Abstract
Purpose:
The impact of the BRCA1-3’UTR-variant on BRCA1 gene expression and altered responses to external stimuli was previously tested in vitro using a luciferase reporter assay. Its ability to predict breast cancer risk in women was also assessed but the conclusions were inconsistent. The present study concerns the relationship between the BRCA1-3’UTR germline variant rs8176318G>T and susceptibility to Breast cancer in an ethnic population of Saudi Arabia.
Methodology:
The study included 100 breast cancer patients and 100 sex matched healthy controls from the northwestern region (Tabuk) and Dammam of Saudi Arabia were investigated for the BRCA1-3’UTR germline variant rs8176318G>T using an allele specific PCR technique. Genotype distributions were then compared.
Results:
The frequencies of the three genotypes GG, TT and GT in our Saudi Arabian patients were 26%, 8% and 66% and in healthy controls were 45%, 5% and 50%, respectively (p=0.03). Risk of developing breast cancer was found to be significantly associated with the GT variant (OR 2.28, 1.24-4.191; RR 1.47, 1.11-1.93; P=0.007), GT+TT (OR, 2.32, 1.28-4.22; RR 1.48, 1.13-1.94; P=0.005) and the T allele (OR 1.62, 1.072- 2.45; RR 1.28, 1.02-1.60: P=0.020). There were 2.76 and 2.28 fold increase risks of developing breast cancer associated with the TT and GT genotypes in our cases. A significant correlation was also found between the BRCA1 3’UTR variants with the stage of the disease and distant metastasis but not with age, grade, and ER, PR and her2/neu status.
Conclusion :
The rs8176318G/T in the 3’untranslated region (UTR) of the BRCA1 gene was found to be associatedwith increased susceptibility to breast cancer in our study population, increased risk being noted with the GT and TT genotypes. Further association studies are needed to confirm this finding in other regions of Saudi Arabia.
Keywords: BRCA1 gene, BRCA1 3’UTR variants, rs8176318G/T, ER-estrogen receptor-PR-progesterone receptor
Introduction
Breast cancer is the most frequently diagnosed noncutaneous malignancy in women (Bassam et al., 2017). It is the most common cancer in females and is ranked second in cancer-related deaths all over the world (Albeshan et al., 2017). Saudi Arabia has the lowest rate of breast cancer incidence in the Arab world. The nation wide average of incidence in the Kingdom is 22 patients for every 100,000 women. In the UAE, 23 patients, Kuwait 46 patients, Jordan 49 patients, Qatar 48 patients and Bahrain 53 patients, for every 100,000 women (Chouchane et al., 2014). Despite improvement in diagnosis, the survival rate of this disease has still not improved and also there is a substantial rise in the incidence of breast cancer in Saudi Arabia in recent years, particularly among younger females compared to affected females’ in western countries (AlJohani el al., 2016). The breast cancer 1 (BRCA1) gene encodes a tumor suppressor protein that plays a critical role in the DNA damage response and repair pathways and functions as a negative regulator of tumor growth (Wu et al., 2016). Lot of efforts are being made to identify the genetic basis of familial breast cancer achieved success few years ago, when the breast- cancer-susceptibility genes BRCA1 and BRCA2 were identified through positional cloning (Nathanson et al., 2001). Germline mutations in either of these genes account for 20-60% of breast cancer cases in families where multiple individuals are affected (~ 2-6% of all cases). Women who have an abnormal BRCA1 or BRCA2 gene (or both) can have up to an 80% risk of being diagnosed with breast cancer during their lifetimes (Smith et al., 2007). BRCA1 is a tumor suppressor gene and operates in a series of cellular processes, including DNA repair, chromatin remodeling, protein ubiquitination, regulation of transcription, apoptosis and cell cycle checkpoint control (Ayoub et al., 2011). Epidemiological studies sparked by the discovery of BRCA1 and BRCA2 have made clear several features of inherited mutations in the genes (Ford et al., 1998). The mutations are highly penetrant, carrying a lifetime risk of 30-70% for cancer incidence with variation related to genetic background (Chen et al., 2007). Recently it was reported that the germline variants in the open-reading-frame of BRCA1 confer a mean risk of 54% and 39% for developing hereditary breast cancer by the age of 70 (Easton et al.,1995).
However, BRCA1 open-reading-frame variants were reported in a small portion of hereditary breast cancer cases that occur primarily in young, premenopausal patients (Garcia et al., 2016). Previous studies have reported that the genetic variants in the BRCA1 3’ untranslated region (3’UTR) are significantly associated with breast cancer risk; however, the role of single nucleotide polymorphisms (SNPs) in the BRCA1 3’UTR remains unclear (Pongsavee et al., 2009). The search for additional germline variants, outside of the BRCA1 open-reading-frame predicting increased breast cancer risk has been undertaken. BRCA1 3’UTR variants at rs12516 and rs8176318 were identified in Breast and ovarian cancer in high-risk families and homozygosity for the less derived alleles A at both SNP sites and were found triple the frequency in cancer patients as seen in unaffected women, yielding a significant cancer association (Pelletier et al., 2011). It has been reported that BRCA1 3’ untranslated region SNPs are disrupting the microRNA (miRNA) binding sites therefore can act as genetic markers of cancer risk including breast and ovarian and cancer (Chen et al., 2015).
Previous studies have demonstrated that the genetic variants in the BRCA1 3’ untranslated region (3’UTR), is significantly associated with breast cancer risk. Similarly, the homozygous variants of rs8176318 were found to be associated with increased risk of breast cancer in African American women and were specifically associated with the development of triple negative breast cancer (Fang et al., 2016). Analysis showed reduced translation of BRCA1 with the derived alleles at both sites when present on the same chromosome, i.e., in cis, with the greatest reduction seen with the derived allele at rs8176318 (Brewster et al., 2012). The independent evaluation of the BRCA1-3’UTR-variants rs12516 and rs8176318 revealed significant variation in baseline frequency by ethnicity, with a documented minor allele frequency in Irish populations of approximately 0.28 (Lheureux et al., 2011). Subsequently, the distribution of all BRCA1 genotypes among 11 distinctive populations was calculated and reported that the genotype frequency distribution varied between populations worldwide (Yang et al., 2016). The discrepancies among these studies may due to the ethnic variation and relatively small sample size. To obtain a comprehensive conclusion, we conducted this case control study to evaluate the association between BRCA1 3’UTR variants rs8176318 and risk of Breast cancer in Saudi Arabia.
Materials and Methods
Blood samples were procured from clinically diagnosed Breast cancer patients. The patients were recruited from the different hospitals: Prince Sultan Oncology Center, KSAFH, Tabuk, and Department of Surgery Breast and Endocrine Unit, KSAFH, Tabuk and Department of Surgery, College of Medicine, Imam Abdulrahman Bin Faisal University (IAU), Dammam, Saudi Arabia.
The same number subjects (Sex matched) served as controls. All samples were collected and stored at -70°C and thawed immediately before assay. Complete clinical data and current therapy plan was obtained. The purpose of the sampling was explained to all patients and written informed consent was obtained prior to enrolment. After assessing the clinico-pathological findings, a four (4ml) sample of peripheral blood was collected by venipuncture in EDTA tubes from each patient and healthy control.
DNA extraction
The DNA was extracted by using DNeasy Blood Kit (cat 69506) from Qiagen (Germany) in accordance with the manufacturer’s instructions. The quality and integrity of DNA from these tissues were checked by electrophoresis on 1% agarose gel, quantified spectrophotometrically using NanoDrop™ (Thermo Scientific, USA). The extracted DNA was dissolved in nuclease-free water and stored at 4°C until use.
BRCA1-3’UTR (rs8176318 G>T) genotyping
BRCA1-3’UTR (rs8176318 G>T) genotypes were analyzed using two tube PCR assay which was performed in a reaction volume of 25uL containing 0.25 uL of 25 pmol of each primers (designing by using primer3 software) as depicted in table 1 and 10 uL from GoTaq® Green Master Mix (M7122) (Promega, USA). Final volume of 25 uL was adjusted by adding nuclease free ddH2O. The PCR coattail was prepared as depicted in the Table 2. Finally, 2ul of 50ng DNA was added from each patient.
Table 1.
Allele Specific PCR Primers for BRCA1-3’UTR Gene Variant
| Direction | Primer Sequence | AT | Product size |
|---|---|---|---|
| Allele specific PCR primers for G allele | |||
| Wild reverse Primer R1 | 5’: CCATTGAAGGGTCTGACTCTCTGTC-3 | 58°C | 171bp |
| Common Forward Primer F1 | 5’: GAGCAAGATGCTGATTCATT-3 | ||
| Allele specific PCR primers for T allele | |||
| Mutant reverse Primer R2 | 5’- CCATTGAAGGGTCTGACTCTCTGTA-3 | 171bp | |
| Common Forward Primer F2 | 5’: GAGCAAGATGCTGATTCATT-3 |
Table 2.
Preparation of PCR Cocktail for BRCA1-3’UTR Gene variant
| AS-PCR for G allele | 1x | AS-PCR for T allele | 1x |
|---|---|---|---|
| PCR master mix | 10ul | PCR master mix | 10ul |
| Common Forward primer F | 0.25 ul | Common Forward primer F | 0.25 ul |
| Wild Reverse primer R1 | 0.25 ul | Mutant Reverse R2 | 0.25 ul |
| Nuclease free water | 12.50 ul | Nuclease free water | 12.50 ul |
| DNA (50ng/ul) | 2ul | DNA (50ng/ul) | 2ul |
| Total volume | 25ul | Total volume | 25ul |
Table 3.
Clinical Data of Breast Cancer Cases
| Parameters | N= | % |
|---|---|---|
| Patients | 100 | 100% |
| Controls | 100 | 100% |
| Age Group | ||
| Age<40 | 22 | 22% |
| Age >40 | 78 | 78% |
| Stage status | ||
| Early (I and II) | 37 | 37% |
| Advanced (III and IV) | 63 | 63% |
| Grading | ||
| Grade I | 14 | 14% |
| Grade II | 33 | 33% |
| Grade III | 53 | 53% |
| Estrogen receptor status | ||
| Positive | 67 | 67% |
| Negative | 33 | 33% |
| Progesterone Receptor status | ||
| Positive | 64 | 64% |
| Negative | 36 | 36% |
| Her2/neu status | ||
| Positive | 60 | 60% |
| Negative | 40 | 40% |
| Distant Metastasis status | ||
| Positive | 65 | 65% |
| Negative | 35 | 35% |
Table 4.
Allelic Frequencies of BRCA1 rs8176318 G/T Gene Variation in Cases and Controls
| Subjects | N= | GG | TT | GT | P-value | G | T |
|---|---|---|---|---|---|---|---|
| Cases | 100 | 26 (26%) | 8 (8%) | 66 (66%) | <0.018 | 0.59 | 0.41 |
| Controls | 100 | 45 (45%) | 5 (5%) | 50 (50%) | 0.70 | 0.30 | |
| Significance | The difference is significant at p = 0.05. |
Table 5.
Association Between BRCA1 UTR Varients and Clinic pathological Features in Breast Cancer Cases
| Parameters | n | GG % | TT % | GT % | P-Value |
|---|---|---|---|---|---|
| Age Group | |||||
| Age<40 | 22 (22%) | 05 (22.72%) | 02 (9%) | 15 (68.18) | 0.554 |
| Age >40 | 78 (78%) | 21 (26.92%) | 06 (7.69%) | 51 (65.38%) | |
| Stage status | |||||
| Early stage | 37 (37%) | 11 (29.72%) | 6 (16.21%) | 20 (54%) | 0.03 |
| Advanced stage | 63 (63%) | 15 (23.80%) | 2 (3%) | 46 (73%) | |
| Grade status | |||||
| Grade I | 14 (14%) | 07 (50%) | 01 (10%) | 06 (40%) | 0.07 |
| Grade II | 33 (33%) | 06 (18.25%) | 2 (6%) | 25 (75.75%) | |
| Grade III | 53 (53%) | 13 (24.52%) | 5 (9.43%) | 35 (66%) | |
| Distant metastasis | |||||
| Positive | 65 (65%) | 10 (15.38%) | 05 (7.79%) | 50 (76.92%) | 0.003 |
| Negative | 35 (35%) | 16 (45.71%) | 03 (8.57%) | 16 (45.71%) | |
| Estrogen receptor status | |||||
| Positive | 67 (67%) | 20 (29.85%) | 05 (7.46%) | 42 (62.68%) | 0.45 |
| Negative | 33 (33%) | 06 (18.18%) | 03 (9%) | 24 (72.72%) | |
| Progesterone Receptor status | |||||
| Positive | 64 (64%) | 13 (20.31%) | 05 (7.81%) | 46 (71.87%) | 0.72 |
| Negative | 36 (36%) | 13 (36.11%) | 3 (8.33%) | 20 (55.55%) | |
| Her2/neu status | |||||
| Positive | 60 (60%) | 15 (25%) | 05 (8.33%) | 40 (66.66%) | 0.44 |
| Negative | 40 (40%) | 11 (27.5%) | 03 (7.5%) | 26 (65%) |
Table 6.
The Association of the BRCA1-3’UTR-variant with Breast Cancer Risk
| Genotypes | Healthy controls | Breast cancer | OR (95% CI) | RR (95% CI) | P-Value | ||
|---|---|---|---|---|---|---|---|
| (N=100) | % | (N=100) | % | ||||
| Co dominant | |||||||
| BRCA1-GG | 45 | (45%) | 26 | (26%) | 1 (ref.) | 1 (ref.) | |
| BRCA1-GT | 50 | (50%) | 66 | (66%) | 2.28 (1.24-4.191) | 1.47 (1.11-1.93) | 0.007 |
| BRCA1-TT | 5 | (05%) | 8 | (08%) | 2.76 (0.81- 9.35) | 1.64 (0.81-3.35) | 0.085 |
| Dominant | |||||||
| BRCA1-GG | 45 | 45% | 26 | 26% | 1 (ref.) | 1 (ref.) | |
| BRCA1 (GT+ TT) | 55 | 55% | 74 | 74% | 2.32 (1.28- 4.22) | 1.48 (1.13-1.94) | 0.005 |
| Recessive | |||||||
| BRCA1-(GG+GT) | 100 | 95% | 100 | 92.60% | 1 (ref.) | 1 (ref.) | |
| BRCA1-TT | 05 | 5% | 8 | 7.40% | 1.6 (0.506-5.05) | 1.02 (0.96-0.10) | 0.41 |
| Allele | |||||||
| BRCA1-G | 140 | 70% | 118 | 59% | 1 (ref.) | 1 (ref.) | |
| BRCA1-T | 60 | 30% | 82 | 41% | 1.62 (1.072- 2.45) | 1.28 (1.028- 1.60) | 0.020 |
*OR, Odd ratio; #RR, Risk ratio
Table 7.
Genotype Frequency of the BRCA1 Gene rs8176318 Polymorphism in Different Populations (Yang et al.,2016)
| Populations | Sample | Genotype frequency, n (%) | Allele frequency, % | |||
|---|---|---|---|---|---|---|
| N | GG | GT | TT | G | T | |
| CEU | 226 | 100 (44.2) | 102 (45.1) | 24 (10.6) | 66.8 | 33.2 |
| CHB | 82 | 38 (46.3) | 32 (39.0) | 12 (14.6) | 65.9 | 34.1 |
| GIH | 176 | 52 (29.5) | 92 (52.3) | 32 (18.2) | 55.7 | 44.3 |
| CHD | 170 | 48 (28.2) | 96 (56.5) | 26 (15.3) | 56.5 | 43.5 |
| YRI | 226 | 180 (79.6) | 42 (18.6) | 4 (1.8) | 88.9 | 11.1 |
| LWK | 180 | 152 (84.4) | 26 (14.4) | 2 (1.1) | 91.7 | 8.3 |
| JPT | 172 | 88 (51.2) | 72 (41.9) | 12 (7.0) | 72.1 | 27.9 |
| MEX | 98 | 62 (63.3) | 28 (28.6) | 8 (8.2) | 77.6 | 22.4 |
| ASW | 98 | 56 (57.1) | 42 (42.9) | 0 (0) | 78.6 | 21.4 |
| MKK | 286 | 206 (72.0) | 72 (25.2) | 8 (2.8) | 84.6 | 15.4 |
| TSI | 176 | 68 (38.6) | 88 (50.0) | 20 (11.4) | 63.6 | 36.4 |
| Saudi Arabia | 100 | 45(45) | 50(50%) | 5(5%) | 70.5 | 30.3 |
CEU, Utah residents with Northern and Western European ancestry from the Center for the Study of Human Polymorphisms collection; JPT, Japanese individuals in Tokyo, Japan; YRI, members of the Yoruba tribe in Ibadan, Nigeria; CHB, Han Chinese individuals in Beijing, China; LWK, members of the Luhya tribe in Webuye, Kenya; MEX, individuals of Mexican ancestry in Los Angeles, California; GIH, Gujarati Indians in Houston, Texas; CHD, Chinese individuals in Metropolitan Denver, Colorado; ASW, individuals of African ancestry in Southwest, USA; MKK, members of the Maasai tribe in Kinyawa, Kenya; TSI, Tuscan individuals in Italy; HWP, Hardy-Weinburg probability; T, thymine; G, guanine.
Thermo cycling conditions
The amplification conditions used were at 95 oC for 10 minutes followed by 40 cycles of 94°C for 35sec, 58 °C for 40 sec, 72 °C for 45 sec followed by the final extension at 72 °C for 10 minutes. The amplification products were separated by electrophoresis through 2% agarose gel stained with ethidium bromide. BRCA1-3’UTR wild (GG) as well as mutant genotype (TT) yielded 171bp band size as depicted in Figure 1.
Figure 1.
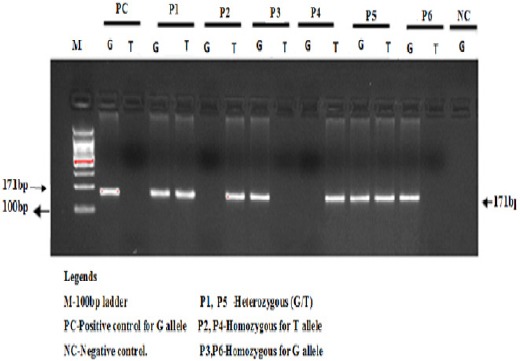
Gel Elec Trophoresis of Allele Specific PCR Amplification for the Detection of BRCA1 3’UTR (rs8176318G>T) Polymorphism in Breast Cancer Patients
Statistical Analysis
Statistical analysis Genotype frequencies between the cases and controls were evaluated using the Chi square test, Hardy-Weinberg equilibrium test used to check the allele frequency and values below 5 were analyzed by Fisher exact test. The associations between BRCA1-3’UTR genotypes and risk of breast cancer were estimated by computing the odds ratios (ORs), risk ratios (RRs) and risk differences (RDs) with 95 % confidence intervals (CIs). Allele frequencies among cases as well as controls were evaluated by using the Chi–square Hardy-Weinberg equilibrium test. A p value < 0.05 was considered significant. All statistical analyses were performed using Graph Pad Prism 6.0 or SPSS 16.0.
Ethical Approval
This study was approved by the Research Ethics committee, University of Tabuk. (HAP07/TU-001).
Results
Study population
Patient clinicopathological characteristics are shown in table 3. At the time of analysis, out of 100 breast cancer patients 22 (22%) were just below or equal to 40 years of age and 78 (78%) were above 40. Thirty seven of 100 (37%) breast cancer cases were in early (I &IIA) stage whereas 63/100 (63%) in advanced stages (IIB, III & IV). Histological grading of the patients’ tumor showed that 14(14%), 33(33%) and 53 (53%) were in grade I, II and III respectively. Of the total cases of Breast cancer 60 (60%) were positive for Her2/neu, 67 (67%) for estrogen receptor and 64 (64%) for progesterone receptor.
BRCA1 rs8176318 variants detection in breast cancer patients and healthy controls
The study included 200 subjects with 100 breast cancer cases and 100 healthy controls. In breast cancer patients, the GG, TT and GT genotype frequencies was 26%, 8% and 66% respectively, compared to 45%, 5% and 50% in healthy controls respectively as depicted in table 4. Higher frequencies of BRCA1 3’UTR (GT) heterozygosity was reported in breast cancer cases (66%) compared with healthy controls (50%). The distribution of BRCA1 3’UTR variants observed between patients and controls was highly significant (p = 0.018).
The frequency of T allele (fT) was found to be higher among breast cancer patients (0.41) than the healthy controls (0.30). However frequency of G allele (fG) was found to be lower among breast cancer patients (0.59) than the healthy controls (0.70) Table 4. It was observed that more than 2.76 fold increase risk of developing breast cancer was associated with T allele of BRCA1 3’UTR variant in Saudi population with more predominant in cases with advanced stage and distant metastasis. This is a comprehensive identification that indicated BRCA1 3’UTR variant is associated with increased risk of developing Breast cancer among our patient group.
Correlation BRCA1 rs8176318 variants with clinicopathological features in breast cancer
This study observed that BRCA1 rs8176318 variants GT genotype was associated with increased breast cancer risk (P= 0.007) when stage, metastasis were adjusted. Age, Grades, receptors status were not associated with BRCA1 rs8176318 polymorphism as depicted in Table no 4 and 5).
Association between BRCA1rs8176318 variants and Stage status in breast cancer
The genotype distribution GG, TT and TT of BRCA1 3’UTR variants (rs8176318G>T) in Breast cancer cases with respect to stage is summarized in table 5. We observed a statistically significant association between the BRCA1 3’UTR variant with the early/advanced stage of cancer (X2 =6.54, P=0.03). Early stage group included Ductal Carcinoma in Situ (DICS) patients in stage I and II of breast cancer. On the other hand, advanced stage group included breast cancer patients in stage III and IV. The frequency of GT was found to be higher among advanced breast cancer patients (73%) than early diseases cases (54%) whereas the higher frequency of GG genotype was observed among early stage cases (30%) than the advanced stage cases (24%) than the early stage cases as depicted in Table 5.
Association between BRCA1rs8176318 variants and Grades of breast cancer cases
The genotype distribution GG, TT and TT of BRCA1 3’UTR variants (rs8176318) in Breast cancer cases with respect to grades is summarized in table 5. We did not find any statistically significant difference in the frequencies of BRCA1 3’UTR variants of GG, TT and GT genotypes between the different grades of breast patients. The frequency of T allele (fT) was found to be higher in grade II and grade III breast cancer patients (0.41) than the grade I cases (0.30) however the higher frequency of G allele (fG) was observed in grade I cases (0.70) than the grade II and grade III (0.59).
Association between BRCA1rs8176318 variants and metastasis status of breast cancer cases
The genotype distribution GG, TT & TT of BRCA1 3’UTR variants (rs8176318) in Breast cancer cases with respect to metastasis is summarized in Table 5. Metastasis is a complex process in which malignant cancer cells from the breast spread into other regions of the body. Once metastasis has occurred, it is much more difficult to effectively treat breast cancer. Breast cancer can metastasize in three general areas called local recurrence, regional recurrence and metastatic or distant recurrence. Sixty five of 100 breast cancer patients (65/100) (65%) were having metastatic status of the disease and 35/100 (35%) did not have metastatic status of disease. We observed a strong significance difference in the frequency of BRCA1 3’UTR variants during the comparison of metastasis status (X2=11.43, p=0.003). The frequency of T allele (fT) was found to be higher in breast cancer patients with metastasis (0.45) than the cases without metastasis (0.28).
The association of the BRCA1-3’UTR-variant with breast cancer risk
To evaluate if there were clinical and biological impacts of the BRCA1-3’UTR-variant, we studied a genetically and environmentally homogeneous population,(ethnic population of Saudi Arabia) to best control for “context” effects on variant function. We used our case–control analysis of 100 cases and 100 controls from Saudi Arabia.
A multivariate analysis based on logistic regression like odds ratio and risk ratio with 95% confidence intervals were calculated for each group to estimate the association between the BRCA1-3’UTR genotypes (rs8176318 G>T) and risk of breast cancer in our study group as depicted in Table 6. The BRCA1-3’UTR-variant (TG or TT) is associated with a modest increased risk for developing breast cancer in the Saudi cohort OR 2.28 95% CI 1.24-4.191) RR 1.47 95% CI 1.11-1.93 p=0.007).
During the allelic comparison, the G allele was compared with the T allele and we found a highly significant association with odd ratio (OR) 1.62 (1.072- 2.45) and risk ratio (RR) 1.28 (1.02- 1.60) < (P=0.20) suggesting a possible dominant effect of this polymorphism on Breast cancer risk. The dominant model was predictive of breast cancer risk compared to controls for all breast cancer patients (OR 2.28, 95% CI 1.24-4.19). During the comparison of GG homozygous genotype with the TT homozygous genotype we did not find a significant association with the OR 2.76(0.81- 9.35) RR 1.64 (0.81-3.35) P=0.08. BRCA1 rs8176318G>T was the important risk factor in our study population. The comparison between genotype, OR and P value revealed that the BRCA1 rs8176318 GT/TT genotype was a most important risk factor in Saudi population.
Discussion
Breast cancer gene 1(BRCA1) encodes a multifunctional protein that operates in a series of cellular processes, including DNA repair, chromatin remodeling, protein ubiquitination, regulation of transcription, apoptosis and cell cycle checkpoint control. BRCA1 is well-established that its mutations result in a significantly increased lifetime risk for the development of breast cancer (Ralhan et al., 2007). Thus, BRCA1, functioning as a tumor suppressor, may be regarded as a strong candidate gene for breast cancer susceptibility. It is indicated that BRCA1 mutations has gained increasing attention due to its high risk and is one of the most important factors responsible for breast cancer cases (Ayoub et al.,2011). About 5%–50% of familial breast cancers could be explained by inherited mutations of BRCA1 in different populations (Yang et al., 2011). BRCA1 open-reading-frame variants is reported to be associated with a small number of hereditary breast cancer patients that occur primarily in young patients (Sedghi et al., 2016). Some variants in the BRCA1 3’UTR have been recently reported and were first implicated in ovarian and breast cancer susceptibility in high-risk families (Newman et al., 1998). BRCA1-3’UTR-(rs8176318G>T) polymorphism is a functional SNP whose occurrence leads to decreased BRCA1 mRNA expression in breast cancer risk patients (Dorairaj et al., 2014). BRCA1-3’UTR polymorphism (rs8176318G>T) has been recently studied in different ethnic populations of the globe and displayed differences in genotype frequency distribution among populations worldwide (Yang et al., 2016) as shown in Table 7.
Distribution of BRCA1-3’UTR-variant G/T genotypes in different populations of the world
Our study observed high percentage of GT (66%) and TT (8%) genotype in patients compared to controls GT (50%) and TT (5%) genotype while lower GG (26%) genotype in patients compared to control CC (45%) genotype. Furthermore, our study reported a statistically significant difference of BRCA1 3’UTR genotypes (GG, GT and TT) between patients and healthy controls (p=0.018). The distribution of BRCA1-3’UTR-variant (rs8176318G>T) genotypes varies among different ethnic groups as depicted in Table 7. Comparing the frequencies of BRCA1-3’UTR-G>T heterozygosity in our study group with other studies, we found GT heterozygosity generally comparable with Italy (TSI) 50%, African ancestry in Southwest USA (ASW) 43%, Japan in Tokyo (JPT) 42%, Chinese in Colorado (CHD) 56%, Gujarati Indians in Houston, Texas (GIH) 52%, European ancestry in Utah, USA (CEU) 45% (26) and 50% in our study population.
Distribution of BRCA1-3’UTR-variant TT genotypes in different populations
Similarly comparing the frequencies of BRCA1-3’UTR-TT genotype in our study with other studies, we found TT homozygosity generally comparable with Nigeria (YRI) (1.8%), Kenya (LWK) (1.1%), Japanese (JPT) (7%), Tuscan (TSI) (11%), (Kinyawa, Kenya) MKK (2.8%) and, (Mexican ancestry in Los Angeles) MEX (8.2) (Brewster BL et al.,2012) as depicted in Table 7.
There have been previous studies conducting haplotype analysis in the BRCA1 region to determine their association with sporadic breast cancer, with little success. However, this data suggests that the BRCA1-3’UTR-variant rs8176318G>T confers an increased risk of developing breast cancer in our group of patients. One could hypothesize from these findings that the BRCA1-3’UTR-variant functions similarly to that of canonical BRCA1 open-reading-frame variants, which are more commonly associated with development of breast cancer as opposed to the other subtypes. Evidence is fast becoming available to support the theory that BRCA1-3’UTR-variant increase susceptibility to cancer through gene expression control (Freedman et al., 2005). Previous study demonstrated that the BRCA1 polymorphism, rs799917, was associated with breast cancer risk (Holm et al., 2010). However, in another study, it was reported that rs799917 played no significant association with breast cancer in Chinese women (Huo et al., 2009). Therefore, the role of genetic variants in the BRCA1 3’UTR and its post-transcriptional regulation remains unclear. Such differences may be due to reproductive patterns, in addition to exposure to particular environmental carcinogens, different lifestyles or different genetic backgrounds.
The present study was conducted on the BRCA1 3’UTR gene polymorphism (rs8176318G>T) in an ethnic population from Saudi Arabia. Furthermore, BRCA1 3’UTR gene polymorphism (rs8176318G>T) exhibited differences in genotype frequency distribution among populations worldwide. This SNP may have a significant association with BRCA1 mRNA expression, therefore implying that these SNPs may partially contribute to BRCA1 post-transcriptional regulation.
Our findings suggest that women with BRCA1 3’UTR gene polymorphism (rs8176318G>T), their BRCA1 gene may become less functional therefore can develop more DNA damage, and perhaps leads for a more aggressive breast cancer genotype. In addition, this findings could indicate that the BRCA1-3’UTR-variant is an important genetic factor for developing breast cancer in Saudi Arabia.
In conclusion, the rs8176318 in the 3’untranslated region (UTR) of the BRCA1 gene was found to be responsible for the susceptibility to Breast cancer in Saudi population. BRCA1 GT and TT genotype was associated with breast cancer risk. However, more association studies are needed to support this finding in different regions of Saudi Arabia.
Consent
All authors hereby declare that all experiments have been examined and approved by the Research ethics committee, University of Tabuk, and have therefore been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki.
Acknowledgement
We acknowledge the support from the Deanship of Scientific Research, University of Tabuk for funding this research (S-1437-0331). We are grateful to the patients with whose cooperation this study was possible. We are also thankful to Mr Imran for his technical support Mr Omar for his help in sample collection.
References
- 1.Albeshan SM, Mackey MG, Hossain SZ, et al. Breast cancer epidemiology in Gulf cooperation council countries: A regional and international comparison. Clin Breast Cancer. 2017;13:1526–8209. doi: 10.1016/j.clbc.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 2.AlJohani B, AlMalik O, Anwar, et al. Impact of surgery on survival in stage IV breast cancer. Breast J. 2016;22:678–82. doi: 10.1111/tbj.12662. [DOI] [PubMed] [Google Scholar]
- 3.Ayoub N, Lucas C, Kaddoumi K. Genomics and pharmacogenomics of breast cancer: Current knowledge and trends. Asian Pac J Cancer Prev. 2011;12:1127–40. [PubMed] [Google Scholar]
- 4.Bassam AA, Rakan FA, Ahmed AA, et al. Breast cancer in Saudi Arabia and its possible risk factors. J Cancer Policy. 2017;12:83–9. [Google Scholar]
- 5.Brewster BL, Rossiello F, French JD, et al. Identification of fifteen novel germline variants in the BRCA1 3'UTR reveals a variant in a breast cancer case that introduces a functional miR-103 target site. Hum Mutat. 2012;33:1665–75. doi: 10.1002/humu.22159. [DOI] [PubMed] [Google Scholar]
- 6.Brewster BL, Rossiello F, French JD, et al. Identification of fifteen novel germline variants in the BRCA1 3'UTR reveals a variant in a breast cancer case that introduces a functional miR-103 target site. Hum Mutat. 2012;33:1665–75. doi: 10.1002/humu.22159. [DOI] [PubMed] [Google Scholar]
- 7.Chen S, Parmigiani G. Meta-analysis of BRCA1 and BRCA2 penetrance. J Clin Oncol. 2007;25:1329–33. doi: 10.1200/JCO.2006.09.1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X, Paranjape T, Stahlhut C, et al. Targeted resequencing of the microRNAome and 3'UTRome reveals functional germline DNA variants with altered prevalence in epithelial ovarian cancer. Oncogene. 2015;34:2125–37. doi: 10.1038/onc.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chouchane L, Boussen H, Sastry KSR. Breast cancer in Arab populations: molecular characteristics and disease management implications. Lancet Oncol. 2013;14:417–24. doi: 10.1016/S1470-2045(13)70165-7. [DOI] [PubMed] [Google Scholar]
- 10.Dorairaj JJ, Salzman DW, Wall D, et al. A germline mutation in the BRCA1 3'UTR predicts Stage IV breast cancer. BMC Cancer. 2014;14:421. doi: 10.1186/1471-2407-14-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Easton DF, Ford D, Bishop DT. Breast and ovarian cancer incidence in BRCA1-variant carriers. Breast cancer linkage consortium. Am J Hum Genet. 1995;56:265–71. [PMC free article] [PubMed] [Google Scholar]
- 12.Fang Y, Fengxia C, Jin X, et al. Identification and frequency of the rs12516 and rs8176318 BRCA1 gene polymorphisms among different populations. Oncol Lett. 2016;11:2481–6. doi: 10.3892/ol.2016.4252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ford T, Easton G, Stratton R, et al. Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The breast cancer linkage consortium. Am J Hum Genet. 1998;62:676–89. doi: 10.1086/301749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freedman ML, Penney KL, Stram DO. A haplo-type-based case-control study of BRCA1 and sporadic breast cancer risk. Cancer Res. 2005;65:7516–22. doi: 10.1158/0008-5472.CAN-05-0132. [DOI] [PubMed] [Google Scholar]
- 15.Garcia AI, Buisson M, Damiola F, et al. Mutation screening of MIR146A/B and BRCA1/2 3'-UTRs in the GENESIS study. Eur J Hum Genet. 2016;24:1324–9. doi: 10.1038/ejhg.2015.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holm K, Melum E, Franke A, et al. SNPexp - A web tool for calculating and visualizing correlation between HapMap genotypes and gene expression levels. BMC Bioinformatics. 2010;11:600. doi: 10.1186/1471-2105-11-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huo X, Lu C, Huang X, et al. Polymorphisms in BRCA1, BRCA1-interacting genes and susceptibility of breast cancer in Chinese women. J Cancer Res Clin Oncol. 2009;135:1569–75. doi: 10.1007/s00432-009-0604-6. [DOI] [PubMed] [Google Scholar]
- 18.Lheureux S, Lambert B, Krieger, et al. Two novel variants in the 3'UTR of the BRCA1 gene in familial breast and/or ovarian cancer. Breast Cancer Res Treat. 2011;125:885–91. doi: 10.1007/s10549-010-1165-8. [DOI] [PubMed] [Google Scholar]
- 19.Nathanson F, Wooster R, Weber Y. Breast cancer genetics: what we know and what we need. Nat Med. 2001;7:552–6. doi: 10.1038/87876. [DOI] [PubMed] [Google Scholar]
- 20.Newman B, Mu H, Butler LM, et al. Frequency of breast cancer attributable to BRCA1 in a population-based series of American women. JAMA. 1998;279:915–21. doi: 10.1001/jama.279.12.915. [DOI] [PubMed] [Google Scholar]
- 21.Pelletier C, Speed WC, Paranjape T, et al. Rare BRCA1 haplotypes including 3′UTR SNPs associated with breast cancer risk. Cell Cycle. 2011;10:90–9. doi: 10.4161/cc.10.1.14359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pongsavee M, Yamkamon V, Dakeng SPOc, et al. The BRCA13'-UTR 5711 + 421 T/T_ 5711 + 1286 T/T genotype is a possible breast and ovarian cancer risk factor. Genet Test Mol Biomarkers. 2009;13:307–17. doi: 10.1089/gtmb.2008.0127. [DOI] [PubMed] [Google Scholar]
- 23.Ralhan R, Kaur J, Kreienberg, et al. Links between DNA double strand break repair and breast cancer: accumulating evidence from both familial and non familial cases. Cancer Lett. 2007;248:1–17. doi: 10.1016/j.canlet.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 24.Sedghi M, Esfandiari E, Fazel-Najafabadi E, et al. Genomic rearrangement screening of the BRCA1 from seventy Iranian high-risk breast cancer families. J Res Med Sci. 2016;21:95. doi: 10.4103/1735-1995.193167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith A, Moran A, Boyd M, et al. Phenocopies in BRCA1 and BRCA2 families: evidence for modifier genes and implications for screening. J Med Genet. 2007;44:10–5. doi: 10.1136/jmg.2006.043091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Q, Paul A, Su D, et al. Structure of BRCA1-BRCT/abraxas complex reveals phosphorylation-dependent BRCT dimerization at DNA damage sites. Mol Cell. 2016;61:434–8. doi: 10.1016/j.molcel.2015.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang ES, Xia F. BRCA1 16 years later: DNA damage-induced BRCA1 shuttling. FEBS J. 2010;277:3079–85. doi: 10.1111/j.1742-4658.2010.07734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang F, Chen F, Xu J, Guan X. Identification and frequency of the rs12516 and rs8176318 BRCA1 gene polymorphisms among different populations. Oncol Lett. 2016;11:2481–6. doi: 10.3892/ol.2016.4252. [DOI] [PMC free article] [PubMed] [Google Scholar]


