Abstract
We recently isolated the AtBI-1 (Arabidopsis Bax Inhibitor-1) gene, the expression of which suppressed Bax-induced cell death in yeast. To determine whether the same is true in the plant system, transgenic Arabidopsis plants overexpressing Bax protein under a dexamethasone (DEX)-inducible promoter were generated. On DEX treatment, such transgenic plants exhibited marked cell death at the whole-plant level, cell shrinkage, membranous destruction, and other apoptotic phenotypes. Transgenic Bax plants were retransformed with a vector containing the AtBI-1 gene (tagged with green fluorescent protein) under the control of the cauliflower mosaic virus 35S promoter. Plants expressing both Bax and AtBI-1 were able to maintain growth on DEX-treatment by sustaining intracellular integrity. Thus, we present here direct genetic evidence that the plant antiapoptotic protein AtBI-1 is biologically active in suppressing the mammalian Bax action in planta.
Although relatively little is known about the mechanistic details of cell death in plants, some aspects of the molecular machinery are conserved between plants and animals (1). It has been demonstrated that overexpression of Bax, which encodes a mammalian proapoptotic protein, is lethal in the budding yeast Saccharomyces cerevisiae (2–5), even though yeasts have neither Bcl-2-related proteins nor caspases. Lacomme and Santa Cruz (6) demonstrated that expression of Bax by using a tobacco mosaic virus (TMV) vector triggered cell death in tobacco leaf cells, which closely resembled the hypersensitive response (HR) induced by TMV in tobacco plants carrying the N gene. Conversely, overexpression of human Bcl-XL in transgenic tobacco suppressed HR and conferred stress tolerance (7). It was also reported that caspase-specific peptide inhibitors could abolish bacteria-induced plant programmed cell death (8). These observations clearly suggest some common features of animal and plant cell death processes.
Xu and Reed (9) transformed yeast cells containing a galactose-inducible Bax plasmid by using a human cDNA library (in which cDNAs were fused to a constitutively active yeast promoter) and isolated cDNAs that prevented Bax-induced lethality in response to galactose. This resulted in the identification of a gene, termed BI-1 (Bax Inhibitor-1), which is identical to a previously isolated human gene of unknown function called TEGT (testis enhanced gene transcript; refs. 10 and 11). We have previously cloned plant BI-1 cDNAs (Arabidopsis AtBI-1, and rice OsBI-1) (12). Interestingly, Sanchez et al. (13) reported that AtBI-1, the same gene isolated by our group, was obtained by differential screening of genes from plants challenged with the phytopathogen Pseudomonas syringae. Expression of AtBI-1 was rapidly up-regulated in plants during wounding or pathogen challenge. Furthermore, accumulation of the AtBI-1 transcript is significantly delayed in coi1 plants, indicating that reduced AtBI-1 mRNA levels may contribute to the enhanced susceptibility exhibited by coi1 plants to infection by various fungal pathogens.
To investigate in vivo cross-talk between Bax and AtBI-1, we established the system where, on dexamethasone (DEX) treatment, dynamic cell death could be induced in transgenic plants harboring the Bax gene. Here, we present direct genetic evidence that Bax-induced cell death can be down-regulated by ectopically expressed AtBI-1 protein in planta.
Materials and Methods
Plasmid Construction and Plant Transformation.
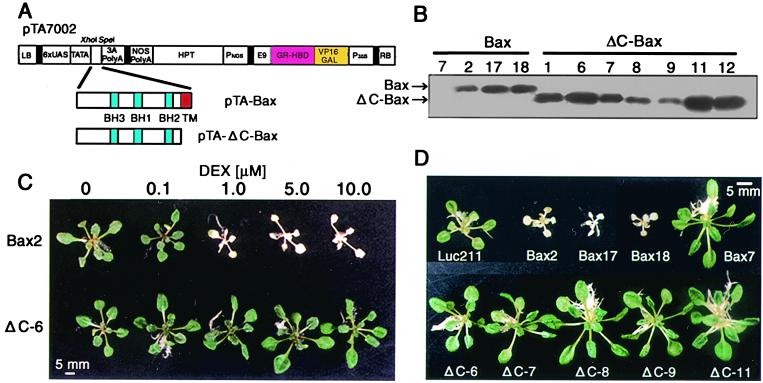
The ORF of mouse Bax (14) was inserted into the DEX-inducible vector pTA7002 (15) (Fig. 1A). The XhoI and SpeI fragment of pTA7002, which contains sequences encoding the VP16 transactivation domain and a portion of the regulatory region of the rat glucocorticoid receptor, was ligated to the XhoI–SpeI-tagged Bax cDNA. To examine the effect of the C-terminal transmembrane (TM) region of Bax protein, pTA-ΔC-Bax, in which the C-terminal 21 aa were deleted, was also constructed. The resulting constructs, pTA-Bax and pTA-ΔC-Bax, were used for Agrobacterium-mediated transformation of Arabidopsis thaliana ecotype Col-0, as described by Bechtold et al. (16). Transformants were selected on Murashige–Skoog medium containing hygromycin (20 μg/ml).
Figure 1.
Phenotypes associated with expression of Bax and ΔC-Bax in transgenic Arabidopsis. (A) Schematic diagrams of the DEX-inducible vector constructs carrying Bax (pTA-Bax) or ΔC-Bax lacking transmembrane (TM) region (pTA-ΔC-Bax). LB, left T-DNA border; 6×UAS, glucocorticoid-regulated transcription factor (GVG)-regulated promoter; 3A PolyA, pea rbcS-3A polyadenylation sequence; NOS PolyA, nopaline synthetase polyadenylation sequence; HPT, hygromycin phosphotransferase II coding sequence; PNOS, nopaline synthetase promoter; E9, pea rbcS-E9 polyadenylation sequence; GR-HBD-VP16GAL, the transcription activation domain of VP16 and the regulatory region of the human glucocorticoid receptor; P35S, cauliflower mosaic virus (CaMV) 35S promoter; RB, right T-DNA border. (B) Immunological detection of Bax and ΔC-Bax proteins under the control of a DEX-inducible system. Three-week-old seedlings germinated and grown in the absence of DEX were transferred onto the inductive medium containing 1 μM DEX and cultured at 23°C for 24 h. Ten micrograms of total protein was used for Western blot analysis. (C) Dose dependency of DEX-induced cell death in transgenic lines. Three-week-old seedlings of Bax2 and ΔC-6 lines grown on noninductive medium were transferred onto medium containing each concentration of DEX and cultured at 23°C for 3 days. (D) Phenotypes of Bax and ΔC-Bax transgenic plants. Three-week-old seedlings of transgenic lines were treated with 1 μM DEX for 4 days. The Bax7, which did not express Bax protein, and Luc211 (containing the luciferase gene with the same promoter) lines were also treated with 1 μM DEX as negative controls.
Immunological Detection.
Protein extraction and Western blot analysis were performed as described (17). Briefly, total protein (10 μg), electrophoresed on SDS/polyacrylamide gels (15%), was blotted to a poly(vinylidene difluoride) (PVDF) membrane (Immobilon, Millipore), which was then treated with anti-Bax antibody (06-499, Upstate Biotechnology). Bax protein was visualized with the enhanced chemiluminescence (ECL) system (Amersham Pharmacia) according to the manufacturer's instructions.
DNA Fragmentation Analysis.
Seedlings treated with or without 1 μM DEX for 3 days were ground in a mortar and pestle with liquid N2 to a fine powder, which was then subjected to the DNA extraction as described (18, 19). Ten micrograms of DNA treated with RNase was electrophoresed on a 1.5% agarose gel, stained with ethidium bromide, and photographed on a UV light box.
Yeast Strain and Expression.
Expression vector pYX112-AtBI-GFP (GFP, green fluorescent protein) was constructed by using the SalI-tagged coding sequence of AtBI-1 cDNA (12) by ligation in the SalI site of cassette vector pTS910-GFP, possessing a 2-μm replicon. The S. cerevisiae strain QX95001, BF264–15Dau (MATα ade1 his2 leu2-3,112 trp1-1a ura3; ref. 20) containing the LEU2-marked mouse Bax-encoding plasmid YEp51-Bax (9), was transformed with the plasmids pYX112-GFP and pYX112-AtBI-GFP by the lithium acetate method. The control plasmids, pYX112 and pYX112-AtBI, were also introduced into the yeast as described (12). Ura+ Leu+ transformants were streaked either on a synthetic dropout (SD)-glucose plate or on a SD-galactose plate, and incubated at 30°C.
Transformation of Plant Cells with GFP-Constructs.
To express the GFP-tagged AtBI-1 in plant cells, the coding sequence of AtBI-1 (12) amplified by PCR was cloned into the SalI site of a GFP cassette plasmid containing the cauliflower mosaic virus (CaMV) 35S promoter and nopaline synthase terminator (21). Subsequently, EcoRI–HindIII fragments were ligated into the EcoRI/HindIII-digested binary vector Bin19, which resulted in Bin-AtBI-GFP. Transformation of tobacco BY2 cells was performed as described by Shaul et al. (22) by using Agrobacterium tumefaciens strain EHA105.
To obtain Arabidopsis plants overexpressing both Bax and AtBI-1, the Bax2 transgenic lines were retransformed with the same Agrobacterium possessing Bin-AtBI-GFP. Transformants were selected on Murashige–Skoog medium containing kanamycin (50 μg/ml).
Northern Blot Analysis.
Total RNAs were isolated from transgenic lines by using guanidinium thiocyanate as described (12). Total RNA (10 μg), electrophoresed on a denaturing 1.2% agarose gel, was transferred to a nylon membrane (Biodyne B, Pall), which was then hybridized to the 32P-labeled cDNA fragments (Bax, GFP to detect AtBI-GFP, 3′-untranslated region of AtBI-1 for detection of endogenous AtBI-1, PR1 and PR2) in 10% dextran sulfate/1 M NaCl/1% SDS/100 μg/ml heat-denatured salmon sperm DNA. Washing was performed at high stringency (0.1× standard saline citrate and 0.1% SDS at 65°C). The membrane was analyzed with an imaging plate scanner BAS 1500 (Fuji Film, Tokyo).
Microscopic Examination.
GFP fluorescence was examined at a 488-nm excitation wavelength under a confocal laser scanning microscope (BX50, Olympus) or under a fluorescence microscope (DMRD, Leica, Wetzlar, Germany). Mitochondria were stained with the fluorescent probe MitoTracker Red (650 nM, Molecular Probes). Stained cells were examined by using excitation light at a wavelength of 568 nm.
For electron microscopic analysis, leaf sections obtained from 3-week-old plants treated with 1 μM DEX were fixed in 2.5% glutaraldehyde in cacodylate buffer (pH 7.2), and treated with OsO4. The substituted samples were embedded in Spurr's resin, and thin serial sections were prepared with an Ultracut N or Ultracut S microtome (Reichert, Vienna, Austria). Sections were stained with uranylacetate and lead citrate (UA/Pb) and observed under an electron microscope (Zmodel 2010; JEOL, Akishima-shi, Japan).
Results
A Transgenic Plant System in Which Expression of Mammalian Bax Triggered Lethality.
We obtained several transgenic Arabidopsis plants expressing either Bax or ΔC-Bax (lacking the C-terminal transmembrane domain) under a DEX-inducible promoter (Fig. 1A). Bax (21.2 kDa) and ΔC-Bax (19.0 kDa) proteins were detected immunologically in transgenic lines (Fig. 1B). The same protein was not detected in either wild-type or control plants (Luc211), in which a luciferase gene was overexpressed under the same promoter (data not shown). When transgenic 3-week-old Bax2 plants were transferred to a medium containing variable concentration of DEX (0 to 10 μM), etiolation at the whole-plant level was observed at 1 μM DEX and higher, whereas the same was not true for the ΔC–6 line (Fig. 1C). As shown in Fig. 1D, within 4 days of Bax2, -17, and -18 lines (3-week-old) being transferred to a medium containing 1 μM DEX, severe discoloration and growth retardation were observed, whereas no such abnormalities were evident in other plants (Luc211 and Bax7, where Bax protein was not produced; see Fig. 1B). The expression of ΔC-Bax protein in transgenic lines did not affect plant growth. These results suggest that expression of full-length Bax protein is essential for induction of plant cell death.
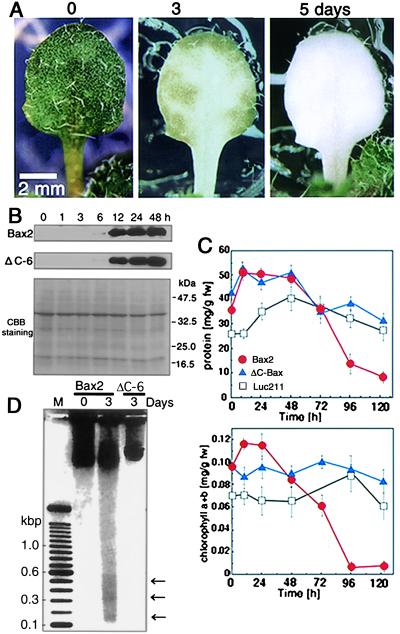
Time course analysis of leaf discoloration after DEX treatment using Bax2 and ΔC-6 lines (3-week-old plants) was also performed. Immunoblot analysis revealed that a small amount of Bax or ΔC-Bax protein was detected as early as 6 h after DEX treatment (Fig. 2B). As time progressed, Bax protein levels in DEX-treated plants rose and remained high until day 2. Color changes of leaves were apparent in the vicinity of veins and extended to the periphery within 3 days (Fig. 2A). An extensive fall in total protein level was evident by 2–3 days after DEX treatment. Chlorophyll breakdown was noted until day 4 (Fig. 2C). A DNA-laddering profile was confirmed in leaves of DEX-treated Bax2 plants after 3 days, whereas no such phenomenon was observed in ΔC-6 plants (Fig. 2D).
Figure 2.
Progression of Bax-induced cell death in DEX-treated transgenic Arabidopsis. (A) Chlorosis in Bax2 plants after DEX treatment. Three-week-old seedlings were transferred to medium containing 1 μM DEX and cultured at 23°C. The selected single leaf was observed for 5 days continuously. (B) Time course analysis of Bax or ΔC-Bax protein accumulation in transgenic lines (Bax2 and ΔC-6). Proteins were isolated at each time point (0, 1, 3, 6, 12, 24, and 48 h) after treatment as described above and used for Western blot analysis. (C) Amounts of total protein (Upper) and chlorophyll a + b (Lower) in leaves at each time point after DEX treatment. Chlorophyll estimation was performed from total leaf extracts according to Porra et al. (31); fw, fresh weight. Values are the mean ± SD of triplicate assays. (D) Detection of DNA laddering in Bax-induced cell death in Arabidopsis. Total DNA was extracted at 0 and 3 days after treatment with 1 μM DEX and separated by electrophoresis. DNA (10 μg) treated with RNase was loaded into each lane. Molecular sizes in bases (kbp) are indicated. A negative image of a photograph is presented. Arrows indicate possible DNA ladders. M, 100-bp DNA ladder (GIBCO/BRL, catalog no. 10380-D12).
Expression of GFP-Tagged AtBI-1 in Yeast and Tobacco Cells.
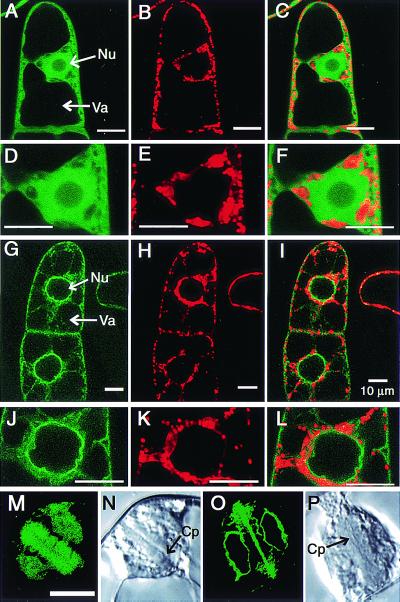
To monitor the intracellular localization of AtBI-1 protein, GFP fused to the C-terminal region of AtBI-1 (AtBI-GFP) was expressed in yeast, and the ability of the fusion protein to suppress cell death was studied. The constructed plasmids pYX112-AtBI-GFP and pYX112-GFP were transformed into the yeast strain QX95001 harboring a galactose-inducible Bax. In addition, plasmids pYX112 and pYX112-AtBI (described in ref. 12) were also transformed into the same yeast strain. As shown in Fig. 3A, when these were plated on the galactose medium, cells transformed with pYX112-AtBI or pYX112-AtBI-GFP survived, whereas cells transformed with empty vectors (pYX112 and pYX112-GFP) did not, confirming that GFP-tagged AtBI-1 conferred sufficient activity to suppress Bax-induced lethality. As shown in Fig. 3B, AtBI-GFP fluorescence was associated with the perinuclear region.
Figure 3.
Effects of GFP-tagged AtBI-1 (AtBI-GFP) on Bax-induced lethality and intracellular distribution in yeast. (A) Suppression of Bax-induced yeast cell death by AtBI-1 tagged with GFP (AtBI-GFP). Expression vectors (pYX112-AtBI-GFP and pYX112-AtBI) or control plasmids (pYX112-GFP and pYX112) were transformed into yeast stain QX95001 harboring Yep51-Bax (9). Transformants were streaked on glucose (Glu)- or galactose (Gal)-containing medium lacking uracil and leucine. Photographs were taken 3 days after incubation at 30°C. (B) Distribution of GFP and AtBI-GFP fluorescence in yeast. Fluorescence (Left) and bright-field images (Right) are displayed. The yeast cells cultured for 16 h in glucose-containing medium were imaged at 488-nm excitation wavelength to detect GFP. Arrows indicate nuclei. v, Vacuole.
To express AtBI-GFP in plant cells, the vector Bin-AtBI-GFP was constructed and introduced into tobacco suspension cells (BY2). Transgenic cells expressing the AtBI-GFP fusion protein displayed a distinct fluorescence localization pattern in both perinuclear and reticulate cortical areas, indicative of endoplasmic reticulum (ER; Fig. 4 G and J), which resembled the image obtained in yeast. Treatment of cells with MitoTracker Red disclosed that AtBI-GFP did not always colocalize with mitochondria (Fig. 4I, more obviously in L). As apparent in Fig. 4O, AtBI-GFP appeared to be associated with the cell plate and the nuclear membrane during mitosis, where ER is known to be distributed.
Figure 4.
Distribution of AtBI-GFP fluorescence in plant cells. To facilitate expression of GFP (A–F, M, and N) and GFP-tagged AtBI-1 (G–L, O, and P) in plant cells, plasmids were constructed and used in Agrobacterium-mediated transformation of tobacco suspension cells as described in Materials and Methods. The transformed cells were imaged at 488-nm excitation wavelength to detect GFP (A, D, G, J, M, and O) and 568-nm excitation wavelength to detect the mitochondrial marker MitoTracker Red (B, E, H, and K) under a confocal laser scanning microscope (Olympus BX50). Merged images are shown in C, F, I, and L. For observation of mitosis, BY2 cells at 3 days after transfer were treated with aphidicolin as described by Hasezawa and Nagata (32) and telophase cells were observed (M–P). Arrows indicate nuclei (Nu), vacuole (Va), and cell plate (Cp).
Bax-Induced Plant Lethality Can Be Down-Regulated by Overexpression of AtBI-1.
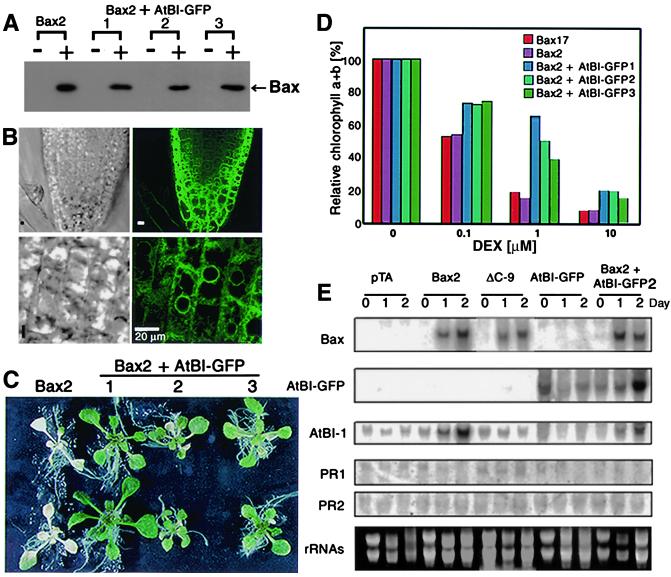
The transgenic plant line (Bax2) was retransformed by an Agrobacterium harboring Bin-AtBI-GFP, and new transgenic lines expressing both AtBI-1 and Bax were selected on a medium containing both kanamycin and hygromycin. Bax proteins were detected immunologically in the Bax2 transgenic lines expressing AtBI-GFP at 24 h after 1 μM DEX treatment (Fig. 5A). Furthermore, these lines exhibited GFP fluorescence in perinuclear and reticulate compartments in the cells (Fig. 5B), which was identical to the pattern in yeast or BY2 cells. The distribution of AtBI-GFP did not change even after 3 days of 1 μM DEX treatment (data not shown). Accordingly, we were prompted to perform experiments to evaluate the effects of AtBI-1 expression on DEX-induced cell death. As shown in Fig. 5C, leaf discoloration was suppressed in plants expressing both Bax and AtBI-1, which was also supported by the quantitative estimation of chlorophyll content (Fig. 5D).
Figure 5.
Resistance to Bax-induced cell death in transgenic Arabidopsis expressing AtBI-1. (A) Western blot analysis of Bax proteins 1 day after 1 μM DEX treatment in transgenic lines 1, 2, and 3, which expressed both pTA-Bax and Bin-AtBI-GFP. (B) Localization pattern of AtBI-GFP in transgenic Arabidopsis grown on Murashige–Skoog medium. The root tip (Upper) was imaged at 488-nm excitation wavelength to detect AtBI-GFP protein (Right) or bright-field (transmission mode, Left). Magnified images are shown in the lower panels. (C) Plants of Bax2 or Bax2 + AtBI-GFP transformants 3 days after 1 μM DEX treatment. (D) Resistance to Bax-induced leaf chlorosis. Transgenic plants were treated for 3 days with several concentrations of DEX. The chlorophyll a + b (%) was compared with corresponding non-DEX-treated leaves (control). Values were calculated from triplicate assays. (E) Analysis of mRNA accumulation in Bax, ΔC-Bax, and AtBI-GFP transgenic lines. Total RNA was isolated from 3-week-old transgenic plants containing pTA7002 (vector control, pTA), pTA-Bax (Bax2), pTA-ΔC-Bax (ΔC-9), Bin-AtBI-GFP (AtBI-GFP), and both pTA-Bax and Bin-AtBI-GFP (Bax2 + AtBI-GFP2). Plants were grown for 3 weeks on Murashige–Skoog medium, and then treated with 1 μM DEX for 0, 1, or 2 days. Coding regions of Bax and GFP, the 3′-untranslated region (UTR) of AtBI-1 (12), PR1 (33), and PR2 (34) were used as hybridization probes. A signal obtained by hybridization with the 3′-UTR of AtBI-1 indicated endogenous AtBI-1 expression.
Causal factors associated with plant cell death are considered to be orchestrated by a variety of key genes engaged in defense responses (23). We examined the possibility that such coordination may exist in Bax-induced plant cell death in relation to pathogen-related (PR) gene expression as well as the endogenous AtBI-1 level. As shown in Fig. 5E, accumulation of Bax mRNA was observed in Bax2, ΔC-9, and Bax2 + AtBI-GFP2 transgenic plants after 1 μM DEX treatment for 1 day. A mRNA for AtBI-GFP driven by the CaMV 35S promoter was detected in the transgenic lines, AtBI-GFP and Bax2 + AtBI-GFP2. Interestingly, the level of endogenous AtBI-1 mRNA was enhanced in Bax2 and Bax2 + AtBI-GFP2 lines. In contrast, we did not observe any apparent differences in the mRNA levels of pathogen-inducible genes (PR1 and PR2). These results indicate that endogenous AtBI-1 mRNA level coordinate with Bax over-expression, however, this could be insufficient to warrant the survival of Bax-expressing plants.
Observation of Intracellular Morphology.
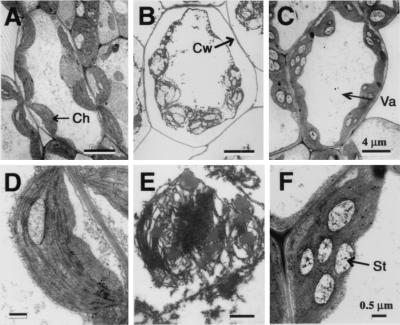
Electron microscopic analysis was performed to examine the intracellular morphology of Bax-expressing cells. As shown in Fig. 6, cytoplasmic shrinkage was apparent in the mesophyll cells of plants expressing Bax (Fig. 6B), compared with those lacking such expression (Fig. 6A). The former was characterized by destruction of chloroplasts with impaired lamella structure (Fig. 6 B and E). As shown in Fig. 6 C and F, no such cellular alteration was noted in Bax transgenic plants expressing AtBI-GFP, except an apparent accumulation of starch granules in chloroplasts.
Figure 6.
Intracellular morphology of Bax2 (A, B, D, and E) and Bax2 + AtBI-GFP2 (C and F) transgenic plants. Mesophyll cells obtained from plants treated with 0 (A and D) or 1 μM DEX (B, C, E, and F) for 3 days were examined by an electron microscope. Arrows indicate chloroplast (Ch), cell wall (Cw), vacuole (Va), and starch granule (St). (Scale bars, 4 μm in A–C and 0.5 μm in D–F.)
Discussion
Bax Conferred Lethality at the Whole-Plant Level.
We previously reported that the AtBI-1 gene suppressed Bax-induced cell death in yeast (12). Before examining the cross-talk between Bax and AtBI-1 genes in planta, we were prompted to establish a stable transgenic system, in which ectopically expressed Bax protein would cause lethality at the whole-plant level. In our experiments, every plant overexpressing Bax protein displayed severe discolored lesions on rosette leaves. Such tissue vulnerability, in mesophylls, was visible at 24 h after DEX treatment, and the discolored lesions in leaves were continued to extend until 3 days. In contrast, transgenic plants containing luciferase (Luc211) or Bax without the C-terminal transmembrane domain (ΔC-Bax) under the same inducible system exhibited normal growth, confirming that only full-length Bax could trigger cell lethality in Arabidopsis leaves.
A plausible model to explain the lethality of Bax in plants or yeast proposes a direct effect of Bax on mitochondria through its ability to form channels in the outer membrane of organelles in animals (24). Bax protein has been proposed to disturb cellular functions, including photosynthesis and the maintenance of membrane integrity (6). The TM region is known to target the Bax molecule to the mitochondrial outer membrane, and is thus required to induce lethality in yeast (4) and in TMV-infected plant cells (6). We have now also proved this to be true in a stable transgenic plant system expressing ΔC-Bax. Apoptotic DNA ladders have been reported in processes of plant cell death such as senescence, HR, and the stress response (18, 19, 25, 26). In this regard, we also detected the oligonucleosomal DNA ladder in Bax-expressing plants, suggesting that Bax-induced lethality might mimic common mechanisms between animal and plant cell death programs.
AtBI-1 Suppressed Bax-Induced Lethality in Plants.
The main objective of this investigation was to evaluate whether AtBI-1 mimics the biological activity of a Bax suppressor in a transgenic plant system. Because of the lack of appropriate antibody, we performed a feasibility study using a GFP-tagged AtBI-1 protein before pursuing these experiments. We demonstrated here that a GFP-tagged AtBI-1 protein also possessed cell-death suppression activity in yeast, and its localization was restricted to the perinuclear region. We then established transgenic Arabidopsis plants and tobacco BY2 cells that overexpressed the AtBI-GFP. Confocal laser scanning microscopy revealed that the GFP-AtBI signal was evident as a perinuclear and reticulate pattern, indicative of ER. Indeed, AtBI-GFP was found to be associated with the cell plate in mitotic BY2 cells.
The evidence that AtBI-1 can function as a suppressor of Bax-induced cell death in higher plants might support the notion that some mechanisms of cell death have been evolutionarily conserved throughout metazoa and plants. In animal, an antiapoptotic protein Bcl-2 localizes with mitochondria and ER (27). In the latter case, the possibility that Bcl-2 might alter ER ionic homeostasis has been reported (28, 29). It is intriguing to find out the mechanism of Bax effects on chloroplast membranes. What we can anticipate at present is that the action of Bax through mitochondria subsequently destroyed chloroplast membrane. Discrepancy in terms of intracellular localization of Bax and AtBI-1 may imply that there would be at least one factor mediating the cross-talk between AtBI-1 and Bax.
Of particular interest is that cell death induced by UV-B irradiation, paraquat treatment, or the HR to TMV infection was suppressed in plants expressing Bcl-XL (7). Similarly enhanced resistance was also observed by overexpression of ced-9 in tobacco cells. Thus, it appears that functional conservation of genes regulating a common cell death pathway may exist in multicellular organisms. We observed enhancement of the endogenous AtBI-1 mRNA level in transgenic plants expressing Bax and Bax + AtBI-GFP, which may indicate that the AtBI-1 gene is inducible by intracellular stresses. Similarly, Sanchez et al. (13) reported that AtBI-1 expression was rapidly up-regulated during wounding or pathogen challenge.
It is widely known that expression of PR genes correlates with cell death by HR (30). However, caspase-specific peptide inhibitors abolished bacteria-induced plant programmed cell death, but did not significantly affect induction of other aspects of HR, such as expression of defense genes (8). Accumulation of PR gene mRNAs was noted in tobacco mesophyll cells on transfection with TMV vectors harboring Bax and other genes encoding elicitor, movement protein, and virus coat protein (6). In regard to this, we have demonstrated that Bax was sufficient to induce plant cell death, independent of expression of defense-related genes.
As demonstrated in this study, the ectopically expressed AtBI-1 protein suppressed the lethal action of the mammalian protein Bax in plant cells. Because the BI-1 gene exists in a wide range of higher organisms, the evolutionary role of this protein in sustaining cell viability remains to be explored.
Acknowledgments
We thank Drs. J. Reed, N. H. Chua, T. Aoyama, Y. Kikuchi, T. Sasaki, Y. Niwa, A. Hasezawa, and F. Kumagai and Ms. H. Suzuki for their help and gift of materials. This research was supported by Research for the Future from the Japan Society for the Promotion of Science.
Abbreviations
- AtBI-1
Arabidopsis Bax inhibitor-1
- CaMV
cauliflower mosaic virus
- DEX
dexamethasone
- HR
hypersensitive response
- PR
pathogenesis-related protein gene
- TMV
tobacco mosaic virus
- ER
endoplasmic reticulum
References
- 1.Pennell R I, Lamb C. Plant Cell. 1997;9:1157–1168. doi: 10.1105/tpc.9.7.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sato T, Harada M, Bodrug S, Irie S, Iwama N, Boise L H, Thompson C B, Golemis E, Fong L, Wang H G, Reed J C. Proc Natl Acad Sci USA. 1994;91:9238–9242. doi: 10.1073/pnas.91.20.9238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harada M, Aimé-Sempé C, Sato T, Reed J C. J Biol Chem. 1995;270:11962–11969. doi: 10.1074/jbc.270.20.11962. [DOI] [PubMed] [Google Scholar]
- 4.Zha H, Fisk H A, Yaffe P, Mahajan N, Herman B, Reed J C. Mol Cell Biol. 1996;16:6494–6508. doi: 10.1128/mcb.16.11.6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenhalf W, Stephan C, Chaudhuri B. FEBS Lett. 1996;380:169–175. doi: 10.1016/0014-5793(96)00044-0. [DOI] [PubMed] [Google Scholar]
- 6.Lacomme C, Santa Cruz S. Proc Natl Acad Sci USA. 1999;96:7956–7961. doi: 10.1073/pnas.96.14.7956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitsuhara I, Malik K A, Miura M, Ohashi Y. Curr Biol. 1999;9:775–778. doi: 10.1016/s0960-9822(99)80341-8. [DOI] [PubMed] [Google Scholar]
- 8.del Pozo O, Lam E. Curr Biol. 1998;8:1129–1132. doi: 10.1016/s0960-9822(98)70469-5. [DOI] [PubMed] [Google Scholar]
- 9.Xu Q, Reed J C. Mol Cell. 1998;1:337–346. doi: 10.1016/s1097-2765(00)80034-9. [DOI] [PubMed] [Google Scholar]
- 10.Walter L, Dirks B, Rothermel E, Heyens M, Szpirer C, Levan G, Gunther E. Mamm Genome. 1994;5:216–221. doi: 10.1007/BF00360548. [DOI] [PubMed] [Google Scholar]
- 11.Walter L, Marynen P, Szpirer J, Levan G, Gunther E. Genomics. 1995;28:301–304. doi: 10.1006/geno.1995.1145. [DOI] [PubMed] [Google Scholar]
- 12.Kawai M, Pan L, Reed J C, Uchimiya H. FEBS Lett. 1999;464:143–147. doi: 10.1016/s0014-5793(99)01695-6. [DOI] [PubMed] [Google Scholar]
- 13.Sanchez P, de Torres Zabala M, Grant M. Plant J. 2000;21:393–399. doi: 10.1046/j.1365-313x.2000.00690.x. [DOI] [PubMed] [Google Scholar]
- 14.Oltvai Z N, Milliman C L, Korsmeyer S J. Cell. 1993;74:609–619. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 15.Aoyama T, Chua N H. Plant J. 1997;11:605–612. doi: 10.1046/j.1365-313x.1997.11030605.x. [DOI] [PubMed] [Google Scholar]
- 16.Bechtold N, Ellis J, Pelletier G. C R Acad Sci Ser III. 1993;316:1194–1199. [Google Scholar]
- 17.Kawai M, Uchimiya H. Plant Mol Biol. 1995;27:943–951. doi: 10.1007/BF00037022. [DOI] [PubMed] [Google Scholar]
- 18.Kawai M, Uchimiya H. Annal Bot. 2000;86:405–414. [Google Scholar]
- 19.Kawai M, Kobayashi Y, Hirata A, Oono Y, Watanabe H, Uchimiya H. Plant Biotech. 2000;17:305–308. [Google Scholar]
- 20.Leu D J, Dulic V, Reed D I. Cell. 1991;66:1197–1206. doi: 10.1016/0092-8674(91)90042-w. [DOI] [PubMed] [Google Scholar]
- 21.Niwa Y, Hirano T, Yoshimot K, Shimizu M, Kobayashi H. Plant J. 1999;18:455–463. doi: 10.1046/j.1365-313x.1999.00464.x. [DOI] [PubMed] [Google Scholar]
- 22.Shaul O, Nironov V, Burssens S, Montague M V, Inze D. Proc Natl Acad Sci USA. 1996;93:4868–4872. doi: 10.1073/pnas.93.10.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Alvarez M E, Pennell R I, Meije P J, Ishikawa A, Dixon R A, Lamb C. Cell. 1998;92:773–784. doi: 10.1016/s0092-8674(00)81405-1. [DOI] [PubMed] [Google Scholar]
- 24.Antonsson B, Conti F, Ciavatta A M, Montessuit S, Lewis S, Martinou I, Bernascoti L, Bernard A, Mermod J J, Mazzei G, et al. Science. 1997;277:370–372. doi: 10.1126/science.277.5324.370. [DOI] [PubMed] [Google Scholar]
- 25.Orzäez D, Granell A. FEBS Lett. 1997;404:275–278. doi: 10.1016/s0014-5793(97)00133-6. [DOI] [PubMed] [Google Scholar]
- 26.Yen C H, Yang C H. Plant Cell Physiol. 1998;39:922–927. [Google Scholar]
- 27.Jacobson M D, Burne J F, King M P, Miyashita T, Reed J C, Raff M C. Nature (London) 1993;361:365–369. doi: 10.1038/361365a0. [DOI] [PubMed] [Google Scholar]
- 28.Pinton P, Ferrari D, Magalhâes P, Schulze-Osthoff K, Vrgilio F, Pozzan T, Rizzuto R. J Cell Biol. 2000;148:857–862. doi: 10.1083/jcb.148.5.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foyouzi-Youssefi R, Arnaudeau S, Borner C, Kelley L W, Tschopp J, Lew D P, Demaurex N, Krause K H. Proc Natl Acad Sci USA. 2000;97:5723–5728. doi: 10.1073/pnas.97.11.5723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Greenberg J T, Guo A, Klessig D F, Ausubel F M. Cell. 1994;77:551–563. doi: 10.1016/0092-8674(94)90217-8. [DOI] [PubMed] [Google Scholar]
- 31.Porra R J, Thompson W A, Kriedemann P E. Biochim Biophys Acta. 1989;975:384–394. [Google Scholar]
- 32.Hasezawa A, Nagata T. Bot Acta. 1991;104:206–211. [Google Scholar]
- 33.Metzler M C, Cutt J R, Klessig D F. Plant Physiol. 1991;96:346–348. doi: 10.1104/pp.96.1.346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uknes S, Mauch-Mani B, Moyer M, Potter S, Williams S, Dincher S, Chandler D, Slusarenko, Ward E R, Ryals J A. Plant Cell. 1992;4:645–656. doi: 10.1105/tpc.4.6.645. [DOI] [PMC free article] [PubMed] [Google Scholar]