Abstract
Purpose
To quantify the precision of in vivo cardiac DTI (cDTI) acquired with a spin echo, first- and second-order motion-compensated (M1M2), convex optimized diffusion encoding (CODE) sequence.
Methods
Free-breathing CODE-M1M2 cDTI were acquired in healthy volunteers (N = 10) at midsystole and diastole with 10 repeated acquisitions per phase. 95% confidence intervals of uncertainty in reconstructed diffusion tensor eigenvectors (E⃗1, E⃗2, E⃗3), mean diffusivity (MD), fractional anisotropy (FA), and tensor Mode were measured using a bootstrapping approach. Trends in observed tensor metric uncertainty were evaluated as a function of scan duration, image SNR, cardiac phase, and bulk motion artifacts.
Results
For midsystolic scans including 5 signal averages (scan time: ~5min), the median myocardial 95% confidence intervals of uncertainties were: E⃗1: 15.5 ± 1.2°, E⃗2: 31.2 ± 3.5°, E⃗3: 21.8 ± 3.1°, MD: 0.38 ± 0.02 × 10−3mm2/s, FA: 0.20 ± 0.01, and Mode: 1.10 ± 0.08. Uncertainty in all parameters increased for diastolic scans: E⃗1: 31.9 ± 7.1°, E⃗2: 59.6 ± 6.8°, E⃗3: 40.5 ± 6.4°, MD: 0.52 ± 0.09 × 10−3 mm2/s, FA: 0.23 ± 0.01, and Mode: 1.57 ± 0.11. Diastolic cDTI also reported higher MD (MDDIA = 1.91 ± 0.34 × 10−3 mm2/s vs. MDSYS = 1.58 ± 0.09 × 10−3 mm2/s, P = 8 × 10−3) and lower FA values (FADIA = 0.32 ± 0.06 vs. FASYS = 0.37 ± 0.03, P = 0.03).
Conclusion
cDTI precision improved with increasing nondiffusion-weighted (b = 0) image SNR, but gains were minimal for SNR ≥ 25 (~10 averages). cDTI precision was also sensitive to intershot bulk motion artifacts, which led to better precision for midsystolic imaging.
Keywords: cardiac diffusion, diffusion tensor imaging, cardiac MRI
1 | INTRODUCTION
Cardiac DTI (cDTI) is an emerging contrast-free technique for quantifying microstructure in healthy1,2 and diseased myocardium.3–5 cDTI provides quantitative maps of local cardiomyocyte orientation, the organization of myolaminar sheetlets, and microstructural anisotropy by probing the diffusion of water molecules contained in myocardial tissue. Increases in the mean diffusivity (MD) have been linked to the presence of fibrosis in myocardial infarction as well as changes in extracellular volume in hypertrophic cardiomyopathy. 6,7 Changes in myocardial sheetlet dynamics have also been demonstrated in hypertrophic and dilated cardiomyopathies using cDTI.3–5
As with any quantitative measure, several sources of error lead to uncertainty for in vivo cDTI measurements. These include, but are not limited to, noise, image distortions, and physiological variability. Noise can have a large impact on cDTI-derived parameters due to the low SNR of the acquired diffusion-weighted images that stems from the diffusion-induced signal decay and the general reliance on single-shot EPI (SS-EPI) readouts. Image distortions can also be substantial in cDTI due to eddy currents and sharp magnetic susceptibility gradients at the heart–lung interface that are exacerbated by the SS-EPI readout.8 Physiological variability is also significant in cDTI due to the presence of bulk cardiac and respiratory motion as well as pulsatile blood flow. This variability can lead to both corrupted individual measurements (i.e., intrashot motion effects) and mismatches between measurements (i.e., intershot variability between encoding directions or repetitions).
The propagation of these errors from the acquired image data through diffusion tensor reconstruction and on to derived tensor metrics is a complex problem that is not easily modeled. As a result, a nonparametric bootstrapped approach has been proposed for quantifying uncertainty in diffusion tensor orientation9 and shape10,11 in neurological DTI. This approach provides insight into the uncertainty underlying neurological DTI experiments,12,13 but these results do not directly apply to in vivo cDTI for several reasons. Namely, cardiomyocytes are approximately 10 times thicker in diameter than neuronal fibers,14,15 which leads to diffusion tensors with substantially lower fractional anisotropy (FA) and consequently larger uncertainties in measures of fiber orientation. 9 Cardiac motion also necessitates the use of highly specialized pulse sequences, which employ either motion-compensated diffusion encoding gradients16–19 or a STEAM with diffusion encoding spread across multiple heart beats.20,21 Furthermore, even when using motion-compensated diffusion encoding schemes, slight variations in heart rate and breathing patterns throughout an acquisition contribute a degree of uncertainty to the measured tensor metrics. The impact of bulk physiological motion also depends on the pulse sequence timing within the cardiac cycle17,19,22; thus, uncertainty is expected to depend on the cardiac phase selected for triggering, although this has not yet been characterized.
Currently, the many sources of variability and moderate SNR present in cDTI acquisitions result in the standard practice of acquiring multiple repetitions of the protocol and averaging the repeated data to increase SNR and thereby improve measurement precision. This, of course, increases the already long cDTI scan times that arise from the need for multiple diffusion encoding directions and both cardiac and respiratory triggering (or multiple repeated breath holds). Therefore, it is useful to quantify the precision of cDTI-derived tensor metrics made within clinically viable scan durations.
The first objective of this study was to use bootstrapped uncertainty measurements to characterize the precision of in vivo spin echo EPI (SE-EPI) cDTI measurements made using first- and second-order motion-compensated (M1M2) convex optimized diffusion encoding (CODE)19 at 2 points in the cardiac cycle (midsystole and diastole). The second objective was to quantify the precision of diffusion tensor orientation (i.e., tensor eigenvectors) and shape (i.e., tensor invariants) for cDTI acquisitions with scan times between 1 and 5min per slice to identify practical guidelines for efficiently measuring high-quality microstructural information.
2 | THEORY
Diffusion tensor uncertainty can be measured for a cDTI acquisition using bootstrapped resampling to generate a distribution of tensors that reflects the uncertainty inherent in the measurement. This was previously described for calculating uncertainty in the tensor primary eigenvector, which was referred to as the “cone of uncertainty.”9 This technique is briefly described herein and extended to quantify other aspects of diffusion tensor uncertainty, including the secondary and tertiary eigenvectors as well as several tensor invariants.
This bootstrapping technique requires the cDTI acquisition to be repeated twice to generate 2 independent but matched datasets. From these, a composite dataset can be generated by randomly selecting 1 image for each diffusion encoding direction from the 2 sets. This composite dataset, which includes 1 randomly chosen image for all diffusion encoding directions, can then be used to reconstruct a diffusion tensor at each voxel with a least squares linear regression. This process can then be repeated a large number of times (Nbootstrap samples) to generate a distribution of diffusion tensors at each voxel, which is much more efficient than actually acquiring Nboot repetitions. Eigensystem decomposition can then be performed on each diffusion tensor to determine their eigenvectors (E1, E2, and E3) and eigenvalues (λ1, λ2, λ3). Tensor invariant quantities such as MD, FA, and tensor Mode23 can also be extracted from each diffusion tensor.
The precision of each eigenvector, Ej, can then be measured by first calculating the dyadic mean, ψj of the Nboot Ej vectors, and then measuring the angle, θj between each vector Ej and ψj. Ej precision (dEj) can then characterized by the 1-sided 95% confidence interval (95CI) of the resultant θj distribution. A 1-sided 95CI is used because by definition θj ≥ 0. Note that dE1 as described here is exactly the cone of uncertainty as described by Jones.9
Analogous measures of tensor invariant precision can be determined by analyzing their distributions across the Nboot repetitions (as done for FA in Pajevic and Basser10). Unlike the θj distributions describing vector uncertainties, which by definition are bounded between 0° and 90°, the invariants form 2-sided distributions and can meaningfully exhibit negative deviations from the median value. As such, their uncertainty should be represented by a 2-sided 95CI, which need not be symmetric about the median. The width of this 95CI (i.e., the upper bound minus the lower bound) can then be used to represent the underlying uncertainty (dMD, dFA, dMode).
3 | METHODS
3.1 | cDTI acquisition
cDTI were acquired in healthy volunteers (N = 10) on a 3.0T scanner (Siemens Prisma, Siemens HealthCare, Erlangen, Germany) after obtaining informed consent under an institutional review board-approved study using a SE-EPI DWI pulse sequence with convex optimized diffusion encoding with first- and second-order moment compensation (CODE-M1M2) to impart bulk motion compensation and reduce the TE, thereby increasing SNR.19 The acquisition included a single midventricular slice, 1 b value (b = 350 s/mm2) along 12 diffusion encoding directions plus 1 nondiffusion-weighted image (b = 0), and 2.0 × 2.0 × 5.0mm spatial resolution with inner volume excitation to reduce the FOV in the phase encode direction24 (FOV = 200 × 160 mm, GRAPPA factor 2,25 full-Fourier, water-only excitation, 90° flip angle, TE = 65 ms). There was a small b-value increase (<1 s/mm2) due to the imaging gradients and to use of a previously described concomitant field correction,19 but only the nominal b value was used for reconstruction. For each scan, 3 dummy scans were performed prior to acquiring images in order to reach a steady state. Local second-order B0 shimming was performed in a focused shim box containing only the left ventricular (LV) myocardium to minimize susceptibility artifacts in the posterior wall. Imaging was timed to an end-expiratory respiratory phase using a liver-dome navigator trigger, which resulted in a TR of 1 respiratory cycle (average TR = 4.2 ± 1.4 s). Separate acquisitions were performed with ECG trigger delays timed to: 1) midsystole (fixed trigger delay = 100 ms); and 2) late diastole (subject-specific trigger delay determined by identifying diastasis from a balanced steady-state free precession cine image, which resulted in trigger delays of 705 ± 63 ms across all subjects). Ten signal repetitions were acquired in a single free-breathing scan at each cardiac phase to facilitate boot-strapped uncertainty measurement for datasets containing up to 5 averages (scan time: ~10 min per cardiac phase). Prior to reconstruction, all images were coregistered using a rigid transformation to correct for differences in cardiac position between scans.
3.2 | Image quality evaluation
Whereas SNR generally increases with additional signal averages ( ), it can vary substantially in space and between subjects due to differences in subject geometry and coil positioning. It is thus useful to quantify voxel-wise image SNR to more completely observe the relationship between SNR and cDTI uncertainty. Because 10 repetitions of each image were acquired, it was possible to map SNR for each reference DWI (b = 0) acquisition. In order to estimate SNR from images that included several averaged repetitions, a bootstrapped approach similar to that described above was used. For example, for Navg = 5 averages, 5 b = 0 images were randomly chosen (with replacement) from the acquired set of 10 and magnitude averaged together (complex averaging was not performed because image intensities were sufficiently above the noise floor with respect to the moderate b value used26. This process was repeated to generate a set of 300 images that each included 5 averages. SNR was then calculated at each voxel by dividing the mean signal value across the 300 images by their SD (no SNR differences were observed when the image sample size was increased beyond 300). SNR maps were generated in this manner for Navg = 1–10.
Because all pulse sequence parameters and setup conditions were held constant between acquisitions at midsystole and diastole, we can assume that any differences observed between the acquisitions arise predominantly from physiological variability. Furthermore, if cardiac bulk motion effects are present in the images, it is expected that they will vary subtly from beat to beat27 and lead to voxel-wise signal fluctuation between repetitions. To quantify this effect, the coefficient of variation (CoV) (i.e., the SD normalized by the mean) of the voxel-wise signal intensity across all 10 repetitions of each direction (CoVDTI) was measured for both cardiac phases.
3.3 | cDTI precision measurement
Subsets of the set of 10 cDTI repetitions were first randomly selected and averaged together to reflect acquisitions with scan times ranging from approximately 1 to 5 min (Navg = 1 to Navg = 5). A second subset was then randomly generated for each case to facilitate bootstrapping. The uncertainty in tensor eigenvectors (dE1, dE2, dE3) and invariants (dMD, dFA, dMode) was then measured from the 2 subsets as described above. These calculations used Nboot = 1000 boot-strapped samples, which was determined to be sufficient to generate stable statistical measures of uncertainty (i.e., increasing Nboot did not alter the measured tensor uncertainties) and is in line with previous studies.2
Histograms of the uncertainty in each tensor quantity were generated for all LV voxels within each subject for Navg = 1 to Navg = 5 at both midsystole and diastole. Median and maximum likelihood values were then extracted from each histogram. Global LV histograms of each quantity’s uncertainty were also generated from data pooled across all subjects at each cardiac phase.
Uncertainty was then characterized as a function of b = 0 image SNR on a voxel-wise basis. This was done by first binning all voxels by SNR (bin size: 2) and then generating histograms of uncertainty across all voxels contained in each SNR bin. The median uncertainty was then measured within each bin, and the 95CI of the median was measured using bootstrapping. Significant differences between midsystole and diastolic median uncertainties were identified by nonoverlapping 95CIs.
In order to examine the impact of variation in bulk motion-induced signal attenuation on precision, the same process was then carried out to measure the median uncertainty of each parameter as a function of CoVDTI (bin size: 2%). The impact of tensor size and shape on precision was also evaluated by measuring the median uncertainties as functions of MD and FA (as reconstructed with 5 averages, MD bin size: 8 × 10−5mm2/s, FA bin size: 0.02).
3.4 | Tensor evolution with varying signal averages
The dependence of eigenvector and tensor invariant parameters was also evaluated as a function of Navg. To do so, diffusion tensors were reconstructed from cDTI image subsets, including the full range of acquired signal averages (Navg = 1–10; note that all 10 averages could be used for this analysis, which did not require bootstrapped resampling). Maps of E⃗1, E⃗2, E⃗3, MD, FA, and tensor Mode were then generated for each subset.
Helix angle (HA) maps were also generated by measuring the elevation angle of the projection of E⃗1 relative to the local circumferential tangent vector, C⃗.1 C⃗ was defined by a b-spline vector fit to the endocardial and epicardial surfaces, as defined by 10 manually defined seed points on each endocardial and epicardial surface.1 The average HA slope across the wall, HApitch, was calculated using a linear regression between HA and percent wall depth for each subject. Percent wall depth values were calculated at each LV voxel by linearly interpolating between the endo- and epicardial splines (where percent wall depth = 0% at the epicardial surface and 100% at the endocardial surface). Myocardial sheet angles (SA), SA2 and SA3 were then defined by the angles between the circumferential-long axis plane and E⃗2 and E⃗3, respectively.1 Median myocardial HA, |SA2|, and |SA3|were then calculated for each subject for Navg = 1–10. The median (m) SD (σ) of myocardial MD, FA, and Mode were also calculated for each subject for Navg = 1–10 using the region between the endo- and epicardial splines described above to form the LV mask. Statistical differences between each median and SD across all subjects as a function of Navg were identified using 1-way analysis of variation. If analysis of variation yielded significant differences, medians and SDs from Navg = 1 were compared with Navg ≥ 2 using paired t tests at a P = 0.05 significance level.
3.5 | Cardiac phase dependence of tensor parameters
Median myocardial HA, HApitch, |SA2|, |SA3|, MD, FA, and tensor Mode were also compared between midsystole and diastole with Navg = 10 using paired t tests.
4 | RESULTS
4.1 | Image quality evaluation
cDTIs from all imaging experiments were included in analysis (no data was discarded). The average heart beat duration (R-R interval) was 1006 ± 101 ms, and the ECG trigger delays (TD) used for systolic (SYS) and diastolic imaging (DIA) were TDSYS = 100 ± 0 ms, which corresponded with 35.4% ± 2.6% of peak systolic contraction and TDDIA = 705 ± 63 ms. The average scan time was 9.1 ± 3.1 min per cardiac phase (54.7 ± 18.5 s per cDTI average).
The mean nondiffusion-weighted (b = 0) image SNR in the LV increased from 8.0 ± 1.9 with Navg = 1 to 22.3 ± 6.1 with Navg = 10. No significant differences in SNR were observed between midsystolic and diastolic b = 0 images (8.2 ± 1.1 vs. 7.9 ± 2.6, P = NS). However, median myocardial CoVDTI was significantly lower in midsystole than in diastole (16.7% ± 2.2% vs. 25.8% ± 9.1%, P = 0.006), which indicates higher signal variability during diastole.
4.2 | cDTI precision measurement
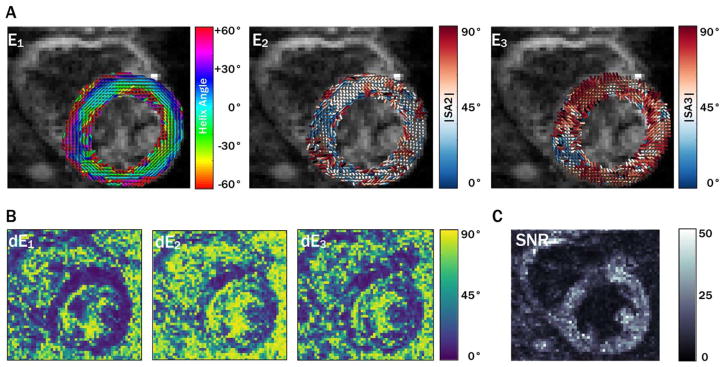
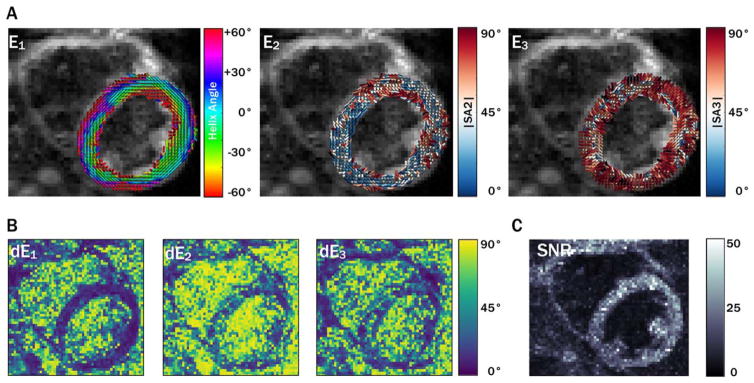
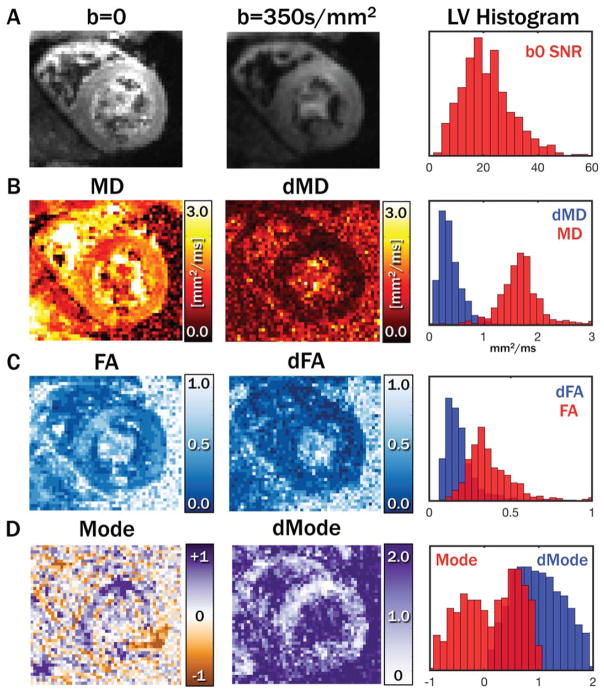
Primary, secondary, and tertiary eigenvector (E⃗1, E⃗2, and E⃗3) maps, along with eigenvector uncertainty maps (dE1, dE2, and dE3) and myocardial uncertainty histograms, are shown for a midsystolic acquisition with 5 averages from a single subject in Figure 1. Diastolic maps for the same subject are shown in Figure 2. Qualitatively, regions with increased uncertainty corresponded with regions with less coherent eigenvectors, and uncertainty was generally higher in the diastolic phase. cDTI images, along with reconstructed maps of MD, FA, and tensor Mode, as well as maps of each parameter’s uncertainty and histograms of myocardial uncertainty, are shown for the same subject (also with 5 averages) in the midsystolic cardiac phase in Figure 3. Analogous maps are shown for a different subject in the diastolic phase in Supporting Information Figure S1.
FIGURE 1.
Maps of eigenvector orientations (A) for 1 subject with 5 averages from a midsystolic cardiac phase with corresponding eigenvector orientation 95CI uncertainty maps (B) and b = 0 image SNR maps (C). Qualitatively, regions with increased uncertainty correspond with regions of eigenvector incoherence. Overall, uncertainty in E2 was greater than uncertainty in E1 and E3, a trend that was observed in all subjects.
95CI, 95% confidence interval; E, eigenvector.
FIGURE 2.
Maps of eigenvector orientations (A) for the same subject shown in Figure 1 with 5 averages from a diastolic cardiac phase with corresponding eigenvector orientation 95CI uncertainty maps (B) and b = 0 image SNR maps (C). Overall, greater uncertainty was observed in diastole than in systole.
FIGURE 3.
Example systolic images with b = 0 and 350 s/mm2 with 5 averages and a histogram of LV b = 0 SNR(A). Maps of tensor MD(B), FA (C), and Mode (D), with corresponding uncertainties and LV histograms. dMD was in general low compared with myocardial MD values, whereas dFA was somewhat closer to myocardial FA. Mode was not well separated from dMode.
FA, fractional anisotropy; LV, left ventricular; MD, mean diffusivity.
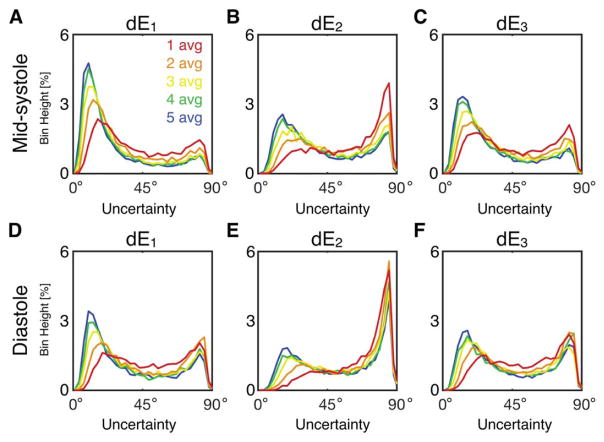
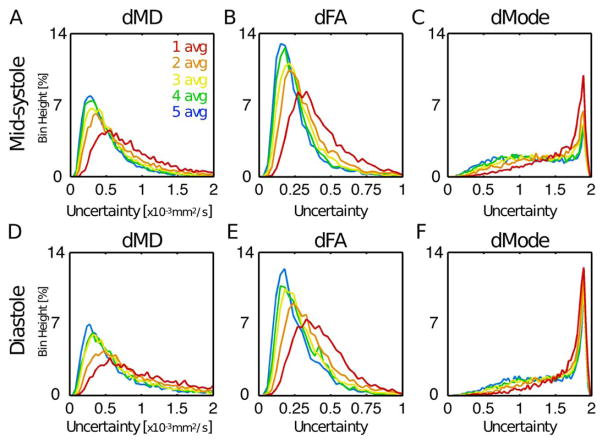
Histograms of tensor eigenvector uncertainty are shown in Figure 4 for midsystolic and diastolic cDTI, with Navg = 1–5 pooled across all subjects at systole (Figures 4A–4C) and diastole (Figures 4D–4F). Uncertainty decreased with increasing signal averages; however, differences were minimal for Navg ≥ 4 in systole (small differences between Navg = 4 and 5 were observed in diastole). Uncertainty was generally larger for cDTI tensor metrics acquired in diastole than in midsystole. Analogous histograms of tensor invariant uncertainty (MD, FA, and Mode) are shown in Figure 5. Invariant uncertainty also decreased with increasing Navg, however, with minimal differences for Navg ≥ 4 in systole (small differences were again observed between Navg = 4 and 5 in diastole). Differences in tensor invariant uncertainty histograms between midsystolic and diastolic cDTI were minimal.
FIGURE 4.
Histograms of uncertainty in tensor eigenvectors pooled from all LV voxels from all subjects at midsystole (A, B, C) and diastole (D, E, F) from acquisitions with Navg = 1–5. Uncertainty decreased with additional signal averages, but differences were minimal for Navg ≥ 4. Uncertainty was generally larger for diastolic cDTI.
FIGURE 5.
Histograms of uncertainty in tensor invariants pooled from all LV voxels from all subjects at midsystole (A, B, C) and diastole (D, E, F) from acquisitions with Navg = 1–5. Differences between midsystolic and diastolic cDTI were minimal.
The median and maximum likelihood values from the uncertainty histograms generated for each subject are shown for Navg = 5 in Table 1. Median and maximum likelihood uncertainties were lower in midsystole than in diastole for all parameters.
TABLE 1.
Maximum likelihood and median 95% confidence intervals for Navg = 5 acquired at 2 cardiac phases (population mean ± SD)
| dE1 (degree) | dE2 (degree) | dE3 (degree) | dMD (x10−3mm2/s) | dFA (unitless) | dMode (unitless) | ||
|---|---|---|---|---|---|---|---|
| Midsystole | Maximum likelihood | 9.7 ± 1.0 | 15.7 ± 1.0 | 12.1 ± 1.5 | 0.25 ± 0.02 | 0.13 ± 0.01 | 1.62 ± 0.47 |
| Median | 15.5 ± 1.2 | 31.2 ± 3.5 | 21.8 ± 3.1 | 0.38 ± 0.02 | 0.20 ± 0.01 | 1.10 ± 0.08 | |
|
| |||||||
| Diastole | Maximum likelihood | 17.8 ± 21.6 | 84.1 ± 1.5 | 42.7 ± 33.2 | 0.28 ± 0.07 | 0.16 ± 0.003 | 1.88 ± 0.02 |
| Median | 31.9 ± 7.1 | 59.6 ± 6.8 | 40.5 ± 6.4 | 0.52 ± 0.09 | 0.23 ± 0.01 | 1.57 ± 0.11 | |
95CI, 95% confidence interval.
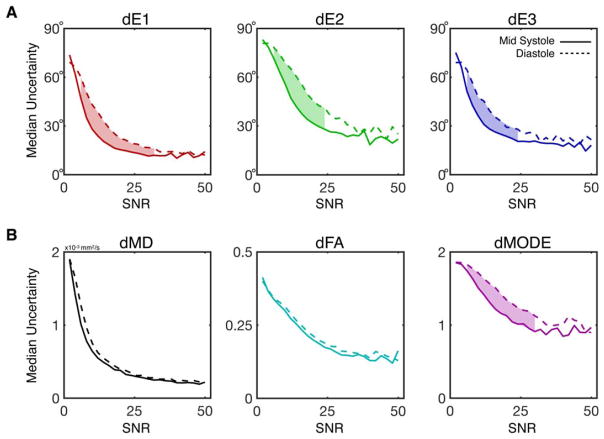
Median eigenvector and invariant uncertainties are plotted as functions of b = 0 image SNR in Figure 6. Uncertainty in all measured tensor metrics dropped consistently with increasing SNR. Uncertainty dropped faster with respect to SNR for midsystolic data, particularly in dE1, dE2, dE3, and dMode, for which significant differences were observed between phases for intermediate SNRs as highlighted in Figure 6. Whereas the maximum observed SNR across all subjects and voxels was 95, histograms were only generated for SNR ≤ 50 due to a lack of sufficient data for meaningful statistics— as seen in the histograms of b = 0 SNR pooled across all subjects and cardiac phases, which are plotted for Navg = 1–5 in Supporting Information Figure S2.
FIGURE 6.
Plots of median uncertainty in tensor eigenvalues (A) and invariants (B) as a function of b = 0 image SNR at midsystole (solid lines) and diastole (dotted lines). Median uncertainties and 95CI of the medians (not shown) were calculated from uncertainty distributions across all voxels and subjects with a particular b = 0 SNR (binned in SNR increments of 2) across images with Navg = 1–5. Significant differences between midsystole and diastole were identified by nonoverlapping 95CI and are indicated by shaded regions between the plots. dE1, dE2, dE3, and dMode were significantly lower at midsystole for moderate SNRs. Although differences in dMD or dFA were observed between phases, these differences did not reach statistical significance for any SNR bin.avg, average.
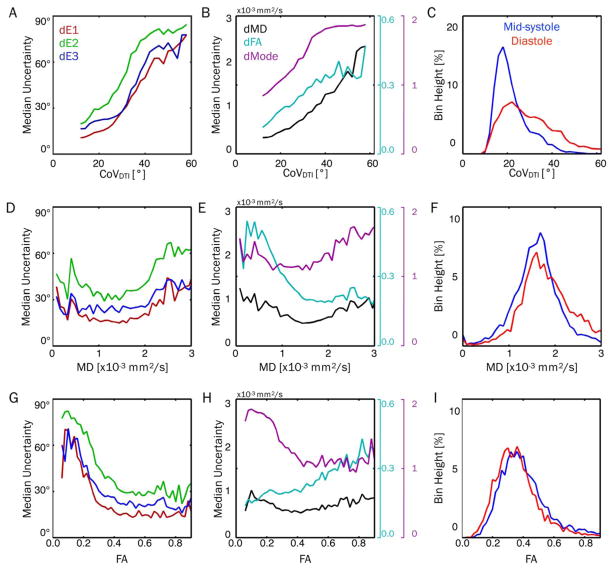
Median uncertainty values were plotted as functions of CoVDTI, MD, and FA in Figure 7. There were clear increases in uncertainty for all parameters with increasing CoVDTI (Figures 7A, 7B). Figure 7C shows global histograms of CoVDTI, which demonstrates the increased CoVDTI in the diastolic phase. Uncertainty in all parameters decreased with increasing MD up to approximately 1.5 × 10−3mm2/s and then increased at larger MDs for all parameters other than FA, which leveled off (Figures 7D, 7E). With increasing FA, uncertainty in all 3 tensor eigenvectors, MD, and Mode generally decreased, whereas FA uncertainty increased (Figures 7G, 7H). Histograms of myocardial MD and FA did not vary substantially between cardiac phases, although a larger population of MD values >2.5 × 10−3mm2/s were observed in diastole (Figures 7F, 7I).
FIGURE 7.
Plots of median uncertainty in tensor eigenvalues (left column) and invariants (center column) as functions of CoVDTI (top row), MD (middle row), and FA (bottom row), with Navg = 5 and pooled across all subjects and cardiac phases. Histograms of CoVDTI, MD, and FA are also shown (right column) for midsystole and diastole. Median values were calculated from uncertainty distributions across all voxels and subjects with a particular CoVDTI, MD, or FA. Uncertainty in all parameters increased with increasing CoVDTI, indicating a strong sensitivity to bulk-motion– induced signal variations. This likely explains the increased uncertainty observed in diastolic cDTI, which had significantly higher CoVDTI. Uncertainty in all parameters decreased with increasing MD up to approximately 1.5 × 10−3 mm2/s and then increased (aside from FA, which leveled off). With increasing FA, uncertainty in all 3 tensor eigenvectors, MD, and Mode generally decreased, whereas FA uncertainty increased. CoV, coefficient of variation; cDTI, cardiac DTI.
4.3 | Tensor evolution with varying signal averages
Measures of HA, HApitch, |SA2|, or |SA3| as a function of signal averages at either midsystole or diastole were not significantly different. Median myocardial values are tabulated for each parameter in Table 2.
TABLE 2.
Median and maximum likelihood eigenvector orientation metrics for two cardiac phases (population mean ± SD)
| Navg = 1 | Navg = 5 | Navg = 10 | |||||
|---|---|---|---|---|---|---|---|
| Median | ML | Median | ML | Median | ML | ||
| Midsystole | HA | 3.8 ± 3.5° | 11.0 ± 22.1° | 1.9 ± 3.8° | 5.5 ± 8.6° | 1.6 ± 3.1° | 8.8 ± 22.3° |
| HApitch | −1.2 ± 0.2°/% | NA | −1.2 ± 0.2°/% | NA | −1.2 ± 0.2°/% | NA | |
| |SA2| | 40.9 ± 5.7° | 24.6 ± 30.6° | 38.2 ± 5.5° | 22.7 ± 20.1° | 36.6 ± 6.0° | 23.7 ± v26.8° | |
| |SA3| | 61.7 ± 4.3° | 79.4 ± 12.6° | 65.2 ± 5.5° | 82.4 ± 9.4° | 66.5 ± 4.8° | 81.7 ± 10.2° | |
|
| |||||||
| Diastole | HA | 4.2 ± 4.5° | 32.5 ± 38.2° | 1.9 ± 3.4° | 18.9 ± v35.6° | 1.1 ± 4.1° | 11.0 ± v27.9° |
| HApitch | −1.3 ± 0.2°/% | NA | −1.3 ± 0.3°/% | NA | −1.2 ± 0.1°/% | NA | |
| |SA2| | 41.2 ± 7.5° | 36.0 ± 39.0° | 37.8 ± 9.9° | 23.0 ± 36.3° | 35.0 ± 10.7° | 13.9 ± 30.0° | |
| |SA3| | 64.7 ± 7.1° | 88.5 ± 3.2° | 67.8 ± 7.7° | 86.9 ± 8.0° | 68.9 ± 8.1° | 87.4 ± 6.0° | |
HA, helix angle; ML, maximum likelihood. NA, not applicable.
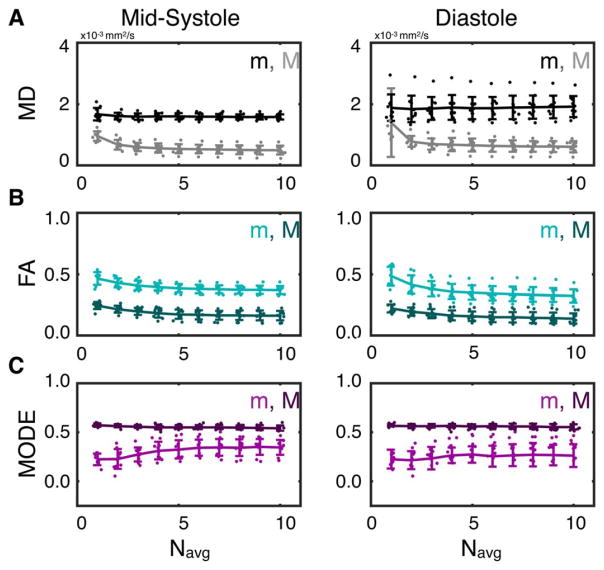
Median (m) and SD (σ) of MD, FA, and Mode across the LV myocardium are plotted for all subjects as functions of Navg in Figure 8. No significant changes in mMD were observed between any number of averages at either midsystole or diastole. Compared with Navg = 1, σMD decreased significantly with Navg ≥ 2 in midsystole (σMD,1avg = 0.95 ± 0.2 × 10−3 mm2/s vs. σMD,2avg = 0.67 ± 0.2 × 10−3 mm2/s, P = 8 × 10−4) and with Navg ≥ 6 in diastole (σMD,1avg = 1.39 ± 1.1 × 10−3 mm2/s vs. σMD,6avg = 0.63 ± 0.2 × 10−3 mm2/s, P = 0.05).
FIGURE 8.
LV medians (m) and SDs (σ) of MD (A), FA (B), and Mode (C) from midsystolic (left) and diastolic (right) cDTI with Navg = 1–10. Lines and error bars represent the population means and SDs across all subjects, and dots represent individual subject values. LVSDs decreased with increasing Navg for all parameters but with diminishing changes with Navg ≥ 4. Median FA values decreased with increasing Navg, whereas median Mode values increased. Median MD values did not change with increasing Navg.
Compared with Navg = 1, mFA decreased significantly with Navg ≥ 3 at both midsystole (mFA,1avg = 0.46 ± 0.05 vs. mFA,3avg = 0.41 ± 0.04, P = 0.01) and diastole (mFA,1avg = 0.48 ± 0.08 vs. mFA,3avg = 0.38 ± 0.06, P = 4 × 10−3). Compared with Navg = 1, σFA decreased significantly with Navg ≥ 2 in midsystole (σFA,1avg = 0.24 ± 0.03 vs. σFA,2avg = 0.21 ± 0.03, P = 0.03) and with Navg ≥ 3 in diastole (σFA,1avg = 0.22 ± 0.03 vs. σFA,3avg = 0.18 ± 0.04, P = 0.01).
A significant increase in mMode was observed in midsystole with Navg ≥ 4 compared with Navg = 1 (mMode,1avg = 0.22 ± 0.6 vs. mMode,4avg = 0.31 ± 0.9, P = 0.02). Compared with Navg = 1, σMode decreased significantly with Navg ≥ 3 in midsystole (σMode,1avg = 0.57 ± 0.02 vs. σMode,1avg = 0.56 ± 0.01, P = 0.03). No corresponding significant differences were observed in diastole.
4.4 | Cardiac phase dependence of tensor parameters
With Navg = 10, median myocardial MD was significantly lower in midsystole than in diastole (MDSYS = 1.58 ± 0.09 × 10−3 mm2/s vs. MDDIA = 1.91 ± 0.34 × 0−3mm2/s, P = 8 × 10−3), whereas FA was significantly higher in midsystole (FASYS = 0.37 ± 0.03 vs. FADIA = 0.32 ± 0.06, P = 0.03). No significant differences were observed between cardiac phases for any of the other parameters examined.
5 | DISCUSSION
The observed increase in bulk motion artifacts (higher CoV-DTI) in diastolic cDTI compared with midsystolic cDTI is consistent with previous reports of the second-order motion-compensated diffusion encoding scheme.19 M1M2 nulled diffusion encoding relies on an assumption that cardiac motion can be well described by a second-order polynomial (i.e., only velocity and acceleration terms). Physiologic deviations from this assumption lead to bulk motion artifacts that appear as localized myocardial signal loss. If these artifacts are not reproducible from beat to beat, this leads to intershot signal fluctuations across cDTI images and consequently to elevated CoVDTI measurements. The lower CoVDTI values observed during systole thus indicate that systolic motion is more reproducible from beat to beat than diastolic motion. It is important to note, however, that CoVDTI is not sensitive to systematic bias caused by higher order motion that is consistent from beat to beat. This means that the low CoVDTI observed in systole does not necessarily imply complete motion compensation. However, the low level of intersubject variability in median midsystolic myocardial MD (Figure 8A) suggests that systolic motion is well characterized by second-order motion and thus is largely compensated by the CODE M1M2 nulled gradient waveform.
Diastolic motion, although smaller in magnitude than contractile motion, accords with a larger CoVDTI than systolic motion. This indicates that diastolic motion does not meet the intrashot second-order assumption as closely (i.e., it is not fully compensated by the M1M2 nulling) and is more dependent on intershot (beat to beat) variations and changes in heart rate. These effects combine to cause deviations in the cDTI signal during an acquisition that result in elevated CoVDTI. This is also reflected in an increase in intersubject variability in median myocardial MD in the diastolic phase (Figure 8B).
The strong positive correlation between CoVDTI and median uncertainty (Figures 7A, 7B) indicates that bulk motion artifacts are a major driver of diffusion tensor uncertainty and explains the greater degree of uncertainty observed in diastolic cDTI.
The negative correlation between eigenvector uncertainty and MD for MD up to approximately 1.5 × 10−3 mm2/s (Figures 7D, 7E) indicates that sufficient diffusion weighting should be achieved to minimize uncertainty, echoing previous reports.13,23,28 On the other hand, the increasing uncertainty with increasing MD beyond 1.5 × 10−3 mm2/s is likely caused by bulk motion effects. The negative correlation between eigenvector uncertainty and FA (Figures 7G, 7H) indicates that the achievable precision will likely vary in patients with conditions that reduce microscopic anisotropy such as diffuse fibrosis.29
The uncertainty in diastolic diffusion tensor metrics could have been reduced by employing specialized image postprocessing algorithms to remove bulk-motion image artifacts such as constrained reconstruction30 or maximum intensity projection.27,31 These correction strategies would likely reduce the effects of physiological variability but limit the SNR gains from acquiring repeated acquisitions by omitting some of the acquired data. It is worth noting that this effect can be mitigated through postprocessing denoising techniques such as principal component analysis,31 which will have an unknown impact on tensor accuracy, precision, and optimal scan efficiency. It is also possible that a TR correction to account for heart rate variability, which was not implemented in this work, could reduce uncertainty.
Uncertainty varied substantially between tensor parameters and the 95CIs were the smallest for E⃗1, MD, and FA; and they were the largest for E⃗2, E⃗3, and Mode. This indicates that SNR and scan time requirements vary depending on the parameters of interest in a given study. To contextualize these uncertainties, it is useful to consider examples of expected physiological variability. For example, the global HApitch value of 1.2% ± 0.2°/% observed in this work (which is in line with other in vivo studies5,32 indicates an inherent range of myofiber orientations present in each imaging voxel. Assuming an LV wall thickness of 10mm and 2.0mm in-plane spatial resolution, each voxel will occupy approximately 20% of wall depth. Consequently, each voxel should contain a range of myofiber orientations, which vary by as much as 20° to 25°, thus placing a reasonable upper bound on the expected precision of E⃗1. For a 5-min acquisition (5 averages, SNR~18), the maximum likelihood and median E⃗1 precision were both well within this bound for midsystolic cDTI: dE1 = 9.7 ± 1.0° and dE1 = 15.4 ± 1.2°, respectively (Table 1).
In comparison to our observed eigenvector uncertainties, an ex vivo study of cDTI in rat hearts reported mean dE1, dE2, and dE3 values of 3.7° ± 0.2°, 10.9° ± 0.4°, and 10.6° ± 0.5°, respectively.2 However, this study was performed at high field (9.4T), employed gradients capable of amplitudes >10 × of standard clinical hardware (1T/m), and did not contend with physiological motion. Furthermore, the 0.1mm spatial resolution protocol resulted in substantially lower range of intravoxel myofiber orientations and thus less physiological variability within each voxel.
Our observed midsystolic uncertainties are consistent with Stoeck et al., who reported comparable E1 uncertainties and lower E2 uncertainties from 2 repeated measurements, with a similar M1M2 compensated SE-cDTI method in in vivo swine but with longer scans (14 signal averages).33
Another example of expected physiological diffusion tensor variability is the 0.9 × 10−3 mm2/s MD increase observed in myocardial infarcts compared with healthy tissue observed by Nguyen et al.16 For the purpose of identifying infarcts, MD precision should thus be sufficiently small to detect this difference. The median MD uncertainty was within this difference for even just 1 average in midsystole (median dMD = 0.73 ± 0.04 × 10−3 mm2/s). However, the uncertainty histogram for 1 average (Figure 5) shows that nearly 40% of voxels exhibited MD uncertainties ≥ 0.9 × 10−3 mm2/s, a number that reduced to 11% with 5 averages (median dMD = 0.38 ± 0.02 × 10−3 mm2/s).
Our results indicate that little benefit in precision is achieved by increasing SNR beyond approximately 25 (Figure 6), which in this study corresponded to an approximately 10 min acquisition (i.e., 10 averages). Note that this study was conducted on a 3.0T clinical system with high-performance gradient hardware (Maximum gradient amplitude (Gmax) = 76 mT/m), which reduces the minimum TE (65 ms) for the cDTI protocol compared with more commonly available gradient hardware sets (Gmax = 40 mT/m and TEMin = 85 ms). The baseline SNR on these systems will consequently be lower than those reported here—and will thus require longer scans to achieve the same degree of uncertainty. Further scan time increases will also be required for imaging at 1.5T or for nonoptimized M1M2-compensated diffusion encoding schemes with longer TEs.
Conversely, no significant changes in median myocardial tensor parameters or their SDs were achieved by increasing scan time beyond approximately 4 min, which corresponded with b = 0 image SNR = 16.0 ± 3.8 (i.e., 4 averages) (Figure 8). With low SNR (Navg<4), we observed an upward bias in FA and a downward bias in Mode, which are both consistent with published reports based on numerical simulations.23 Notably, no significant differences in median MD were observed between any number of signal averages. This indicates that MD measurement accuracy is very robust to variability and does not require long scan times. Whereas all reconstructions were performed using linear least squares, it has been shown that this can introduce bias in the presence of noise, and that this bias can be mitigated by using nonlinear least squares.34 It is thus possible that nonlinear fitting could reduce the scan time requirements for accurate reconstruction.
The only significant differences observed between cardiac phases were an increase in MD and a decrease FA in diastole relative to midsystole. It is likely that the increase in MD in diastole stems from both the increased sensitivity to bulk motion in this phase19 and the increased variability of diastolic motion from beat to beat. The FA decrease may be due to an increase in myocardial blood volume in diastole, which contributes an isotropic diffusive compartment. This comes in contrast to the increase in FA in diastole observed with STEAM cDTI.35 However, this increase was observed in comparison to peak systolic measurements as opposed to the midsystolic phase examined in this work.
The steepening of the secondary sheet angle in diastole compared with peak systole previously observed was not observed in this study. This may arise, in part, because the midsystolic phase imaged (TDSYS = 100 ms corresponded with 35% of peak systole) does not correspond with a fully contracted myocardium, which limits the observable differences in sheetlet mobility. Furthermore, the relatively large E⃗2 uncertainties indicate that the CODE-M1M2 method may not be as sensitive to sheetlet mobility, possibly because of the short diffusion times of the CODE-M1M2 sequence. This may also lead to sorting errors that can flip the order of eigenvectors, which could explain the secondary peaks in E⃗1, E⃗2, and E⃗3 uncertainty observed at 90°.
This study only examines the precision in SE-EPI cDTI. Substantial differences may be observed in STEAM cDTI, which generally reports higher FA (FASTEAM~0.5–0.635,36 vs. FASE~0.3–0.4 reported here and elsewhere36 values than spin echo data, possibly due to the longer diffusion “mixing times” that allow diffusing molecules to probe larger length scales during diffusion encoding. The higher effective FA will likely reduce the inherent uncertainty in E1. This may also reduce uncertainty in tensor Mode measurements, which can distinguish planar and linear diffusion anisotropy and have shown sensitivity to microstructural changes in pacing-induced heart failure37 but were relatively imprecise in this study. However, STEAM cDTI has half the baseline SNR of SE-EPI cDTI and also double the scan time due to the need for 2 heartbeats per diffusion encode, which results in lower SNR efficiency.36 Currently, however, it is unclear how the tradeoffs between FA and SNR efficiency affect the relative tensor uncertainty for the STEAM approach.
The number of unique diffusion encoding gradient orientations sampled in a DTI acquisition also impacts measurement precision.12 Although studies in neurological and skeletal muscle tissue provide guidance in this regard (the 12-direction sampling scheme was used based on such guidance12,38, it remains an open question whether additional signal averages or diffusion encoding directions more efficiently improve the precision of in vivo cDTI. Furthermore, it is unclear how to best employ EPI acceleration techniques such as parallel imaging and partial Fourier, which have complex relationships with image SNR in that they reduce SNR efficiency but often permit shorter TEs. Partial Fourier also has the added effect of increasing bulk motion sensitivity.39 The b value of 350 s/mm2 used in this study is lower than the optimal values proposed by Jones13 based on noise propagation (~700 s/mm2 for the cardiac diffusivities observed in this study) or by Scott et al.28 for STEAM cDTI (750 s/mm2). The b value of 350 s/mm2 was chosen for this spin echo cDTI protocol to empirically balance bulk motion sensitivity, diffusion-weighted contrast, and SNR. However, the optimal b value and its effect on optimal scan efficiency for spin echo cDTI precision have yet to be determined and should be the focus of future study.
Although it would be interesting to observe the progression of tensor precision with Navg>5, the bootstrapped technique used in this work requires a repeated acquisition and thus can only quantify uncertainties for datasets containing half of the acquired 10 averages. Model-based techniques such as the wild bootstrap,40 however, can quantify precision without a repeated acquisition but require assumptions of the underlying uncertainties. It is unclear whether these approaches can be successfully applied to in vivo cDTI, and they have not been examined in this work.
6 | CONCLUSION
Acceptable levels of precision in E⃗1, MD, and FA were achieved in a 5-min (per slice) free-breathing scan when using the CODE-M1M2 SE-EPI cDTI approach in vivo. Precision improved with increasing b = 0 image SNR, but gains were minimal for SNR ≥ 25, which corresponded with a ~10-min scan with this protocol. Lower uncertainty was observed in midsystolic compared to diastolic acquisitions, indicating that bulk cardiac motion effects are more consistently compensated by the M1M2 approach during midsystole.
Supplementary Material
Figure S1. Example diastolic images with b=0 and 350s/mm2 with 5 averages and a histogram of LV b=0 SNR (A). Maps of tensor MD (B), FA (C), and Mode (D) with corresponding uncertainties and LV histograms. Uncertainty was generally higher in diastolic than in mid-systolic cDTI.
Figure S2. Histograms of b=0 image SNR within the LV with Navg=1–5 for all subjects and phases. Median (±95CI) SNR increased from 7.4 ± 0.3 with Navg = 1 to 16.7 ± 0.7 with Navg = 5.
Acknowledgments
Funding information Grant sponsors: supported, in part, by the graduate program in bioscience at UCLA; the department of radiological sciences at UCLA; and the National Institute of Health (NIH) grant number NIH R01HL131975 and R01HL131823 and the American Heart Association (AHA) grant number AHA16PRE27380023
This work was supported, in part, by the graduate program in Bioscience at UCLA, the department of radiological Sciences at UCLA, NIH R01HL131975, and AHA 16PRE27380023.
Footnotes
Additional supporting information may be found in the online version of this article.
References
- 1.Kung GL, Nguyen TC, Itoh A, Skare S, Ingels NB, Miller DC, Ennis DB. The presence of two local myocardial sheet populations confirmed by diffusion tensor MRI and histological validation. J Magn Reson Imaging. 2011;34:1080–1091. doi: 10.1002/jmri.22725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Teh I, McClymont D, Burton RA, Maguire ML, Whittington HJ, Lygate CA, Kohl P, Schneider JE. Resolving Fine cardiac structures in rats with high-resolution diffusion tensor imaging. Sci Rep. 2016;6:30573. doi: 10.1038/srep30573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nielles-Vallespin S, Khalique Z, Ferreira PF, et al. Assessment of myocardial microstructural dynamics by in vivo diffusion tensor cardiac magnetic resonance. J Am Coll Cardiol. 2017;69:661–676. doi: 10.1016/j.jacc.2016.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferreira PF, Kilner PJ, McGill LA, et al. In vivo cardiovascular magnetic resonance diffusion tensor imaging shows evidence of abnormal myocardial laminar orientations and mobility in hypertrophic cardiomyopathy. J Cardiovasc Magn Reson. 2014;16:87. doi: 10.1186/s12968-014-0087-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.von Deuster C, Sammut E, Asner L, Nordsletten D, Lamata P, Stoeck CT, Kozerke S, Razavi R. Studying dynamic myofiber aggregate reorientation in dilated cardiomyopathy using in vivo magnetic resonance diffusion tensor imaging. Circ Cardiovasc Imaging. 2016:9. doi: 10.1161/CIRCIMAGING.116.005018. https://doi.org/10.1161/circimaging.116.005018. [DOI] [PMC free article] [PubMed]
- 6.Nguyen C, Fan Z, Xie Y, Dawkins J, Tseliou E, Bi X, Sharif B, Dharmakumar R, Marban E, Li D. In vivo contrast free chronic myocardial infarction characterization using diffusion-weighted cardiovascular magnetic resonance. J Cardiovasc Magn Reson. 2014;16:68. doi: 10.1186/s12968-014-0068-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nguyen C, Lu M, Fan Z, Bi X, Kellman P, Zhao S, Li D. Contrast- free detection of myocardial fibrosis in hypertrophic cardiomyopathy patients with diffusion-weighted cardiovascular magnetic resonance. J Cardiovasc Magn Reson. 2015;17:107. doi: 10.1186/s12968-015-0214-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Le Bihan D, Poupon C, Amadon A, Lethimonnier F. Artifacts and pitfalls in diffusion MRI. J Magn Reson Imaging. 2006;24:478–488. doi: 10.1002/jmri.20683. [DOI] [PubMed] [Google Scholar]
- 9.Jones DK. Determining and visualizing uncertainty in estimates of fiber orientation from diffusion tensor MRI. Magn Reson Med. 2003;49:7–12. doi: 10.1002/mrm.10331. [DOI] [PubMed] [Google Scholar]
- 10.Pajevic S, Basser PJ. Parametric and non-parametric statistical analysis of DT-MRI data. J Magn Reson. 2003;161:1–14. doi: 10.1016/s1090-7807(02)00178-7. [DOI] [PubMed] [Google Scholar]
- 11.Heim S, Hahn K, Samann PG, Fahrmeir L, Auer DP. Assessing DTI data quality using bootstrap analysis. Magn Reson Med. 2004;52:582–589. doi: 10.1002/mrm.20169. [DOI] [PubMed] [Google Scholar]
- 12.Jones DK. The effect of gradient sampling schemes on measures derived from diffusion tensor MRI: a Monte Carlo study. Magn Reson Med. 2004;51:807–815. doi: 10.1002/mrm.20033. [DOI] [PubMed] [Google Scholar]
- 13.Jones DK. Precision and accuracy in diffusion tensor magnetic resonance imaging. Top Magn Reson Imaging. 2010;21:87–99. doi: 10.1097/RMR.0b013e31821e56ac. [DOI] [PubMed] [Google Scholar]
- 14.Olivetti G, Cigola E, Maestri R, Corradi D, Lagrasta C, Gambert SR, Anversa P. Aging, cardiac hypertrophy and ischemic cardiomyopathy do not affect the proportion of mononucleated and multinucleated myocytes in the human heart. J Mol Cell Cardiol. 1996;28:1463–1477. doi: 10.1006/jmcc.1996.0137. [DOI] [PubMed] [Google Scholar]
- 15.Liewald D, Miller R, Logothetis N, Wagner HJ, Schuz A. Distribution of axon diameters in cortical white matter: an electron-microscopic study on three human brains and a macaque. Biol Cybern. 2014;108:541–557. doi: 10.1007/s00422-014-0626-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen C, Fan Z, Sharif B, He Y, Dharmakumar R, Berman DS, Li D. In vivo three-dimensional high resolution cardiac diffusion-weighted MRI: a motion compensated diffusion-prepared balanced steady-state free precession approach. Magn Reson Med. 2013;72:1257–1267. doi: 10.1002/mrm.25038. [DOI] [PubMed] [Google Scholar]
- 17.Stoeck CT, von Deuster C, Genet M, Atkinson D, Kozerke S. Second- order motion-compensated spin echo diffusion tensor imaging of the human heart. Magn Reson Med. 2016;75:1669–1676. doi: 10.1002/mrm.25784. [DOI] [PubMed] [Google Scholar]
- 18.Welsh C, Di Bella E, Hsu E. Higher-order motion-compensation for in vivo cardiac diffusion tensor imaging in rats. IEEE Trans Med Imaging. 2015;34:1843–1853. doi: 10.1109/TMI.2015.2411571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aliotta E, Wu HH, Ennis DB. Convex optimized diffusion encoding (CODE) gradient waveforms for minimum echo time and bulk motion compensated diffusion weighted MRI. Magn Reson Med. 2017;77:717–729. doi: 10.1002/mrm.26166. [DOI] [PubMed] [Google Scholar]
- 20.Reese TG, Weisskoff RM, Smith RN, Rosen BR, Dinsmore RE, Wedeen VJ. Imaging myocardial fiber architecture in vivo with magnetic resonance. Magn Reson Med. 1995;34:786–791. doi: 10.1002/mrm.1910340603. [DOI] [PubMed] [Google Scholar]
- 21.Nielles-Vallespin S, Mekkaoui C, Gatehouse P, et al. In vivo diffusion tensor MRI of the human heart: reproducibility of breath-hold and navigator-based approaches. Magn Reson Med. 2013;70:454–465. doi: 10.1002/mrm.24488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Froeling M, Strijkers GJ, Nederveen AJ, Luijten PR. Whole heart DTI using asymmetric bipolar diffusion gradients. J Cardiovasc Magn Reson. 2015;17(suppl 1):P15. [Google Scholar]
- 23.Gahm JK, Kindlmann G, Ennis DB. The effects of noise over the complete space of diffusion tensor shape. Med Image Anal. 2014;18:197–210. doi: 10.1016/j.media.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 24.Feinberg DA, Hoenninger JC, Crooks LE, Kaufman L, Watts JC, Arakawa M. Inner volume MR imaging: technical concepts and their application. Radiology. 1985;156:743–747. doi: 10.1148/radiology.156.3.4023236. [DOI] [PubMed] [Google Scholar]
- 25.Griswold MA, Jakob PM, Heidemann RM, Nittka M, Jellus V, Wang J, Kiefer B, Haase A. Generalized autocalibrating partially parallel acquisitions (GRAPPA) Magn Reson Med. 2002;47:1202–1210. doi: 10.1002/mrm.10171. [DOI] [PubMed] [Google Scholar]
- 26.Scott AD, Nielles-Vallespin S, Ferreira PF, McGill LA, Pennell DJ, Firmin DN. The effects of noise in cardiac diffusion tensor imaging and the benefits of averaging complex data. NMR Biomed. 2016;29:588–599. doi: 10.1002/nbm.3500. [DOI] [PubMed] [Google Scholar]
- 27.Rapacchi S, Wen H, Viallon M, Grenier D, Kellman P, Croisille P, Pai VM. Low b-value diffusion-weighted cardiac magnetic resonance imaging: initial results in humans using an optimal time-window imaging approach. Invest Radiol. 2011;46:751–758. doi: 10.1097/RLI.0b013e31822438e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott AD, Ferreira PF, Nielles-Vallespin S, Gatehouse P, McGill LA, Kilner P, Pennell DJ, Firmin DN. Optimal diffusion weighting for in vivo cardiac diffusion tensor imaging. Magn Reson Med. 2015;74:420–430. doi: 10.1002/mrm.25418. [DOI] [PubMed] [Google Scholar]
- 29.Abdullah OM, Drakos SG, Diakos NA, et al. Characterization of diffuse fibrosis in the failing human heart via diffusion tensor imaging and quantitative histological validation. NMR Biomed. 2014;27:1378–1386. doi: 10.1002/nbm.3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aliotta E, Rapacchi S, Hu P, Ennis D. Increased maximum gradient amplitude improves robustness of spin-echo cardiac diffusion-weighted MRI. J Cardiovasc Magn Reson. 2015;17(suppl 1):P388. [Google Scholar]
- 31.Pai VM, Rapacchi S, Kellman P, Croisille P, Wen H. PCATMIP: enhancing signal intensity in diffusion-weighted magnetic resonance imaging. Magn Reson Med. 2011;65:1611–1619. doi: 10.1002/mrm.22748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nguyen C, Fan Z, Xie Y, Pang J, Speier P, Bi X, Kobashigawa J, Li D. In vivo diffusion-tensor MRI of the human heart on a 3 Tesla clinical scanner: an optimized second order (M2) motion compensated diffusion-preparation approach. Magn Reson Med. 2016;76:1354–1363. doi: 10.1002/mrm.26380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stoeck CT, von Deuster C, Fleischmann T, Lipiski M, Cesarovic N, Kozerke S. Direct comparison of in vivo versus postmortem second-order motion-compensated cardiac diffusion tensor imaging. Magn Reson Med. 2017 doi: 10.1002/mrm.26871. https://doi.org/10.1002/mrm.26871. [DOI] [PubMed]
- 34.Jones DK, Basser PJ. “Squashing peanuts and smashing pumpkins”: how noise distorts diffusion-weighted MR data. Magn Reson Med. 2004;52:979–993. doi: 10.1002/mrm.20283. [DOI] [PubMed] [Google Scholar]
- 35.McGill LA, Ferreira PF, Scott AD, Nielles-Vallespin S, Giannakidis A, Kilner PJ, Gatehouse PD, de Silva R, Firmin DN, Pennell DJ. Relationship between cardiac diffusion tensor imaging parameters and anthropometrics in healthy volunteers. J Cardiovasc Magn Reson. 2016;18:2. doi: 10.1186/s12968-015-0215-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Deuster C, Stoeck CT, Genet M, Atkinson D, Kozerke S. Spin echo versus stimulated echo diffusion tensor imaging of the in vivo human heart. Magn Reson Med. 2016;76:862–872. doi: 10.1002/mrm.25998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kung GL, Ouadah S, Hsieh Y-C, Garfinkel A, Chen P-S, Ennis D. Transmural heterogeneity of microstructural remodeling in pacing induced heart failure measured by diffusion tensor MRI. J Cardiovasc Magn Reson. 2013;15(suppl 1):P119. [Google Scholar]
- 38.Froeling M, Nederveen AJ, Nicolay K, Strijkers GJ. DTI of human skeletal muscle: the effects of diffusion encoding parameters, signal-to-noise ratio and T2 on tensor indices and fiber tracts. NMR Biomed. 2013;26:1339–1352. doi: 10.1002/nbm.2959. [DOI] [PubMed] [Google Scholar]
- 39.Storey P, Frigo FJ, Hinks RS, Mock BJ, Collick BD, Baker N, Marmurek J, Graham SJ. Partial k-space reconstruction in single-shot diffusion-weighted echo-planar imaging. Magn Reson Med. 2007;57:614–619. doi: 10.1002/mrm.21132. [DOI] [PubMed] [Google Scholar]
- 40.Whitcher B, Tuch DS, Wisco JJ, Sorensen AG, Wang L. Using the wild bootstrap to quantify uncertainty in diffusion tensor imaging. Hum Brain Mapp. 2008;29:346–362. doi: 10.1002/hbm.20395. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Example diastolic images with b=0 and 350s/mm2 with 5 averages and a histogram of LV b=0 SNR (A). Maps of tensor MD (B), FA (C), and Mode (D) with corresponding uncertainties and LV histograms. Uncertainty was generally higher in diastolic than in mid-systolic cDTI.
Figure S2. Histograms of b=0 image SNR within the LV with Navg=1–5 for all subjects and phases. Median (±95CI) SNR increased from 7.4 ± 0.3 with Navg = 1 to 16.7 ± 0.7 with Navg = 5.