Abstract
Conventional ubiquitination regulates key cellular processes by catalyzing the ATP-dependent formation of an isopeptide bond between ubiquitin (Ub) and primary amines in substrate proteins1. Recently, SidE family of bacterial effector proteins (SdeA, SdeB, SdeC and SidE) of pathogenic Legionella pneumophila were shown to utilize NAD+ to mediate phosphoribosyl-linked ubiquitination (PR-ubiquitination) of serine residues in host proteins2,3. Yet, the molecular architecture of the catalytic platform enabling such a complex multistep process remained unknown. Here, we describe the structure of the catalytic core of SdeA composed of the mono-ADP-ribosyltransferase (mART) and the phosphodiesterase (PDE) domains and shed light on the activity of two distinct catalytic sites for serine ubiquitination. The mART catalytic site is composed of an α-helical lobe (AHL) that together with the mART-core creates a chamber for NAD+ binding and ADP-ribosylation of Ub. The catalytic site in the PDE domain cleaves ADP-ribosylated Ub to phosphoribosyl Ub (PR-Ub) and mediates a two-step PR-Ub transfer reaction: first to a catalytic histidine 277 (forming a transient SdeA:H277-PR-Ub intermediate) and subsequently to a serine residue in host proteins. Structural analysis revealed a substrate binding cleft in the PDE domain juxtaposing the catalytic site that is essential for serine positioning for ubiquitination. Using degenerate substrate peptides and newly identified ubiquitination sites in RTN4B, we show that disordered polypeptides with hydrophobic residues surrounding the target serine residues are preferred substrates for SdeA ubiquitination. Infection studies with L. pneumophila expressing substrate-binding mutants of SdeA revealed that substrate ubiquitination rather than modification of the cellular Ub pool determines the pathophysiological effect of SdeA during acute bacterial infection.
Keywords: Legionella, SdeA, ubiquitin ligase, PDE, mART, phosphoribose, Phosphoribosyl-ubiquitination, Histidine intermediate
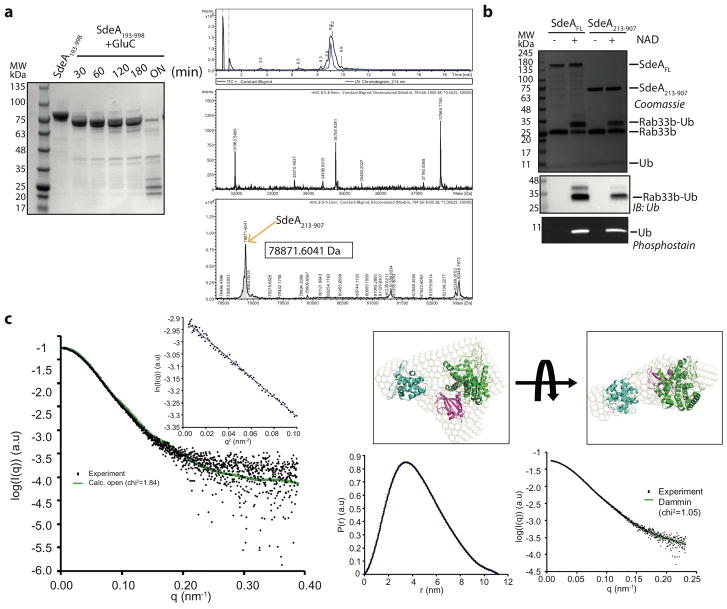
To understand the novel mode of ubiquitination by SdeA, we aimed to obtain structural insights into the function of this enzyme. First, we identified SdeA residues 213 to 907 comprising both PDE and mART domains as the minimal stable fragment that can ubiquitinate the known SdeA substrate Rab33b2,3, albeit less efficiently than full-length SdeA (SdeAFL) (Extended Data Fig. 1a, b). We crystallized SdeA213-907 and determined the structure at 2.8Å (Table S1, supplementary information). In the structure, each asymmetric unit contained one molecule of SdeA213-907 comprising three distinct domains (Fig. 1a). The PDE domain spans residues 222-593, which is α-helical in nature. Structure comparison analysis revealed that the PDE domain of SdeA is most similar to that of the Legionella effector protein lpg1496 (PDB: 5BU2) (r.m.s.d. of 2.3Å over 239 Cα atoms)4. The closest structural mammalian homologue of the SdeA PDE domain is human SAMHD1, a dNTP hydrolase with functions in the innate immune response (r.m.s.d. of 4.1Å over 165 Cα-atoms)5. The mART domain is situated at the C-terminus (residues 594-907) and comprises two distinct and spatially separated lobes, namely the α-helical lobe (residues 594-758, AHL) and the mART-core (residues 759-907). The mART-core interacts strongly with the PDE domain and is composed mostly of β-strands with a couple of α-helices. Surprisingly, in our crystal structure the AHL has no physical proximity to the mART-core unlike in the structures of other bacterial ADP-ribosylating enzymes where it is an integral part of the mART domain and contributes to NAD+ binding and ADP-ribosylation of the substrate6. The solution structure of SdeA213-907 that was determined using small-angle X-ray scattering (SAXS) revealed a similar orientation of AHL in solution as seen in the crystal (Extended Data Fig. 1c, Table S2, Supplementary information). Superimposition of the AHLs of SdeA and Vis toxin, a bacterial ADP-ribosyl transferase from Vibrio splendidus (PDB: 4Y1W), revealed a proximal conformation of the AHL, which differs substantially from that seen in the crystal structure (Fig. 1b). We hypothesize that the AHL of SdeA213-907 could transiently adopt a conformation proximal to the mART-core for NAD+ binding and processing (Fig. 1b). Consistent with this hypothesis, deletion of the AHL (residues 599-758) led to a complete loss of ADP-ribosylation of ubiquitin and ε-NAD+ hydrolysis7 (Fig. 1c, Extended Data Fig. 2a). Mutating residues in the two flexible loops flanking the AHL affected substrate ubiquitination in SdeA213-907 but not in SdeAFL, suggesting that the dynamic conformational shift of AHL only occurs in the context of SdeA213-907, whilst the position of AHL in SdeAFL is fixed to the proximal, active form by the C-terminal region (CTR, residues 909-1499) (Extended Data Fig. 2b,c). Accordingly, SdeAFL exhibited a much greater NAD+ sensitivity in our in vitro ubiquitination experiments, resulting in complete modification of 10 μM Ub with 20μM NAD+, whereas the activity of SdeA213-907 gradually increased proportional to the increase in the NAD+ concentration (Fig. 1d). Similarly, SdeAFL exhibited a marked increase in activity compared to SdeA213-907 with respect to the ε-NAD+ hydrolysis kinetics measured in vitro (Fig. 1e). SdeA213-907 failed to detectably ubiquitinate Rab33b in HEK293T cells perhaps due to insufficient cellular NAD+ concentration (Extended Data Fig. 2d). Moreover, limited proteolysis experiments with SdeA constructs containing different C-terminal extensions revealed that the construct ending at residue 1233 is indigestible while shorter constructs collapse to SdeA213-907, indicating that the CTR induces a compact/closed state of the SdeA structure (Extended Data Fig. 2e). Mixing purified CTR (residues 909-1499) or shorter CTR (residues 909-1233) with SdeA213-907 increased the catalytic activity of SdeA213-907 to the same level as SdeAFL (Fig. 1e). These results are in accordance with the crystal structure of a longer construct of SdeA whereby CTR is stabilizing the proximal orientation of AHL (Yue Feng, accompanying manuscript). Detailed analysis of the structure of the mART domain and its interaction with the PDE domain is presented in the supplemental information (Extended Data Fig. 3–4).
Figure 1. Crystal structure of the SdeA catalytic core.
a) Crystal structure and domain organization of SdeA213-907 stable fragment identified by limited proteolysis. Loops connecting the AHL to PDE and mART domains are partially disordered in the crystal structure and are depicted with dotted lines for clarity. b) Superimposition of mART-core and SdeA AHL in proximal orientation with the mART-core of Vis toxin from Vibrio splendidus (PDB: 4Y1W). Inset shows the NAD+ binding pocket between mART-core and AHL in proximal orientation. c) In vitro Rab33b ubiquitination assays comparing the activity of SdeA213-907 and SdeA213-907ΔAHL. ProQ-Diamond phosphostain was used to monitor PR-Ub. d) NAD+ sensitivity of SdeAFL and SdeA213-907. Abcam Ub antibody was used to monitor the levels of unmodified Ub. e) ε-NAD+ hydrolysis assay with various constructs of SdeA. Experiments were repeated independently three times with similar results (1c–1e). For gel source data, see Supplementary Figure 1.
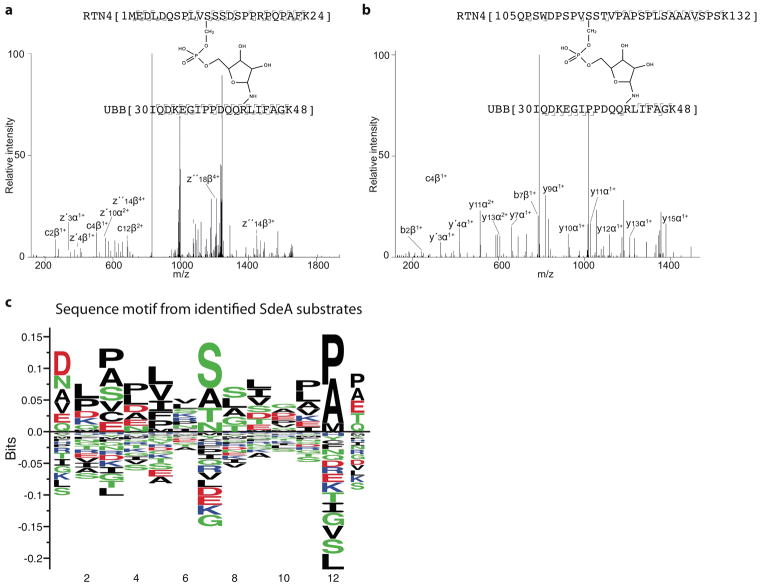
The PDE domain of SdeA hydrolyses ADP-ribosylated Ub (ADPR-Ub) and catalyses transfer of Ub to serine residues of the substrate protein via a phosphoribose linker3. The catalytic pocket of PDE in SdeA is lined by several conserved histidines (Fig. 2a) and mutation of H277 or H407 was previously shown to completely abolish the activity of the PDE domain8. We hypothesized that the PR-ubiquitination reaction by SdeA might take place via a transient intermediate involving covalent attachment of phosphate to a catalytic histidine residue9. Phosphohistidine intermediates are difficult to observe due to their extreme lability10,11. Therefore, we introduced a few key mutations in the catalytic pocket of the PDE domain to stabilize a potential intermediate. Surprisingly, we observed a SdeA-Ub intermediate that is sensitive to heat treatment only in the H407N mutant (Extended Data Fig. 5a, Fig. 2b). Using mass-spectrometry analysis of the tryptic digest of the intermediate reaction, we could identify an ion with the exact mass of Ub 34–48 bridged to SdeA 275-284 by phosphoribose. We applied low energy HCD (High-energy collisional dissociation) fragmentation to specifically cleave the phosphoramidate bond. Indeed, the generated fragment ion corresponded to SdeA 275-284 and phosphoribosylated Ub 34–48 (Fig. 2c). This analysis revealed that H277 of SdeA is linked by phosphoribose to Ub through a phosphoramidate bond. We further validated the identity of the histidine-bridged intermediate by high-energy fragmentation generated ion series of the peptide backbones (Extended Data Fig. 5b). Utilizing rhodamine-labelled Ub and HA-Ub, we observed that both Ub variants were attached to SdeA H407N in a heat-dependent manner (Extended Data Fig. 5c, d). Upon heating, the levels of intermediate decreased with a concomitant increase of PR-Ub, indicating that PR-Ub is indeed attached to a catalytic histidine of SdeA (Extended Data Fig. 5e). Interestingly, residue E340 forms a hydrogen bond with H277 potentially activating this histidine to be a strong nucleophile (Fig. 2a)12. Both double mutants H277A_H407N and E340A_H407N failed to form the intermediate, supporting our notion that H277 is the intermediate forming residue and that E340 plays a critical role in the activation of H277 (Extended Data Fig. 5f). Based on the observed stabilization of the intermediate in H407 mutants, we propose a general role for H407 in orienting and activating a water molecule or the substrate serine for a nucleophilic attack on H277-PR-Ub (Fig. 2d). Accordingly, SdeA213-907 H407N is defective in substrate ubiquitination and only partially active in producing PR-Ub (Extended Data Fig. 6a). Together, we propose a two-step phosphoryl transfer reaction scheme for the PDE-mediated PR-ubiquitination of substrates (Fig. 2d). This model is also supported by the crystal structure of ADPR-Ub in complex with the PDE domain of SdeD (Yuxin Mao, accompanying manuscript). In addition to revealing critical roles for the two histidine residues, the structure of PDE also showed that side chains of Y347 and R413 are inserted into the catalytic centre and could be potentially involved in either the phosphoryl transfer activity or the binding of ADPR-Ub to SdeA (Fig. 2a). Indeed, mutating these residues into alanine inhibited PR-ubiquitination by SdeA (Extended Data Fig. 6b, c).
Figure 2. PDE catalytic mechanism.
a) Active site of SdeA PDE domain depicting residues important for catalysis. Distances between important catalytic residues are indicated in Å. b) In vitro ubiquitination assays with PDE histidine mutations. This experiment was repeated independently three times with similar results. c) Stabilized intermediate was analyzed by targeted LC-MS/MS after tryptic digestion. By low energy HCD, the phosphoramidate bond was targeted specifically for partial fragmentation, creating, besides the intact precursor, marker ions for the tryptic peptides of the active site of SdeA’s PDE domain and PR-Ub. This experiment was repeated independently two times with similar results. d) Proposed catalytic cycle in SdeA PDE domain mediated by E340, H277 and H407. Electron transfer is indicated by curved arrows. Detailed description of the mechanism can be found in supplementary information. For gel source data, see Supplementary Figure 1.
To gain insights into substrate recognition by SdeA, we set out to identify ubiquitination sites within the recently described SdeA substrate RTN4B using a mass-spectrometry based approach3. We discovered two ubiquitination sites in the cytoplasmic part of RTN4B, where each site contained two serine residues that are targeted by SdeA for ubiquitination (Extended Data Fig. 7a,b). RTN4B peptides of ~13 residues containing the target serine residues served as substrates for SdeA (Fig. 3a). Alignment of all peptide sequences containing known SdeA target serines using Seq2logo server13 generated a sequence motif (Fig. 3b, Extended Data Fig. 7c), in which the target serine is in the vicinity of hydrophobic residues and is flanked by proline residues. Furthermore, we produced 47 degenerate peptides based on one of the RTN4B ubiquitination sites and performed ubiquitination assays with all of them (Table S3, supplementary information, Fig. 3c). Individual peptide sequences and their fractional activity was given as input for NNalign server to generate a sequence motif14. The resulting sequence motif confirms the importance of hydrophobic residues surrounding the target serine sites for SdeA ubiquitination (Fig. 3d). Importantly, we have identified an AMP analogue, Adenosine 5′-O-thiomonophosphate (AMPS) that acts as a low affinity inhibitor of substrate ubiquitination by the SdeA PDE domain via affecting substrate positioning (Supplementary information, Extended Data Fig. 8). AMPS also inhibited the ubiquitination activity of SdeC, a paralogue of SdeA (Extended data Fig. 8g) indicating the potential implications of this lead compound in developing novel inhibitors against this class of Legionella toxins.
Figure 3. Substrate recognition by SdeA.
a) In vitro ubiquitination of Rtn4 peptides and Rab33b by SdeAFL. b) Alignment of target serine sequences of SdeA identified thus far. c) In vitro ubiquitination of 47 degenerate peptides designed with Rtn4 substrate peptide as template. d) Sequence motif generated by NNalign software, resulting from analysis of in vitro ubiquitination data of the peptides. Experiments were repeated independently two times with similar results (3a, 3c). For gel source data, see Supplementary Figure 1.
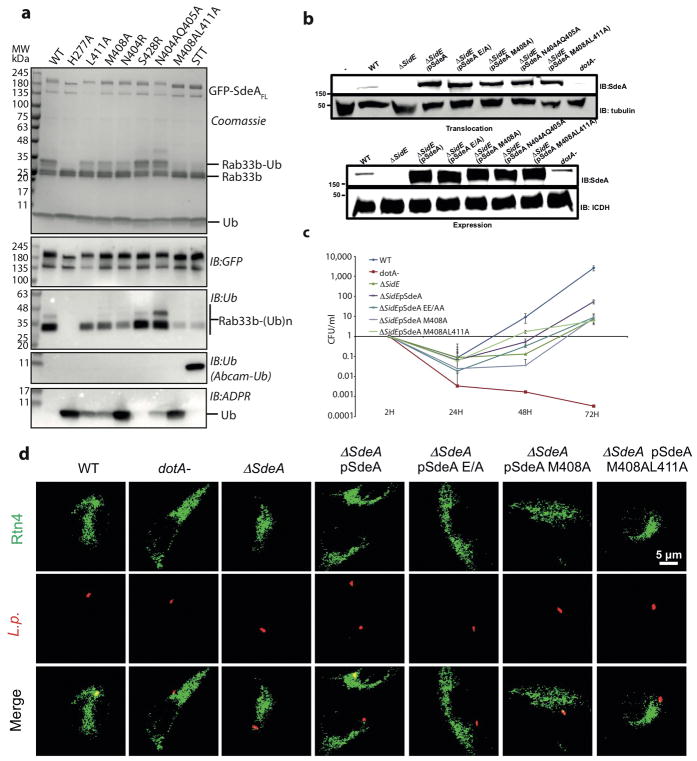
Further inspection of the SdeA PDE domain structure revealed the existence of a cleft on the surface that leads to the active site of the PDE containing the catalytic residue H277 (Fig. 4a). We hypothesized that this cleft could bind and position polypeptides containing substrate serines. We identified several SdeA cleft mutants (Fig. 4b) that are associated with a substantial decrease in substrate ubiquitination, whilst showing negligible effect on phosphoribosylation of Ub (Fig. 4c, Extended Data Fig. 9a). Among the mutants, M408A, L411A and M408A_L411A had the biggest effect on Rab33b ubiquitination in both SdeA213-907 and SdeAFL. These mutations also affected ubiquitination of RTN4B indicating that SdeA recognizes multiple structurally different substrates via this region (Fig. 4d).
Figure 4. Substrate-binding site in SdeA PDE domain.
a) SdeA PDE domain in surface representation with the catalytic site colored in orange. b) Amino acid residues in the PDE active site (orange) and the putative substrate-binding cleft (magenta) are indicated. c) In vitro Rab33b ubiquitination assays with SdeA213-907 substrate-binding mutants. d) In vitro RTN4B ubiquitination assays with SdeA213-907 substrate-binding mutants. e) Fold change in colony forming units in wild type L. pneumophila and the ΔsidEs strain complemented with mutants defective in substrate ubiquitination. SdeA catalytic dead mutant EE/AA (E860_E862A) was used as a control (n= 3 biological replicates). Exact p-values are: (ΔsidEs: pSdeA=0.024), (pSdeA: pSdeA EE/AA=0.032), (pSdeA: pSdeA M408A=0.028), (pSdeA: pSdeA M408AL411A=0.023) as analysed by two-tailed test. f) Percentage of RTN4 positive vacuoles containing relevant L. pneumophila strains (n= 3 biological replicates). Exact p values are (ΔsidEs: pSdeA=0.0015), (pSdeA: pSdeA EE/AA=0.0015), (pSdeA: pSdeA M408A=0.0034), (pSdeA: pSdeA M408AL411A=0.0051) as analysed by two-tailed t-test. Error bars represent s.e.m. Centre is mean. * and ** represents p < 0.05 and p <0.01, respectively (4e, 4f). Experiments shown in panels c and d were repeated independently two times with similar results. For gel source data, see Supplementary Figure 1.
The identification of substrate-binding mutants of SdeA enabled us to investigate which of its two functions: phosphoribosylation of Ub or substrate ubiquitination3 is physiologically relevant. The L. pneumophila ΔsidEs mutant2 was complemented with either SdeAFL wt or various substrate-binding mutants (Extended Data Fig. 9b). Both M408A and M408A_L411A mutants lost the ability to complement the growth of the bacterial strain lacking the sidE effector family in the amoebae host Dictyostelium discoideum (Fig. 4e, Extended Data Fig. 9c) and also failed to restore the ΔsidEs mutant in the recruitment of RTN4 to the Legionella-containing vacuole during infection in primary murine macrophages (Fig. 4f, Extended Data Fig. 9d). Together, our results indicate that targeting of specific substrates for ubiquitination rather than the modification of Ub is the central function of SdeA in acute bacterial pathogenicity.
The SdeA structure and the detailed biochemistry presented here give us a first-hand glimpse into the atomic details of the PR-ubiquitination catalysed by the SidE family of bacterial enzymes and allowed us to pin down substrate ubiquitination as the pathogenic principle of SdeA. PR-ubiquitination by the PDE domain progresses through a transient intermediate in the form of SdeA:H277-PR-Ub, which is subsequently attacked by the OH group of the target serine of the substrate for successful Ub transfer. This suggests a double displacement mechanism for PDE catalysis where the binding of PR-ubiquitination substrates at the PDE active site may first require release of AMP generated during the intermediate formation. This is consistent with the juxtaposed catalytic groove of PDE domain and the substrate binding cleft. Interestingly, the active sites of mART and PDE are facing opposite sides of the molecule (Fig. 1a–b), hinting that there may not be a direct transfer of ADP-ribosylated Ub between the catalytic centres of mART and PDE domain. However, potential dimerization of SdeA as observed with purified proteins in solution (Extended Data Fig. 10) may enable these two catalytic sites to face each other in trans. Sequence analysis of SdeA substrates revealed that the target serine residues could occur in disordered regions in line with the limiting size of the substrate-binding cleft in SdeA PDE domain (Fig. 4b). Based on the substrate recognition motif identified in this study, we propose that SdeA could be a broad specificity ligase targeting disordered serine residues in multiple substrates. Therefore, the specificity of SdeA-mediated ubiquitination during Legionella infection could rather be conferred by its recruitment to the endoplasmic reticulum, where currently identified in vivo substrates of SdeA reside2,8. Importantly, the dissection of PR-ubiquitination catalysis by SdeA presented here may also aid the future discovery of related mammalian enzymes.
Methods
Expression and Purification
Expression and purification of SdeA and Rab33b have been described previously2. Briefly, T7 express cells were transformed with SdeA wildtype (UniProt accession code: Q5ZTK4) and mutant constructs cloned in a modified pET21a vector with a C-terminal CPD (Cysteine Protease Domain)- His tag. Rab33b was cloned into pET21a vector with a C-terminal His tag. For Selenomethionine labeled SdeA213-907, the plasmid was transformed into B834 competent cells and was expressed using Selenomethionine containing minimal media (Molecular Dimensions). T7 express transformed cells were grown in LB media at 37°C to an OD of 0.6–0.8, induced with 0.5 mM IPTG (Isopropyl β-D-1-thiogalactopyranoside), grown overnight at 18°C and harvested. The cell pellet was resuspended in a buffer containing 50 mM Tris-HCl pH 7.5, 300 mM NaCl, 10% (v/v) Glycerol, 1 mM PMSF, Dnase and Protease inhibitor cocktail tablets (Roche). The cells were lysed by sonication. Clarified supernatant (12,000 rpm, 4°C, 40 minutes) was incubated for 1 hour with pre-equilibriated Talon beads before washing three times with 50 mM Tris-HCl pH 7.5, 300 mM NaCl, 10% (v/v) Glycerol. For RTN4B purification, the clarified supernatant was spun again (40,000 rpm, 4°C, 90 min) to collect membrane fractions. The membranes were homogenized and solubilized in the presence of 1% n-Dodecyl β-D-Maltoside (DDM). For the rest of RTN4B purification, 0.05% (w/v) DDM was maintained in the buffer. For Rab33b purification, the protein was eluted using 200 mM Imidazole after binding to Talon beads. The CPD-His tag of SdeA and RTN4B were cleaved off the protein while it was still bound to Talon beads using 100 μM Phytic acid. For biochemical assays, the buffer was exchanged to a final buffer of 10 mM HEPES pH 7.5, 150 mM NaCl and 1 mM TCEP. For crystallization, SdeA213-907 was loaded onto a Q-Sepharose column after CPD tag cleavage. SdeA213-907 eluted in flowthrough, while most of the impurities were bound to the column. The fractions containing the protein was then concentrated before injecting on a Superdex 75 16/60 size exclusion chromatography column pre-equilibrated with 10 mM HEPES pH 7.5, 150 mM NaCl, 1 mM TCEP. The protein eluted in a single peak and fractions were pooled together and concentrated to 25 mg/ml before setting up crystallization screens.
Limited proteolysis
1 mg of SdeA193-998 was incubated with 50 μg of GluC in 20 mM HEPES pH 7.5, 50 mM NaCl and 10 mM MgSO4 for 1 hour on ice. This was followed by size exclusion chromatography of the reaction on a Superdex 75 16/60 column equilibrated with 10 mM HEPES pH 7.5, 150 mM NaCl. The fractions were pooled and protein MALDI mass spectrometry was performed. The protein fragment of interest was then identified based on the exact mass using the Expasy server tool Findpept. Various constructs of SdeA were also similarly proteolyzed by GluC and analyzed by coomassie stained SDS gel.
Crystallization
The purified protein was clarified by centrifugation (10,000 rpm, 10 min, 4 °C) before setting up crystallization plates. Sitting drop and hanging drop sparse matrix screens in 96-well format were set up with 125 nl protein + 125 nl precipitant solution. The protein crystallized in 100 mM Bis Tris Propane pH 7.0–8.0, 0.1–0.2 M sodium citrate tribasic dihydrate, 20–30% (w/v) PEG 3350 at 20°C. The morphology of the thin plate like crystals was improved using 0.5–1% (v/v) ethylene glycol and 50–200 mM Non-detergent sulfobetaine-201 (NDSB-201) as additives. The crystals were cryoprotected using mother liquor supplemented with 15% (v/v) glycerol and 10% (v/v) ethylene glycol and flash frozen in liquid nitrogen. For inhibitor bound structure, the purified protein was incubated with 5 mM 5′-AMPS before setting up crystallization plates. SeMet labelled SdeA crystallized in similar conditions as wildtype SdeA.
Data collection, data processing and structure solution
Both SeMet and native diffraction data were collected at PXIII beamline of the Swiss Light Source, Villigen. Native data were collected at wavelength 1.00003 Å. The data were processed using XDS15. SAD (Single Anomalous Dispersion) data from SeMet crystals were collected at wavelength 0.97927 Å and were processed using XDS and the phases were calculated using Phenix autosol16. Using these initial phases, buccaneer was used to build most of PDE domain and segments of mART and AHL that were well-ordered 17. After manual correction of the output model of buccaneer, MR-SAD was performed using PHENIX autosol to further improve the phases. Further building was done manually in Coot using SeMet positions as a guide18. Using the model obtained from experimental phases, molecular replacement was performed for the native data of SdeA extending to 2.8 Å. Further iterative cycles of manual building and refinement were performed using coot and refinement programs phenix refine and Buster (Global phasing)16,19. Ramachandran statistics for the refined SdeA core structure are favored: 93.6%, allowed: 6.4%, outliers: none.
In vitro ubiquitination assays
SdeA ubiquitination experiments were done as described before3. Briefly, 2.5 μg of purified untagged ubiquitin and 2 μg Rab33b was incubated with 1 μg of SdeA (FL, 213-907 wildtype and variants and SdeA PDE domain) at 37 °C for 30 minutes in the presence or absence of 200 μM NAD+ in a buffer containing 50 mM Tris-HCl pH 7.5, 50 mM NaCl in a final reaction volume of 30 μl. For reactions involving SdeA PDE domain, purified ADP ribosylated ubiquitin (2.5 μg) was used instead of NAD+. ADP ribosylated ubiquitin was generated using SdeA H277A mutant and purified using size exclusion chromatography. The reaction mixture was subjected to SDS-PAGE followed by Coomassie staining. Alternatively, reaction mixture was subjected to SDS-PAGE followed by western blotting using ubiquitin antibody and pan-ADP ribose antibody (Millipore) or to phosphostaining procedure to identify PR-UB (Pro-Q diamond phosphostaining protocol, ThermoFischer). Ubiquitination assays with bacterial lysates were performed in a similar manner except that the clarified bacterial lysate containing over-expressed SdeA213-907 or mutants were used in the reaction instead of purified SdeA proteins. For ubiquitination assays using FL GFP tagged SdeA constructs, we transfected 2 μg of GFP tagged SdeA constructs into HEK293T cells cultured in 6-well plate. Cells were harvested after 24 hours and lysed in 150 μL of lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1% Triton X-100, 10% glycerol, protease inhibitors cocktail and 1mM PMSF). Using 15μL GFP-trap beads we purified the GFP tagged SdeA proteins from the clarified lysate. After extensive washing of the beads, an in vitro ubiquitination reaction was set up in 30 μL reaction buffer (50 mM Tris-HCl, 50 mM NaCl, pH 7.4) with 3 μg purified Rab33b, 2.5 μg ubiquitin and 0.2 μM NAD+. Reaction was performed at 37 °C in a thermomixer while shaking. After 30 mins, the reaction was stopped by adding SDS loading buffer and the samples were analyzed using Coomassie staining of SDS gel and western blotting. The reaction mixture was analyzed similar to the assays using bacterially purified SdeA213-907 except for the use of complimentary ubiquitin antibodies described in our previous paper (CS-Ub and abcam-Ub) to monitor ubiquitin modification3. Where indicated ADPR-Ub was used as co-factor in reactions instead of Ub and NAD. Peptide ubiquitinations were carried out with 0.5mM of each peptide as substrate with SdeAFL. All experiments were repeated at least twice.
ε-NAD+ hydrolysis
For measuring ubiquitin ADP-ribosylation kinetics of SdeA213-907 and mutants, ε-NAD hydrolysis assay was performed. The method has been previously described 7. 5 μg of SdeA wt or mutant protein was incubated in a buffer containing 50 mM Tris pH 7.5, 50 mM NaCl with 100 μg of ubiquitin in a 100 μL reaction buffer. ε-NAD was added to final concentration of 1 mM to start the reaction. Fluorescence of ε-adenine (excitation wavelength: 300 nm, emission wavelength: 410 nm) was monitored using a plate reader at 25 °C at every 1 minute interval. Constructs of C-terminal region (909-1499 and 909-1233) were added in 1.5 molar excess of the SdeA213-907 to test their effect on the activity. All experiments were repeated at least twice.
Histidine intermediate analysis
This analysis was performed as described in Fuhs et al., with slight modifications11. Briefly, 4 μg of SdeA213-907 wt and mutant proteins are incubated with 10 μg Ub or labeled Ub and 2 mM NAD+ for 5 minutes at 37 °C. Reaction was stopped by transferring the contents onto ice and adding 5X SDS loading buffer (pH 8.8). Gel electrophoresis and transfer were conducted at 4 °C and analyzed by either Coomassie staining or fluorescence scanning or western blotting. All experiments were repeated at least twice.
Small-angle X-ray scattering
SdeA213-907 was purified in 10 mM HEPES pH 7.5, 150 mM NaCl, 1mM TCEP by size exclusion chromatography (Superdex 200 increase). Each fraction was collected and concentrated with centrifugal concentration devices (50 kDa cut-off, Table S2, supplementary information). Scattering profile of flow through buffer from size-exclusion column was recorded as reference buffer scattering. SAXS data were collected at beamline P12, EMBL-DESY (Table S2, supplementary information). Primary data analysis was performed by PRIMUS from ATSAS package24. Bead modeling was conducted with DAMMIF25 and DAMMIN26 ATSAS package. SAXS curves from atomic model were generated and fitted to experimental data by CRYSOL27 and SUPCOMB28 from ATSAS 2.8.3 package.
Mass-spectrometry
For analysis of the Histidine intermediate, an in vitro ubiquitination reaction of SdeA213-907 H407N was stopped after 5 minutes by denaturation on ice in 5.3 M Urea pH 8.8 for 10 minutes. The sample was diluted to 2 M Urea with 50 mM ABC pH 8.8 and was loaded on a 30 kDa filter (Amicon Ultra, 0.5 ml, Merck). (Ub-)SdeA was trypsinised according to an adapted FASP-protocol as described previously3. In brief, the proteins were washed four times with 100 μl ABC and after adding Trypsin Gold (Promega) in an enzyme to protein ratio of 1:2 tryptic digestion was performed for 20 minutes at 22 °C. Tryptic peptides in 50 mM Ammonium bicarbonate pH 8.8 were loaded onto a 15 cm self-packed C18 column and separated with a short gradient (12 minutes from 10–38% Buffer B (80% Acetonitrile, 0.1% Formic Acid)) on an easy nLC2 system and injected online in Q Exactive HF mass-spectrometer. Targeted MS2 scans were used to specifically fragment the bridged intermediate with different collision energies. For partial and specific fragmentation of the phosphoramidate bond a normalized collision energy (NCE) of 15 was applied and for fragmentation of the peptide backbone NCE 30 was used. Spectra were annotated manually and StavroX 3.6 was used for additional identification of the peptide backbone fragments20. Identification of Phosphoribose bridged Ubiquitination sites in RTN4 was done as described before by HCD and targeted ETD fragmentation after a modified FASP digest3.
L. pneumophila strains and host infections
L. pneumophila strains used in this study were derivatives of the Philadelphia 1 strain Lp021 and were grown and maintained on CYE (charcoal-yeast extract) plates or in N-(2-Acetamido)-2-aminoethane (ACES) buffered yeast extract (AYE) broth as previously described21. The sidE family in-frame deletion strain and complementation strains were described previously2. sdeAM408A and sdeAM408A_L411A mutant genes were cloned into pZL5073 for complementation. Raw264.7 cells were cultured in RPMI 1640 medium supplemented with 10% FBS. Dictyostelium discoideum AX4 cells were cultured in HL-5 medium, cells were maintained in MB medium for infection as described2. Infection experiments were performed in triplicates.
Infection
L. pneumophila strains were grown to the post-exponential phase (OD600=3.0–3.6) in AYE broth. Complementation strains were induced with 0.2 mM IPTG for 4 hours at 37°C before infection. Raw 264.7 cells were infected with L. pneumophila strains at an MOI of 10 for 2 h to detect the translocation of SdeA and its mutants. Raw cells were collected and lysed with 0.2% Saponin on ice for 30 minutes. Cleared cell lysates were resolved by SDS-PAGE and followed by immunoblotting with antibodies specific for SdeA and tubulin, respectively. Total L. pneumophila proteins were resolved by SDS-PAGE to evaluate the expression of SdeA by immunoblotting with SdeA-specific antibodies, and isocitrate dehydrogenase (ICDH) was probed as loading control with antibodies previously described22. For intracellular growth in Dictyostelium discoideum, infection was performed at an MOI of 0.1 and the total bacterial counts were determined at 24 hour intervals as described23. The enrichment of RTN4 by bacterial phagosomes was assessed by immunostaining in primary murine macrophages infected with the relevant L. pnuemophila strains for 2 hours at an MOI of 1.0. Immunostaining with anti-RTN4 (Lsbio cat#LS-B6516-50) (1:500) was performed as described8. Infection experiments were performed in triplicates.
Antibodies and Immunoblotting
For immunoblotting, samples resolved by SDS-PAGE were transferred onto 0.2 μm nitrocellulose membranes (Pall Life Sciences cat#66485). Membranes were blocked with 5% non-fat milk, incubated with the appropriate primary antibodies: anti-SdeA2, 1:10,000, anti-ICDH3, 1:10,000, anti-tubulin (DSHB, E7) 1:10,000. Membranes were then incubated with an appropriate IRDye infrared secondary antibody (dilution: 1:20,000) and scanned using an Odyssey infrared imaging system (Li-Cor’s Biosciences).
Extended Data
Extended data Figure 1. Catalytic core - SdeA213-907.
a) Limited proteolysis of SdeA fragment 193–998 and subsequent analysis of the fragments by Coomassie stained SDS gel and total mass analysis by mass spectrometry. b) In vitro ubiquitination of Rab33b by SdeAFL and SdeA213-907. c) Scattering profile of SdeA213-907 with calculated scattering curve from crystal structure. Gunier region is shown in inset. (right upper) Ab initio bead model from DAMMIN was superimposed with crystal structure and shown in two orientations. (right lower) Pair distance distribution plot and DAMMIN model fitting results were shown. Experiments were repeated independently two times with similar results (Extended data Figure 1a–1b). For gel source data, see Supplementary Figure 1.
Extended data Figure 2. Role of AHL in SdeA.
a) ε-NAD+ hydrolysis assay in the presence of SdeA (SdeA213-907 and SdeA213-907ΔAHL) and ubiquitin (wildtype or R42A_R72A). b) In vitro ubiquitination assay with mutations in loops connecting AHL to PDE and mART catalytic core in SdeA213-907 and c) in SdeAFL. d) Substrate ubiquitination and ubiquitin modification by SdeAFL and SdeA213-907 in HEK293T cells. Abcam Ub and Cell signaling Ub antibodies were used to monitor the levels of unmodified Ub and total Ub, respectively. e) Limited proteolysis analysis of various SdeA constructs. All experiments were repeated independently two times with similar results. For gel source data, see Supplementary Figure 1.
Extended data Figure 3. Characterization of mART domain.
a) Superimposition of the mART-core of SdeA with that of NAD+ bound iota toxin structure (PDB: 4H0Y). Residues in SdeA that are predicted to be important for NAD+ binding and hydrolysis are labeled b) In vitro ubiquitination assay with NAD+ binding site mutants in the mART-core of SdeA213-907. c) Residues at the interface between mART-core and AHL in proximal conformation (AHLprox). d) In vitro ubiquitination assays with the mutants of SdeA213-907 mART-core-AHLprox interface residues indicated in 3c. e) Comparison of ε-NAD+ hydrolysis by SdeA213-907 and NAD+ binding site mutants and mutants disrupting the predicted mART-core-AHLprox interaction. f) In vitro ubiquitination assays with the mutants of SdeAFL mART-core-AHLprox interface residues indicated in 3c. Experiments were repeated independently two times with similar results. For gel source data, see Supplementary Figure 1.
Extended data Figure 4. Interaction between PDE and mART-core.
a) Cartoon diagram showing the details of interaction between SdeA PDE and mART core. Important residues mediating the interaction are indicated in inset. b) Testing in vitro substrate ubiquitination and ubiquitin modification by PDE-mART core interaction mutants in SdeA213-907 and c) in SdeAFL. Experiments were repeated two times independently with similar results. For gel source data, see Supplementary Figure 1.
Extended data Figure 5. Histidine intermediate in SdeA catalysis.
a) In vitro ubiquitination reactions by various SdeA213-907 PDE site histidine mutants probed by coomassie stained SDS-PAGE. b) High-energy HCD fragmentation was used to generate fragments of the peptide backbone. We could identify multiple fragments of the SdeA 275–284 as well as of the Ubiquitin 34–48 peptide, to further validate the identity of the bridged active site. c) In vitro ubiquitination reaction by SdeA213-907 H407N mutant using Rhodamine labeled ubiquitin. d) In vitro ubiquitination reaction using HA tagged ubiquitin. e) In vitro ubiquitination reaction by SdeA213-907H407N without and with heating probed by phosphostain. f) In vitro ubiquitination reaction using HA-Ubiquitin by various PDE mutants. These experiments were repeated independently two times with similar results. For gel source data, see Supplementary Figure 1.
Extended data Figure 6. PDE domain catalytic site.
a) In vitro Rab33b ubiquitination by SdeA PDE mutants. b) In vitro ubiquitination assays with PDE catalytic site mutants. c) In vitro Rab33b ubiquitination assays with GFP-SdeAFL PDE catalytic site mutants purified from HEK293T cells. These experiments were repeated independently two times with similar results. For gel source data, see Supplementary Figure 1.
Extended data Figure 7. Substrate specificity of SdeA.
a,b) Fragmentation spectra of the bridged peptide indicating RTN4B ubiquitination sites. These experiments were done once c) Sequence motif of target serine sequences of SdeA as computed by Seq2Logo server.
Extended data Figure 8. Chemical inhibition of SdeA.
a) Chemical structure of adenosine-5′-thio-monophosphate (5′-AMPS). b) 5′-AMPS inhibition of Rab33b PR-ubiquitination by SdeAFL, SdeA213-597 and SdeA213-907 in the presence of ADPR-Ub. c) 5′-AMPS inhibition of RTN4B PR-ubiquitination by SdeAFL d) In vitro ubiquitination by SdeAFL in the presence of increasing concentration of 5′-AMPS and AMP. e) Apparent inhibition constants of AMP and 5′-AMPS against SdeAFL calculated from quantification of substrate ubiquitination (panel-d). f) PDE domain of SdeC ubiquitinates Rab33b and RTN4B. g) Effect of 5′-AMPS on the ubiquitination activity by SdeC PDE. These experiments were done two times independently with similar results. For gel source data, see Supplementary Figure 1.
Extended data Figure 9. Effect of SdeA substrate-binding mutations in vivo.
a) In vitro Rab33b ubiquitination assays with GFP-SdeAFL substrate-binding mutants purified from HEK293T cells. b) Expression and translocation of SdeA using wildtype and various mutant strains of Legionella. c) CFU fold change monitored in wildtype Legionella and ΔsidEs strain complemented with substrate ubiquitination defective mutant plasmids (n=3 biological replicates). Error bars represent s.e.m. Centre is mean. d) Co-localization of Legionella-containing vacuole and RTN4 network in primary murine macrophage cells. These experiments (panels a,b and d) were repeated two times independently with similar results. For gel source data, see Supplementary Figure 1.
Extended data Figure 10. Size-exclusion chromatography profile of SdeAFL.
SdeAFL shows dimeric behavior in Size-exclusion chromatography column (Superdex 200 16/60). This experiment was repeated two times independently with similar results. For the inset n=1. For gel source data, see Supplementary Figure 1.
Supplementary Material
Acknowledgments
We thank Joseph Vogel for the kind gift of anti-SidC serum, Rajan Prabu for help with total mass analysis of proteolysed SdeA, Özkan Yildiz for sharing synchrotron time and Apirat Chaikuad for initial construct design of SdeA. We are grateful to Tony Hunter, Chris Lima for advice on histidine intermediate protocols and Thomas Hanke for the PDE mechanism scheme and discussion. We thank Thomas Colby and Ivan Matic for their help with identifying ubiquitination sites of RTN4B by LC-MS/MS. We also thank Brenda Schulman and Danny Scott for crystallography advices. Swiss Light Source beamtime was part of the proposal 20161958. We thank the staff of SLS for their assistance in data collection as well as Elena Veshkova, Sofia Rodriguez Gomez, Stela Jelenic and Frane Miljkovic for technical assistance. We thank Kerstin Koch, Daniela Höller, Volker Dötsch and Stefan Knapp for critical comments on the paper. We thank Dmitri Svergun’s group at beamline P12, PETRA III, EMBL-DESY for SAXS data collection. This work was supported by the iNEXT (PID:3515). This work was supported by the DFG-funded Collaborative Research Centre on Selective Autophagy (SFB 1177), by the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement No 742720), by the DFG-funded Cluster of Excellence “Macromolecular Complexes” (EXC115), by the DFG-funded SPP 1580 program “Intracellular Compartments as Places of Pathogen-Host-Interactions” (I.D.) and by the LOEWE program Ubiquitin Networks (Ub-Net) and the LOEWE Center for Gene and Cell Therapy Frankfurt (CGT), both funded by the State of Hesse/Germany. NIH-NIAID grant R01AI127465 (ZQL). The work of S.B. is also funded by Goethe University Nachwuchsforscher grant.
Footnotes
Author contributions
S.K, S.B and I.D conceived the project. S.K. performed initial crystallization. S.K. and S.B performed crystal optimization, structure solution, protein purification and biochemistry. J.B. contributed to crystallization and structure solution. F.B. performed mass-spectrometry. D.S. performed SAXS and contributed to protein purification. Y.L. and P.G. performed experiments with SdeAFL. N.G. performed bacterial infection experiments. S.K., S.B, Z.Q.L. and I.D. analysed the data. S.K., S.B., and I.D. wrote the manuscript. I.D. supervised the project.
Structure coordinates are deposited and available from Protein Data Bank under the accession code: 6G0C. Small-angle X-ray scattering data and models will be deposited and available from SASBDB (SASBDB ID:SASDD65).
The authors declare no competing financial interests.
Data availability:
Structure coordinates are deposited and available from Protein Data Bank under the accession code: 6G0C. Small-angle X-ray scattering data and models are available from SASBDB (SASBDB ID:SASDD65). Full gel source data can be found in SI Figure 1.
The data that support the findings of this study are available from the corresponding author upon request.
Ethical Compliance:
Animal protocols used in the study were approved by Purdue Animal Care and Use Committee. We complied with all the relevant ethical regulations.
References
- 1.Hershko A, Ciechanover A, Varshavsky A. Basic Medical Research Award. The ubiquitin system. Nature medicine. 2000;6:1073–1081. doi: 10.1038/80384. [DOI] [PubMed] [Google Scholar]
- 2.Qiu J, et al. Ubiquitination independent of E1 and E2 enzymes by bacterial effectors. Nature. 2016;533:120–124. doi: 10.1038/nature17657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhogaraju S, et al. Phosphoribosylation of Ubiquitin Promotes Serine Ubiquitination and Impairs Conventional Ubiquitination. Cell. 2016;167:1636–1649.e13. doi: 10.1016/j.cell.2016.11.019. [DOI] [PubMed] [Google Scholar]
- 4.Wong K, Kozlov G, Zhang Y, Gehring K. Structure of the LegionellaEffector, lpg1496, Suggests a Role in Nucleotide Metabolism. Journal of Biological Chemistry. 2015;290:24727–24737. doi: 10.1074/jbc.M115.671263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ji X, et al. Mechanism of allosteric activation of SAMHD1 by dGTP. Nat Struct Mol Biol. 2013;20:1304–1309. doi: 10.1038/nsmb.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simon NC, Aktories K, Barbieri JT. Novel bacterial ADP-ribosylating toxins: structure and function. Nature reviews Microbiology. 2014;12:599–611. doi: 10.1038/nrmicro3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrio JR, Secrist JA, Leonard NJ. A Fluorescent Analog of Nicotinamide Adenine Dinucleotide. Proc Natl Acad Sci USA. 1972;69:2039–2042. doi: 10.1073/pnas.69.8.2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kotewicz KM, et al. A Single Legionella Effector Catalyzes a Multistep Ubiquitination Pathway to Rearrange Tubular Endoplasmic Reticulum for Replication. Cell Host & Microbe. 2017;21:169–181. doi: 10.1016/j.chom.2016.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matte A, Tari LW, Delbaere LT. How do kinases transfer phosphoryl groups? Structure. 1998;6:413–419. doi: 10.1016/s0969-2126(98)00043-4. [DOI] [PubMed] [Google Scholar]
- 10.Kee JM, Muir TW. Chasing Phosphohistidine, an Elusive Sibling in the Phosphoamino Acid Family. ACS Publications. 2011;7:44–51. doi: 10.1021/cb200445w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuhs SR, et al. Monoclonal 1- and 3-Phosphohistidine Antibodies: New Tools to Study Histidine Phosphorylation. Cell. 2015;162:198–210. doi: 10.1016/j.cell.2015.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lima CD, Klein MG, Hendrickson WA. Structure-Based Analysis of Catalysis and Substrate Definition in the HIT Protein Family. Science. 1997;278:286–290. doi: 10.1126/science.278.5336.286. [DOI] [PubMed] [Google Scholar]
- 13.Thomsen MCF, Nielsen M. Seq2Logo: a method for construction and visualization of amino acid binding motifs and sequence profiles including sequence weighting, pseudo counts and two-sided representation of amino acid enrichment and depletion. Nucleic Acids Research. 2012;40:W281–W287. doi: 10.1093/nar/gks469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nielsen M, Andreatta M. NNAlign: a platform to construct and evaluate artificial neural network models of receptor–ligand interactions. Nucleic Acids Research. 2017;45:W344–W349. doi: 10.1093/nar/gkx276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kabsch WIUCrXDS. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adams, P. D. et al. in F, 539–547 (International Union of Crystallography, 2012).
- 17.Cowtan K. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr Sect D Biol Crystallogr. 2006;62:1002–1011. doi: 10.1107/S0907444906022116. [DOI] [PubMed] [Google Scholar]
- 18.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bricogne G, Blanc E, Brandl M, Flensburg C, Keller P, Paciorek W, Roversi P, Sharff A, Smart OS, Vonrhein C, Womack TO. BUSTER version 2.10.2. Cambridge: United Kingdom Global Phasing Ltd; 2017. [Google Scholar]
- 20.Götze M, et al. StavroX—A Software for Analyzing Crosslinked Products in Protein Interaction Studies. J Am Soc Mass Spectrom. 2011;23:76–87. doi: 10.1007/s13361-011-0261-2. [DOI] [PubMed] [Google Scholar]
- 21.Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 22.Xu L, et al. Inhibition of Host Vacuolar H+-ATPase Activity by a Legionella pneumophila Effector. PLoS Pathog. 2010;6:e1000822. doi: 10.1371/journal.ppat.1000822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci USA. 2004;101:841–846. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Franke D, et al. ATSAS 2.8: a comprehensive data analysis suite for small-angle scattering from macromolecular solutions. J Appl Crystallogr. 2017;50:1212–1225. doi: 10.1107/S1600576717007786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J Appl Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Svergun D, Barberato C, Koch MHJIUCr. CRYSOL – a Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. J Appl Crystallogr. 1995;28:768–773. [Google Scholar]
- 28.Kozin MB, Svergun DIIUCr. Automated matching of high- and low-resolution structural models. J Appl Crystallogr. 2001;34:33–41. [Google Scholar]
- 29.Tsuge H, Tsurumura T. Endogenous ADP-Ribosylation. Vol. 384. Springer International Publishing; 2014. pp. 69–87. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.