Abstract
Atmospheric nitrogen (N) deposition has enhanced soil carbon (C) stocks in temperate forests. Most research has posited that these soil C gains are driven primarily by shifts in fungal community composition with elevated N leading to declines in lignin degrading Basidiomycetes. Recent research, however, suggests that plants and soil microbes are dynamically intertwined, whereby plants send C subsidies to rhizosphere microbes to enhance enzyme production and the mobilization of N. Thus, under elevated N, trees may reduce belowground C allocation leading to cascading impacts on the ability of microbes to degrade soil organic matter through a shift in microbial species and/or a change in plant–microbe interactions. The objective of this study was to determine the extent to which couplings among plant, fungal, and bacterial responses to N fertilization alter the activity of enzymes that are the primary agents of soil decomposition. We measured fungal and bacterial community composition, root–microbial interactions, and extracellular enzyme activity in the rhizosphere, bulk, and organic horizon of soils sampled from a long-term (>25 years), whole-watershed, N fertilization experiment at the Fernow Experimental Forest in West Virginia, USA. We observed significant declines in plant C investment to fine root biomass (24.7%), root morphology, and arbuscular mycorrhizal (AM) colonization (55.9%). Moreover, we found that declines in extracellular enzyme activity were significantly correlated with a shift in bacterial community composition, but not fungal community composition. This bacterial community shift was also correlated with reduced AM fungal colonization indicating that declines in plant investment below-ground drive the response of bacterial community structure and function to N fertilization. Collectively, we find that enzyme activity responses to N fertilization are not solely driven by fungi, but instead reflect a whole ecosystem response, whereby declines in the strength of belowground C investment to gain N cascade through the soil environment.
Keywords: arbuscular mycorrhizal fungi, belowground carbon allocation, extracellular enzymes, microbial community, nitrogen fertilization, plant–microbial interactions
1 INTRODUCTION
Atmospheric nitrogen (N) deposition has enhanced the carbon (C) sink in both the soils and vegetation of temperate forests (Frey et al., 2014; Janssens et al., 2010; Thomas, Canham, Weathers, & Goodale, 2010). Aboveground responses to N are relatively straightforward with additional N inputs relieving N limitation to tree growth (Thomas et al., 2010). Belowground, a synthesis of data across a suite of N addition experiments showed that soil C stocks in forests are typically enhanced by N fertilization due to declines in soil respiration (Janssens et al., 2010). While this response appears robust across multiple forest types, the proximate mechanism driving these declines in soil respiration remains unclear. Given that the world’s soils contain more C than the atmosphere and vegetation combined, understanding the key mechanisms driving the response of forest soil C to shifts in N input regimes is critical to predicting future rates of C sequestration (Reay, Dentener, Smith, Grace, & Feely, 2008).
Classically, shifts in fungal community composition have been viewed as the main driver of decomposition responses to enhanced N inputs (Frey, Knorr, Parrent, & Simpson, 2004; Morrison et al., 2016; Wallenstein, McNulty, Fernandez, Boggs, & Schlesinger, 2006). According to this fungal-driven view, elevated N inputs reduce the abundance of white-rot fungi (phylum Basidiomycota), one of the primary fungal guilds including both saprotrophs and ectomycorrhizal species that can produce oxidative enzymes capable of degrading lignin (Dix & Webster, 1995). The inability of Basidiomycota to compete when N limitation is removed, then leads to declines in decomposition and increased soil organic matter accumulation (Carreiro, Sinsabaugh, Repert, & Parkhurst, 2000; Fog, 1988; Waldrop, Zak, Sinsabaugh, Gallo, & Lauber, 2004; Zak, Holmes, Burton, Pregitzer, & Talhelm, 2008). In support, many studies report reduced ligninolytic enzyme activity in response to N fertilization (DeForest, Zak, Pregitzer, & Burton, 2004; Fog, 1988; Frey et al., 2014; Saiya-Cork, Sinsabaugh, & Zak, 2002; Zak et al., 2008). However, these enzymatic declines are not always coupled with a corresponding decline in Basidomycota abundance (DeForest et al., 2004; Hassett, Zak, Blackwood, & Pregitzer, 2009). This apparent disconnect between enzyme activity and fungal community composition suggests that soil C responses to N fertilization may not solely be driven by elevated N directly inhibiting fungal growth and activity.
Emerging research suggests that enzyme activity declines in response to N fertilization may reflect an integrated ecosystem response that is driven by interactions among fungi, bacteria, and plants. Temperate forest trees allocate nearly 20% of net primary production belowground to mycorrhizae and free-living microbes in the rhizosphere which can influence microbial community composition, stimulate extracellular enzyme production and enhance plant access to N (Brzostek, Fisher, & Phillips, 2014; Hobbie, 2006; Paterson, Gebbing, Abel, Sim, & Telfer, 2007; Yin et al., 2013). When forest ecosystems are fertilized with N, trees likely reduce the C subsidies they send to rhizosphere microbes, which can alter the structure and function of not only fungal communities but also bacterial communities as well. These potential shifts in bacterial community composition are consequential given recent evidence that bacteria can synthesize enzymes to degrade simple and complex C (i.e., polyphenols, lignin) in soil organic matter (SOM; Datta et al., 2017; De Gonzalo, Colpa, Habib, & Fraaije, 2016; Jackson, Couger, Prabhakaran, Ramachandriya, & Canaan, 2017) and that shifts in bacterial function can reduce enzyme activities under N fertilization (Freedman, Upchurch, Zak, & Cline, 2016). As such, enzyme activity declines in response to N fertilization may result from parallel shifts in plant, bacterial, and fungal function; whereby reductions in plant C allocation to rhizosphere microbes indirectly alter fungal and bacterial community composition, decrease the energy available to synthesize enzymes, and ultimately reduce the fungal and bacterial community’s ability to degrade SOM.
While fungi have been shown to be important drivers of SOM increases with N fertilization (DeForest et al., 2004; Fog, 1988; Frey et al., 2014; Saiya-Cork et al., 2002; Zak et al., 2008), focusing solely on fungal responses may mask important interactions between other components of the ecosystem that can alter enzyme activity responses. Thus, the objective of this study was to examine the extent to which couplings among plant, fungal, and bacterial responses to N fertilization change extracellular enzyme activity (the proximate agent of SOM decomposition; Schimel & Weintraub, 2003) in a long-term (>25 year), watershed-scale, N fertilization experiment in West Virginia, United States. To meet this objective, we linked assays of soil enzyme activities with measurements of fungal and bacterial community composition and belowground C allocation across replicate plots in a reference and fertilized watershed, and used these measurements to parameterize a simple microbial enzyme decomposition model to examine long-term effects of N fertilization on soil C stocks.
2 MATERIALS AND METHODS
2.1 Site description
The Fernow Experimental Forest (herein Fernow) is located in the central Appalachian Mountains near the town of Parsons, West Virginia (39.03°N, 79.67°W). Mean annual precipitation is ~1460 mm and mean annual temperature is 8.9°C. (Kochenderfer, 2006). Ambient wet N deposition at the site has been declining over time from 10.4 kg N ha−1 year−1 in 1989 (with an additional 4–5 kg N ha−1 year−1 dry deposition) to 5.0 kg N ha−1 year−1 in 2014 (Clean Air Status and Trends Network (Gilliam, Yurish, & Adams, 1996). Soils are derived from sandstone and shale bedrock and are classified as coarse textured Inceptisols (loamy-skeletal, mixed mesic Typic Dystrochrept) of the Berks and Calvin Series (Gilliam, Turrill, Aulick, Evans, & Adams, 1994).
To examine the impacts of N deposition on plant–microbial interactions, we sampled soils and roots from a reference watershed receiving only ambient N deposition inputs and an N-fertilized watershed at the Fernow. Both watersheds have stands of similar age (~45 years old) and tree species assemblages. Trees >2.5 cm in diameter at breast height (DBH) were cut in the N-fertilized watershed (Watershed 3) in 1970 and recovered naturally until 1989 when N treatments began (Peterjohn, Adams, & Gilliam, 2017). N is added by aerial fertilization via helicopter at a rate of 35 kg N ha−1 year−1 in the form of solid pellet (NH4)2 SO4 (Adams, Edwards, Wood, & Kochenderfer, 1993). The N-fertilized watershed is 34.3 ha with a south-facing aspect and elevation ranging from 735 to 860 m (Gilliam et al., 2016). The reference watershed (Watershed 7) was clear cut from 1963 to 1967 (the upper half in 1963/1964 and the lower half in 1966/1977) and kept barren with herbicides until 1969 when it began natural recovery (Kochenderfer, 2006). This watershed is 24.0 ha with an east-facing aspect and elevation ranging from 731–850 m (Gilliam et al., 2016). The watersheds are dominated by arbuscular mycorrhizal associated (AM) tree species that include tulip poplar (Liriodendron tulipifera), red maple (Acer rubrum), black cherry (Prunus serotina), and sugar maple (Acer saccharum), which together account for 59% of the basal area in the reference watershed and 72% in the N-fertilized watershed. Less abundant, ectomycorrhizal associated (ECM) tree species include sweet birch (Betula lenta), red oak (Quercus rubra), and American beech (Fagus grandifolia) which together account for 25% and 16% of the total basal area in the reference and N-fertilized watershed, respectively.
2.2 Soil sampling protocol
We sampled soils on three dates in June, July, and August of 2015 in order to capture early, peak, and late growing season dynamics. In each watershed, we sampled soils from an existing network of 10 randomly placed 25 × 25 m plots that have similar tree species composition (Gilliam et al., 1996). Three 10 × 10 cm organic horizon (OH) samples were collected from each plot. One OH sample was used for root metrics and the other two for measures of soil biogeo-chemistry. After OH removal, three 5-cm diameter soil cores were extracted from each plot to a depth of 15 cm. All samples were stored on ice and returned to West Virginia University for further processing.
Within 24 hr of sampling, mineral soil samples were separated into bulk and rhizosphere fractions. Rhizosphere soil was operationally defined as soil that adhered to roots upon removal from the soil matrix, and was carefully subsampled using forceps and homogenized (sensu Phillips & Fahey, 2005). The remaining soil was considered mineral bulk soil. After root and rhizosphere separation, the bulk and OH soil was homogenized by sieving through a 2-mm mesh. A subsample of each soil sample and fraction was stored at −80°C for assays of enzyme activity. All fine roots (<2 mm) were washed in deionized water and subsequently dried and weighed to determine standing root biomass. Subsamples of roots from each plot from the final sampling date were used to examine AM colonization and root morphology.
2.3 Arbuscular Mycorrhizal colonization and root morphology
To determine the extent to which N fertilization altered AM colonization, we analyzed a subsample comprising ~1/3 of all roots from the mineral and OH horizons for % AM colonization. Roots were cleared by incubating in 10% potassium hydroxide for 48–96 hr at room temperature depending on the pigmentation of the roots. Excess pigment was leached from roots by incubating in 85% ethanol for 24–48 hr. This step was necessary due to the dark pigments present in the woody tissue of our samples. Any remaining pigment was removed by soaking roots in ammonia/hydrogen peroxide solution for 15 min. Roots were prepared for staining by acidifying them in 5% hydrochloric acid, and then stained for 5 min in 0.05% trypan blue (Comas, Callahan, & Midford, 2014).
We calculated AM colonization for each sample using the grid-intersect method (Giovannetti & Mosse, 1980; McGonigle, Miller, Evans, Fairchild, & Swan, 1990). Each sample was suspended in water on a 1 × 1 cm gridded Petri dish and examined under a stereoscopic microscope. At each root-gridline intersect, colonization was determined by the presence of arbuscules or hyphae. Percent AM colonization was then calculated as the percent of total intersects examined that had visible AM structures. Given that roots with high levels of AM colonization are likely to be more involved in releasing C to rhizosphere microbes (Kaiser et al., 2015), we used the level of colonization by AM fungi as a metric of the degree to which roots and microbes were actively coupled.
We created digital images of root morphology using an LA2400 desktop scanner (WinRhizo; Regent Instruments Inc), and then used WinRhizo software to determine root length, surface area, average diameter, and number of forks of each sample. All measurements were expressed per g of root mass and scaled to a per m2 basis based on total standing root biomass (g/m2) at the plot level (Table 1).
TABLE 1.
Fine root and mycorrhizal metrics in bulk and organic horizon soils under ambient and long-term N-fertilized conditions
| Fine root metric | Control | +N | % Change |
|---|---|---|---|
| Organic horizon | |||
| Root biomass (g/m2) | 30.83 ± 4.07 | 52.13 ± 8.50 | 69.1* |
| Mycorrhizal colonization (%) | 72.8 ± 3.6 | 60.9 ± 0.01 | −16.3* |
| Specific root length (cm/g) | 2038.69 ± 260.99 | 1992.99 ± 179.60 | −2.24 |
| Surface area (cm2/g) | 51.88 ± 5.32 | 85.63 ± 28.22 | 25.1 |
| Average diameter (mm) | 0.44 ± 0.02 | 0.43 ± 0.02 | −4.2 |
| Root volume (cm3/g) | 1.85 ± 0.10 | 1.85 ± 0.17 | 0.24 |
| Forks/g | 6027 ± 739 | 5967 ± 910 | −0.98 |
| Mineral soil | |||
| Root biomass (g/m2) | 319.23 ± 26.31 | 240.51 ± 48.20 | −24.7* |
| Mycorrhizal colonization (%) | 75.3 ± 5.8 | 33.2 ± 6.7 | −55.9* |
| Specific root length (cm/g) | 1240.57 ± 95.98 | 976.58 ± 93.93 | −21.3 |
| Surface area (cm2/g) | 167.13 ± 8.81 | 125.25 ± 8.47 | −25.1* |
| Average diameter (mm) | 0.44 ± 0.02 | 0.43 ± 0.02 | −6.5 |
| Root volume (cm3/g) | 1.84 ± 0.12 | 1.30 ± 0.57 | −29.4* |
| Forks/g | 2598 ± 351 | 2091 ± 183 | −19.5* |
Mean values of all sampling dates are accompanied by standard error.
Significant differences between treatments (p < .05).
2.4 Extracellular enzyme activity
For all soil fractions at each sampling date, we assayed the potential activity of hydrolytic enzymes that release nitrogen (N-Acetylglucosaminidase; NAG), phosphorus (acid phosphatase; AP), and simple carbon (β-glucosidase; BG). Given extracellular enzyme activity is the rate-limiting step of the decomposition of organic matter, we consider these measurements a metric of decomposition (Schimel & Weintraub, 2003). Soil extracellular enzyme activity has been linked to mass-loss rates of wood (Sinsabaugh et al., 1992, 1993), and soil C-mineralization (Brzostek, Greco, Drake, & Finzi, 2013; Hernández & Hobbie, 2010). After thawing, 1 g of each sample was homogenized in 50 mM sodium acetate buffer (pH 5.0). Hydrolytic enzyme activities were determined using a fluorometric microplate assay with methylumbelliferone-linked substrates. In addition, we assayed for activities of liginolytic oxidative enzymes, phenol oxidase and peroxidase, using a colorimetric microplate assay based on the hydrolysis of L-3,4-dihydroxyphenylalanine linked substrates (Saiya-Cork et al., 2002). These assays were performed under saturated substrate conditions wherein the concentration of enzyme limits activity.
We measured ambient proteolytic activity of each soil sample following a protocol adapted from Brzostek and Finzi (2011). Each subsample (2–3 g) received 10 ml of 0.05 M sodium acetate buffer (pH 5.0) and 400 μl toluene to inhibit microbial reuptake of amino acids. Initial samples were immediately terminated by adding 3 ml of trichloroacetic acid. Incubated samples were shaken at room temperature for 4 hr prior to termination. Proteolysis was expressed as the difference between incubated and initial amino acid concentrations. Amino acid concentrations were measured following the fluorometric OPAME method (Jones, Owen, & Farrar, 2002) where fluorescence of each sample was a function of amino acid concentration calculated from a glycine standard curve. In contrast to the other enzyme assays described above, this proteolytic enzyme assay is run at ambient substrate conditions and reflects the limitation of activity by both enzyme and substrate concentrations.
2.5 Bacterial and fungal community composition
Soil microbial DNA was extracted from samples collected in July 2015 using the Zymo soil microbe mini prep kit (Zymo Research Corporation, Irvine, CA, USA) and quantified on a Nanodrop 3300 (Thermo Scientific, Wilmington, DE, USA). Due to logistical constraints, we chose the July sampling date in order to examine microbial community composition at the peak of the growing season. The fungal ITS1 and bacterial 16S loci were chosen for amplification via polymerase chain reaction (PCR) in order to delineate microbial community composition and the corresponding shifts associated with N amendment. All PCR reactions contained 1–5 ng template DNA, 1× Mg-included Taq buffer, 0.8 mM dNTPs, 0.05 U of Taq polymerase (Denville Scientific, South Plainfield, NJ, USA), and 0.4 pM each of either ITS1-F_KYO1 (Toju, Tanabe, Yamamoto, & Sato, 2012) and ITS2 primers (Gardes & Bruns, 1993; White, Bruns, Lee, & Taylor, 1990) for fungal amplification, or S-D-Bact-0341-b-D-17 and S-D-Bact-0785-a-A-21 (Klindworth et al., 2013) for bacterial analysis. Primers were modified to include 197 unique 8 bp index sequences (for individual sample “bar-coding”) in addition to Illumina adapters and Nextera sequencing motifs (sequences available upon request). Reaction conditions were as follows: 95°C denaturation for 3 min, 30 cycles of 98°C for 20 s, 61°C for 30 s, 72°C for 30 s, followed by a final extension at 72°C for 5 min. All amplification products were resolved on a 1% agarose gel, extracted with a sterile razor, and purified using the PureLink Quick Gel Extraction Kit (Invitrogen, Carlsbad, CA, USA). Purified products were quantified via Qubit, pooled in equimolar amounts, and sequenced in concert on the Illumina MiSeq at the West Virginia University Genomics Core Facility (Morgantown, WV).
Paired end sequences were merged and barcodes and primers trimmed using the Quantitative insights into Microbial Ecology (QIIM) pipeline, version 1.9.0 (Caporaso et al., 2010). Using a Phred score cut-off of 30, reads with less than 75% high quality base calls were excluded from further analysis. We also discarded sequences with three or more adjacent low-quality base calls. Sequences were clustered into operational taxonomic units (OTUs) using the USEARCH algorithm with an open reference OTU picking strategy using a 97% sequence similarity threshold (Edgar, 2010). Bacterial 16S sequences were clustered against the Greengenes reference database (DeSantis et al., 2006), and fungal sequences were clustered against the UNITE ITS database (Kõljalg et al., 2013). Taxonomy was assigned using the RDP × 2.2 classifier (Wang, Garrity, Tiedje, & Cole, 2007) and associated Greengenes and UNITE reference databases. To determine the effect of N fertilization on overall species composition, the top four abundant fungal phyla (Ascomycota, Basidiomycota, Chytridiomycota, and Zygotmycota as well as ‘unclassified’) were examined for shifts in relative abundance.
To further analyze fungal and bacterial community composition, each OTU table was proportionally normalized, such that the per OTU sequence count within a sample was divided by the total number of sequences detected in a given sample generating a proportion on the interval [0,1]. This proportional normalization has been demonstrated to be sufficiently accurate when using the Bray–Curtis similarity metric (Weiss et al., 2017). We then used normalized OTU tables to calculate Bray–Curtis similarity using the vegdist function within the vegan package for R statistical software (Oksanen et al., 2015; R Core Team, 2017). Community similarity was analyzed as a function of watershed, soil horizon, and their interaction using the adonis function within the vegan package. If interactions were not significant then they were removed and we only analyzed main effects.
To understand how microbial community composition related to variation in soil enzyme profiles, we created similarity matrices of soil enzymes. We calculated Bray–Curtis similarity of enzyme profiles using all enzymes using the vegdist function in vegan. We then performed nonmetric multidimensional scaling (NMDS) to generate NMDS scores for both microbial and enzymatic profiles. We tested for relationships between microbial communities and enzymatic profiles in bulk and rhizosphere soil separately by comparing the first NMDS axes of microbial and enzymatic profiles using linear regression in SAS JMP vs. 10.0.2 (SAS Institute, Cary, NC, USA). We further examined the relationship between microbial communities and enzymatic profiles across all soil horizons by comparing the first NMDS axes of microbial and enzymatic profiles using linear regression across both watersheds.
To assess the relative importance of belowground C allocation (i.e., % AM colonization) on fungal and bacterial community composition we explored whether there were correlations between % AM colonization and the first NMDS axis of bacterial and fungal community composition in bulk and rhizosphere soil separately using SAS JMP vs. 10.0.2 (SAS Institute, Cary, NC, USA).
2.6 Net mineralization and nitrification
For all soil fractions at each sampling date, we measured the rate of net N mineralization and net nitrification within 48 hours of collection. Net N mineralization and nitrification rates were expressed as the difference in the 1 M KCl extractable pool sizes of and between an initial sample and a sample that was aerobically incubated in the lab for 14 days at room temperature and under constant soil moisture (Finzi, Van Breemen, & Canham, 1998). To perform the extractions, 50 ml of 1 M KCl was added to a jar containing 5 g of each soil sample. Samples were sealed and shaken for 30 min and allowed to settle for 2 hr. Samples were filtered using Pall Supor® 450 filters via syringe into 20-ml vials and stored at −20°C prior to analysis by flow injection (Lachat QuikChem® 8500 series 2).
2.7 Statistical analysis
To determine the extent to which N fertilization altered enzyme activities, N mineralization, nitrification, and root metrics, we performed a three-way analysis of variance (ANOVA) in SAS JMP vs. 10.0.2 (SAS Institute, Cary, NC, USA) and used date, watershed, and soil horizon as factors. Although we included date in our statistical models, we present growing season means in the results reflecting our focus on treatment rather than seasonal effects. Post hoc multiple comparisons were also made among watersheds, dates, and soil fractions using the Tukey–Kramer HSD test.
2.8 Modeling potential impacts of plant–microbial linkages on soil decomposition
To further examine the extent to which linkages between plants and soil microbes drive soil enzyme responses to N fertilization as well as to estimate potential impacts on soil C stocks, we used a microbial enzyme decomposition model (Allison, Wallenstein, & Bradford, 2010) that was modified in Cheng et al. (2014) to include seasonal differences in soil temperature, the inputs of root and leaf litter, and root transfers of C to the rhizosphere. For leaf and root litter inputs, we used the mean observed litterfall rate from 1998 to 2015 for each watershed (Kochenderfer, 2006; Peterjohn et al., 2017) and our own measurements of root biomass with the assumption that roots in both watersheds turnover once each year (McCormack, Adams, Smithwick, & Eissenstat, 2017). For both roots and leaves, we assumed that turnover primarily occurred during the leaf senescence in the fall. It is important to note there are no significant differences in leaf litterfall rates between the two watersheds (WS3 = 143 g C/m2; WS7 = 146 g C/m2; Kochenderfer, 2006). Finally, we used published values of AM root exudation rates on a per g root basis (Yin, Wheeler, & Phillips, 2014) to estimate root C transfers, assuming that these inputs occurred only during the growing season. Using these model inputs, we then ran two scenarios: (1) the fertilized and control watersheds had similar root C exudation rates per g of root, and (2) N fertilization reduced exudations rates by 25%. We chose this reduction in exudation rates to be conservative as it represents around half of the decline in AM colonization we observed in the fertilized watershed. All model simulations were run for 30 year to achieve steady state.
3 RESULTS
3.1 Fine root metrics
Fine root biomass and the extent of mycorrhizal symbiosis in the bulk mineral soil layer were significantly lower in the N-fertilized watershed (Table 1). Overall, the bulk mineral soil in the N-fertilized watershed had 24.7% less standing fine root biomass than the reference watershed. This reduction in fine root biomass was accompanied by decreases in fine root surface area, volume, and the amount of root forks per g. Additionally, N fertilization reduced AM colonization of fine roots by 55.9%. Given the inherent limitations of measuring root production using minirhizotron tubes or ingrowth cores, we use fine root biomass and AM colonization as proxies for plant C allocation to gain nutrients. While root turnover may be enhanced under N fertilization, we base the assumption that fine root biomass is synonymous with belowground C allocation to fine roots on our observations that fine root biomass was consistently lower in the fertilized than control watersheds over all three of the sequentially cored soil sampling dates during the growing season. In addition, AM fungi have been shown to act as conduits for photosynthate C to rhizosphere microbes and as such, represent an important belowground flux of C (Kaiser et al., 2015). Thus, reduced investment in AM colonization may also lead to reduced transfers of photosynthate C to the rhizosphere resulting in an overall decline in belowground C allocation.
By contrast, in the OH, standing fine root biomass was larger in the N-fertilized watershed although root biomass in the OH horizon is markedly lower than in the bulk mineral soil (Table 1). However, like the bulk mineral soil, AM colonization of OH roots was significantly higher in the reference watershed (72.8% colonized) than the N-fertilized watershed (60.9% colonized).
3.2 Extracellular enzyme activity
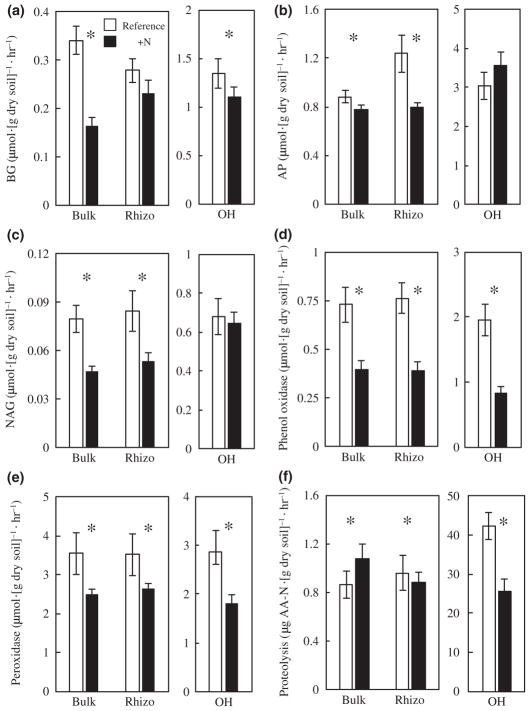
Nearly all hydrolytic enzyme activities were lower under elevated N conditions across all three soil fractions. BG activity was significantly lower in the N-fertilized watershed in both the bulk mineral (52% decrease) and OH (17% decrease) soil fractions (p < .05, Figure 1a). N fertilization significantly lowered AP activity in the bulk (12%) and rhizosphere (36%) fractions of the mineral soil (p < .05, Figure 1b). Similarly, N amendment significantly lowered NAG activity in both bulk and rhizosphere fractions of the mineral soil by 41 and 37%, respectively (p < .05, Figure 1c). However, BG activity in the rhizosphere of the mineral soil was not significantly different between watersheds. In addition, AP and NAG activities in the OH horizon did not vary by treatment.
FIGURE 1.
N fertilization reduces hydrolytic and oxidative enzyme activities. Values are the overall seasonal mean enzyme activities (mean ± SE) of (a) BG, (b) AP, (c) NAG, (d) phenol oxidase, (e) peroxidase, and (f) proteolysis for each soil fraction (i.e., bulk, rhizosphere, and organic horizon) measured in June, July, and August 2015 across all plots (n = 10 plots per watershed). Asterisks indicate significant differences in enzyme activity between watersheds within soil fractions (p < .05). Note difference in scale between OH vs. rhizosphere and bulk soil fractions. AA-N is amino acid nitrogen
Liginolytic oxidative enzyme activities were also generally lower in the N-fertilized watershed. Phenol oxidase activity was significantly lower in bulk (45% decrease), rhizosphere (49% decrease), and OH horizons (57% decrease) of the fertilized watershed relative to the reference watershed (p < .05 Figure 1d). Peroxidase activity was lower in the fertilized watershed in the bulk, rhizosphere, and OH soil fractions with reductions of 30, 25, and 36%, respectively (p < .05 Figure 1e). Proteolytic enzyme activity was consistently lower in each soil fraction under N fertilization, with a 48% decrease in bulk, 56% decrease in rhizosphere, and 40% decrease in OH proteolysis (Figure 1f).
3.3 Microbial community composition
When fungal taxonomic units were aggregated to the phylum level, there were no significant changes in the relative abundance of the four most common fungal phyla or the unclassified group in any soil horizon (Figure S1). When bacterial taxonomic units were aggregated to the phylum level, there were limited shifts in the relative abundance of the seven most common bacterial phyla. Relative abundance of Actinobacteria was higher in the OH soil of the fertilized watershed (Figure S2a). Relative abundance of Proteobacteria was lower in the fertilized bulk soil and relative abundance of Firmicutes was higher in both rhizosphere and bulk soil in the fertilized watershed (Figure S2b,c).
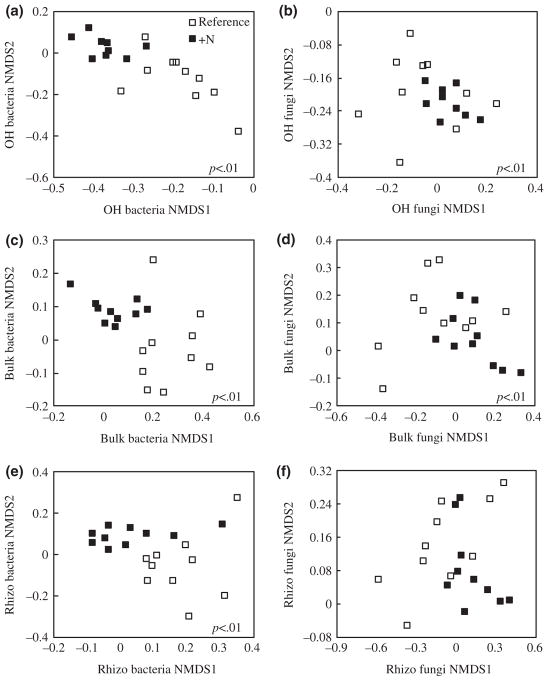
Adonis analysis of bacterial communities revealed significant effects for watershed, soil horizon (p < .001), and their interaction (p = .02, total model R2 = .32). Within watersheds, post hoc comparisons showed OH communities were different than bulk and rhizosphere communities (Figure S3a,b). There was no difference between bulk and rhizosphere communities (Figure S3a,b). Across watersheds, bacterial community composition differed in all soil fractions such that OH, rhizosphere and bulk soil exhibited unique communities in the N-fertilized watershed compared to the reference (Figure 2a,c,e).
FIGURE 2.
N fertilization altered bacterial community composition in OH (a), bulk (c), and rhizosphere (e) soils and fungal community composition in OH (b) and bulk (d) soils, but not rhizosphere (f). All community data were obtained for each soil fraction in July 2015 (n = 10 plots per watershed). p values indicate significant differences between the N-fertilized and reference community
Adonis analysis of fungal communities revealed significant effects of watershed and soil horizon (p < .001, total model R2 = .12), but not their interaction. Within watersheds, post hoc comparisons of fungal communities showed OH communities were different than bulk and rhizosphere communities in both watersheds, but there was no difference between bulk and rhizosphere communities (Figure S3c,d). Across watersheds, fungal community composition within OH and bulk soil fractions was significantly different (p < .01; Figure 2b,d). However, there was no significant difference in fungal communities between watersheds in rhizosphere soil (Figure 2f).
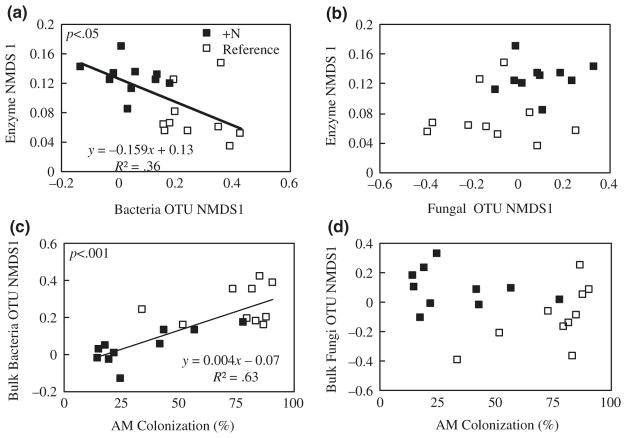
Comparison of bacterial and enzymatic NMDS scores across all soil horizons showed a positive relationship between bacterial communities and enzyme profiles (R2 = .48, p < .01; Figure S4a). Comparison of fungal and enzymatic NMDS scores across all soil horizons and both watersheds showed no significant relationship between fungal communities and enzyme profiles (Figure S4b). Across watersheds, comparison of bacterial and enzymatic NMDS scores showed a negative relationship between bacterial communities and enzyme profiles in the bulk soil (R2 = .36, p < .05; Figure 3a), but no significant relationship in rhizosphere soil. Comparison of fungal and enzymatic NMDS scores across watersheds showed no significant relationships in either bulk or rhizosphere soil (Figure 3b). Linear regression of % AM colonization and the first bacterial NMDS axis resulted in a positive linear relationship in bulk soil (R2 = 0.63, p < .001; Figure 3c), but not rhizosphere soil. There were no significant linear relationships between % AM colonization and the first fungal NMDS axis in either soil fraction (Figure 3d).
FIGURE 3.
Bacterial, but not fungal community composition is correlated with the first NMDS axis of enzyme activity (a & b). Percent AM colonization is correlated with bacterial community composition (c), but not fungal community composition (d). Data presented are bulk soil community composition data from July 2015 (n = 10 plots per watershed). p values indicate significance of correlation
3.4 Net N mineralization and nitrification
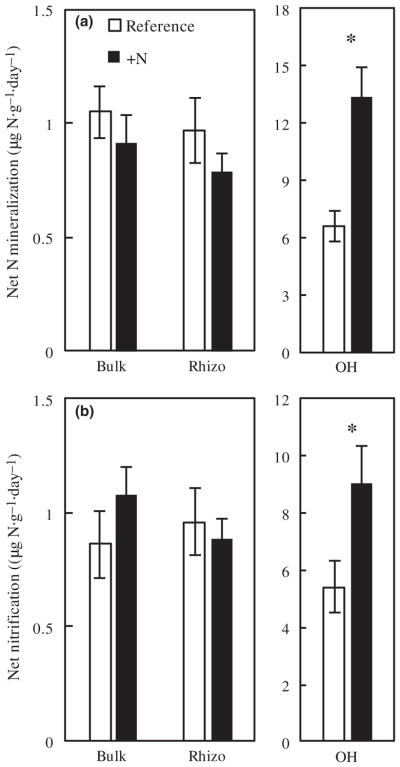
The only significant difference we found in rates of nitrogen cycling was that N mineralization and net nitrification in the OH were 40% and 51% higher in the fertilized watershed than the reference. N transformation rates did not significantly differ between watersheds in either the bulk or rhizosphere soil fractions (Figure 4a,b).
FIGURE 4.
N fertilization increases (a) N mineralization and (b) nitrification in the OH but not the mineral soil. Values are the overall watershed-level mean rates (mean ± SE) measured in June, July, and August 2015 for each soil fraction (n = 10 plots per watershed). Asterisks indicate significant differences between treatments (p < .05)
3.5 Modeling potential impacts of plant–microbial linkages on soil decomposition
When we ran the model to steady state, we found that reductions in root biomass and root C transfers to the rhizosphere in the N-fertilized watershed reduced microbial enzyme pools by ~16% and enhanced soil C by ~3% compared to the reference watershed (Table 2). Even though exudation rates on a per g root basis were the same, the fertilized watershed had lower overall exudations rates at the ecosystem scale than the control watershed because of its lower root biomass (Table 2). When we reduced exudation rates by 25% on a per g root basis in the fertilized watershed, there was a further exacerbation in the reduction in microbial enzyme activity to a ~28% decline and a larger increase in soil C by ~20% (Table 2).
TABLE 2.
At model steady state, fertilization reduced microbial enzyme pools by ~16% and enhanced soil C by ~3%
| Watershed | Exudation | Enzyme pool (mg/cm3) | Soil C pool (mg/cm3) |
|---|---|---|---|
| Control | No change | 0.0136 | 184.73 |
| Fertilized | No change | 0.0114 | 190.92 |
| 25% reduction | 0.0097 | 221.16 |
When root C exudation was reduced 25% on a per g root basis in the fertilized watershed, there was a further exacerbation in the reduction in microbial enzyme activity to a ~28% decline and a larger increase in soil C by ~20%.
4 DISCUSSION
Here we provide evidence that coupled interactions among plants, fungi, and bacteria play an important role in enzyme activity responses to N fertilization. For fungi, we observed distinct shifts in fungal community structure in response to N fertilization (Figure 2b, d), but we found no evidence for a link between these shifts and extracellular enzyme activity (Figure 3b). By contrast, we found that bacterial community composition shifts under N fertilization are correlated with declines in enzyme activity (Figure 3a) and that these compositional shifts are tightly coupled to reductions in plant C allocation to roots and mycorrhizal fungi (Figure 3c). Overall, these results suggest that N fertilization drives an integrated ecosystem response whereby reductions in plant C allocation to roots and AM fungi feedback on bacterial community structure and function.
While whole-watershed fertilization at the Fernow results in a pseudoreplicated experimental design (Hurlbert, 1984), we conclude that the effects we measured are driven by N fertilization, rather than pre-existing differences between these adjacent watersheds, for four main reasons. First, soil chemistry (i.e., soil pH, cation exchange capacity, nutrient content, etc.) were similar at the beginning of the experiment (Adams & Angradi, 1996). Second, the amount of N added to the watershed yearly was originally chosen in 1989 to approximately double ambient N deposition rates, but is now more than quadruple current rates and as such, it seems unlikely that this does not incur a biogeochemical response. Third, the results from this watershed study are consistent with measurements we made during the same year in a replicated N fertilization study <2 km away from these watersheds (the Fork Mountain Long-Term Soil Productivity Plots; Adams, Burger, Zelazny, & Baumgras, 2004). In this small-scale replicated study, N fertilization reduced fine root biomass and ligninolytic enzyme activity (Figure S5). Finally, this work builds upon other research at the Fernow that has found the N-fertilized watershed had lower rates of litter decomposition, (Adams & Angradi, 1996), reduced understory richness (Gilliam et al., 1994, 2016; Walter, Adams, Gilliam, & Peterjohn, 2017), and altered N cycling (Adams et al., 1993; Burnham, Cumming, Adams, & Peter-john, 2017; Gilliam, Yurish, & Adams, 2001; Gilliam et al., 1996, 2016) compared to the reference watershed.
While we did observe significant declines in enzyme activity across all three soil fractions, particularly for the liginolytic enzymes (Figure 1a–f), these declines were not correlated with significant shifts in fungal community composition (Figure 3b). The lack of a clear link between enzyme declines and changes in the fungal community does not support the prevailing paradigm that white-rot Basidiomycota, are the dominant cause of a decline in enzyme activities following N additions (Edwards, Zak, Kellner, Eisenlord, & Pregitzer, 2011; Fog, 1988; Freedman, Romanowicz, Upchurch, & Zak, 2015; Morrison et al., 2016). It is possible that fungal enzyme activity response to N fertilization is independent of community composition (i.e., investment in enzyme activity declines with no change in community structure); however, microbial community composition has been linked to catabolic functioning and enzyme activities across N gradients and seasons (Fierer et al., 2011; Vorıskova, Brabcova, Cajthaml, & Baldrian, 2014). Furthermore, our data indicate that shifts in bacterial community composition under elevated N are correlated with reduced enzyme activity at the Fernow (Figure 3a). While changes in bacterial community may be influenced by overall fungal community composition, these shifts in bacterial community composition are tightly coupled to %AM colonization (a metric of belowground C allocation) suggesting that plant responses to N fertilization feedback on bacterial community structure and function (Figure 3c). While we cannot rule out that N-induced declines in total fungal biomass led to reductions in liginolytic enzyme activity (DeForest et al., 2004; Frey et al., 2004; Treseder, 2008; Wallenstein et al., 2006), the dominance of AM trees in our plots, whose inorganic nutrient economy is largely driven by bacteria, suggests that free-living fungi may not be an important driver of N deposition responses in AM-dominated systems (Cheeke et al., 2017; Phillips, Brzostek, & Midgley, 2013).
In contrast, it appears that the declines in enzyme activity we observed appear to be the result of a cascade of ecosystem responses affecting both microbial community composition and plant C allocation belowground. The N-induced declines in fine root biomass, AM colonization, and root morphology all indicate that there was a reduction in the investment of C belowground by trees to gain nutrients (Table 1). While previous research has shown that below-ground C allocation is inversely correlated with N availability (Bae, Fahey, Yanai, & Fisk, 2015; Haynes & Gower, 1995; Phillips & Fahey, 2005), the tight linkage between the first bacterial NMDS axis and %AM colonization suggest that N-induced declines in belowground C allocation caused a shift in the bacterial community composition and subsequent reductions in enzyme activity (Figure 3a,c). This result also supports the emerging view that roots are active engineers of microbial activity (Brzostek et al., 2013; Finzi et al., 2015; Yin et al., 2014), and highlights the need to integrate root-microbial interactions into our conceptual and predictive modeling frameworks of how forest ecosystems respond to global change (Johnson, Gehring, & Jansa, 2016; Terrer, Vicca, Hungate, Phillips, & Prentice, 2016).
An additional impact of the N fertilization treatment is that it reduced soil pH (as measured in a 0.01 M calcium chloride buffer) of the upper 5 cm of mineral soil in the fertilized watershed to 3.4 compared to 3.8 in the control watershed (Peterjohn, unpublished data). Soil pH is an important control on microbial community diversity (Fierer et al., 2012; Kaiser et al., 2016; Lauber, Hamady, Knight, & Fierer, 2009), microbial biomass, and enzyme activities (Rousk & Bååth, 2011; Sinsabaugh, 2010). Thus, the decline in soil pH may account for a portion of the enzyme reductions we observed under N fertilization. To address this, we assayed the sensitivity of phenol oxidase and peroxidase enzyme activity in organic horizon, bulk and rhizosphere soils from the control watershed to three different levels of pH that spanned a 1 unit shift (data not shown). Peroxidase activity was insensitive to pH (slope = −0.74, r2 = .05, p > .05). While phenol oxidase activity did significantly increase as pH increased (slope = 0.10, r2 = 0.53, p < .05), this sensitivity would only account for a 10% decline in activity. Given that we observed a 50% decline in phenol oxidase in the treatment watershed coupled with the greater overall importance of peroxidase enzymes in our study (i.e., nearly an order of magnitude higher activity), it appears that pH is an important, but secondary driver of the enzyme activity responses we observed.
Over longer time scales, the reductions in root and microbial activity we observed at the Fernow may have important implications for soil C stocks. In our model simulation, we found that feedbacks between reductions in root biomass and enzyme production have the potential to drive nearly a 3% increase in soil C stocks (Table 2). When these were coupled with a 25% reduction in specific root C exudation rates, soil C in the fertilized watershed increased by nearly 20% over the 30-yr simulation. This model was designed to be theoretical and as such, it is used here to show the sensitivity of the system to the perturbations in plant inputs and is not meant to be quantitatively predictive. However, this simple modeling effort provides compelling evidence that N-induced shifts in root–microbial interactions have the potential to alter not only microbial production of soil enzymes but also the size of the soil C pool. To generate the necessary data for fully integrating root-microbial interactions into more sophisticated ecosystem models, future research should couple direct measurements of belowground C allocation responses to N fertilization (e.g., total belowground C allocation, root exudation) with the resulting impacts on microbial activity.
Given the prevailing paradigm that free-living fungi drive soil enzyme activity and subsequent decomposition responses to N fertilization (Carreiro et al., 2000; Fog, 1988; Waldrop et al., 2004; Zak et al., 2008), our results raise an important question of why shifts in plant C allocation, bacterial community structure and enzyme activities were tightly coupled at the Fernow. The divergent results between the Fernow and other long-term fertilization sites may be the result of differences in ambient N deposition and tree species composition. The Fernow is in a region that has historically received some of the highest levels of N deposition in the United States (Driscoll et al., 2001), which may have enhanced bacterial dominance before treatment began in 1989. By contrast, N fertilization experiments that have linked reductions in soil C decomposition to fungal community composition and activity tend to be located in areas with lower historical N deposition rates (Edwards et al., 2011; Freedman et al., 2015; Frey et al., 2004; Morrison et al., 2016; Wallenstein et al., 2006). Second, the Fernow is predominantly dominated by AM trees; whereas most N fertilization experiments have tended to occur in ecosystems dominated by ECM trees, with ECM-dominated sites comprising ~80% of the studies included in the Janssens et al. (2010) meta-analysis. AM trees show greater growth enhancements with N addition (Thomas et al., 2010), are characterized by lower fungal to bacterial ratios (Cheeke et al., 2017), and are less dependent on plant–microbial interactions to access N than ECM trees (Brzostek, Dragoni, Brown, & Phillips, 2015). As such, in AM-dominated systems like the Fernow, bacteria may outweigh fungi in controlling soil decomposition responses to N fertilization. Additionally, evidence that ECM trees rely more on roots and rhizosphere microbes to access N than AM trees suggests that N fertilization may lead to greater declines in belowground C allocation and enzyme activity than observed at the Fernow (Brzostek et al., 2015; Yin et al., 2014). Moving forward, the equal distribution of ECM and AM trees across the temperate forest landscape highlights a critical need to investigate ECM and AM stand responses to N fertilization within the same ecosystem (Midgley & Phillips, 2014).
Although enzyme declines were observed across all three soil fractions (Figure 1a–f), N cycling shifts were only observed in the OH, where rates were elevated by nearly 50% and 40% for mineralization and nitrification, respectively (Figure 4a,b). OH and mineral soils differ in key biogeochemical traits and this may contribute to these divergent N cycling responses. In temperate forests, OH typically have rapid N cycling rates reflecting greater organic matter content and root densities than mineral soils (Brzostek & Finzi, 2011). Shifts in the ratio of gross immobilization to gross mineralization may also play a role. Net N mineralization rates increased in the OH despite N fertilization induced declines in proteolytic and chitinolytic enzyme activity that produce N monomers for microbial uptake. While we do not have data on gross N cycling, we hypothesize that declines in gross microbial N immobilization due to reduced N demand may have outpaced declines in gross N mineralization in the fertilized watershed. Regardless of the exact mechanism, it is important to put the OH results into context: on a mass per unit area basis, the OH is less important to total ecosystem N cycling than the underlying mineral soil.
Much of the research on soil N fertilization has focused on aboveground responses such as litter input and quality or fungal community shifts to explain widely observed reductions in enzyme activities and subsequent soil decomposition (Edwards et al., 2011; Frey et al., 2004; Morrison et al., 2016; Sinsabaugh et al., 2008; Waldrop et al., 2004; Wallenstein et al., 2006). Our results provide evidence that enzyme activity declines under long-term N fertilization at the Fernow are driven by a cascade of ecosystem responses whereby reductions in belowground plant C investment lead to shifts in bacterial community structure and a decline in their ability to degrade soil organic matter. Thus, there is a need to integrate plant–microbial interactions into our current conceptual and predictive models of N deposition impacts on temperate forests in order to forecast soil C responses to shifting N deposition regimes.
Supplementary Material
Acknowledgments
Funding information
Division of Graduate Education, Grant/Award Number: DGE-1102689; Division of Environmental Biology, Grant/Award Number: DEB-0417678, DEB-1019522; National Science Foundation Graduate Research Fellowship; Long-Term Research in Environmental Biology (LTREB) program at the National Science Foundation; NOAA Climate and Global Change Postdoctoral Fellowship Program, administered by Cooperative Programs for the Advancement of Earth System Science (CPAESS); University Corporation for Atmospheric Research (UCAR)
We acknowledge Mary Beth Adams, Tom Schuler, and the US Forest Service staff at the Fernow Experimental Forest for logistical assistance and access to the experimental watersheds and the LTSP site. This work was also supported by the National Science Foundation Graduate Research Fellowship to Joseph Carrara under Grant No. DGE-1102689, and by the Long-Term Research in Environmental Biology (LTREB) program at the National Science Foundation (Grant Nos. DEB-0417678 and DEB-1019522) to William Peterjohn. Colin Averill was supported by the NOAA Climate and Global Change Postdoctoral Fellowship Program, administered by Cooperative Programs for the Advancement of Earth System Science (CPAESS), University Corporation for Atmospheric Research (UCAR), Boulder, Colorado, USA. We acknowledge the WVU Genomics Core Facility, Morgantown WV for support provided to help make this publication possible. We also thank Leah Baldinger, Brittany Carver, Mark Burn-ham, Hannah Hedrick, Jennifer Mangano, and Catherine Sesa for assistance in the field and in the laboratory.
Footnotes
ORCID
Joseph E. Carrara http://orcid.org/0000-0003-0597-1175 Colin Averill http://orcid.org/0000-0003-4035-7760
Additional Supporting Information may be found online in the supporting information tab for this article.
References
- Adams MB, Angradi TR. Decomposition and nutrient dynamics of hardwood leaf litter in the Femow Whole-Watershed Acidification Experiment. Forest Ecology and Management. 1996;1127:61–69. https://doi.org/10.1016/0378-1127(95)03695-4. [Google Scholar]
- Adams MB, Burger J, Zelazny L, Baumgras J. Description of the fork mountain long-term soil productivity study: Site characterization. USDA Forest Service General Technical Report NE. 2004;43:323. [Google Scholar]
- Adams MB, Edwards PJ, Wood F, Kochenderfer JN. Artificial watershed acidification on the Fernow Experimental Forest, USA. Journal of Hydrology. 1993;150:505–519. https://doi.org/10.1016/0022-1694(93)90123-Q. [Google Scholar]
- Allison SD, Wallenstein MD, Bradford MA. Soil-carbon response to warming dependent on microbial physiology. Nature Geoscience. 2010;3:336–340. https://doi.org/10.1038/ngeo846. [Google Scholar]
- Bae K, Fahey TJ, Yanai RD, Fisk M. Soil nitrogen availability affects belowground carbon allocation and soil respiration in northern hardwood forests of new hampshire. Ecosystems. 2015;18:1179–1191. https://doi.org/10.1007/s10021-015-9892-7. [Google Scholar]
- Brzostek ER, Dragoni D, Brown ZA, Phillips RP. Mycorrhizal type determines the magnitude and direction of root-induced changes in decomposition in a temperate forest. New Phytologist. 2015;206:1274–1282. doi: 10.1111/nph.13303. https://doi.org/10.1111/nph.13303. [DOI] [PubMed] [Google Scholar]
- Brzostek ER, Finzi AC. Substrate supply, fine roots, and temperature control proteolytic enzyme activity in temperate forest soils. Ecology. 2011;92:892–902. doi: 10.1890/10-1803.1. https://doi.org/10.1890/10-1803.1. [DOI] [PubMed] [Google Scholar]
- Brzostek ER, Fisher JB, Phillips RP. Modeling the carbon cost of plant nitrogen acquisition: Mycorrhizal trade-offs and multipath resistance uptake improve predictions of retranslocation. Journal of Geophysical Research: Biogeosciences. 2014;119:1684–1697. [Google Scholar]
- Brzostek ER, Greco A, Drake JE, Finzi AC. Root carbon inputs to the rhizosphere stimulate extracellular enzyme activity and increase nitrogen availability in temperate forest soils. Biogeochemistry. 2013;115:65–76. https://doi.org/10.1007/s10533-012-9818-9. [Google Scholar]
- Burnham MB, Cumming JR, Adams MB, Peterjohn WT. Soluble soil aluminum alters the relative uptake of mineral nitrogen forms by six mature temperate broadleaf tree species: possible implications for watershed nitrate retention. Oecologia. 2017;185:1–11. doi: 10.1007/s00442-017-3955-8. [DOI] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, … Huttley GA. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. https://doi.org/10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carreiro MM, Sinsabaugh RL, Repert DA, Parkhurst DF. Microbial enzyme shifts explain litter decay responses to simulated nitrogen deposition. Ecology. 2000;81:2359–2365. https://doi.org/10.1890/0012-9658(2000)081[2359:MESELD]2.0.CO;2. [Google Scholar]
- Cheeke TE, Phillips RP, Brzostek ER, Rosling A, Bever JD, Fransson P. Dominant mycorrhizal association of trees alters carbon and nutrient cycling by selecting for microbial groups with distinct enzyme function. New Phytologist. 2017;214:432–442. doi: 10.1111/nph.14343. https://doi.org/10.1111/nph.14343. [DOI] [PubMed] [Google Scholar]
- Cheng W, Parton WJ, Gonzalez-Meler MA, Phillips R, Asao S, McNickle GG, … Jastrow JD. Synthesis and modeling perspectives of rhizosphere priming. New Phytologist. 2014;201:31–44. doi: 10.1111/nph.12440. https://doi.org/10.1111/nph.12440. [DOI] [PubMed] [Google Scholar]
- Comas LH, Callahan HS, Midford PE. Patterns in root traits of woody species hosting arbuscular and ectomycorrhizas: Implications for the evolution of belowground strategies. Ecology and Evolution. 2014;4:2979–2990. doi: 10.1002/ece3.1147. https://doi.org/10.1002/ece3.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta R, Kelkar A, Baraniya D, Molaei A, Moulick A, Meena RS, Formanek P. Enzymatic degradation of lignin in soil: A review. Sustainability. 2017;9:1163. https://doi.org/10.3390/su9071163. [Google Scholar]
- De Gonzalo G, Colpa DI, Habib MHM, Fraaije MW. Bacterial enzymes involved in lignin degradation. Journal of Biotechnology. 2016;236:110–119. doi: 10.1016/j.jbiotec.2016.08.011. https://doi.org/10.1016/j.jbiotec.2016.08.011. [DOI] [PubMed] [Google Scholar]
- DeForest JL, Zak DR, Pregitzer KS, Burton AJ. Atmospheric nitrate deposition, microbial community composition, and enzyme activity in northern hardwood forests. Soil Science Society of America Journal. 2004;68:132–138. https://doi.org/10.2136/sssaj2004.1320. [Google Scholar]
- DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, … Andersen GL. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Applied and Environmental Microbiology. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. https://doi.org/10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dix NJ, Webster J. Colonization and decay of wood. In: Dix NJ, editor. Fungal ecology. Dordrecht, The Netherlands: Springer; 1995. pp. 145–171. https://doi.org/10.1007/978-94-011-0693-1. [Google Scholar]
- Driscoll CT, Lawrence GB, Bulger AJ, Butler TJ, Cronan CS, Eagar C, … Weathers KC. Acidic deposition in the Northeastern United States : Sources and inputs, ecosystem effects, and management strategies. AIBS Bulletin. 2001;51:180–198. [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. https://doi.org/10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- Edwards IP, Zak DR, Kellner H, Eisenlord SD, Pregitzer KS. Simulated atmospheric N deposition alters fungal community composition and suppresses ligninolytic gene expression in a Northern Hardwood forest. PLoS One. 2011;6:e20421. doi: 10.1371/journal.pone.0020421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Lauber CL, Ramirez KS, Zaneveld J, Bradford MA, Knight R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. The ISME Journal. 2011;6:1007–1017. doi: 10.1038/ismej.2011.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Leff JW, Adams BJ, Nielsen UN, Bates ST, Lauber CL, … Caporaso JG. Cross-biome metagenomic analyses of soil microbial communities and their functional attributes. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:21390–21395. doi: 10.1073/pnas.1215210110. https://doi.org/10.1073/pnas.1215210110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finzi AC, Abramoff RZ, Spiller KS, Brzostek ER, Darby BA, Kramer MA, Phillips RP. Rhizosphere processes are quantitatively important components of terrestrial carbon and nutrient cycles. Global Change Biology. 2015;21:2082–2094. doi: 10.1111/gcb.12816. https://doi.org/10.1111/gcb.12816. [DOI] [PubMed] [Google Scholar]
- Finzi AC, Van Breemen N, Canham CD. Canopy tree soil interactions within temperate forests: Species effects on soil carbon and nitrogen. Ecological Applications. 1998;8:440–446. [Google Scholar]
- Fog K. The effect of added nitrogen on the rate of decomposition of organic matter. Biological Reviews. 1988;63:433–462. https://doi.org/10.1111/j.1469-185X.1988.tb00725.x. [Google Scholar]
- Freedman ZB, Romanowicz KJ, Upchurch RA, Zak DR. Differential responses of total and active soil microbial communities to long-term experimental N deposition. Soil Biology and Biochemistry. 2015;90:275–282. https://doi.org/10.1016/j.soilbio.2015.08.014. [Google Scholar]
- Freedman ZB, Upchurch RA, Zak DR, Cline LC. Anthropogenic N deposition slows decay by favoring bacterial metabolism: Insights from metagenomic analyses. Frontiers in Microbiology. 2016;7:1–11. doi: 10.3389/fmicb.2016.00259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey SD, Knorr M, Parrent JL, Simpson RT. Chronic nitrogen enrichment affects the structure and function of the soil microbial community in temperate hardwood and pine forests. Forest Ecology and Management. 2004;196:159–171. https://doi.org/10.1016/j.foreco.2004.03.018. [Google Scholar]
- Frey SD, Ollinger S, Nadelhoffer K, Bowden R, Brzostek E, Burton A, … Finzi A. Chronic nitrogen additions suppress decomposition and sequester soil carbon in temperate forests. Biogeochemistry. 2014;121:305–316. https://doi.org/10.1007/s10533-014-0004-0. [Google Scholar]
- Gardes M, Bruns TD. ITS primers with enhanced specificity for basidiomycetes - application to the identification of mycorrhizae and rusts. Molecular Ecology. 1993;2:113–118. doi: 10.1111/j.1365-294x.1993.tb00005.x. https://doi.org/10.1111/j.1365-294X.1993.tb00005.x. [DOI] [PubMed] [Google Scholar]
- Gilliam FS, Turrill NL, Aulick SD, Evans DK, Adams MB. Herbaceous layer and soil response to experimental acidification in a central Appalachian hardwood forest. Journal of Environmental Quality. 1994;23:835–844. https://doi.org/10.2134/jeq1994.00472425002300040032x. [Google Scholar]
- Gilliam FS, Welch NT, Phillips AH, Billmyer JH, Peterjohn WT, Fowler ZK, … Adams MB. Twenty-five-year response of the herbaceous layer of a temperate hardwood forest to elevated nitrogen deposition. Ecosphere. 2016;7:1–16. [Google Scholar]
- Gilliam FS, Yurish BM, Adams MB. Ecosystem nutrient responses to chronic nitrogen inputs at Fernow Experimental Forest, West Virginia. Canadian Journal of Forest Research. 1996;26:196–205. https://doi.org/10.1139/x26-023. [Google Scholar]
- Gilliam FS, Yurish BM, Adams MB. Temporal and spatial variation of nitrogen transformations in nitrogen-saturated soils of a central Appalachian hardwood forest. Canadian Journal of Forest Research. 2001;1785:1768–1785. https://doi.org/10.1139/x01-106. [Google Scholar]
- Giovannetti M, Mosse B. An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytologist. 1980;84:489–500. https://doi.org/10.1111/j.1469-8137.1980.tb04556.x. [Google Scholar]
- Hassett JE, Zak DR, Blackwood CB, Pregitzer KS. Are basidiomycete laccase gene abundance and composition related to reduced lignolytic activity under elevated atmospheric NO3- Deposition in a northern hardwood forest? Microbial Ecology. 2009;57:728–739. doi: 10.1007/s00248-008-9440-5. https://doi.org/10.1007/s00248-008-9440-5. [DOI] [PubMed] [Google Scholar]
- Haynes BE, Gower ST. Belowground carbon allocation in unfertilized and fertilized red pine plantations in northern Wisconsin. Tree Physiology. 1995;15:317–325. doi: 10.1093/treephys/15.5.317. https://doi.org/10.1093/treephys/15.5.317. [DOI] [PubMed] [Google Scholar]
- Hernández DL, Hobbie SE. The effects of substrate composition, quantity, and diversity on microbial activity. Plant and Soil. 2010;335:397–411. https://doi.org/10.1007/s11104-010-0428-9. [Google Scholar]
- Hobbie EA. Carbon allocation to ectomycorrhizal fungi correlates with belowground allocation in culture studies. Ecology. 2006;87:563–569. doi: 10.1890/05-0755. https://doi.org/10.1890/05-0755. [DOI] [PubMed] [Google Scholar]
- Hurlbert SH. Psuedoreplication and the design of ecological experiments. Ecological Monographs. 1984;54:187–211. https://doi.org/10.2307/1942661. [Google Scholar]
- Jackson CA, Couger MB, Prabhakaran M, Ramachandriya KD, Canaan P. Isolation and characterization of Rhizobium sp. strain YS-1r that degrades lignin in plant biomass. Journal of Applied Microbiology. 2017;122:940–952. doi: 10.1111/jam.13401. https://doi.org/10.1111/jam.13401. [DOI] [PubMed] [Google Scholar]
- Janssens IA, Dieleman W, Luyssaert S, Subke JA, Reichstein M, Ceulemans R, … Papale D. Reduction of forest soil respiration in response to nitrogen deposition. Nature Geoscience. 2010;3:315–322. https://doi.org/10.1038/ngeo844. [Google Scholar]
- Johnson NC, Gehring C, Jansa J. Mycorrhizal mediation of soil: Fertility, structure, and carbon storage. Cambridge, MA: Elsevier; 2016. [Google Scholar]
- Jones DL, Owen AG, Farrar JF. Simple method to enable the high resolution determination of total free amino acids in soil solutions and soil extracts. Soil Biology and Biochemistry. 2002;34:1893–1902. https://doi.org/10.1016/S0038-0717(02)00203-1. [Google Scholar]
- Kaiser C, Kilburn MR, Clode PL, Fuchslueger L, Koranda M, Cliff JB, … Murphy DV. Exploring the transfer of recent plant photosynthates to soil microbes: Mycorrhizal pathway vs direct root exudation. New Phytologist. 2015;205:1537–1551. doi: 10.1111/nph.13138. https://doi.org/10.1111/nph.13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser K, Wemheuer B, Korolkow V, Wemheuer F, Nacke H, Schoning I, … Daniel R. Driving forces of soil bacterial community structure, diversity, and function in temperate grasslands and forests. Scientific Reports. 2016;6:1–12. doi: 10.1038/srep33696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klindworth A, Pruesse E, Schweer T, Peplies J, Quast C, Horn M, Glockner FO. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Research. 2013;41:1–11. doi: 10.1093/nar/gks808. https://doi.org/10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochenderfer JN. Fernow and the appalachian hardwood region. In: Adams MB, DeWalle DR, Horn JL, editors. The Fernow watershed acidification study. Dordrecht, The Netherlands: Springer; 2006. pp. 17–39. [Google Scholar]
- Kõljalg U, Nilsson RH, Abarenkov K, Tedersoo L, Taylor AF, Bahram M, … Douglas B. Towards a unified paradigm for sequence-based identification of fungi. Molecular Ecology. 2013;22:5271–5277. doi: 10.1111/mec.12481. https://doi.org/10.1111/mec.12481. [DOI] [PubMed] [Google Scholar]
- Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Applied and Environmental Microbiology. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. https://doi.org/10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack ML, Adams TS, Smithwick EA, Eissenstat DM. Variability in root production. Phenology. 2017;95:2224–2235. doi: 10.1890/13-1942.1. [DOI] [PubMed] [Google Scholar]
- McGonigle TP, Miller MH, Evans DG, Fairchild GL, Swan JA. A new method which gives an objective measure of colonization of roots by vesicular - Arbuscular Mycorrhizal Fungi. New Phytologist. 1990;115:495–501. doi: 10.1111/j.1469-8137.1990.tb00476.x. https://doi.org/10.1111/j.1469-8137.1990.tb00476.x. [DOI] [PubMed] [Google Scholar]
- Midgley MG, Phillips RP. Mycorrhizal associations of dominant trees influence nitrate leaching responses to N deposition. Bio-geochemistry. 2014;117:241–253. https://doi.org/10.1007/s10533-013-9931-4. [Google Scholar]
- Morrison EW, Frey SD, Sadowsky JJ, van Diepen LTA, Thomas WK, Pringle A. Chronic nitrogen additions fundamentally restructure the soil fungal community in a temperate forest. Fungal Ecology. 2016;23:48–57. https://doi.org/10.1016/j.funeco.2016.05.011. [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R, Legendre P, O’hara RB, Simpson GL, … Wagner H. vegan: Community Ecology Package 2015 [Google Scholar]
- Paterson E, Gebbing T, Abel C, Sim A, Telfer G. Rhizode-position shapes rhizosphere microbial community structure in organic soil. The New Phytologist. 2007;173:600–610. doi: 10.1111/j.1469-8137.2006.01931.x. [DOI] [PubMed] [Google Scholar]
- Peterjohn WT, Adams MB, Gilliam FS. Symptoms of nitrogen saturation in two central appalachian hardwood forest ecosystems. Biogeochemistry. 2017;35:507–522. [Google Scholar]
- Phillips RP, Brzostek E, Midgley MG. The mycorrhizal-associated nutrient economy: A new framework for predicting carbon-nutrient couplings in temperate forests. New Phytologist. 2013;199:41–51. doi: 10.1111/nph.12221. https://doi.org/10.1111/nph.12221. [DOI] [PubMed] [Google Scholar]
- Phillips RP, Fahey TJ. Patterns of rhizosphere carbon flux in sugar maple (Acer saccharum) and yellow birch (Betula allegheniensis) saplings. Global Change Biology. 2005;11:983–995. https://doi.org/10.1111/j.1365-2486.2005.00959.x. [Google Scholar]
- R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2017. [Google Scholar]
- Reay DS, Dentener FJ, Smith P, Grace J, Feely RA. Global nitrogen deposition and carbon sinks. Nature Geoscience. 2008;1:430–437. https://doi.org/10.1038/ngeo230. [Google Scholar]
- Rousk J, Bääth E. Growth of saprotrophic fungi and bacteria in soil. FEMS Microbiology Ecology. 2011;78:17–30. doi: 10.1111/j.1574-6941.2011.01106.x. https://doi.org/10.1111/j.1574-6941.2011.01106.x. [DOI] [PubMed] [Google Scholar]
- Saiya-Cork KR, Sinsabaugh RL, Zak DR. The effects of long term nitrogen deposition on extracellular enzyme activity in an Acer saccharum forest soil. Soil Biology and Biochemistry. 2002;34:1309–1315. https://doi.org/10.1016/S0038-0717(02)00074-3. [Google Scholar]
- Schimel JP, Weintraub MN. The implications of exoenzyme activity on microbial carbon and nitrogen limitation in soil: A theoretical model. Soil Biology and Biochemistry. 2003;35:549–563. https://doi.org/10.1016/S0038-0717(03)00015-4. [Google Scholar]
- Sinsabaugh RL. Phenol oxidase, peroxidase and organic matter dynamics of soil. Soil Biology and Biochemistry. 2010;42:391–404. https://doi.org/10.1016/j.soilbio.2009.10.014. [Google Scholar]
- Sinsabaugh RL, Antibus RK, Linkins AE, McClaugherty CA, Ray-burn L, Repert D, Weiland T. Wood decomposition over a first-order watershed: Mass loss as a function of lignocellulase activity. Soil Biology and Biochemistry. 1992;24:743–749. https://doi.org/10.1016/0038-0717(92)90248-V. [Google Scholar]
- Sinsabaugh RL, Antibus RK, Linkins AE, McClaugherty CA, Rayburn L, Repert D, Weiland T. Wood decomposition: Nitrogen and phosphorus dynamics in relation to extracellular enzyme activity. Ecology. 1993;74:1586–1593. https://doi.org/10.2307/1940086. [Google Scholar]
- Sinsabaugh RL, Lauber CL, Weintraub MN, Ahmed B, Allison SD, Crenshaw C, Gartner TB. Stoichiometry of soil enzyme activity at global scale. Ecology Letters. 2008;11:1252–1264. doi: 10.1111/j.1461-0248.2008.01245.x. https://doi.org/10.1111/j.1461-0248.2008.01245.x. [DOI] [PubMed] [Google Scholar]
- Terrer C, Vicca S, Hungate BA, Phillips RP, Prentice IC. Mycorrhizal association as a primary control of the CO2 fertilization effect. Science. 2016;353:72–74. doi: 10.1126/science.aaf4610. https://doi.org/10.1126/science.aaf4610. [DOI] [PubMed] [Google Scholar]
- Thomas RQ, Canham CD, Weathers KC, Goodale CL. Increased tree carbon storage in response to nitrogen deposition in the US. Nature Geoscience. 2010;3:13–17. https://doi.org/10.1038/ngeo721. [Google Scholar]
- Toju H, Tanabe AS, Yamamoto S, Sato H. High-coverage ITS primers for the DNA-based identification of ascomycetes and basidiomycetes in environmental samples. PLoS One. 2012;7:e40863. doi: 10.1371/journal.pone.0040863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treseder KK. Nitrogen additions and microbial biomass: A meta-analysis of ecosystem studies. Ecology Letters. 2008;11:1111–1120. doi: 10.1111/j.1461-0248.2008.01230.x. https://doi.org/10.1111/j.1461-0248.2008.01230.x. [DOI] [PubMed] [Google Scholar]
- Vorıskova J, Brabcova V, Cajthaml T, Baldrian P. Seasonal dynamics of fungal communities in a temperate oak forest soil. New Phytologist. 2014;201:269–278. doi: 10.1111/nph.12481. https://doi.org/10.1111/nph.12481. [DOI] [PubMed] [Google Scholar]
- Waldrop MP, Zak DR, Sinsabaugh RL, Gallo M, Lauber C. Nitrogen deposition modifies soil carbon storage through changes in microbial enzymatic activity. Ecological Applications. 2004;14:1172–1177. https://doi.org/10.1890/03-5120. [Google Scholar]
- Wallenstein MD, McNulty S, Fernandez IJ, Boggs J, Schlesinger WH. Nitrogen fertilization decreases forest soil fungal and bacterial biomass in three long-term experiments. Forest Ecology and Management. 2006;222:459–468. https://doi.org/10.1016/j.foreco.2005.11.002. [Google Scholar]
- Walter CA, Adams MB, Gilliam FS, Peterjohn WT. Non-random species loss in a forest herbaceous layer following nitrogen addition. Ecology. 2017;98:2322–2332. doi: 10.1002/ecy.1928. https://doi.org/10.1002/ecy.1928. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Applied and Environmental Microbiology. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. https://doi.org/10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss S, Xu ZZ, Peddada S, Amir A, Bittinger K, Gonzalez A, … Hyde ER. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome. 2017;5:27. doi: 10.1186/s40168-017-0237-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns TD, Lee SB, Taylor JW. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. New York, NY: Academic Press; 1990. pp. 315–322. [Google Scholar]
- Yin H, Li Y, Xiao J, Xu Z, Cheng X, Liu Q. Enhanced root exudation stimulates soil nitrogen transformations in a subalpine coniferous forest under experimental warming. Global Change Biology. 2013;19:2158–2167. doi: 10.1111/gcb.12161. https://doi.org/10.1111/gcb.12161. [DOI] [PubMed] [Google Scholar]
- Yin H, Wheeler E, Phillips RP. Root-induced changes in nutrient cycling in forests depend on exudation rates. Soil Biology and Biochemistry. 2014;78:213–221. https://doi.org/10.1016/j.soilbio.2014.07.022. [Google Scholar]
- Zak DR, Holmes WE, Burton AJ, Pregitzer KS, Talhelm AF. Simulated atmospheric NO 3 – deposition increases organic matter by slowing decomposition. Ecological Applications. 2008;18:2016–2027. doi: 10.1890/07-1743.1. https://doi.org/10.1890/07-1743.1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.