Abstract
Background:
DNA methylation plays a vital role in the pathogenesis of the myelodysplastic syndrome (MDS), a heterogeneous group of clonal hematopoietic stem cell (HSC) disorders. It is reported to be an independent prognostic factor affecting overall survival (OS). Our aim was to analyze the role of global DNA methylation using an anti-5-methylcytosine (5-MC) antibody by immunohistochemistry (IHC) of bone marrow biopsy (BM Bx) specimens in MDS patients, assessing correlations with various clinical and biological prognostic factors.
Material and methods:
A total of 59 MDS cases, classified as per the World Health Organization (WHO) 2008 guidelines, were evaluated over a period of 4 years. Clinical data were retrieved from departmental case records and anti-5-MC expression was analyzed with formalin fixed paraffin embedded sections of BM Bx specimens of MDS patients and controls.
Results:
The median age at diagnosis was 52 years (15-85years). Patients were categorized into low risk (59%) and high risk (41%) according to International Prognostic Scoring System (IPSS). The median follow-up time was 10 months (1 to 37 months). We generated a methylation score (M-score) using anti-5-MC and with the derived cut-off of 30.5 from the receiver operator curve (ROC), there was a significant difference between the two groups in the percentage of BM blasts (p=0.01), WHO sub-type (p=0.01), IPSS (p=0.004), progression to AML (p=0.04) on univariate analysis. Interestingly, patients showing a high M-score (M-score ≥ 30.5) demonstrated a significantly shorter OS and progression to AML. However, on multivariate analysis, only BM blasts (p=0.01) and IPSS (p=0.02) remained independent variables for progression to AML and OS respectively.
Conclusion:
Immunostaining with anti-5-MC antibody with BM Bx samples is a simple and cost effective technique to detect global methylation, a powerful tool to predict overall survival in patients with MDS.
Keywords: DNA methylation, 5, methylcytosine, myelodysplastic syndrome
Introduction
Myelodysplastic syndrome (MDS) is a clonal hematopoietic stem cell disorder characterized by cytopenia(s), dysplasia in one or more cell lineages, ineffective hematopoiesis, and carries an increased risk of progression to acute myeloid leukemia (AML) (Swedlow et al., 2008). The natural history of MDS varies considerably; while some patients experience an indolent course, others show short overall survival (OS) and rapid transformation to AML.
DNA methylation is an epigenetic mechanism by which methyl groups are covalently added at the 5-carbon of the cytosine ring by enzyme DNA methyltransferase (DNMT), resulting in 5-methylcytosine (5-MC) (Jones and Baylin, 2007). DNA methylation was studied in the context of CpG-rich regions called CpG islands, which are often found within promoter regions. Nearly 60% of mammalian gene promoters contain CpG islands and methylation of CpG-rich promoters lead to silencing of gene expression by enabling binding of specific proteins (called methyl binding proteins). These proteins recruit transcriptional co-repressors and change the accessibility of DNA to transcription factors (Jones and Liang, 2009). DNA methylation has been associated with silencing of important tumor suppressor genes (Jones and Baylin, 2007). It plays an important role during early embryogenesis, genomic imprinting, X-chromosome inactivation, repression of transposable elements, aging and carcinogenesis (Issa, 2000). There is considerable evidence that DNA methylation is altered in MDS and affects the expression of genes important in regulating hematopoietic stem cell proliferation and differentiation. DNA methylation is an independent prognostic factor affecting OS in MDS patients (Uchida et al., 1997; Jiang et al., 2009; Vosa et al,. 2010; Shen et al., 2010). The relationship between prognosis and DNA methylation has been evaluated in single genes or combination of multiple genes in MDS patients (Lin et al., 2008; Quesnel et al., 1998; Tien et al., 2001). Hypomethylating agents have a central role in therapy of higher-risk MDS patients and demonstrated a significant prolongation of survival. Therefore, it is crucial to analyse the impact of DNA methylation in the outcome of MDS patients (Fenaux et al., 2009).
Several molecular techniques are available for assessment of global methylation; however, these techniques are laborious, cost intensive and requiring molecular laboratory with trained staff. 5-MC is a marker for global DNA methylation, immunostaining for anti-5-MC in tumor tissues has been previously explored in several tumor types including cervical, colon and oral cavity (Piyathilake et al., 2000; Kim et al., 1994; Hernandez-Blazquez et ai., 2000; Piyathilake et al., 2005). Recently, Poloni et al, evaluated the role of 5-MC by immunohistochemical method (anti-5-MC antibodies) on bone marrow biopsy (BM Bx) of MDS and found that high levels of global DNA methylation to be an independent poor prognostic factor (Poloni et al., 2013).
This study was conducted to establish the role of global DNA methylation using an anti-5-MC antibody by immunohistochemistry (IHC) on BM Bx in MDS patients and its correlation with various clinical and biological prognostic factors in MDS.
Materials and Methods
This was a prospective and retrospective study conducted in the Department of Hematology, All India Institute of Medical Sciences (AIIMS), New Delhi, India over a period of four years from June 2012 to May 2016. A total of 59 MDS cases were obtained during this period. Clinical data were retrieved from the departmental case records. All these cases were classified as per World Health Organization (WHO) 2008 classification of MDS (Swedlow et al., 2008). Concurrent conventional cytogenetic results were obtained from patient’s medical records. Clinico-hematological profiles were analyzed and International Prognostic Scoring System (IPSS) for MDS was calculated for determining the prognosis of these patients. Patients were categorized in low risk (low and intermediate-1) and high risk (intermediate-2 and high) groups according to the IPSS. The study was approved by the Institutional Ethics Committee.
Immunohistochemistry (IHC) for anti-5-MC
Anti-5-methylcytosine (anti-5-MC) expression was detected on formalin fixed paraffin embedded sections of bone marrow biopsy of 59 cases of MDS. The expression was also analyzed on 10 BM Bx each of positive controls (acute myeloid leukemia) and normal controls (immune thrombocytopenia purpura and lymphoma with uninvolved bone marrow).
IHC was performed on 4µm paraffin embedded BM biopsy section on poly-L-lysine coated slides. Sections were dewaxed in xylene and rehydrated by passing through graded alcohol. Antigen retrieval was done using 0.01 M citrate buffer, pH 6 using microwave at high heat for 30 minutes. The sections were treated with peroxidase blocking for 5 minutes and then incubated with serum blocking agent for 15 minutes to reduce the non specific staining. The sections were incubated first with primary antibody [anti- 5-MC Ab (AMM99021, clone 33D3, diluted 1:150; Aviva System, San Diego, CA, USA)] in moist chamber at room temperature for 1 hour and then incubated with biotinylated secondary antibody for 30 minutes. The antibody was visualized by incubated with 3,3’-diaminobenzidine and counter stained with Mayer’s hematoxylin.
Evaluation of anti-5-MC expression
Immunostaining for anti-5-MC antibody was assessed using light microscopy with blinding to the clinical characteristics of the patients. Initially, the entire bone marrow section was systematically scanned at 100x and 400x magnification. The percentage of positive cells stained with the specific antibody (anti-5-MC) within cellular areas was estimated. Nuclear positivity of anti-5-MC antibody was taken as positive and was assessed in mononuclear cells. Expression of anti-5-MC was determined by calculating the percentage of nuclear positivity in the stained cells. For each patient, the cell counts for methylation score (M-score) were repeated three times, and the mean values were used to calculate a final M-score. The results were expressed in percentage.
Statistical Analysis
Descriptive statistical analysis of main characteristics of patients was performed. Categorical and continuous data were presented in frequency (%), mean ± SD/ median (minimum – maximum). The association between two categorical variables was seen by Chi-square/ Fisher’s exact test. Comparison of continuous variables between two independent groups was evaluated by using Wilcoxon rank sum test. The receiver operating characteristic (ROC) curve analysis was done to determine an appropriate cut off of continuous variable. Survival curve was constructed by Kaplan-Meier method using the interval from the date of diagnosis to the date of last follow-up or death (overall survival - OS). The Cox regression method was used for multivariate survival analysis and to identify the independent prognostic factors for OS and progression to AML. Univariate and multivariate survival analysis were done to see unadjusted and adjusted hazard ratio with 95% confidence interval (CI), respectively. All the p-value < 0.05 were taken as statistically significant. All the statistical analysis was done using Stata 12.1.
Results
Patient characteristics
A total of 59 MDS cases were analyzed during this period. The median age at diagnosis was 52 years (15-85 years). Twenty-seven patients were younger than 50 years, and 32 were more than 50 years. There was male preponderance with the male to female ratio of 1.8:1. The median duration of symptoms before diagnosis was Six months (1-72 months). The median hemoglobin (Hb), absolute neutrophil count (ANC) and platelet count were 60 g/L (30-130 g/L), 1.6 x 109/L (0.08-7.5 x 109/L) and 61x 109/L (5-640 x 109/L) respectively. Most of the patients (62.7%) had bicytopenia or pancytopenia. These patients were categorized in following groups according to 2008 WHO classification as RA/RCUD (n=8), RCMD (n=19), RAEB-1 (n=10), RAEB-2 (n=17), and 5q deletion syndrome (n=5). Cytogenetic analysis was available in 54 patients out of which 24 patients showed cytogenetic abnormalities. IPSS was calculated in the patients where cytogenetic analysis was available. Fifty nine percentage of these patients were in low risk (Low and intermediate-1) while 41% were in high risk (intermediate-2 and high) category. The majority of our patients received only supportive care (n=43), eight patients received hypomethylating agents, seven patients received lenalidomide and one patient underwent allogeneic stem cell transplantation. During the period of study, nine patients progressed to AML and median time to progression was three months (1-8 months). Nineteen patients died during the study period. The median follow-up time was 10 months (1 to 37 months).
Global methylation score (M-score) in patients with MDS
On analysis, it was found that the patients with MDS have significantly higher M-score than in the normal controls (p<0.001). We compared M-score between the high-risk and low-risk MDS, there was a significantly difference between the two groups (p<0.001) (Table 1). The pattern of staining was also different in our patients as compared to normal controls. Normal controls had few scattered positivity in mononuclear cells and constituting on an average 7% (3-15%) of all marrow nucleated cells. Neutrophils and erythroid cells were anti-5-MC negative; however majority of megakaryocyte were weakly positive. The intensity of the anti-5-MC stain was weak to moderate in MDS cases and normal as well as positive control. In MDS patients the percentage of positive cells were increased, especially in high-risk MDS cases (Figure 1).
Table 1.
Comparison of M-Score in Normal Controls and MDS Patients
| M-score (median) | Range | P <0.001 | Overall | |
|---|---|---|---|---|
| Normal controls (a) | 7 | 2.5-15 | a-b | <0.001 |
| MDS | 27.5 | 9-67.5 | ||
| Low risk (b) | 23.2 | 9-39.5 | b-c | |
| High risk (c) | 41 | 17.5-67.5 | a-c |
Figure 1.
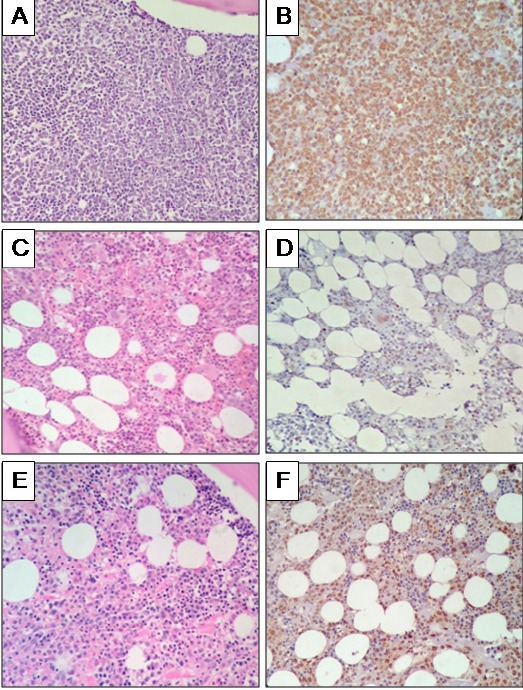
Immunohistochemistry (IHC) for Anti-5-MC (400 X). In AML (A), blasts cells are diffuse and strongly positive for anti-5-MC (B), in normal bone marrow (C) few cells are positive for anti-5-MC (D), in RAEB-2 bone marrow (E), immature cells show positivity for anti-5-MC (F). H and E- Hematoxylin and eosin.
Prognostic role of anti-5-MC in patients with MDS
For evaluating the prognostic role of global DNA methylation by anti-5-MC in patients with MDS, we performed ROC analysis to derive an optimal cut-off for M-score (30.5). On the basis of this cutoff, patients were divided into two groups (M-score< 30.5 and M-score ≥ 30.5). As depicted in Table 2, there was significant difference between two groups for ANC (p=0.04), percentage of bone marrow blasts (p=0.01), WHO sub-type of MDS (p=0.01), IPSS (p=0.003), progression to AML (0.04) and OS (p=0.02). However, there was no difference between two groups for age, gender, duration of symptoms, hemoglobin, platelet count, cytopenias, trilineage dysplasia in bone marrow and abnormal cytogenetics. Interestingly, patients showing a high M-Score (M-score ≥ 30.5) showed a significantly shorter OS and progression to AML (Figure 2).
Table 2.
Baseline Characteristics of MDS Patients according to M Score < 30.5 and ≥30.5
| Parameters | M-score < 30.5 (n=31) n (%) |
M-score ≥ 30.5 (n=28) n (%) |
‘p’ value |
|---|---|---|---|
| Age | 0.09 | ||
| <50 years | 11 (35.4) | 16 (57.1) | |
| ≥50 years | 20 (64.5) | 12 (47.9) | |
| Male | 21 (67.7) | 17 (60.7) | 0.57 |
| Female | 10 (32.3) | 11 (39.3) | |
| Median duration of symptoms before diagnosis in months (range) | 6 (1-72) | 5 (2-36) | 0.65 |
| Hemoglobin < 100 (g/L) | 29 (93.5) | 28 (100) | 0.49 |
| ANC < 1.8 (x 109/L) | 13 (41.9) | 19 (67.8) | 0.04 |
| Platelets < 100 (x 109/L) | 19 (61.3) | 17 (60.7) | 0.96 |
| Bicytopenia/pancytopenia | 18 (58.0) | 19 (67.8) | 0.43 |
| Trilineage dysplasia | 13 (41.9) | 11 (39.2) | 0.83 |
| % of bone marrow blasts | 0.01 | ||
| 0-4 | 23 (74.2) | 10 (35.8) | |
| 5-9 | 3 (9.7) | 7 (25.0) | |
| 10-14 | 3 (9.7) | 5 (17.8) | |
| 15-19 | 2 (6.4) | 12 (21.4) | |
| Abnormal karyotype | 11 (40.7) | 13 (50.0) | 0.49 |
| WHO 2008 classification | 0.01 | ||
| RCUD/RA | 5 (16.1) | 3 (10.7) | |
| RCMD | 15 (48.4) | 4 (14.3) | |
| RAEB-1 | 3 (9.7) | 7 (25.0) | |
| RAEB-2 | 5 (16.1) | 12 (42.9) | |
| 5q deletion | 3 (9.7) | 2 (7.1) | |
| IPSS | 0.003 | ||
| Low risk | 22 (78.6) | 10 (38.5) | |
| High risk | 6 (21.4) | 16 (61.5) | |
| Progression to AML | 2 (6.5) | 7 (25.0) | 0.04 |
| Patients died during study | 6 (19.3) | 13 (46.4) | 0.02 |
IPSS, International prognostic scoring system; WHO, World Health organization; RCUD, Refractory cytopenia with unilineage dysplasia; RA, Refractory anemia; RCMD, Refractory cytopenia with multilineage dysplasia; RAEB-1, Refractory anemia with excess blast-1; RAEB-2, Refractory anemia with excess blasts-2; AML-Acute myeloid leukemia.
Figure 2.
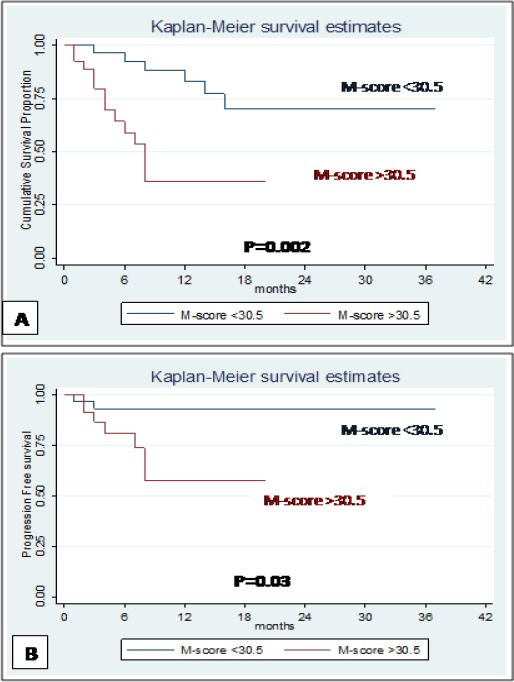
A: Overall survival in patients with MDS, M-score <30.5 and M-score >30.5, B: Progression to AML in patients with MDS, M-score <30.5 and M-score >30.5
Overall survival (OS) and progression to AML in patients with MDS
Nineteen patients died during the study period. We evaluated the impact of various clinical and biological factors for OS in the patients with MDS. Survival analysis was carried out using Kaplan-Meier method by log-rank tests. We observed that BM blasts percentage (p<0.001), WHO sub-type (p<0.001), IPSS (p=0.001), progression to AML (p=0.001) and M-score (p=0.002) were significantly correlated with OS in univariate analysis (Figure 2A). However, in multivariate analysis using Cox regression, only IPSS (p=0.02) was the significant independent prognostic factor for OS (Table 3). The M-score showed significance (p=0.002) on univariate analysis for OS in our patients but not in multivariate analysis.
Table 3.
Univariate and Multivariate Analysis of Various Clinical and Biological Factors Related to Overall Survival (OS) in Patients with MDS
| Variables | Parameters | Total patient | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|---|---|
| P value | Hazard ratio | 95% CI | P value | Hazard ratio | 95% CI | |||
| Age | <50 years | 27 | 0.37 | 0.66 | 0.27-1.64 | - | - | - |
| >50 years | 32 | |||||||
| Sex | Male | 38 | 0.56 | 0.74 | 0.26-2.06 | - | - | - |
| Female | 21 | |||||||
| Hemoglobin | <100 | 57 | 0.78 | 1.32 | 0.17-9.94 | - | - | - |
| (g/L) | >100 | 2 | ||||||
| ANC | <1.8 | 32 | 0.16 | 0.5 | 0.19-1.33 | - | - | - |
| (x 109/L) | >1.8 | 27 | ||||||
| Platelet count | <100 | 36 | 0.41 | 0.67 | 0.26-1.72 | - | - | - |
| (x 109/L) | >100 | 23 | ||||||
| Cytopenia | Unilineage | 37 | 0.11 | 2.27 | 0.81-6.34 | - | - | - |
| Bi/Trilineage | 22 | |||||||
| BM Blasts | <5% | 33 | <0.001 | 7.74 | 2.53-23.69 | - | - | - |
| >5% | 26 | |||||||
| Trilineage dysplasia | Absent | 35 | 0.14 | 1.95 | 0.78-4.83 | - | - | - |
| Present | 24 | |||||||
| Cytogenetics | Normal | 29 | 0.43 | 1.45 | 0.56-3.78 | - | - | - |
| Abnormal | 24 | |||||||
| WHO sub-type | Low risk | 32 | <0.001 | 11.06 | 3.17-38.52 | - | - | - |
| High risk | 27 | |||||||
| IPSS | Low risk | 32 | 0.001 | 5.18 | 1.92-13.98 | 0.02 | 3.578 | 1.21-10.59 |
| High risk | 22 | |||||||
| Progression to AML | No | 50 | 0.001 | 5.29 | 2.02-13.84 | - | - | - |
| Yes | 9 | |||||||
| M-score | Low | 31 | 0.002 | 4.59 | 1.71-12.34 | - | - | - |
| High | 28 | |||||||
ANC, Absolute neutrophil count; BM, Bone marrow; IPSS, International prognostic scoring system; M-score, Methylation score
Nine patients progressed to AML with median progression period of three months (1-8 months). We observed that BM blasts percentage (p=0.01), WHO diagnosis (p=0.01), IPSS (p=0.03) and M-score (p=0.03) significantly correlated with progression to AML in univariate analysis (Figure 2B). However, on multivariate analysis using Cox regression, only BM blasts percentage (p=0.01) was the significant independent prognostic factor for progression to AML (Table 4).
Table 4.
Univariate and Multivariate Analysis of Various Clinical and Biological Factors Related to Progression to AML in Patients with MDS
| Variables | Parameters | Total patient | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|---|---|
| P value | Hazard ratio | 95% CI | P value | Hazard ratio | 95% CI | |||
| Age | <50 years | 27 | 0.36 | 0.54 | 0.14-2.02 | - | - | - |
| >50 years | 32 | |||||||
| Sex | Male | 38 | 0.94 | 0.95 | 0.23-3.81 | - | - | - |
| Female | 21 | |||||||
| Hemoglobin | <10 | 57 | 1 | 5.93 | 0 | - | - | - |
| (g/L) | >10 | 2 | ||||||
| ANC | <1.8 | 32 | 0.73 | 1.25 | 0.33-4.68 | - | - | - |
| (x 109/L) | >1.8 | 27 | ||||||
| Platelet count | <100 | 36 | 0.92 | 1.06 | 0.28-3.98 | - | - | - |
| (x 109/L) | >100 | 23 | ||||||
| Cytopenia | Unilineage | 22 | 0.62 | 1.4 | 0.35-5.65 | - | - | - |
| Bi/Trilineage | 37 | |||||||
| BM Blasts | <5% | 33 | 0.01 | 13.71 | 1.71-109.99 | 0.01 | 12.45 | 1.55-99.99 |
| >5% | 26 | |||||||
| Trilineage dysplasia | Absent | 35 | 0.57 | 1.45 | 0.38-5.47 | - | - | - |
| Present | 24 | |||||||
| Cytogenetics | Normal | 29 | 0.83 | 0.85 | 0.20-3.60 | - | - | - |
| Abnormal | 24 | |||||||
| WHO sub-type | Low risk | 32 | 0.01 | 13.63 | 1.69-109.37 | - | - | - |
| High risk | 27 | |||||||
| IPSS | Low risk | 32 | 0.03 | 4.51 | 1.10-18.48 | - | - | - |
| High risk | 22 | |||||||
| M-score | Low | 31 | 0.03 | 5.78 | 1.18-28.13 | - | - | - |
| High | 28 | |||||||
ANC, Absolute neutrophil count; BM, Bone marrow; IPSS, International prognostic scoring system; M-score, Methylation score
Discussion
Several studies showed that DNA methylation is an independent adverse prognostic factor in MDS patients (Jiang et al., 2009; Shen et al., 2010; Poloni et al., 2013; Calvo et al., 2014). The majority of previous studies showed that the role of methylation in single or combination of multiple genes; however, only few studies assessed the impact of global methylation on clinical outcome (Lin et al., 2008; Quesnel et al., 1998; Tien et al., 2001). Quillen et al., (2012) compared analytical performances and predictive values of five techniques (methylation-specific polymerase chain reaction, methylight, pyrosequencing, methylation-sensitive high-resolution melting, and IHC) to analyze DNA methylation in a series of 100 glioblastoma patients. They observed that by combining pyrosequencing and IHC improved predictive power for OS, but not for progression free survival. In this study, we assessed the global DNA methylation in 59 MDS patients, detected by anti-5-MC IHC and its impact on clinical outcome.
We observed that M-score in MDS patients was significantly higher than the normal controls. The M-score increased in higher risk MDS group or as BM blast percentage increased. Solomon et al made a similar observation in their study that the clinical severity of MDS was directly proportional to hypermethylation and determined the progression of MDS to AML (Solomon et al., 2008). Similar results were also found by Jiang et al., (2009) who observed that aberrant DNA methylation in MDS played an important role in silencing of tumor suppressor gene and determined progression to AML.
Our data showed that a significant difference between two groups (M-score <30.5 and ≥30.5) for ANC, BM blasts percentage, IPSS, progression to AML and OS in univariate analysis. However, in the multivariate analysis, the role of M-score as an independent prognostic factor for OS could not be established. Our results were similar to Rommerman et al., (2008) who observed that global DNA methylation was significantly higher in patients with de-novo MDS compared with the normal controls and was correlated with karyotype, BM blast percentage and IPSS. However, they could not confirm its association with the OS. Similarly, Shen et al., (2010) observed no correlation between methylation at baseline and that following response to decitabine therapy. However, they noted reduction of methylation over the course of treatment and a better clinical response.
Poloni et al., (2013) detected global DNA methylation by IHC (anti 5-MC antibody) on the BM Bx of MDS patients and found high level of DNA methylation was an independent adverse prognostic factor. Similar results was also observed by Calvo et al., (2014) who detected global DNA methylation and hydroxymethylation using ELISA and determined that high levels of global DNA methylation were independent adverse prognostic factor in MDS (Clavo et al., 2014).
The risk of MDS increases with age, and approximately 86% of MDS cases are diagnosed in individuals aged more than 60 years (Cheson et al., 2006; Rollison et al., 2008). On analyzing the basic demographic profile of our patients, we found that the median age (52 years) of our patients was one to two decade lower than the western population (70 years), which is in concordance with reports from India and Southeast Asia (Chaubey et al., 2011; Gupta et al., 2017). In our study 45% patients were less than 50 years of age. The median age of presentation of MDS patients in India and Southeast countries are generally younger than those from Western and the reason for this difference is not clear.
It was also noticed that 45% of patients in our study were diagnosed as higher risk WHO sub-type like RAEB-1 and RAEB-2. This may be attributed to the fact that, being a tertiary care and referral center, most of our patients presented late in advance stage after being worked up for cytopenias and not responding to conventional therapy. This could be the one reason that more number of patients in the present study were in high risk group.
A number of studies showed that BM blasts, WHO sub-type, karyotype, IPSS, transfusion dependency, and cytopenias were independent prognostic factors in MDS (Sole et al., 2000; Voso et al., 2013; Malcovati et al., 2005; Bejar et al., 2014; Sanz et al., 1998). However, in our study, we observed that BM blasts, WHO sub-type, IPSS, progression to AML and M-score were significantly correlated with OS in MDS patients on univariate analysis. But only BM blasts and progression to AML was found to be significant independent prognostic factors affecting OS.
The major limitations of our study were small sample size, short follow up period, which could be the possible reason for not obtaining a statistical significance in multivariate analysis. We assessed global DNA methylation using IHC anti-5-MC antibody and could not compare to the gold standard method such as pyrosequencing due to unavailability of same in our institute. Using fully quantitative techniques and its comparison with anti-5-MC antibody would give a better assessment of global DNA methylation. A larger prospective study with a longer follow-up would give a better evaluation of global DNA methylation and various prognostic factors for OS and progression-free survival in MDS patients.
In conclusion, global DNA methylation can be detected using simple and inexpensive anti-5-MC antibody by IHC technique on BM Bx. The global methylation may predict OS in patients with MDS. However, proper validation of this technique needs to be done using gold standard quantitative technique.
References
- Bejar R. Clinical and genetic predictors of prognosis in myelodysplastic syndromes. Haematologica. 2014;99:956–64. doi: 10.3324/haematol.2013.085217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo X, Nomdedeu M, Navarro A, et al. High levels of global DNA methylation are an independent adverse prognostic factor in a series of 90 patients with de novo MDS. Leuk Res. 2014;38:874–81. doi: 10.1016/j.leukres.2014.04.015. [DOI] [PubMed] [Google Scholar]
- Chaubey R, Sazawal S, Dada R, Mahapatra M, Saxena R. Cytogenetic profile of Indian patients with de novo myelodysplastic syndromes. Indian J Med Res. 2011;134:452–7. [PMC free article] [PubMed] [Google Scholar]
- Cheson BD, Bennett JM, Kantarjian H, et al. Report of an international working group to standardize response criteria for myelodysplastic syndromes. Blood. 2000;96:3671–4. [PubMed] [Google Scholar]
- Fenaux P, Mufti GJ, Hellstrom-Lindberg E, et al. Efficacy of azacitidine compared with that of conventional care regimensin the treatment of higher-risk myelodysplastic syndromes:a randomised, open-label, phase III study. Lancet Oncol. 2009;10:223–32. doi: 10.1016/S1470-2045(09)70003-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Rahman K, Singh MK, et al. Clinico-pathological spectrum and novel karyotypic findings in myelodysplastic syndrome:Experience of tertiary care center in India. Mediterr J Hematol Infect Dis. 2017;16:e2017048. doi: 10.4084/MJHID.2017.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Blazquez FJ, Habib M, Dumollard JM, et al. Evaluation of global DNA hypomethylation in human colon cancer tissues by immunohistochemistry and image analysis. Gut. 2000;47:689–93. doi: 10.1136/gut.47.5.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa JP. CpG-island methylation in aging and cancer. Curr Top Microbiol Immunol. 2000;249:101–18. doi: 10.1007/978-3-642-59696-4_7. [DOI] [PubMed] [Google Scholar]
- Jiang Y, Dunbar A, Gondek LP, et al. Aberrant DNA methylation is a dominant mechanism in MDS progression to AML. Blood. 2009;113:1315–25. doi: 10.1182/blood-2008-06-163246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–92. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805–11. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YI, Giuliano A, Hatch KD, et al. Global DNA hypomethylation increases progressively in cervical dysplasia and carcinoma. Cancer. 1994;74:893–9. doi: 10.1002/1097-0142(19940801)74:3<893::aid-cncr2820740316>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- Lin J, Yao D, Qian J, et al. Methylation status of fragilehistidine triad (FHIT) gene and its clinical impact on prognosis of patients withmyelodysplastic syndrome. Leuk Res. 2008;32:1541–5. doi: 10.1016/j.leukres.2008.02.008. [DOI] [PubMed] [Google Scholar]
- Malcovati L, Porta MG, Pascutto C, et al. Prognostic factors and life expectancy in myelodysplastic syndromes classified according to WHO criteria:a basis for clinical decision making. J Clin Oncol. 2005;23:7594–603. doi: 10.1200/JCO.2005.01.7038. [DOI] [PubMed] [Google Scholar]
- Piyathilake CJ, Bell WC, Jones J, et al. Pattern of nonspecific (or global) DNA methylation in oral carcinogenesis. Head Neck. 2005;27:1061–7. doi: 10.1002/hed.20288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piyathilake CJ, Johanning GL, Frost AR, et al. Immunohistochemical evaluation of global DNA methylation:comparison with in vitro radiolabeled methyl incorporation assay. Biotech Histochem. 2000;75:251–8. doi: 10.3109/10520290009085128. [DOI] [PubMed] [Google Scholar]
- Poloni A, Goteri G, Zizzi A, et al. Prognostic role of immunohistochemical analysis of 5 mc in myelodysplastic syndromes. Eur J Haematol. 2013;91:219–27. doi: 10.1111/ejh.12145. [DOI] [PubMed] [Google Scholar]
- Quesnel B, Guillerm G, Vereecque R, et al. Methylation of the p15(INK4b) gene in myelodysplastic syndromes is frequentand acquired during disease progression. Blood. 1998;91:2985–90. [PubMed] [Google Scholar]
- Quillien V, Lavenu A, Karayan-Tapon L, et al. Comparative assessment of 5 methods (methylation-specific polymerase chain reaction, MethyLight, pyrosequencing, methylation-sensitive high-resolution melting, and immunohistochemistry) to analyze O6-methylguanine-DNA-methyltranferase in a series of 100 glioblastoma patients. Cancer. 2012;118:4201–11. doi: 10.1002/cncr.27392. [DOI] [PubMed] [Google Scholar]
- Rollison DE, Howlader N, Smith MT, Strom SS, et al. Epidemiology of myelodysplastic syndromes and chronic myeloproliferative disorders in the United States 2001-2004, using data from the NAACCR and SEER programs. Blood. 2008;112:45–52. doi: 10.1182/blood-2008-01-134858. [DOI] [PubMed] [Google Scholar]
- Romermann D, Hasemeier B, Metzig K, et al. Global increase in DNA methylation in patients with myelodysplastic syndrome. Leukemia. 2008;22:1954–6. doi: 10.1038/leu.2008.76. [DOI] [PubMed] [Google Scholar]
- Sanz GF, Sanz MA, Greenberg PL. Prognostic factors and scoring systems in myelodysplastic syndromes. Haematologica. 1998;83:358–68. [PubMed] [Google Scholar]
- Shen L, Kantarjian H, Guo Y, et al. DNA methylation predicts survival and response to therapy in patients with myelodysplastic syndromes. J ClinOncol. 2010;28:605–13. doi: 10.1200/JCO.2009.23.4781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sole F, Espinet B, Sanz GF, et al. Incidence, characterization and prognostic significance of chromosomal abnormalities in 640 patients with primary myelodysplastic syndromes. Grupo cooperativo sspanol de citogenetica hematologica. Br J Haematol. 2000;108:346–56. doi: 10.1046/j.1365-2141.2000.01868.x. [DOI] [PubMed] [Google Scholar]
- Solomon PR, Munirajan AK, Tsuchida N, et al. Promoter hypermethylation analysis in myelodysplastic syndromes:Diagnostic & prognostic implication. Indian J Med Res. 2008;127:52–7. [PubMed] [Google Scholar]
- Swedlow SH, Campo E, Harris NL, et al. WHO classification of tumours of haematopoietic and lymphoid tissues. Lyon, France: IARC Press; 2008. [Google Scholar]
- Tien HF, Tang JH, Tsay W, et al. Methylation of thep15(INK4B) gene in myelodysplastic syndrome:it can be detected early at diagnosis or during disease progression and is highly associated with leukaemictransformation. Br J Haematol. 2001;112:148–54. doi: 10.1046/j.1365-2141.2001.02496.x. [DOI] [PubMed] [Google Scholar]
- Uchida T, Kinoshita T, Nagai H, et al. Hypermethylation of the p15INK4B gene in myelodysplastic syndromes. Blood. 1997;90:1403–9. [PubMed] [Google Scholar]
- Voso MT, D'Alo F, Greco M, et al. Epigenetic changes in therapy- related MDS/AML. Chem Biol Interac. 2010;184:46–9. doi: 10.1016/j.cbi.2009.10.013. [DOI] [PubMed] [Google Scholar]
- Voso MT, Fenu S, Latagliata R, et al. Revised international prognostic scoring system (IPSS) predicts survival and leukemic evolution of myelodysplastic syndromes significantly better than IPSS and WHO prognostic scoring system:validation by the gruppo romano mielodisplasie Italian regional database. J Clin Oncol. 2013;31:2671–7. doi: 10.1200/JCO.2012.48.0764. [DOI] [PubMed] [Google Scholar]


