15 field-of-views for each sample aggregated from 3 independent tissues). **P < 0.001 by two-tailed Student’s t-test.
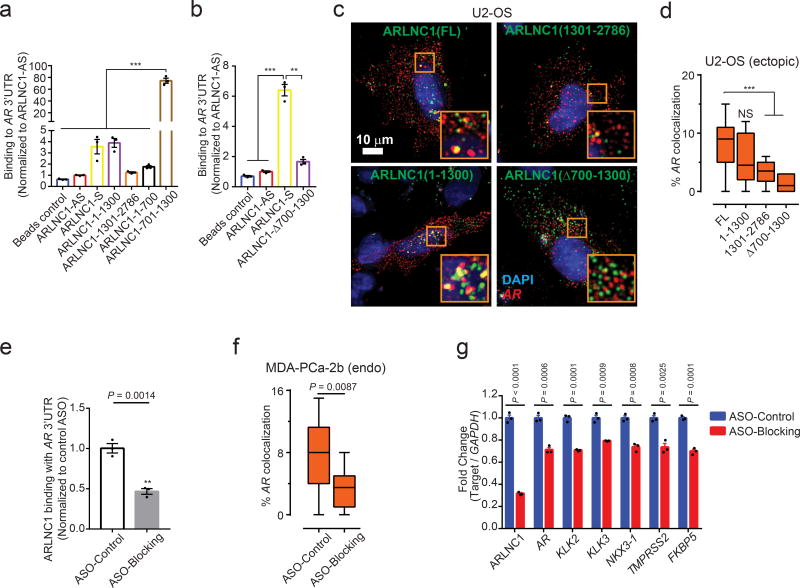
Using an in vitro RNA-RNA binding assay, we identified nucleotides (nt) 700–1300 of ARLNC1 to be critical for binding to the AR 3’UTR (Fig. 6a–b). To confirm this observation within the cellular context, we ectopically overexpressed different fragments of ARLNC1 together with AR in U2OS osteosarcoma cells. In this exogenous system, colocalization between AR and ARLNC1 was once again demonstrated, wherein colocalization was dependent on the presence of 700–1300 nt of ARLNC1 (Fig. 6c–d, Supplementary Fig. 6c). Furthermore, incubation with antisense oligos (ASOs) that blocked the interaction site led to a significant reduction in ARLNC1-AR interaction in vitro and in situ (Fig. 6e–f, Supplementary Fig. 6d–e). Decreased AR signaling was also observed following blocking of this interaction (Fig. 6g, Supplementary Fig. 6f).
Fig. 6. Identification of ARLNC1 fragment mediating RNA-RNA interaction with AR mRNA.
a, In vitro RNA-RNA interaction assay identifies nucleotides 700–1300 on ARLNC1 as critical binding sites to AR 3’UTR-1-980. ARLNC1 fragments covering nucleotides 700–1300 display comparable or higher AR 3’UTR-binding affinity compared to ARLNC1-S, with ARLNC1-700–1300 exhibiting the highest binding affinity. Data were normalized to ARLNC1-AS control. Mean ± s.e.m. are shown, n = 3. ***Adjusted P = 0.0001, determined by ANOVA with Dunnett’s multiple comparisons test. b, Deletion of nucleotides 700–1300 on ARLNC1 results in impaired binding to AR 3’UTR, as shown by in vitro RNA-RNA interaction assay. Data were normalized to ARLNC1-AS control. Mean ± s.e.m. are shown, n = 3. ***P = 0.0001, **P = 0.0003 by two-tailed Student’s t-test. c–d, smFISH shows that 700–1300nt in ARLNC1 is important for colocalization in situc, Representative pseudo-colored images of U2-OS cells stained for DAPI (nucleus, blue), ARLNC1 (green) and AR transcripts (red). Scale bar, 10 µm. Inset, 10×10 µm2 zoomed-in view of orange box in the image. d, Quantification of the percent of AR molecules colocalizing with various ARLNC1 fragments. Center line and whiskers depict the median and range respectively and box extends from 25th to 75th percentiles (n= 50 cells for each sample aggregated from 3 independent experiments). ***P < 0.0001 by two-tailed Student’s t-test. NS, not significant. e, Antisense oligos targeting sites 700–1300 on ARLNC1 transcript (Blocking ASO pool) inhibit ARLNC1 interaction with AR 3’UTR. In vitro RNA-RNA interaction assays were performed using ARLNC1 and AR 3’UTR, with the addition of blocking ASO pool or control ASO. Data were normalized to control ASO. Mean ± s.e.m. are shown, n = 3. P = 0.0014 by two-tailed Student’s t-test. f, smFISH shows that ASOs targeting 700–1300nt on ARLNC1 transcript (ASO-Blocking) inhibit ARLNC1 colocalization with AR, in situ. Quantification of the percent of AR transcripts colocalizing with ARLNC1 after various treatments in MDA-PCa-2b cells. Center line and whiskers depict the median and range and box extends from 25th to 75th percentiles (n= 50 cells for each sample aggregated from 3 independent experiments). P value computed by two-tailed Student’s t-test. g, qPCR analysis of ARLNC1, AR transcript and AR signaling genes (KLK2, KLK3, NKX3-1, TMPRSS2, FKBP5) in MDA-PCa-2b cells transfected with control or blocking ASOs targeting the interaction sites between ARLNC1 and AR 3’UTR. Mean ± s.e.m. are shown, n = 3. Significance was determined by two-tailed Student’s t-test.
ARLNC1 regulates the cytoplasmic levels of AR transcripts
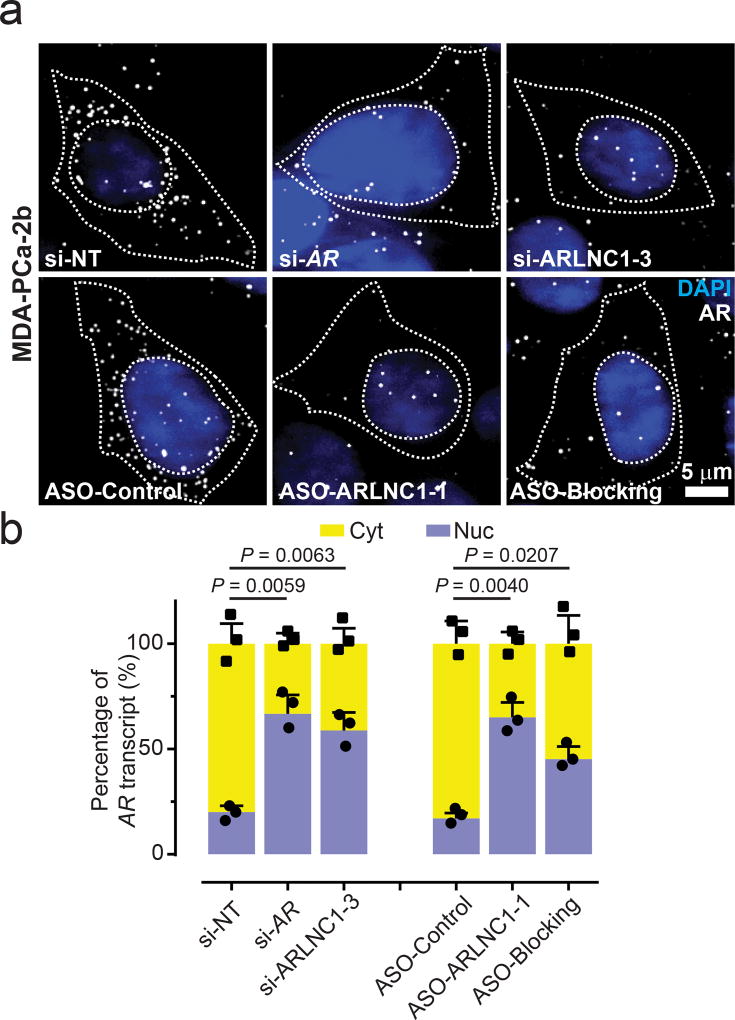
We then sought to delineate the mechanism of ARLNC1-mediated AR regulation. We first monitored the stability of these two transcripts and found that AR and ARLNC1 have similar half-lives of ~9 hours (Supplementary Fig. 6g). Because ARLNC1 depletion resulted in a striking reduction of AR protein levels, much more than that could be explained by AR transcript reduction, we hypothesized that ARLNC1 could affect AR post-transcriptionally. To test this hypothesis, we tracked sub-cellular localization of AR transcripts using smFISH after depleting ARLNC1. We confirmed successful in situ knockdown of ARLNC1 using siRNAs, antisense oligo (ASO), and the blocking oligos that targeted ARLNC1-AR interaction (ASO-blocking) in MDA-PCa-2b cells (Supplementary Fig. 6h–i). Quantification of the sub-cellular distribution of ARLNC1 suggested that the nuclear fraction of ARLNC1 was enriched only in the si-ARLNC1 condition (Supplementary Fig. 6j), a result which was expected for siRNAs that are typically more functional in the cytosol45. Surprisingly, ARLNC1 knockdown or obstruction of the AR-ARLNC1 interaction increased the nuclear AR fraction by dramatically decreasing cytoplasmic levels of AR transcript (Fig. 7a–b, Supplementary Fig. 6k–l). This observation was further supported by BrU-seq and BrU-chase-seq, two high-throughput tools that monitor transcript synthesis and stability. Upon ARLNC1 knockdown, the synthesis rate of the AR transcript remained the same (Supplementary Fig. 6m), while the stability of the transcript decreased, particularly through the 3’UTR region (Supplementary Fig. 6n). Taken together, our data suggest that ARLNC1 regulates the cytoplasmic levels of AR transcripts. Furthermore, the transcriptional coupling between AR and ARLNC1 transcripts is mediated by direct interactions which are encoded in their sequences.
Fig. 7. ARLNC1 regulates cytoplasmic level of AR transcript.
a, ARLNC1 regulates AR post-transcriptionally by specifically affecting cytoplasmic AR mRNA. Representative pseudo-colored images of MDA-PCa-2b cells stained for DAPI (nucleus, blue) and AR (gray) after treatment with siRNA against AR (si-AR), siRNA against ARLNC1 (si-ARLNC1–3), ASO against ARLNC1 (ASO-ARLNC1-1) or blocking ASO against AR-ARLNC1 colocalizing segment (ASO-Blocking). Scale bar, 5 µm. Quantification of knockdown are represented in Supplementary Fig. 6(k–l). b, Fractional column plots depicting the nucleo-cytoplasmic distribution of AR mRNA after various treatment conditions in (a), as computed using smFISH. Mean ± s.e.m. are shown, n = 3 independent experiments and 60 cells analyzed for each sample. P values were computed by comparing to si-NT or ASO-Control treated cells, by two-tailed Student’s t-test.
Inhibition of ARLNC1 delays prostate cancer growth in vitro and in vivo
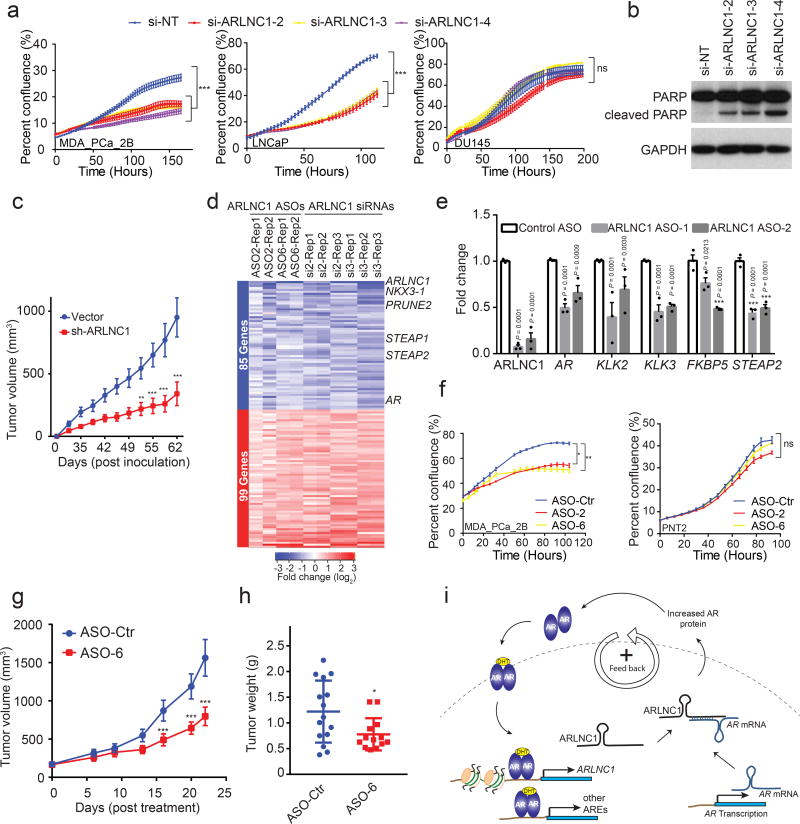
Having established a role for ARLNC1 in the regulation of AR signaling, we further evaluated the biological effects of ARLNC1 in prostate cancer cell lines. GO pathway enrichment analysis of the knockdown microarray data showed that ARLNC1-regulated genes were involved in cell proliferation and apoptosis (Fig. 4a). Knockdown of ARLNC1 had a significant effect on the proliferation of AR-dependent MDA-PCa-2b and LNCaP cells, but had no effect on AR-negative DU145 and PC3 cells (Fig. 8a, Supplementary Fig. 7a–b). Knockdown of ARLNC1 also resulted in increased apoptosis in AR-positive prostate cancer cells (Fig. 8b, Supplementary Fig. 7c). Importantly, these results translated to effects in vivo, as cells expressing shRNA targeting ARLNC1 formed smaller tumors in mice when compared to cells expressing non-targeting shRNA (Fig. 8c), thus suggesting that ARLNC1 is an important survival factor for AR-dependent prostate cancer.
Fig. 8. ARLNC1 as a therapeutic target in AR-positive prostate cancer models.
a, siRNA knockdown of ARLNC1 in vitro in AR-positive prostate cancer cell lines (MDA-PCa-2b and LNCaP) inhibits cell proliferation. The AR-negative prostate cell line DU145 serves as negative control. Mean ± s.d. are shown, n = 6 independent cell cultures per group, **Adjusted P = 0.0001 compared to si-NT treated cells, by one-way ANOVA analysis with Dunnett’s multiple comparison test; ns: not significant. b, ARLNC1 loss leads to increased apoptosis as shown by western blot analysis of PARP and cleaved PARP in LNCaP cells following ARLNC1 knockdown. The experiment was repeated independently for 3 times with similar results. Uncropped images are shown in Supplementary Fig. 9. c, Tumor growth of LNCaP-AR cells expressing shRNA targeting ARLNC1 or shRNA vector. Mean ± s.e.m. are shown. n = 10 independent tumors, ***P < 0.0001 and **P = 0.0007, significance tested by two-tailed Student’s t-test. d, Gene expression profiling for siRNA-mediated or ASO-mediated ARLNC1 knockdown in MDA-PCa-2b cells. Numbers above the heatmap represent the specific microarray replicates. e, qRT–PCR analysis of ARLNC1, AR, and AR targets (KLK2, KLK3, FKBP5, and STEAP2) in MDA-PCa-2b cells transfected with ASOs against ARLNC1. Data were normalized to a housekeeping gene and the levels in control ASO-treated cells were set to 1. Mean ± s.e.m. are shown, n = 3. Adjusted P values determined by one-way ANOVA with Dunnett correction for multiple comparisons. f, Transfection of ASOs targeting ARLNC1 in AR-positive MDA-PCa-2b cells inhibits cell proliferation. AR-negative prostate cell line PNT2 serves as negative control. Mean ± s.e.m. are shown, n = 6 independent cell cultures per treatment group. *Adjusted P = 0.0112, **Adjusted P = 0.0065, ns: not significant; compared to control-ASO group by one-way ANOVA analysis with Dunnett correction for multiple comparisons. g–h, Effect of ASO treatment on the growth of MDA-PCa-2b xenografts in male NOD-SCID mice, with control ASO (n = 15) or ARLNC1 ASO (n = 13) treatment subcutaneously at 50 mg/kg, five times per week for three weeks. Tumors were measured by caliper bi-weekly (g) and tumor weights were measured at end point (h). Mean ± s.d. are shown. *P = 0.0251, ***P < 0.0001; compared to control ASO by two-tailed Student’s t-test. i, Model depicting positive feedback loop between ARLNC1 and AR that is critical for prostate cancer growth.
Since modulation of ARLNC1 expression levels resulted in a striking proliferation phenotype, we hypothesized that ARLNC1 inhibition could be used therapeutically for the treatment of prostate cancer. Antisense oligos have recently been shown to be effective in targeting RNA in vivo46–49, thus, we designed ASOs targeting the ARLNC1 transcript (Supplementary Fig. 7d). Transfection of ASOs exhibited strong knockdown efficiency (Supplementary Fig. 7e), and ASO-mediated knockdown resulted in similar effects on gene expression profiling as siRNA (Fig. 8d–e, Supplementary Fig. 7f). Furthermore, AR-positive cells transfected with ARLNC1 ASOs exhibited retarded growth, similar to those treated with siRNAs (Fig. 8f). To evaluate the therapeutic potential of ARLNC1 ASOs in vivo, we first assessed the cellular free uptake efficiency of ARLNC1 ASOs, a prerequisite for ASO therapeutic use. Importantly, several ASOs significantly reduced ARLNC1 levels through free uptake (Supplementary Fig. 7g). Free uptake of ARLNC1 ASOs led to a significant decrease in the proliferation capacity of MDA-PCa-2b cells in both normal cell culture and 3D sphere conditions (Supplementary Fig. 7h–j). Treatment of mice bearing MDA-PCa-2b xenografts with ARLNC1-targeting ASOs led to significant decreases in tumor growth compared to control ASO (Fig. 8g–h, Supplementary Fig. 8a–e). Taken together, these data, along with the association of ARLNC1 with aggressive androgen signaling (Supplementary Fig. 8f–j), suggest that ARLNC1 plays a critical role in the proliferation of AR-dependent prostate cancer and can be effectively exploited as a therapeutic target.
Discussion
As AR signaling remains a significant driver of CRPC pathogenesis, it is imperative to generate novel strategies to target this pathway. Even with the addition of enzalutamide or abiraterone to CRPC treatment regimens, progression invariably occurs. Exploiting players other than AR itself that are pivotal to maintaining the magnitude of the androgen response is an alternative approach. Our comprehensive profiling of AR-regulated, prostate cancer-associated lncRNAs identified the top-ranking candidate ARLNC1 that we functionally characterized. We identified a positive feedback loop between ARLNC1 and AR that maintains the androgen transcriptional program in AR-positive prostate cancer cells, specifically through regulating the cellular levels of AR (Fig. 8i). The mechanism we identified echoes previous studies on lncRNAs: 1/2-sbsRNAs42, BACE1-AS9, and TINCR41, which highlights the role of lncRNA in increasing or decreasing RNA stability.
As a novel non-coding regulator of AR signaling, ARLNC1 has the potential to be not only a mechanistic biomarker, but also a therapeutic target for advanced prostate cancer. In addition, acting upstream of AR signaling presents the possibility that targeting ARLNC1 may afford an additional option to patients that have de novo or acquired resistance to therapies targeting AR itself (i.e. enzalutamide or abiraterone). Furthermore, specific antisense nucleotides targeting ARLNC1, which we demonstrate to be specifically expressed in the prostate, could circumvent undesirable side effects that occur in other tissues with exposure to androgen synthesis inhibitors or antiandrogens.
Although we have identified a new node of the AR signaling network that can be therapeutically-targeted, the molecular mechanism through which ARLNC1 regulates AR transcript levels remains to be fully characterized. At this time, it is unclear whether the physical interaction between the AR 3’UTR and ARLNC1 functions with the aid of additional RNA-binding proteins (e.g. HuR) and/or RNAs in vivo50,51. Nonetheless, the application of ASOs has ushered in an exciting era that makes it possible to target previously “undruggable” molecules directly at the transcript level, such as ARLNC1, which is likely to yield promising opportunities in cancer treatment.
Methods
Cell lines
Cell lines were purchased from the American Type Culture Collection (ATCC) and maintained using standard media and conditions. All cell lines were genotyped by DNA fingerprinting analysis and tested for mycoplasma infection every two weeks. All cell lines used in this study were mycoplasma-negative. For androgen stimulation experiments, VCaP and LNCaP cells were grown in charcoal-stripped serum containing media for 48 hours and then stimulated with 10 nM DHT (Sigma-Aldrich) for 6 or 24 hours.
RNA-Seq
Total RNA was extracted from LNCaP and VCaP cells following DHT treatment, using the miRNeasy kit (QIAGEN). RNA quality was assessed using the Agilent Bioanalyzer. Each sample was sequenced using the Illumina HiSeq 2000 (with a 100-nt read length) according to published protocols52.
RNA-Seq data analysis to identify AR-regulated genes
RNA-Seq data were analyzed as previously described53. Briefly, the strand-specific paired-end reads were inspected for sequencing and data quality (e.g. insert size, sequencing adapter contamination, rRNA content, sequencing error rate). Libraries passing QC were trimmed of sequencing adapters and aligned to the human reference genome, GRCh38. Expression was quantified at the gene level using the “intersection non-empty” mode54 as implemented in featureCounts55 using the Gencode v2256 and/or MiTranscriptome assemblies10. All pairwise differential expression analyses were carried out using the voom-limma approach57,58 with all default parameters. Relative expression levels (FPKMs, fragments per kilobase of transcript per million mapped reads) were normalized for differences in sequencing depth using scaling factors obtained from the calcNormFactors (default parameters) function from edgeR59.
AR-regulated genes (ARGs) were identified from expression data of VCaP and LNCaP cells treated with DHT after 6 and 24 hours using three linear models: separate models for each of the cell lines treating the two time-points as biological replicates, and a merged model with all treated samples as replicates. ARGs were defined as genes that were significant (P value < 0.1 and absolute log fold-change > 2) in both separate models and/or the merged model.
Identification of prostate cancer associated protein-coding genes and lncRNAs
Raw RNA-Seq data for primary and metastatic patients were obtained from the TCGA/PRAD and PCF/SU2C projects, respectively. External transcriptome samples were re-analyzed using in-house pipelines (see above) to facilitate direct comparisons of expression levels and identification of DEGs. Pan-cancer analyses based on the MiTranscriptome assembly10 were leveraged as FPKMs and enrichment scores (SSEA) were computed as part of that project. To visualize data, fold changes were computed relative to median expression levels estimated across the combined (normal, primary, metastatic) cohorts and subjected to unsupervised hierarchical clustering separately within each cohort. Tissue lineage (prostate) and prostate cancer-specific genes were identified using the sample set enrichment analysis (SSEA) method as previously described10. Briefly, the SSEA test was used to determine whether each gene was significantly associated with a set of samples (e.g. prostate cancer), or cancer progression in a given lineage (e.g. prostate normal to prostate cancer). The genes were ranked according to their strength of association.
Oncomine concept analysis of the ARLNC1 signature
Genes with expression levels significantly correlated with ARLNC1 were separated into positively and negatively correlated gene lists. These two lists were then imported into Oncomine as custom concepts and queried for association with other prostate cancer concepts housed in Oncomine. All the prostate cancer concepts with odds ratio > 2.0 and P-value < 1 × 10−4 were selected. Top concepts (based on odds ratios) were selected for representation. We exported these results as the nodes and edges of a concept association network and visualized the network using Cytoscape version 3.3.0. Node positions were computed using the Edge-weighted force directed layout in Cytoscape using the odds ratio as the edge weight. Node positions were subtly altered manually to enable better visualization of Mode labels60.
Chromatin immunoprecipitation (ChIP)-Seq data analysis
ChIP-Seq data from published external and in-house data sets, GSE56288 and GSE55064, were reanalyzed using a standard pipeline. Briefly, groomed reads (vendor QC, adapter removal) were aligned to the GRCh38 reference genome using STAR settings that disable spliced alignment: outFilterMismatchNoverLmax: 0.05, outFilterMatchNmin: 16, outFilterScoreMinOverLread: 0, outFilterMatchNminOverLread: 0, alignIntronMax: 1. Improperly paired alignments and non-primary alignments were discarded. Peaks were called using MACS2 (callpeak --broad --qvalue 0.05 --broad-cutoff 0.05 and callpeak --call-summits --qvalue 0.05)61 and Q (−n 100000)62. ChIP enrichment plots were computed from alignment coverage files (BigWig63) as trimmed (trim=0.05) smooth splines (spar=0.05). The baseline (non-specific) ChIP signal was estimated from genomic windows furthest from the center of the queried region (peak summit, transcription start site) and subtracted from each signal before plotting.
AR binding motif search
Unsupervised motif search was carried out using MEME64. DNA sequences (GRch38) from the uni-peak ChIP-Seq regions overlapping promoters (5 kb upstream, 1 kb downstream of the assembled or known TSS) of ARGs were used as input to MEME (default parameters).
ChIP-qPCR assay
AR, FOXA1, or NKX3-1 ChIP was performed following our previous protocol32. (Antibodies: AR, Millipore Cat# 06–680; FOXA1, Thermo Fisher Cat# PA5–27157; NKX3-1, CST Cat# 83700S.) Quantitative PCR (qPCR) analysis was performed using primers listed in Supplementary Table 3. Primers targeting CYP2B7 promoter were purchased from CST, Cat #84846.
RNA in situ hybridization (RNA ISH) on tissue microarray
In situ hybridization assays were performed on tissue microarray sections from Advanced Cell Diagnostics, Inc. as described previously7. In total, 133 tissue samples were included (11 from benign prostate, 85 from localized prostate cancer, and 37 from metastatic prostate cancer). ARLNC1 ISH signals were examined in morphologically-intact cells and scored manually by a study pathologist, using a previously described expression value scoring system65. For each tissue sample, the ARLNC1 product score was averaged across evaluable TMA tissue cores. Mean ARLNC1 product scores were plotted in Fig. 2e.
Rapid Amplification of cDNA Ends (RACE)
5’ and 3’ RACE were performed to determine the transcriptional start and termination sites of ARLNC1, using the GeneRacer RLM-RACE kit (Invitrogen), according to the manufacturer’s instructions.
Northern blot analysis
NorthernMax-Gly Kit (Ambion) was used for ARLNC1 detection following the manufacturer’s protocol. Briefly, 20 µg of total RNA was resolved on a 1% agarose glyoxal gel and then transferred to nylon membrane (Roche), cross-linked to the membrane (UV Stratalinker 1800; Stratagene), and the membrane was pre-hybridized. Overnight hybridization was performed with ARLNC1-specific P32-labeled RNA probe. Membranes were exposed to HyBlot CL autoradiography film (Denville Scientific). The primer sequences used for generating the probes are given in Supplementary Table 3.
RNA isolation and cDNA synthesis
Total RNA from cell lines was isolated using QIAzol Lysis reagent (QIAGEN) and miRNeasy kit (QIAGEN) with DNase digestion according to manufacturer’s instructions. cDNA was synthesized using Superscript III (Invitrogen) and random primers (Invitrogen).
qRT–PCR analysis
Relative RNA levels determined by qRT–PCR were measured on an Applied Biosystems 7900HT Real-Time PCR System, using Power SYBR Green MasterMix (Applied Biosystems). All of the primers were obtained from Integrated DNA Technologies (IDT), and gene-specific sequences are listed in Supplementary Table 3. GAPDH, HMBS, or ACTB were used as internal controls for quantification of gene targets. The relative expression of RNAs was calculated using ΔΔCt method.
Cytoplasmic and nuclear RNA purification
Cell fractionation was performed using the NE-PER nuclear extraction kit (Thermo Scientific) according to manufacturer’s instructions. RNA was extracted using the previously mentioned protocol.
siRNA-mediated knockdown
siRNA oligonucleotides targeting ARLNC1, AR, FOXA1, BRD4, NKX3-1, LSD1, IRF1, POU1F1, or EZH2 and a non-targeting siRNA were purchased from Dharmacon. (si-AR-pool, Cat# L-003400-00–0005; si-FOXA1, Cat# LU-010319-00–0005; si-BRD4, Cat# LU-004937-00–0002; si-NKX3-1, Cat# LU-015422-00–0005; si-LSD1, Cat# LU-009223-00–0002; si-IRF1, Cat# LU-011704-00–0005; si-POU1F1, Cat# LU-012546-00–0005; si-EZH2, Cat# L-004218-00–0005; si-NT, Cat# D-001810-01–05.) siRNA sequences for ARLNC1 knockdown are listed in Supplementary Table 3. For AR knockdown, two more siRNAs were purchased from Life Technologies (#HSS179972, #HSS179973). Transfections with siRNA (50 nM) were performed with Lipofectamine RNAiMAX according to the manufacturer’s instructions. RNA and protein were harvested for analysis 72 hours after transfection.
ASO-mediated knockdown
Antisense oligos targeting ARLNC1 were obtained from Ionis Pharmaceuticals. Transfections with ASOs (50 nM) were performed with Lipofectamine RNAiMAX according to the manufacturer’s instructions. RNA and protein were harvested for analysis 72 hours after transfection.
Gene expression profiling
Total RNA was extracted following the aforementioned protocol. RNA integrity was assessed using the Agilent Bioanalyzer. Microarray analysis was carried out on the Agilent Whole Human Oligo Microarray platform, according to the manufacturer’s protocol. siRNA-mediated knockdown experiments were run in triplicates, comparing knockdown samples treated with two independent ARLNC1 siRNAs to samples treated with non-targeting control siRNA. ASO-mediated knockdown experiments were run in replicates, comparing knockdown samples treated with two ARLNC1 ASOs to samples treated with non-targeting control. An AR signature was generated using MDA-PCa-2b cells treated with 10 nM DHT in triplicates.
Analysis of Agilent 44k microarrays was carried out using limma and included background subtraction (bc.method=“half”, offset=100) and within-array normalization (method=“loess”). Between array quantile normalization of average expression levels (but not log-fold changes) was performed using the function normalizeBetweenArrays (method=“Aquantile”). Control probes and probes with missing values were excluded from further analyses. Probes were annotated to Gencode v22 genes using the mapping downloaded from Ensembl (efg_agilent_wholegenome_4×44k_v2). Probes originally annotated as AK093002 were used to detect ARLNC1. Differentially-expressed genes following ARLNC1 knockdown in MDA-PCA-2b cells were identified from triplicate biological repeats using adjusted P value < 0.1 and absolute log fold-change > 0.6 cut-offs. Consensus targets of ARLNC1 knockdown using siRNA and ASOs were identified using a merged linear model (all 10 samples treated replicates) and a P value < 0.001 cut-off.
Gene Set Enrichment Analysis
Enrichment analyses for custom and experimentally-derived signatures (i.e. AR targets, genes up- and down-regulated following DHT treatment) were carried out using the nonparametric GSEA software with all default settings. For Gene Ontology (GO) term enrichment, we applied the parametric randomSet66 enrichment statistic to voom-limma estimated fold-changes (see above).
Overexpression of ARLNC1
Full-length ARLNC1 was amplified from MDA-PCa-2b cells and cloned into the pCDH clone and expression vector (System Biosciences). Insert sequences were validated by Sanger sequencing at the University of Michigan Sequencing Core. Full-length sequence for ARLNC1 expression is listed in Supplementary Table 4.
Single molecule fluorescent in situ hybridization (smFISH)
smFISH and image analysis were performed as described67,68, Probe sequences targeting ARLNC1, PCAT1, DANCR, AR, EZH2 and FOXA1 were designed using the probe design software in https://www.biosearchtech.com/stellaris-designer and are listed in Supplementary Table 5. TERRA probes were designed as described69. Other probes were purchased directly from the LGC-Biosearch. U2-OS cells were seeded in 6-well dishes and transfected with ARLNC1-expression vector alone, or in combination with AR expression vector, using Fugene-HD (Promega) according to the manufacturer’s protocol. Cells were incubated for 24 hours, reseeded into 8-well chambered coverglasses, and were formaldehyde-fixed for smFISH (as described above) after 24 hours.
RNA in vitro transcription
Linearized DNA templates for full-length ARLNC1, ARLNC1 fragments, ARLNC1 deletion, antisense ARLNC1, LacZ, SCHLAP1-AS, THOR, and AR-3’UTR-1–980 were synthesized using T7-containing primers. In vitro transcription assays were performed with T7 RNA polymerase (Promega) according to the manufacturer’s instructions. For BrU-labeled RNA synthesis, 5-Bromo-UTP was added to the transcription mix. At the end of transcription, DNA templates were removed by Turbo DNase (ThermoFisher), and RNA was recovered using RNA Clean and Concentrator Kit (Promega). RNA size and quality was further confirmed by the Agilent Bioanalyzer.
RNA-RNA in vitro interaction assay
25 µl of Protein A/G Magnetic Beads (Pierce) were washed twice with RIP Wash Buffer (Millipore, Cat# CS203177) before incubating with BrU antibody for one hour at room temperature. After antibody conjugation, beads were washed twice with RIP Wash Buffer and then resuspended in Incubation Buffer containing RIP Wash Buffer, 17.5 mM EDTA (Millipore, Cat# CS203175), and RNase Inhibitor (Millipore, Cat# CS203219). Equal amounts (5 pmol) of BrU-labeled RNAs (ARLNC1, ARLNC1-AS, ARLNC1-1–1300, ARLNC1–1301–2786, ARLNC1-1–700, ARLNC1–701–1300, ARLNC1-del-701–1300, LacZ, SCHLAP1-AS, THOR) were incubated with beads in Incubation Buffer for two hours at 4°C. Following incubation, 2.5 pmol of AR 3’UTR-1–980 RNA fragment were added into individual tubes and incubated overnight at 4°C. After incubation, beads were washed six times with RIP Wash Buffer. To recover RNA, beads were digested with proteinase K buffer containing RIP Wash Buffer, 1% SDS (Millipore, Cat# CS203174), and 1.2 µg/µL proteinase K (Millipore, Cat# CS203218) at 55°C for 30 minutes with shaking. After digestion, RNA was extracted from supernatant using the miRNeasy kit (QIAGEN), and reverse transcription was performed using the Superscript III system (Invitrogen). The amount of AR 3’UTR-1–980 recovered in each interaction assay was quantified by qPCR analysis. Data were normalized to ARLNC1-AS control, using ΔCt method. We designed ASOs blocking the AR-ARLNC1 interaction sites (ASO-Blocking, Ionis Pharmaceuticals) and used them in the in vitro interaction assays. Data were normalized to the control ASO, using the ΔCt method.
RNA stability assay
LNCaP cells were treated with 5 µg/mL of actinomycin D for various times as indicated. RNA was extracted and qRT-PCR was carried out as described above. RNA half-life (t1/2) was calculated by linear regression analysis.
Cell proliferation assay
Cells treated with siRNAs or ASOs were seeded into 24-well plates and allowed to attach. Cell proliferation was recorded by IncuCyte live-cell imaging system (Essen Biosciences), following manufacturer’s instructions.
Apoptosis analysis
Cells were grown in 6-well plates and transfected with nonspecific siRNA or siRNAs targeting ARLNC1. Apoptosis analysis was performed 48 hours after transfection, using the Dead Cell Apoptosis Kit (Molecular Probes #V13241) according to manufacturer’s instructions.
Immunoblot analysis
Cells were lysed in RIPA lysis and extraction buffer (Thermo Scientific #89900) supplemented with protease inhibitor cocktail (ROCHE #11836170001). Protein concentrations were quantified using the DC protein assay (BIO-RAD), and protein lysates were boiled in sample buffer. Protein extracts were then loaded and separated on SDS-PAGE gels. Blotting analysis was performed with standard protocols using polyvinylidene difluoride (PVDF) membrane (GE Healthcare). Membranes were blocked for 60 minutes in blocking buffer (5% milk in a solution of 0.1% Tween-20 in Tris-buffered saline (TBS-T) and then incubated overnight at 4°C with primary antibody. After three washes with TBS-T, membranes were incubated with HRP-conjugated secondary antibody. Signals were visualized with an enhanced chemiluminescence system as described by the manufacturer (Thermo Scientific Pierce ECL Western Blotting Substrate). Primary antibodies used were: Androgen Receptor (1:1000 dilution, Millipore, #06–680, rabbit), GAPDH (1:5000 dilution, Cell Signaling, #3683, rabbit), PSA (KLK3) (1:5000 dilution, Dako, #A0562, rabbit), and cleaved PARP (1:1000 dilution, Cell Signaling, #9542, rabbit).
Androgen receptor reporter gene assay
Dual luciferase reporter assays were performed using Cignal Androgen Receptor Reporter Kit (Qiagen) according to the manufacturer’s instructions. Briefly, cells were co-transfected with siRNAs (nonspecific, targeting AR or ARLNC1) and reporter vectors (negative control or AR reporter), using Lipofectamine 2000 transfection reagent (Thermo Fisher Scientific). Forty hours after transfection, DHT (or ethanol vehicle control) was added to induce AR signaling. The Dual Luciferase assay was conducted eight hours after DHT stimulation, using the Dual Luciferase Reporter Assay System from Promega (Cat #1910). Reporter activity was analyzed based on ratio of Firefly/Renilla to normalize for cell number and transfection efficiency.
In vivo experiments
All experiments were approved by the University of Michigan Institutional Animal Care and Use Committee (IACUC). For tumor generation with shRNA-mediated knockdown, shRNA targeting ARLNC1 was cloned in pSIH1-H1-copGFP-T2A–Puro (System Biosciences). Lentiviral particles were generated at the University of Michigan Vector Core. LNCaP-AR cells were infected with lentivirus expressing ARLNC1 shRNA for 48 hours. Knockdown of ARLNC1 was confirmed by qPCR analysis. Male athymic nude mice were randomized into two groups at six to eight weeks of age. 5 million cells expressing sh-ARLNC1 or sh-vector were injected into bilateral flanks of mice. Caliper measurements were taken in two dimensions twice a week by an investigator blinded to the study objective and used to calculate tumor volume. The study was terminated when the tumor volume reached 1000 mm3. For ASO treatment in vivo, six to eight week old male athymic nude mice were inoculated subcutaneously with MDA-PCa-2b cells suspended in matrigel scaffold in the posterior dorsal flank region (5 million cells/site, two sites/animal). When the mean tumor volume reached approximately 150 mm3, mice were randomized into two groups, respectively treated with ARLNC1-specific or control ASO. ASOs, dosed 50 mg/kg, were subcutaneously injected between the scapulae once daily for three periods of five days on/two days off. Tumor size was measured twice per week using a digital caliper by a researcher blinded to the study design. Mouse body weights were monitored throughout the dosing period. When average tumor size in the control group reached 1500 mm3, mice were sacrificed and the primary tumors were excised for weight determination. One-third of the resected specimen was placed in 10% formalin buffer, and the remaining tissue was snap frozen.
BrU-seq and BrUChase-seq
BrU-seq and BrUChase-seq assays were performed as previously described70,71 with MDA-PCa-2b cells treated with either si-NT or si-ARLNC1. BrU-labeling was performed for 30 minutes, and chase experiments were performed for six hours.
Statistical analysis
For in vivo experiments, power analysis (GPOWER software) performed for each tumor type tested to date indicates that the sample size we chose yields a statistical power >90% for detection of tumor size reduction of 40%. Sample sizes were not pre-determined for all other assays. For in vivo experiments, animals were randomized. Randomization was not performed for all other assays. Statistical analyses were performed using Graphpad Prism software or using R. Data were presented as either means ± s.e.m. or means ± s.d. All the experiments were performed in biological triplicates unless otherwise specified. Statistical analyses shown in figures represent two-tailed t-tests, one-way ANOVA, two-way ANOVA, or Kruskal-Wallis rank sum test as indicated. P < 0.05 was considered to be statistically significant. Details regarding the statistical methods employed during microarray, RNA-Seq, and ChIP-Seq data analysis were included in aforementioned methods for bioinformatics analyses.
Data availability
RNA-seq and microarray datasets generated from this study have been deposited into Gene Expression Omnibus, with accession number: GSE110905. Other data supporting the finding of this study are included in the Supplementary Information files.
Code availability
Software for transcriptome meta-assembly and lncRNAs discovery is available at https://tacorna.github.io/.
Supplementary Material
Acknowledgments
We thank A. Poliakov, A. Parolia, V. Kothari, and J. Siddiqui for helpful discussions, the University of Michigan Sequencing Core for Sanger sequencing, H. Johansson (LGC-Biosearch) for initial assistance with smFISH probe design, and S. Ellison, S. Gao, and K. Giles for critically reading the manuscript and submitting documents. This work was supported in part by NCI Prostate SPORE (P50CA186786 to A.M.C.) and EDRN (U01 CA214170 to A.M.C.) grants. A.M.C. is also supported by the Prostate Cancer Foundation and by the Howard Hughes Medical Institute. A.M.C. is an American Cancer Society Research Professor and a Taubman Scholar of the University of Michigan. R. Malik was supported by a Department of Defense Postdoctoral Award (W81XWH-13-1-0284). Y.Z. is supported by a Department of Defense Early Investigator Research Award (W81XWH-17-1-0134). R. Malik, M.C., Y.S.N., J.C-Y.T., and Y.Q. were supported by the Prostate Cancer Foundation Young Investigator Award. R. Mehra was supported by an Idea Development Award (W81XWH-16-1-0314). Y.S.N. is supported by a University of Michigan Cellular and Molecular Biology National Research Service Award Institutional Predoctoral Training Grant. S.P. was supported by AACR-Bayer Prostate Cancer Research Fellowship (16-40-44-PITC). L.X. is supported by U.S. Department of Defense Postdoctoral Fellowship (W81XWH-16-1-0195). M.B. was supported by NIH DP5 grant OD012160. G.C.S. was supported by the Department of Defense awards W81XWH-14-1-0508 and W81XWH-14-1-0509. M.U. was funded by the German Research Foundation (DFG grant BA2168/11-1 SPP 1738).
Footnotes
Author contributions
R. Malik, Y.Z., M.C., S.P. and A.M.C. conceived the study and designed the research. Y.Z. and R. Malik performed most of the cellular and molecular biology experiments with the assistance of Y.H., S.Y., S.S., S.K.S., L.X., X.J., S.M.D., X.C., J.T.W., and F.Y.F. M.C. performed most of the bioinformatics analyses with the help of Y.S.N. and M.K.I. S.P., U.P., and M.B. performed all smFISH work and S.P. performed the mechanistic work-up. J.C-Y.T. and K.M.J. carried out in vivo mouse xenograft studies, and Y.Q. performed 3-D sphere model work. L.P.K. performed histopathological analyses. L.W. and R. Mehra carried out RNA-ISH on tissue microarrays, and T-Y. L., H.J. performed the statistical analysis for this technique. M.U., A.R.G., R.B., and C.S.S. performed the in silico binding predictions. S.M.F., A.T.W., and S.G. provided ASOs. G.C.S. provided AR expression construct. M.T.P. and M.L. performed BrU and BrUChase sample preparation. Y.Z., M.C., R. Malik, S.P. and A.M.C. wrote the manuscript. All authors discussed the results and commented on the manuscript.
Competing Financial Interests
The University of Michigan has filed a patent on lncRNAs as biomarkers of cancer and A.M.C. is named as an inventor. A.M.C. is a co-founder of LynxDx, which is developing lncRNA biomarkers. S.M.F., A.T.W., and S.G. are employees of Ionis Pharmaceuticals, which developed the ASOs against ARLNC1 that were used in this study.
References
- 1.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–9. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 2.Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–14. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012;81:145–66. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell. 2007;129:1311–23. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee N, Moss WN, Yario TA, Steitz JA. EBV noncoding RNA binds nascent RNA to drive host PAX5 to viral DNA. Cell. 2015;160:607–18. doi: 10.1016/j.cell.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wutz A, Rasmussen TP, Jaenisch R. Chromosomal silencing and localization are mediated by different domains of Xist RNA. Nat Genet. 2002;30:167–74. doi: 10.1038/ng820. [DOI] [PubMed] [Google Scholar]
- 7.Prensner JR, et al. The long noncoding RNA SChLAP1 promotes aggressive prostate cancer and antagonizes the SWI/SNF complex. Nat Genet. 2013;45:1392–8. doi: 10.1038/ng.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta RA, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faghihi MA, et al. Expression of a noncoding RNA is elevated in Alzheimer’s disease and drives rapid feed-forward regulation of beta-secretase. Nat Med. 2008;14:723–30. doi: 10.1038/nm1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iyer MK, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208. doi: 10.1038/ng.3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malik R, et al. The lncRNA PCAT29 inhibits oncogenic phenotypes in prostate cancer. Mol Cancer Res. 2014;12:1081–7. doi: 10.1158/1541-7786.MCR-14-0257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shukla S, et al. Identification and Validation of PCAT14 as Prognostic Biomarker in Prostate Cancer. Neoplasia. 2016;18:489–99. doi: 10.1016/j.neo.2016.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu-Yao GL, et al. Fifteen-year survival outcomes following primary androgen-deprivation therapy for localized prostate cancer. JAMA Intern Med. 2014;174:1460–7. doi: 10.1001/jamainternmed.2014.3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huggins C, Hodges CV. Studies on prostatic cancer. I. The effect of castration, of estrogen and of androgen injection on serum phosphatases in metastatic carcinoma of the prostate. 1941. J Urol. 2002;167:948–51. discussion 952. [PubMed] [Google Scholar]
- 15.Treatment and survival of patients with cancer of the prostate. The Veterans Administration Co-operative Urological Research Group. Surg Gynecol Obstet. 1967;124:1011–7. [PubMed] [Google Scholar]
- 16.Chen Y, Sawyers CL, Scher HI. Targeting the androgen receptor pathway in prostate cancer. Curr Opin Pharmacol. 2008;8:440–8. doi: 10.1016/j.coph.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wong YN, Ferraldeschi R, Attard G, de Bono J. Evolution of androgen receptor targeted therapy for advanced prostate cancer. Nat Rev Clin Oncol. 2014;11:365–76. doi: 10.1038/nrclinonc.2014.72. [DOI] [PubMed] [Google Scholar]
- 18.Mukherji D, Pezaro CJ, De-Bono JS. MDV3100 for the treatment of prostate cancer. Expert Opin Investig Drugs. 2012;21:227–33. doi: 10.1517/13543784.2012.651125. [DOI] [PubMed] [Google Scholar]
- 19.Scher HI, et al. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367:1187–97. doi: 10.1056/NEJMoa1207506. [DOI] [PubMed] [Google Scholar]
- 20.Tran C, et al. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324:787–90. doi: 10.1126/science.1168175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Scher HI, et al. Antitumour activity of MDV3100 in castration-resistant prostate cancer: a phase 1–2 study. Lancet. 2010;375:1437–46. doi: 10.1016/S0140-6736(10)60172-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stein MN, Goodin S, Dipaola RS. Abiraterone in prostate cancer: a new angle to an old problem. Clin Cancer Res. 2012;18:1848–54. doi: 10.1158/1078-0432.CCR-11-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reid AH, et al. Significant and sustained antitumor activity in post-docetaxel, castration-resistant prostate cancer with the CYP17 inhibitor abiraterone acetate. J Clin Oncol. 2010;28:1489–95. doi: 10.1200/JCO.2009.24.6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Bono JS, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364:1995–2005. doi: 10.1056/NEJMoa1014618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watson PA, Arora VK, Sawyers CL. Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer. 2015;15:701–11. doi: 10.1038/nrc4016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antonarakis ES, et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028–38. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Attard G, Richards J, de Bono JS. New strategies in metastatic prostate cancer: targeting the androgen receptor signaling pathway. Clin Cancer Res. 2011;17:1649–57. doi: 10.1158/1078-0432.CCR-10-0567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hearn JW, et al. HSD3B1 and resistance to androgen-deprivation therapy in prostate cancer: a retrospective, multicohort study. Lancet Oncol. 2016;17:1435–1444. doi: 10.1016/S1470-2045(16)30227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chan SC, Li Y, Dehm SM. Androgen receptor splice variants activate androgen receptor target genes and support aberrant prostate cancer cell growth independent of canonical androgen receptor nuclear localization signal. J Biol Chem. 2012;287:19736–49. doi: 10.1074/jbc.M112.352930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson D, et al. Integrative clinical genomics of advanced prostate cancer. Cell. 2015;161:1215–28. doi: 10.1016/j.cell.2015.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Visakorpi T, et al. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet. 1995;9:401–6. doi: 10.1038/ng0495-401. [DOI] [PubMed] [Google Scholar]
- 32.Asangani IA, et al. Therapeutic targeting of BET bromodomain proteins in castration-resistant prostate cancer. Nature. 2014;510:278–82. doi: 10.1038/nature13229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roche PJ, Hoare SA, Parker MG. A consensus DNA-binding site for the androgen receptor. Mol Endocrinol. 1992;6:2229–35. doi: 10.1210/mend.6.12.1491700. [DOI] [PubMed] [Google Scholar]
- 34.Pomerantz MM, et al. The androgen receptor cistrome is extensively reprogrammed in human prostate tumorigenesis. Nat Genet. 2015;47:1346–51. doi: 10.1038/ng.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cancer Genome Atlas Research, N. The Molecular Taxonomy of Primary Prostate Cancer. Cell. 2015;163:1011–25. doi: 10.1016/j.cell.2015.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takayama K, et al. Androgen-responsive long noncoding RNA CTBP1-AS promotes prostate cancer. EMBO J. 2013;32:1665–80. doi: 10.1038/emboj.2013.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mele M, et al. Human genomics. The human transcriptome across tissues and individuals. Science. 2015;348:660–5. doi: 10.1126/science.aaa0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rhodes DR, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–80. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Engreitz JM, et al. RNA-RNA interactions enable specific targeting of noncoding RNAs to nascent Pre-mRNAs and chromatin sites. Cell. 2014;159:188–99. doi: 10.1016/j.cell.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kretz M, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–5. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature. 2011;470:284–8. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wright PR, et al. CopraRNA and IntaRNA: predicting small RNA targets, networks and interaction domains. Nucleic Acids Res. 2014;42:W119–23. doi: 10.1093/nar/gku359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mann M, Wright PR, Backofen R. IntaRNA 2.0: enhanced and customizable prediction of RNA-RNA interactions. Nucleic Acids Res. 2017;45:W435–W439. doi: 10.1093/nar/gkx279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lennox KA, Behlke MA. Cellular localization of long non-coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res. 2016;44:863–77. doi: 10.1093/nar/gkv1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Meng L, et al. Towards a therapy for Angelman syndrome by targeting a long non-coding RNA. Nature. 2015;518:409–12. doi: 10.1038/nature13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wheeler TM, et al. Targeting nuclear RNA for in vivo correction of myotonic dystrophy. Nature. 2012;488:111–5. doi: 10.1038/nature11362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hua Y, et al. Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev. 2010;24:1634–44. doi: 10.1101/gad.1941310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Evers MM, Toonen LJ, van Roon-Mom WM. Antisense oligonucleotides in therapy for neurodegenerative disorders. Adv Drug Deliv Rev. 2015;87:90–103. doi: 10.1016/j.addr.2015.03.008. [DOI] [PubMed] [Google Scholar]
- 50.Yeap BB, et al. Novel binding of HuR and poly(C)-binding protein to a conserved UC-rich motif within the 3’-untranslated region of the androgen receptor messenger RNA. J Biol Chem. 2002;277:27183–92. doi: 10.1074/jbc.M202883200. [DOI] [PubMed] [Google Scholar]
- 51.Lebedeva S, et al. Transcriptome-wide analysis of regulatory interactions of the RNA-binding protein HuR. Mol Cell. 2011;43:340–52. doi: 10.1016/j.molcel.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 52.Prensner JR, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–9. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cieslik M, et al. The use of exome capture RNA-seq for highly degraded RNA with application to clinical cancer sequencing. Genome Res. 2015;25:1372–81. doi: 10.1101/gr.189621.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–30. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 56.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–40. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cline MS, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2:2366–82. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hansen P, et al. Saturation analysis of ChIP-seq data for reproducible identification of binding peaks. Genome Res. 2015;25:1391–400. doi: 10.1101/gr.189894.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–7. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–8. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mehra R, et al. A novel RNA in situ hybridization assay for the long noncoding RNA SChLAP1 predicts poor clinical outcome after radical prostatectomy in clinically localized prostate cancer. Neoplasia. 2014;16:1121–7. doi: 10.1016/j.neo.2014.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Newton MA, Quintana FA, Den Boon JA, Sengupta S, Ahlquist P. Random-Set Methods Identify Distinct Aspects of the Enrichment Signal in Gene-Set Analysis. Annals of Applied Statistics. 2007;1:85–106. [Google Scholar]
- 67.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–9. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Niknafs YS, et al. The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1 in breast cancer progression. Nat Commun. 2016;7:12791. doi: 10.1038/ncomms12791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rossiello F, et al. DNA damage response inhibition at dysfunctional telomeres by modulation of telomeric DNA damage response RNAs. Nat Commun. 2017;8:13980. doi: 10.1038/ncomms13980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Paulsen MT, et al. Coordinated regulation of synthesis and stability of RNA during the acute TNF-induced proinflammatory response. Proc Natl Acad Sci U S A. 2013;110:2240–5. doi: 10.1073/pnas.1219192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Paulsen MT, et al. Use of Bru-Seq and BruChase-Seq for genome-wide assessment of the synthesis and stability of RNA. Methods. 2014;67:45–54. doi: 10.1016/j.ymeth.2013.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and microarray datasets generated from this study have been deposited into Gene Expression Omnibus, with accession number: GSE110905. Other data supporting the finding of this study are included in the Supplementary Information files.