Abstract
Objective:
Persistence of HPV infection is the true cause of cervical disorders. It is reported that competition may exist among HPV genotypes for colonization. This survey was designed to establish the multiple HPV genotype status in our community and the probability of multiple HPV infections involvement.
Methods:
All multiple HPV infections were selected for investigation in women suffering from genital infections referred to private laboratories in Tehran, Iran. A total of 160 multi HPV positive specimens from cervical scraping were identified by the HPV genotyping methods, “INNO-LiPA and Geno Array”.
Result:
In present study, HPV 6 (LR), 16 (HR), 53 (pHR), 31 (HR) and 11 (LR) were included in 48.8% of detected infections as the most five dominant genotypes. HPV 16 was detected at the highest rate with genotypes 53, 31 and 52, while HPV 53 appeared linked with HPV 16, 51 and 56 in concurrent infections. It appears that HPV 16 and 53 may have significant tendencies to associate with each other rather than with other genotypes. Analysis of the data revealed there may be some synergistic interactions with a few particular genotypes such as “HPV 53”.
Conclusion:
Multiple HPV genotypes appear more likely to be linked with development of cervical abnormalities especially in patients with genital infections. Since, there are various patterns of dominant HPV genotypes in different regions of world, more investigations of this type should be performed for careHPV programs in individual countries.
Keywords: HPV, multi genotypes, genital infection, Iran
Introduction
Infection with high-risk Human Papillomavirus (HR-HPV) genotypes are responsible for cervical cancer. Frequency and the distribution of HPV in each community have its own dominant types with a specific pattern that may be comparable in some genotypes with other areas. Although, HPV 16 is usually reported as one of the most prevalent types in most countries. Hence, it is necessary to have prevalence evaluated of HR-HPVs and LR-HPVs in cervical cancer prevention and Care HPV programs (Quint et al., 2012; Shayanfar et al., 2013; Sohrabi et al., 2014; Sohrabi et al., 2014). It is reported various parameters such as age of the infected patients, sexual behavior, and number of the partners, environmental conditions, smoke and some other risk factors are the main causes of HPV infection. Some of these influential parameters can cause enhancement of multiple HPV genotypes such as having different sexual partners (Chaturvedi et al., 2011; Mzarico et al., 2015 ;Tran et al., 2015). General tendency has been reported in previous studies for all HPV types being in similar cluster. It is frequently reported cervical concurrent infections with more than one HPV genotype are common, especially among young women (Chaturvedi et al., 2005; Mendez et al., 2005; Vaccarella et al., 2010). It is continuously are discussed these infections occur randomly or the result of interactions between HPV types (Chaturvedi et al., 2011; Chaturvedi et al., 2005 ; Plummer et al., 2012). Some studies have formally evaluated synergistic interactions across multiple HPV genotypes on cervical malignancies and risk of genital infections. In these reports higher rate have mentioned in those patients with abnormal cervical cytology especially those which included in α-9 family (Alpha-PV 9), particularly HPV16, 31 and 35 (Trottier et al., 2006; Wentzensen et al., 2013; Bernard et al., 2010; Dickson et al., 2013; Tommasino et al., 2014). It is emphasized the association between HPV16 and cancer is significantly stronger than that between either HPV31 or HPV35 (Pirog et al., 2014). This survey aims to find out the status of multiple HPV genotypes in our community among identified women suffering from multiple HPVs infections, also to understand whether certain HPV types have the tendency to be involved in multiple HPV infections or not.
Materials and Methods
Patients
Based on our previous study all patients who had multiple HPV genotypes were entered in this study (Sohrabi et al., 2016). These patients were all women referred to private clinical laboratories for diagnosis of genital infections and regular checkups in Tehran, Iran from January 2012 till December 2013. They were totally160 multi HPV positive specimens from cervical scrapping (Liquid Based Cytology) and genital lesions. All clinical data were collected from patients’ medical records.
HPV Genotyping
HPV DNA was extracted by the High Pure PCR Template Preparation Kit (Roche Diagnostics®, Mannheim, Germany). Extracted DNA was stored at −20°C until experiment. HPV genotyping was performed by approved HPV commercial diagnostic technologies as PCR Hybridization methods by either GenoArray Test kit (Hybribio) and INNO-LiPA HPV Genotyping Extra (INNOGENETICS®, Belgium) according to the manufacturer’s instructions. The GenoArray allowed for the detection of the following 21 HPV genotypes: 14 high risk types (HPV16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66 and 68), five low risk genotypes (HPV6, 11, 42, 43, 44and CP8304 [81]), and probably HR types (HPV 53). However, the INNO-LiPA Extra I and Extra II were able to detect high risk types (HPV 16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68, 70, 73, 82), low risk types (HPV6, 11, 40, 42, 43, 44, 54, 61, 62, 67, 81, 83 and 89), and probably HR types (HPV 53).
Definition of Multiple Infections
All specimens with two and more detected HPV genotypes considered as multiple HPV infections.
Statistical Analysis
In order to determine any significant relationship between the HPV genotypes, an independent tests (Chi-Square) were used. All tests were performed at the significant level of 0.05 using SPSS software version 23.
Results
Overall Prevalence of HPV Genotypes
Of the 426 HPV genotypes were identified from 160 multi HPV specimens which 32 HPV genotypes (high risk-HPVs:16, 18, 26, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 66, 68, 70, 73, 82; low risk-HPV: 6, 11, 40, 42, 43, 44, 54, 61, 62, 67, 69/71, 81 and 89 and probably HR-HPVs: 53) were diagnosed by genotyping assays Figure 1. The distribution and frequency rate of Multiple HPV infections were determined for all low and high risk HPV genotypes. Comprehensive HPV genotypes interactions are shown in Figure 1. In present study, HPV 6 (LR), 16 (HR), 53 (pHR), 31 (HR) and 11 (LR) were involved in 48.83% from all detected infections as the most five dominant genotypes with 59, 57, 33, 31, and 29 cases, respectively. None of these dominant genotypes were involved with HPV40, 43, and 62 (Figures 1 and 2).
Figure 1.
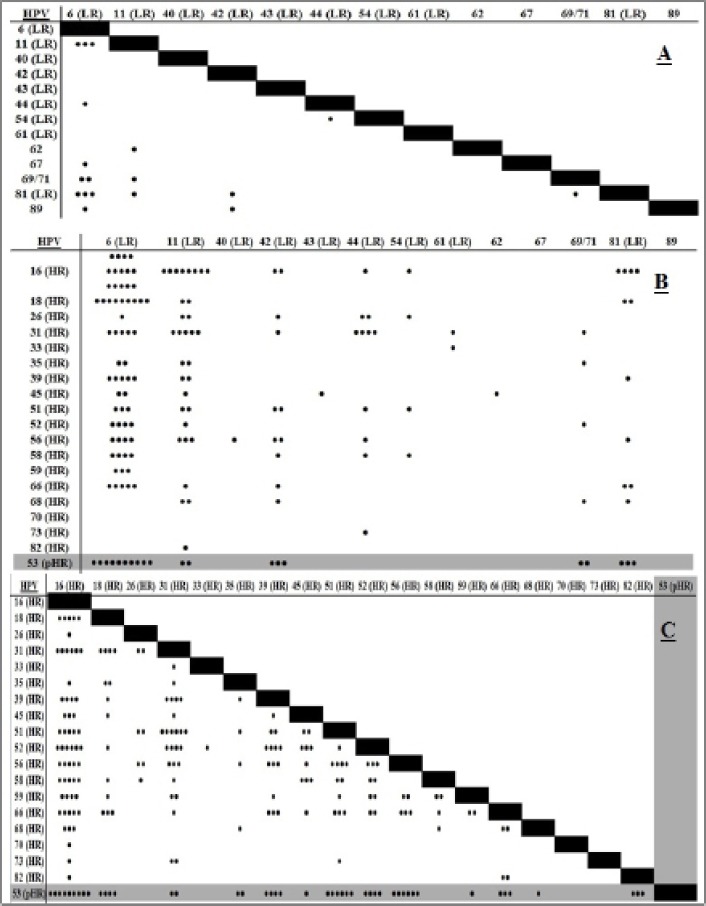
Associations Between LR-LR (A), HR-LR (B) and HR-HR HPV Genotypes Involved in Patients with Multiple Infections
Figure 2.
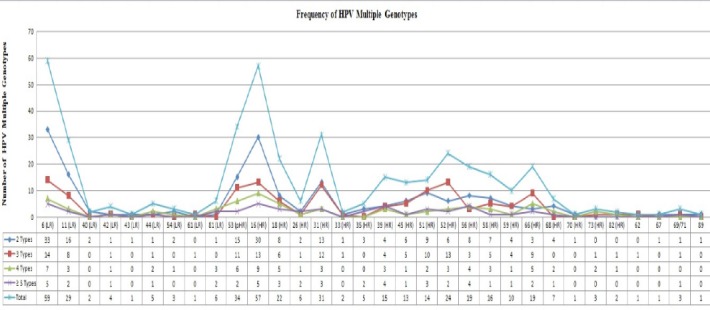
Frequency of Detected HPV Genotypes in Multiple HPV Infections
Frequencies of Multiple HPV Genotypes
All detected genotypes were analyzed regarding their tendencies to the other genotypes in another aspect. In order to find out the rate of multiple HPV genotypes distribution with each other, each detected HPV genotypes were compared with others. The significant directly relationship between HPV genotypes in multiple HPV infections especially in dominant genotypes are shown in Table 1. Frequency rate for the three most dominant HR types 16, 53 and 18 have been compared with each others. HPV 16 was detected at the highest rate with genotypes 53, 31 and 52, while HPV 53 was with HPV 16, 51 and 56 in multiple HPV genotypes (Figure 3). HPV 16, 53 obviously showed higher tendencies to each other rather than other types Figure 3. These detected HPV multiple genotypes were belong to 92, 42, 17, 7, 1 and 1 cases as two, three, four, five, six and seven HPV multiple infections, respectively. Rate of multiple HPV infections were reduced in those specimens contain more than two types. As a result, there were no statistically significant differences between these multiple HPV genotypes (p≥ 0.05).
Table 1.
Comparison Frequency of HPV Genotypes Rates which Involved in Multiple HPV Infections
| HPV Genotypes | No. of Detected HPV Genotypes in Subjects | No. of Involved HPV Genotypes in Multiple Infections |
|---|---|---|
| 16 (HR) | 95 | 23 |
| 18 (HR) | 36 | 13 |
| 26 (HR) | 15 | 10 |
| 31 (HR) | 57 | 22 |
| 33 (HR) | 3 | 3 |
| 35 (HR) | 15 | 11 |
| 39 (HR) | 36 | 14 |
| 45 (HR) | 22 | 14 |
| 51 (HR) | 46 | 18 |
| 52 (HR) | 39 | 15 |
| 56 (HR) | 45 | 17 |
| 58 (HR) | 26 | 14 |
| 59 (HR) | 21 | 11 |
| 66 (HR) | 38 | 16 |
| 68 (HR) | 13 | 9 |
| 70(HR) | 1 | 1 |
| 73 (HR) | 5 | 4 |
| 82 (HR) | 3 | 3 |
| 53 (pHR) | 63 | 17 |
| 6 (LR) | 82 | 20 |
| 11 (LR) | 40 | 18 |
| 40 (LR) | 1 | 1 |
| 42 (LR) | 16 | 11 |
| 43 (LR) | 1 | 2 |
| 44 (LR) | 13 | 9 |
| 54 (LR) | 5 | 5 |
| 61 (LR) | 2 | 1 |
| 81 | 20 | 11 |
| 62 | 2 | 2 |
| 67 | 1 | 1 |
| 69/71 | 10 | 8 |
| 89 | 2 | 2 |
Figure 3.
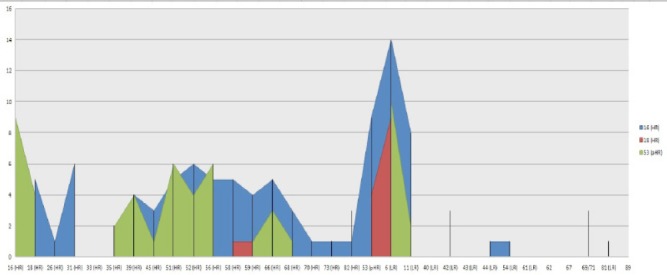
Comparison Frequency of Dominant HR- HPV Genotypes 16, 53,18
Discussion
It is reported multiple HPV genotypes are the major cause of the cell transformations in infected cases for progressing toward the cervical intraepithelial neoplasia (CIN I-IV) and association with abnormal cytology (Beca et al., 2014; Salazar et al., 2015). In this regard, the influence of newly diagnostic HPV technologies have been obviously had an important tools in the increasing of HPV genotypes. Therefore, apply high quality genotyping methods have now been necessarily more than ever (Sohrabi et al., 2015 ; Sorabi et al., 2016; Sorabi et al., 2016). Characterization of HPV genotypes in detected multiple infections had various frequencies in present study. Our results revealed all HPV genotypes are participating in genital infections in our community. HPV genotypes in multiple infections could be random or may be synergistic interactions across concurrent infections for each genotype in cervical disorders. Those HPV scatters in Figure 1 illustrated multi-type infections in some particular types, which supposed of the synergistic interactions between them. On the other hand, some types had interaction with many HPV genotypes but with low frequency rate. It could be meaning and suggesting for a random pattern rather than some specific tendency for these types. However, a specific tendency can be observed in those types having higher frequency rate. Analysis of scatters (Figure 1) demonstrated that LR-HPVs have low association with themselves, while HR-HPVs have strong and high association with themselves and even with LR-HPV types. The pattern of the dominant HPV genotypes are evaluating in most regions of the world in last decade. Furthermore, newly pHR-HPV genotypes have recently been raised in genital infections and also in cervical malignancies (Dickson et al., 2013; Shafaghi et al., 2013; Puryasin et al., 2015). No strong evidence has not been reported for HPV53 in cervical cancer in spite of frequent reports (Beca et al., 2014; Salazar et al., 2015). We observed stronger association of other HPV genotypes (HPV 52, 56, and 16) with HPV 53 in this study. The role of HPV53 and some other pHR types have been studied in cervical cancer and compared their effect with HR carcinogenic types. This type repeatedly reported as dominant type in many countries in the world (Shafaghi et al., 2013; Puryasin et al., 2014; Meloni et al., 2014; Sun et al., 2014). Phylogenic analysis proved HPV 53 has been located in 2B carcinogens with HPV26, 66, 67, 68, 70 and 73 which is belonging to the family of the α-Papillomaviridae the same group as HR-HPV types. It was reported these pHR-HPV genotypes were biologically active when presented as a single infection in cervical cancer affecting the same cellular pathways as any of the fully recognized carcinogenic HR-HPV types (Miller et al., 2012; Gill et al., 2014). Its potential effect has been interested in some reports (Padalko et al., 2015). However, our results demonstrated the rate of interactions were strongest between HR-HPV genotypes. Although, these results illustrated random patterns (multi HPV genotypes) were more likely in genital disorders. It still needs to be worked on: “is the pattern of multiple HPV genotypes, random or synergistic interaction?”. However there may be some synergistic interactions in a few particular types specifically “HPV 53” in those patients with multi HPV infections. It is suggested that more surveys should be emphasized in different populations to draw better conclusion for probability of synergistic interactions and its influence on the rising the rate of multi-type infections.
Disclosure
The authors who have taken part in this study declare that they do not have anything to disclose regarding the conflict of interest with respect to this manuscript.
References
- 1.Beca F, Pinheiro J, Rios E, Pontes P, Amendoeira I. Genotypes and prevalence of HPV single and multiple concurrent infections in women with HSIL. Diagn Cytopathol. 2014;42:919–23. doi: 10.1002/dc.23143. [DOI] [PubMed] [Google Scholar]
- 2.Bernard HU, Burk RD, Chen Z, et al. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virol J. 2010;401:70–9. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chaturvedi AK, Myers L, Hammons AF. Human Papillomavirus infection with multiple types: pattern of coinfection and risk of cervical disease. J Infect Dis. 2011;203:910–20. doi: 10.1093/infdis/jiq139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chaturvedi AK, Myers L, Hammons AF. Prevalence and clustering patterns of human papillomavirus genotypes in multiple infections. Cancer Epidemiol Biomarkers Prev. 2005;14:2439–45. doi: 10.1158/1055-9965.EPI-05-0465. [DOI] [PubMed] [Google Scholar]
- 5.Dickson EL, Vogel R, Bliss R, Downs LS. Multiple-type HPV infections: a cross sectional analysis of the prevalence of specific types in 309,000 women referred for HPV testing at the time of cervical cytology. Int J Gynecol Cancer. 2013;23:1295–302. doi: 10.1097/IGC.0b013e31829e9fb4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gill SS, Jana AM, Shrivastav A, Sharma S, Sharma A. The carcinogenic potential of e6 & e7 genes of high-risk hpv compared with e6-e7 genes of low-risk HPV in human cervical cancer: a review. Int J Pharm Sci Res. 2014;5:703–12. [Google Scholar]
- 7.Meloni A, Pilia R, Campagna M. Prevalence and molecular epidemiology of human papillomavirus infection in Italian women with cervical cytological abnormalities. J Public Health Res. 2014;26:157. doi: 10.4081/jphr.2014.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendez F, Munoz N, Posso H. Cervical coinfection with human papillomavirus (HPV) types and possible implications for the prevention of cervical cancer by HPV vaccines. J Infect Dis. 2005;192:1158–65. doi: 10.1086/444391. [DOI] [PubMed] [Google Scholar]
- 9.Miller DL, Puricelli MD, Stack MS. Virology and molecular pathogenesis of HPV (Human Papillomavirus) associated oropharyngeal squamous cell carcinoma. Biochem J. 2012;443:339–53. doi: 10.1042/BJ20112017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mzarico E, Gómez-Roig MD, Guirado L, Lorente N, Gonzalez-Bosquet E. Relationship between smoking, HPV infection, and risk of cervical cancer. Eur J Gynaecol Oncol. 2015;36:677–80. [PubMed] [Google Scholar]
- 11.Padalko E, Ali-Risasi C, Van Renterghem L. Evaluation of the clinical significance of human papillomavirus (HPV53) Eur J Obstet Gynecol Reprod Biol. 2015;191:7–9. doi: 10.1016/j.ejogrb.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Pirog EC, Lloveras B, Molijin A. HPV prevalence and genotypes in different histological subtypes of cervical adenocarcinoma, a worldwide analysis of 760 cases. Mod Pathol. 2014;27:1559–67. doi: 10.1038/modpathol.2014.55. [DOI] [PubMed] [Google Scholar]
- 13.Plummer M, Vaccarella S, Franceschi S. Multiple human papillomavirus infections: the exception or the rule? J Infect Dis. 2012;203:891–3. doi: 10.1093/infdis/jiq146. [DOI] [PubMed] [Google Scholar]
- 14.Puryasin M, Sharafi H, Mousavi AS. Distribution of Human Papillomavirus Genotypes in Liquid based Samples: Abundance of HPV-53 in Tehran, Iran. Iran J Public Health. 2014;43:1159–60. [PMC free article] [PubMed] [Google Scholar]
- 15.Quint W, Jenkins D, Molijn A. One virus, one lesion-individual components of CIN lesions contain a specific HPV type. J Pathol. 2012;227:62–71. doi: 10.1002/path.3970. [DOI] [PubMed] [Google Scholar]
- 16.Salazar KL, Zhou HS, Xu J. Multiple human papilloma virus infections and their impact on the development of high-risk cervical lesions. Acta Cytol. 2015;59:391–8. doi: 10.1159/000442512. [DOI] [PubMed] [Google Scholar]
- 17.Shafaghi B, Jarollahi A, Yousefzadeh B, et al. Human papillomavirus prevalence and types among Iranian women attending regular gynecological visit. Rep Radiother Oncol. 2015;1:73–9. [Google Scholar]
- 18.Shayanfar N, Hosseini N, Panahi M. Detection of mucosal type human papillomavirus in cutaneous squamous cell carcinoma in Iran. Pathol Res Pract. 2013;209:90–4. doi: 10.1016/j.prp.2012.10.010. [DOI] [PubMed] [Google Scholar]
- 19.Sohrabi A, Rahnmaye-Farzami M, Mirab-Samiee S, Mahdavi S, Babei M. Comparison of in-house multiplex real time PCR, Diagcor GenoFlow HPV Array test and INNO-LiPA HPV genotyping extra assays with LCD- Array kit for human papillomavirus genotyping in cervical liquid based cytology specimens and genital lesions in Tehran, Iran. Clin Lab. 2016;62:615–19. doi: 10.7754/clin.lab.2015.150808. [DOI] [PubMed] [Google Scholar]
- 20.Sohrabi A, Hajia M, Jamali F, Kharazi F. Is incidence of multiple HPV genotypes rising in genital infections? J Public Health Infect Dis. 2017;10:730–3. doi: 10.1016/j.jiph.2016.10.006. [DOI] [PubMed] [Google Scholar]
- 21.Sohrabi A, Hajia M. Cervical cancer and genital infections: assessment of performance and validation in human papillomavirus genotyping assays in Iran, its neighbor countries and Persian Gulf areas. Iran J Pathol. 2016;12:35–44. [PMC free article] [PubMed] [Google Scholar]
- 22.Sohrabi A, RahnamayeFarzami M, MirabSamiee S, Modarressi MH. An overview on papillomaviruses as the main cause of cervical cancer. Iran J Obstet Gynecology Infertil. 2015;18:14–25. [Google Scholar]
- 23.Sohrabi A, Mirab-Samiee S, Rahnmaye-Farzami M. C13orf18 and C1orf166 (MULAN) DNA genes methylation are not associated with cervical cancer and precancerous lesions of human papillomavirus genotypes in Iranian women. Asian Pac J Cancer Prev. 2014;15:6745–6748. doi: 10.7314/apjcp.2014.15.16.6745. [DOI] [PubMed] [Google Scholar]
- 24.Sohrabi A, Mirab-Samiee S, Modarresi MH, et al. Development of in-house multiplex real time PCR for human papillomavirus genotyping in iranian women with cervical cancer and cervical intraepithelial neoplasia. Asian Pac J Cancer Prev. 2014;15:6257–61. doi: 10.7314/apjcp.2014.15.15.6257. [DOI] [PubMed] [Google Scholar]
- 25.Sun B, He J, Chen X, et al. Prevalence and genotype distribution of human papillomavirus infection in Harbin, Northeast China. Arch Virol. 2014;159:1027–32. doi: 10.1007/s00705-013-1886-1. [DOI] [PubMed] [Google Scholar]
- 26.Tran LT, Tran LT, Bui TC. Risk factors for high risk and multi type human papillomavirus infections among women in Ho Chi Minh City, Vietnam: a cross-sectional study. BMC Womens Health. 2015;15:16. doi: 10.1186/s12905-015-0172-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tommasino M. The human papillomavirus family and its role in carcinogenesis. Semin Cancer Biol. 2014;26:13–21. doi: 10.1016/j.semcancer.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 28.Trottier H, Mahmud S, Costa MC. Human papillomavirus infections with multiple types and risk of cervical neoplasia. Cancer Epidemiol Biomarkers Prev. 2006;15:1274–80. doi: 10.1158/1055-9965.EPI-06-0129. [DOI] [PubMed] [Google Scholar]
- 29.Vaccarella S, Franceschi S, Snijders PJ, et al. Concurrent infection with multiple human papillomavirus types: pooled analysis of the IARC HPV Prevalence Surveys. Cancer Epidemiol Biomarkers Prev. 2010;19:503–10. doi: 10.1158/1055-9965.EPI-09-0983. [DOI] [PubMed] [Google Scholar]
- 30.Wentzensen N, Schiffman M, Dunn T. Multiple human papillomavirus genotype infections in cervical cancer progression in the study to understand cervical cancer early endpoints and determinants. Int J Cancer. 2009;125:2151–8. doi: 10.1002/ijc.24528. [DOI] [PMC free article] [PubMed] [Google Scholar]


