Abstract
Nasopharyngeal carcinoma (NPC), although not very common in many parts of the world, is a major concern in some countries, including Iran. Molecular studies are very helpful to provide essential information regarding underlying carcinogenetic mechanisms. Here, considering NPC proteomic approaches, established biomarkers were designated for protein-protein interaction network construction and analysis with corresponding plug-ins. A network of reported protein markers was constructed and topological and biological process features were investigated. Centrality analysis showed that JUN, CALM1, HSB1, and SOD1 are more important than other differentially expressed proteins in an interacting pattern. What is more, by extending the network, Tp53, PRDM10, AKT1, ALB, HSP90AA1, and EGFR achieved the highest values for NPC network strength. It can be concluded that these proteins as well as their contributing processes, particularly in a second network, may be important for NPC onset and development. Targeting these candidate proteins may allow novel treatment approaches following appropriate validation.
Keywords: Nasopharyngeal carcinoma, protein−protein interaction network analysis, topological analysis
Introduction
Nasopharyngeal carcinoma, the cancer of head and neck, has been known as one of the most common type of nasopharyngeal cancers with poor prognosis (Janvilisri, 2015; Safavi-Naini et al., 2015). While the incident of NPC is not very high in the world, it has been reported with an increasing rate in some regions (Safavi et al., 2015). NPC while not occur frequently among population around the world, it has a significant distribution in distinct areas (Luo et al., 2017). Similarly, In Iran, the trend is increasing, particularly for men (Safavi-Naini et al., 2015).
Nasopharyngeal carcinoma is a very complex malignancy, there are many contributing phenomenon to its etiology (Tao and Chan, 2007). That is, genetic, infection such as Epstein-Barr virus (EBV), and environment factors, such as the consumption of salt-preserved fish are correlated to the risk of NPC (Chen et al., 2015). EBV has strong linkage to NPC. In Iran, EBV type 1, is dominant in NPC patients (Shahani et al., 2017). The clinical manifestation of NPC can be lump in the neck region, blood-stained post nasal drip, tinnitus, hearing impairment, and headache that usually appear in late stages. Absence of diagnostic features of NPC in early stages can hamper the treatment goals (Yue et al., 2017). The only practical treatment for NPC is radiotherapy with or without chemotherapy (Ameri et al., 2017); however, 20% of these patients has been reported radioresistant (Chen et al., 2015). Therefore, a need for optimizing and reaching a better treatment options is sensed. Meanwhile, molecular approaches can be favorable in this regard due to better understanding of mechanisms of any type of cancers (Karbalaei et al., 2017).
Recently, molecular pathogenesis of NPC has also been in a great attention (Chou et al., 2008; Ooft et al., 2017). As mentioned earlier, genetic components play indispensable role in different types of malignancies. Many genes may correspond to cancer trigger and development (Lo and Huang, 2002). Moreover, as it is known that proteins are the final functional part of normal and abnormal phenotype of a cell; studying at this scale, may provide more tangible molecular information (Rezaei-Tavirani et al., 2017). In this view, a systematic study at the protein levels by the mean of proteomics can offer more information (Rezaie-Tavirani et al., 2017).
In addition to proteomics, one of the widespread used new disciplines in molecular studies is protein-protein interaction network analysis. This evaluation underlies the importance of some the specific proteins that are more prominent than the others considering their centrality properties. Any changes in these protein may promote abnormal connection between proteins and consequently resulting in a disease state (Safaei et al., 2017). In this study, proteins from proteomic investigations that are extensively applied for biomarker identification of NPC (Li et al., 2008; Chen et al., 2015) were considered for network evaluation. The introduced proteomic studies provided some important proteins related to NPC that may serve as useful biomarkers in diagnostic and therapeutic feature of disease (Chen et al., 2015). These proteins were applied for our network study and analyzed via relevant methods.
Materials and Methods
As known, one of the proper sources for protein-protein interaction network analysis is proteome investigations (Squires et al., 2017). In this study, protein biomarkers identified by proteomic approaches are used for protein-protein interaction network construction and analysis. There are many conducted proteomic evaluation on NPC. A paper published by Ze-Tan Chen et al., (2015) reviewed proteomic investigations on NPC. This study was conducted in 2015 and several proteins related to cell proteomics, mice proteomics, and human body fluids and tissues of NPC were gathered. In the present study the proteins related to human body fluid and tissue from this review article were extracted and combined with the same data from proteomic sources related to NPC from 2015 to 2017. By listing the total extracted proteins, it is possible to organize patterns of proteins expression changes in NPC.
Network construction of these proteins was handled via Cytoscape v, 3.5.1. (Shannon et al., 2003). The source for network query was STRING DB V 10.5 (http://string-db.org/), Plug-in. Four different sources are applicable through String db including protein query, PubMed query, STITCH, and disease query (Szklarczyk et al., 2017). Here, the protein query was the chosen source for interaction analysis. Two networks of differentially expressed proteins with and without neighbor proteins were constructed. The number of additional nodes (as neighbor proteins) was set to 100 and the combined interaction score for both networks was set to 0.5. Furthermore, the network central analysis is conducted by Network Analyzer. This analyzer is a well-integrated plug-in in Cytoscape (Assenov et al., 2007). The centrality analysis was done by considering cut offs for degree (K), betweenness centrality (BC) based on 10% of the highest values. The proteins with the highest values of degree and betweenness are known as hub and bottlenecks, respectively. Those nodes possessing both feature are categorized as hub-bottlenecks, which are very imperative for a PPI network function (Wu et al., 2009).
The enrichment analysis of the central proteins of the network and differentially expressed ones was followed by ClueGO+ CluePedia Cytoscape Plug-in (Bindea et al., 2009; Bindea et al., 2013). The criteria for biological process analysis for differentially expressed proteins and also central proteins of the network are as follow:
Kappa Score: 0.4, Min level of ontology: 3 Max level of ontology: 8, Number of min gene per term: 2 and 3, Percentage of min gene per term: 3 and 4, The correction method: Bonferroni step down, Enrichment/depletion test for the terms: 2-sided enrichment/depletion based on hypergeometric method.
The kappa score expresses how much the terms are grouped together as clusters. Here, it is assigned 0.4 as the default option of CluePedia Panel.
Results
Proteins presented by proteomic studies are gathered from one review article which is discussed about proteomic of NPC by Ze-Tan Chen et al., (2015) published in 2015 and recent proteomic investigations from 2015 to 2017. The list of resulted proteins and their expression pattern is tabulated in Table 1.
Table 1.
The List of Identified NPC Proteins via Proteomic Studies from Human Body Fluid and Tissue. The proteins which are added to the list of Chen et al., 2015 are refered to the correspoded references. If there were several reports about individual protein, the number of documents are shown by superscript numbers
| Row | Gene Name | Protein Name | Up-regulated | Down-regulated |
|---|---|---|---|---|
| 1 | CP | Ceruloplasmin2(Doustjalali et al., 2015) | √ | |
| 2 | HSP70 | Heat shock 70 kDa protein 2 | √ | √ |
| 3 | HSP60, HSPD1 | 60 kDa heat shock protein | √ | |
| 4 | PHB | Prohibitin4 | √ | √ |
| 5 | KRT 19 | Keratin-194 | √ | √ |
| 6 | KRT5 | Keratin-52(Xiao et al., 2017) | √ | |
| 7 | NME1 | nm-23 protein2 | √ | |
| 8 | VIM | Vimentin3 | √ | √ |
| 9 | HSPB1 | Heat shock protein beta-15(Cai et al., 2015) | √ | |
| 10 | STMN1 | Stathmin | √ | |
| 11 | KRT 8 | Keratin-83 | √ | √ |
| 12 | ANXA3 | annexin-A3 | √ | |
| 13 | ANXA1 | annexin-A12 | √ | √ |
| 14 | ENO1 | α-enolase2 | √ | √ |
| 15 | slCAM-1 | Intercellular adhesion molecule 1 | √ | |
| 16 | CTSG | cathepsin G | √ | |
| 17 | HNRNPK | Heterogeneous nuclear ribonucleoprotein Q | √ | |
| 18 | SOD1 | Superoxide dismutase 14 | √ | √ |
| 19 | LGALS1 | Galectin-1 | √ | |
| 20 | STMN1 | Stathmin 1 | √ | |
| 21 | KIT | Mast/stem cell growth factor receptor Kit | √ | |
| 22 | ATP1A1 | Sodium/potassium-transporting ATPase subunit alpha-1 | √ | |
| 23 | KRT31 | Keratin-31 | √ | |
| 24 | SYN1 | Synapsin | √ | |
| 25 | MAP2K4 | SEK1 | √ | |
| 26 | H2AFX | histone H2AX | √ | |
| 27 | KLKB1 | KalliKrein | √ | |
| 28 | POSTN | Periostin | √ | |
| 29 | KRT1 | keratin-1 | √ | |
| 30 | SOD2 | Manganese superoxide dismutase (Mn-SOD) | √ | |
| 31 | GSTO1 | Glutathione S-transferase ω1 (GST ω1) | ||
| 32 | PPIA | Cyclophilin A (CYPA) | √ | |
| 33 | CA2 | Carbonic Anhydrase 2(Luo et al., 2017) | √ | |
| 34 | NPM1 | Nucleophosmin (Cai et al., 2015) | √ | |
| 35 | NCL | Nucleolin (Cai et al., 2015) | √ | |
| 36 | CALM1 | Calmodulin-1(Meng et al., 2017) | √ | |
| 37 | RKIP | Raf kinase inhibitor protein2 | √ | |
| 38 | KRT18 | keratin-18 | √ | |
| 39 | SFN | 14-3-3σ2 | √ | |
| 40 | ANXA2 | annexin-A2 | √ | |
| 41 | GDIA1 | Rho GDP dissociation inhibitor (GDI) Β2 | √ | |
| 42 | TPI1 | triosephosphate isomerase | √ | |
| 43 | NM23 | NM-23-H1 proteins | √ | |
| 44 | C-Jun | Transcription factor AP-1 | √ | |
| 45 | HNRPC | heterogeneous nuclear ribonucleoproteins C1/C2 | √ | |
| 46 | CTSD | Cathepsin D2 | √ | |
| 47 | AHSG | α2-HS glycoprotein(Doustjalali et al., 2015) | √ |
Network construction of the NPC corresponding proteins can provide a new perspective of the malignancy features as indicated in Figures 1 and 2. In the first network (Figure 1), 47 proteins without neighbor proteins are organized in the form of free scale network while in the second network (Figure 2) 50 proteins are added as neighbors. In the first one, the aim is to investigate the manner, which our proteins interact with each other in an interacting system while in the second one; it is objective to show the behavior of the designated proteins in a whole interacting pattern with other adjacent agents. The lists of central proteins in the two constructed of NPC Networks are tabulated in the Tables 2 and 3. There are five and six crucial proteins related to the first and the second networks of NPC. The central nodes were selected based on degree and betweenness centrality parameters.
Figure 1.
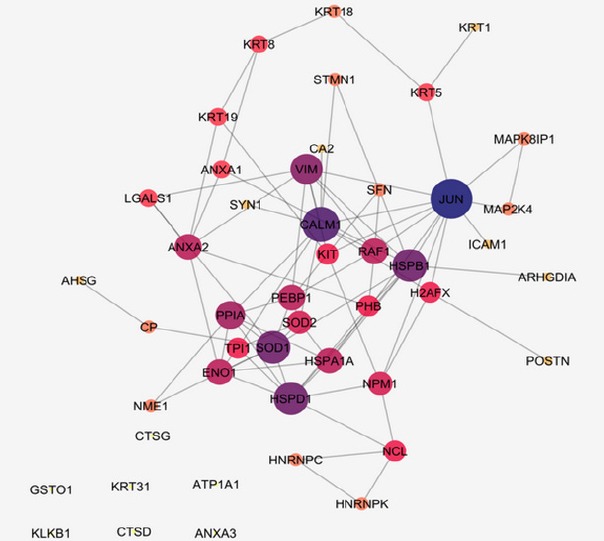
Network Analysis of NPC Proteins, Confidence Score: 0.5. The color changes from dark blue to yellow and the changes of node size reflect degree centrality value reduction. A number of 47 proteins in NPC are shown without addition of neighbor proteins with the number of edges of 82. Six proteins are isolated including GSTO1, KRT31, ATP1A1, KLKB1, CTSD, and ANXA3. These proteins are not in the interaction when designating confidence score of 0.5
Figure 2.
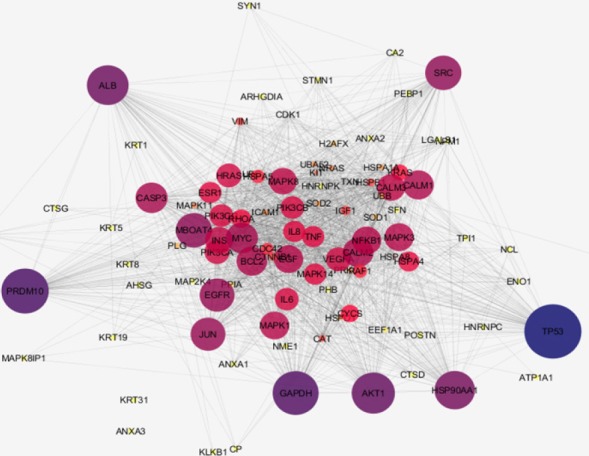
Network Analysis of NPC Proteins with Addition of Neighbor Proteins. Nodes number: 97 Edge number: 1587. Confidence score: 0.5. The color changes from dark blue to yellow and the changes of node size indicate degree centrality value reduction. ANXA3 and KRT31 are remained isolated.
Table 2.
The List of Central Proteins in the First NPC Network. The proteins are ranked based on degree values, which are hubs. The cut offs are designated by above 10% of the highest values of degree and betweenness (BC). Considering 10% of highest values for degree and betweenness includes five nodes for each that are also common. The cut off based on calculation for degree and BC are 9 and 0.12. Here the common nodes above the designated threshold is considered as the hub-bottlenecks.
| Row | Name | Degree | BC |
|---|---|---|---|
| 1 | JUN | 12 | 0.33 |
| 2 | CALM1 | 10 | 0.18 |
| 3 | HSPB1 | 9 | 0.18 |
| 4 | HSPD1 | 9 | 0.12 |
| 5 | SOD1 | 9 | 0.15 |
Table 3.
The List of Hub-Bottlenecks Ranked Based on Highest Value of Degree. The cut off for degree was above 58 and for BC was 0.02 Common proteins are known as hub-bottleneck proteins. Considering 10% of highest values for degree and betweenness includes ten nodes for each that six of which are common.
| Row | Name | Degree | BC |
|---|---|---|---|
| 1 | TP53 | 74 | 0.05 |
| 2 | PRDM10 | 67 | 0.03 |
| 3 | AKT1 | 64 | 0.03 |
| 4 | ALB | 63 | 0.05 |
| 5 | HSP90AA1 | 62 | 0.03 |
| 6 | EGFR | 58 | 0.02 |
The results of biological process analysis of NPC related central proteins (the tabulated proteins in the Table 3) are shown in Pie chart as depicted in Figure 3. The terms were classified based on statistical parameters, which are described in the legend of Figure 3. As it is shown in the Figure 4, the significant biological processes are positive regulation of nitric oxide biosynthetic process, release of cytochrome c from mitochondria, response to antibiotic, positive regulation of reactive oxygen species metabolic process, and protein insertion to membrane. Two proteins including ALB and PRDM10 are not in the result based on the assigned criteria.
Figure 3.
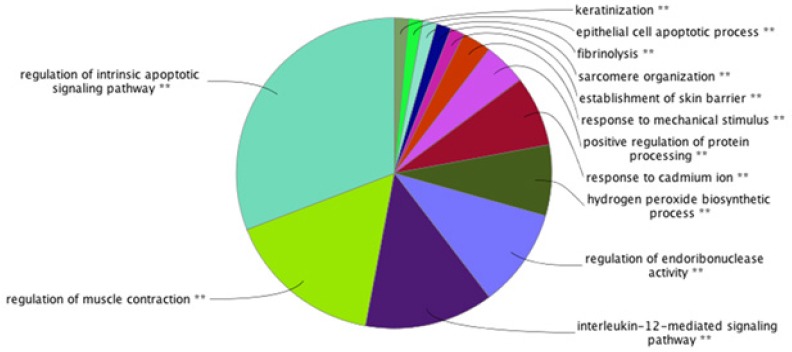
Pie Chart Biological Process Analysis of NPC Related Proteins from Proteomic Studies. As it is clear, the pie chart specify which group is the most highlighted in NPC. These groups are significantly P< 0.05 associated with NPC. The light blue colored area is the most highlighted biological process for our studied proteins, which is regulation of intrinsic apoptotic signaling pathway. The other ranked bps are regulation of muscle contraction, interleukin-12-mediated signaling pathway, regulation of endoribonuclease activity, hydrogen peroxide biosynthetic process, response to cadmium ion, positive regulation of protein processing, establishment of skin barrier, sarcomere organization, fibrinolysis, epithelial cell apoptotic process, and keratinization. Number of genes per term: 3, percentage of genes per term: 4% Kappa score=0.4
Figure 4.
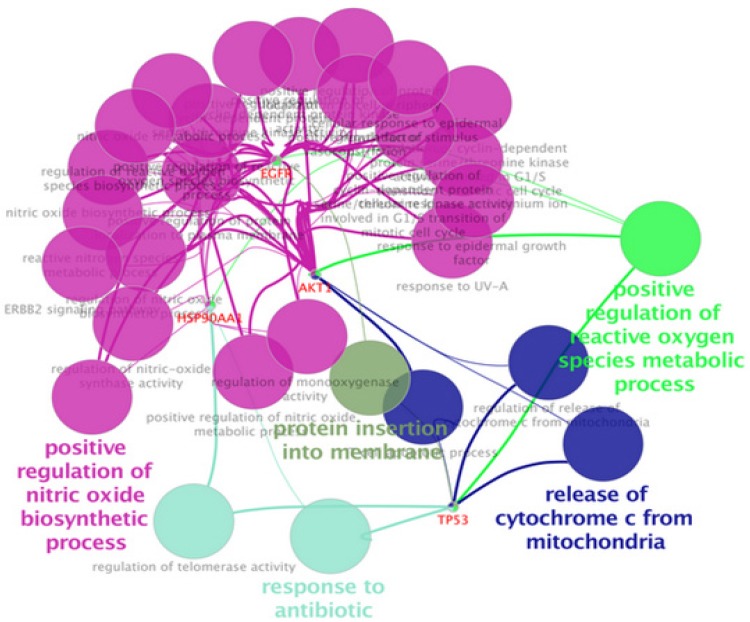
Clusters of Biological Process Identification of Central Proteins in the Second Network. The significant processes are positive regulation of nitric oxide biosynthetic process, release of cytochrome c from mitochondria, response to antibiotic, positive regulation of reactive oxygen species metabolic process, and protein insertion to membrane. Two proteins including ALB and PRDM10 are not in the result based on the assigned criteria. Number of genes per term: 2, percentage of genes per term: 3% Kappa score=0.4
Discussion
As one of the main contributing factors in NPC onset and development is molecular part, the role of these agents particularly proteins and their communications worth more investigations. In this regard, by defining NPC linked biological processes, a wider interpretation of the neoplasm underlying mechanisms can be achievable (Khayyer et al., 2017). Here it is tried to have a comprehensive analysis of interactome portrait of NPC related candidate proteins. At first the chosen candidate proteins were listed based on expression profile as presented in Table 1. As it is shown in the Table 1, there are eight proteins which their expression patterns are reported both up and down regulated by different documents. These proteins are from a paper that reviews NPC proteomic studies published in 2015 and the recent studies after this year. As it is depicted in this table, presence of 16 proteins among studied documents is repeated several times (2 to 5 times). These repeatable reports may entail on their significant linkage to NPC. It can also be interpreted that most of our studied proteins are over-expressed in NPC. In figure 1, network analysis of the reported proteins in NPC, showed that some proteins demonstrate higher centrality values based on degree and betweenness centrality comparing to the other elements. These five proteins are tabulated in table 2. Among them, SOD1, is also referred by many studies that has linkage to NPC (see Table 1). So, here, the central values of this proteins supports additional possible prominent role of this protein in NPC. On the other hand, as it is shown in the Figure 1, there are six isolated genes including GSTO1, KRT31, ATP1A1, KLKB1, CTSD, and ANXA3. After adding the neighbor nodes (see Figure 2) the numbers of isolated nodes decreases to two genes including KRT31 and ANXA3. It can be concluded that these two isolated proteins have not impact in construction of NPC network and their expression changes are happened under regulatory effect of the other genes. Likewise, the other four proteins that now are involved in the network, do not express high centralities values. Therefore, no six individual proteins may have fundamental properties in NPC network strength. The central proteins as our query proteins in network 1, which are shown in the Table 2, are not among the hub-bottlenecks in network 2. This implies on the fact that, there is a group of central proteins aside from the query proteins which conducting our NPC PPI network system. Among ten high ranked proteins regarding degree and betweenness centralities, six were common that were considered as hub-bottlenecks. Evaluating the recognized central proteins including TP53, AKT1, ALB, HSP90AA1, and EGFR through literature denotes some connections of these proteins to NPC via other molecular approaches (Li et al., 2013; Irungu et al., 2015; Ooft et al., 2015; Sahu et al., 2016). In addition, EGFR differentially expression was reported by a cell proteomic study (Ruan et al., 2011). Further evaluation based on gene ontology, explored some biological process corresponding to our up and down regulated proteins. These processes are important about their role in the NPC onset and development (Mirvish, 1995). In other words, alteration and malfunction of them may related to the NPC mechanisms. The dominant process of the central proteins identified as regulation of intrinsic apoptotic signaling pathway as shown in Figure 3 as a part of a pie chart. It has been well-known that apoptosis has crucial associations with cancer events (Ghobrial et al., 2005). The most involved biological process for our differentially expressed proteins as revealed in Figure 4, is positive regulation of nitric oxide biosynthetic process that may play indispensable role in NPC as it is also reported for high participation in progression of other tumors of head and neck (Choudhari et al., 2013).
In conclusion, examining networks of NPC reflects that, apparently, there are central proteins with relevant processes, which may be chief in this malignancy. However, this finding needs to be well-investigated and analyzed by validation tests before considering for prognosis and treatment approaches.
Acknowledgments
This research is derived from Ph.D. thesis of Dr. Mona Zamanian Azodi.
References
- 1.Ameri A, Mortazavi N, Kashi ASY, et al. Clinical outcome and prognostic factors for nasopharyngeal carcinoma: A single institution study in Iran. Int J Cancer Manag. 2017;10:1–5. [Google Scholar]
- 2.Assenov Y, Ramírez F, Schelhorn S-E, et al. Computing topological parameters of biological networks. Bioinformatics. 2007;24:282–4. doi: 10.1093/bioinformatics/btm554. [DOI] [PubMed] [Google Scholar]
- 3.Bindea G, Galon J, Mlecnik B. CluePedia cytoscape plugin: pathway insights using integrated experimental and in silico data. Bioinformatics. 2013;29:661–3. doi: 10.1093/bioinformatics/btt019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bindea G, Mlecnik B, Hackl H, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. 2009;25:1091–3. doi: 10.1093/bioinformatics/btp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cai XZ, Zeng WQ, Xiang Y, et al. iTRAQ-based quantitative proteomic analysis of nasopharyngeal carcinoma. J Cell Biochem. 2015;116:1431–41. doi: 10.1002/jcb.25105. [DOI] [PubMed] [Google Scholar]
- 6.Chen Z-T, Liang Z-G, Zhu X-D. A review: proteomics in nasopharyngeal carcinoma. Int J Mol Sci. 2015;16:15497–530. doi: 10.3390/ijms160715497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chou J, Lin YC, Kim J, et al. Nasopharyngeal carcinoma-review of the molecular mechanisms of tumorigenesis. Head Neck. 2008;30:946–63. doi: 10.1002/hed.20833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choudhari SK, Chaudhary M, Bagde S, et al. Nitric oxide and cancer: a review. World J Surg Oncol. 2013;11:118. doi: 10.1186/1477-7819-11-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doustjalali S, Bhuiyan M, Al-Jashamy K, et al. Differential expression of serum ceruloplasmin and α2-HS glycoprotein among nasopharyngeal carcinoma patients. J Mol Biomark Diagn S. 2015;2:2. [Google Scholar]
- 10.Ghobrial IM, Witzig TE, Adjei AA. Targeting apoptosis pathways in cancer therapy. CA Cancer J Clin. 2005;55:178–94. doi: 10.3322/canjclin.55.3.178. [DOI] [PubMed] [Google Scholar]
- 11.Irungu CW, Oburra HO, Ochola B. Prevalence and Predictors of Malnutrition in Nasopharyngeal Carcinoma Clinical medicine insights. Ear Nose Throat. 2015;8:19. doi: 10.4137/CMENT.S12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Janvilisri T. Omics-based identification of biomarkers for nasopharyngeal carcinoma. Dis Markers. 2015;2015:1–10. doi: 10.1155/2015/762128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karbalaei R, Piran M, Rezaei-Tavirani M, et al. One systems biology analysis protein-protein interaction of NASH and IBD based on comprehensive gene information. Gastroenterol Hepatol Bed Bench. 2017;10:194–201. [PMC free article] [PubMed] [Google Scholar]
- 14.Khayyer N, Azodi MZ, Mansouri V, et al. Oral squamous cell cancer protein-protein interaction network interpretation in comparison to esophagus adenocarcinoma. Gastroenterol Hepatol Bed Bench. 2017;10:118–24. [PMC free article] [PubMed] [Google Scholar]
- 15.Li F, Guan Y, Chen Z. Proteomics in nasopharyngeal carcinoma. Cell Mol Life Sci. 2008;65:1007–12. doi: 10.1007/s00018-008-7444-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li G, Qiu Y, Su Z, et al. Genome-wide analyses of radioresistance-associated miRNA expression profile in nasopharyngeal carcinoma using next generation deep sequencing. PLoS One. 2013;8:e84486. doi: 10.1371/journal.pone.0084486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo K-W, Huang DP. Genetic and epigenetic changes in nasopharyngeal carcinoma. Semin Cancer Biol. 2002;l:451–62. doi: 10.1016/s1044579x02000883. [DOI] [PubMed] [Google Scholar]
- 18.Luo Y, Mok TS, Lin X, et al. SWATH-based proteomics identified carbonic anhydrase 2 as a potential diagnosis biomarker for nasopharyngeal carcinoma. Sci Rep. 2017;7:1–11. doi: 10.1038/srep41191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meng H, Zhu X, Li L, et al. Identification of CALM as the potential serum biomarker for predicting the recurrence of nasopharyngeal carcinoma using a mass spectrometry-based comparative proteomic approach. Int J Mol Med. 2017;40:1152–64. doi: 10.3892/ijmm.2017.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ooft ML, Braunius WW, Heus P, et al. Prognostic significance of the EGFR pathway in nasopharyngeal carcinoma: a systematic review and meta-analysis. Biomarkers. 2015;9:997–1010. doi: 10.2217/bmm.15.68. [DOI] [PubMed] [Google Scholar]
- 21.Ooft ML, van Ipenburg J, van Loo R, et al. Molecular profile of nasopharyngeal carcinoma: analysing tumour suppressor gene promoter hypermethylation by multiplex ligation-dependent probe amplification. J Clin Pathol. 2017;2017:204661. doi: 10.1136/jclinpath-2017-204661. [DOI] [PubMed] [Google Scholar]
- 22.Rezaei-Tavirani M, Okhovatian F, Azodi MZ, et al. Duchenne muscular dystrophy (DMD) protein-protein interaction mapping. Iran J Child Neurol. 2017;11:7–14. [PMC free article] [PubMed] [Google Scholar]
- 23.Rezaie-Tavirani M, Hasanzadeh H, Seyyedi S, et al. Proteomic analysis of the effect of extremely low-frequency electromagnetic fields (ELF-EMF) with different intensities in SH-SY5Y neuroblastoma cell line. Lasers Med Sci. 2017;8:79–83. doi: 10.15171/jlms.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruan L, Li X-H, Wan X-X, et al. Analysis of EGFR signaling pathway in nasopharyngeal carcinoma cells by quantitative phosphoproteomics. J Proteome Sci. 2011;9:35. doi: 10.1186/1477-5956-9-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Safaei A, Tavirani MR, Azodi MZ, et al. Diabetic ?etinopathy and laser therapy in rats: A protein-protein interaction network analysis. Lasers Med Sci. 2017;8:20–1. doi: 10.15171/jlms.2017.s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Safavi-Naini A, Raad N, Ghorbani J, et al. Incidence trends and geographical distribution of nasopharyngeal carcinoma in Iran. Iran J Cancer Prev. 2015;8:24. [PMC free article] [PubMed] [Google Scholar]
- 27.Safavi A, Raad N, Raad N, et al. Epidemiology of nasopharyngeal cancers in Iran: A 6-year report. Asian Pac J Cancer Prev. 2015;16:4447–50. doi: 10.7314/apjcp.2015.16.10.4447. [DOI] [PubMed] [Google Scholar]
- 28.Sahu S, Chakrabarti S, Roy S, et al. Association of p53 codon72 Arg>Pro polymorphism with susceptibility to nasopharyngeal carcinoma: evidence from a case–control study and meta-analysis. Oncogenesis. 2016;5:e225. doi: 10.1038/oncsis.2016.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shahani T, Makvandi M, Samarbafzadeh A, et al. Frequency of epstein barr virus type 1 among nasopharyngeal carcinomas in Iranian patients. Asian Pac J Cancer prev. 2017;18:327. doi: 10.22034/APJCP.2017.18.2.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Squires S, Ewing R, Prugel-Bennett A, et al. A method of integrating spatial proteomics and protein-protein interaction network data. International Conference on Neural Information Processing. 2017:782–90. [Google Scholar]
- 32.Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:362–8. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tao Q, Chan AT. Nasopharyngeal carcinoma: molecular pathogenesis and therapeutic developments. Exp Rev Mol Med. 2007;9:1–24. doi: 10.1017/S1462399407000312. [DOI] [PubMed] [Google Scholar]
- 34.Wu J, Vallenius T, Ovaska K, et al. Integrated network analysis platform for protein-protein interactions. Nature Methods. 2009;6:75–7. doi: 10.1038/nmeth.1282. [DOI] [PubMed] [Google Scholar]
- 35.Yue PY-K, Ha W-Y, Lau C-C, et al. Micro RNA profiling study reveals miR-150 in association with metastasis in nasopharyngeal carcinoma. Sci Rep. 2017;7:12012. doi: 10.1038/s41598-017-10695-2. [DOI] [PMC free article] [PubMed] [Google Scholar]


