Abstract
Objectives:
Genetic markers are crucial fort diagnostic and prognostic investigation of hematological malignancies (HM). The conventional cytogenetic study (CCS) has been the gold standard for more than five decades. However, FISH (Fluorescence in Situ Hybridization) testing has become a popular modality owing to its targeted approach and the ability to detect abnormalities in non-mitotic cells. We here aimed to compare the diagnostic yields of a FISH panel against CCS in HMs.
Methods:
Samples of bone marrow and peripheral blood for a total of 201 HMs were tested for specific gene rearrangements using multi-target FISH and the results were compared with those from CCS.
Results:
Exhibited a greater diagnostic yield with a positive result in 39.8% of the cases, as compared to 17.9% of cases detected by CCS. Cases of chronic lymphocytic leukaemia (CLL) benefited the most by FISH testing, which identified chromosomal aberrations beyond the capacity of CCS. FISH was least beneficial in myelodysplastic syndrome (MDS) where the highest concordance with CCS was exhibited. Acute lymphocytic leukaemia (ALL) demonstrated greater benefit with CCS. In addition, we found the following abnormalities to be most prevalent in HMs by FISH panel testing: RUNX1 (21q22) amplification in ALL, deletion of D13S319/LAMP1 (13q14) in CLL, CKS1B (1q21) amplification in multiple myeloma and deletion of EGR1/RPS14 (5q31/5q32) in MDS, consistent with the literature.
Conclusions:
In conclusion, FISH was found to be advantageous in only a subset of HMs and cannot completely replace CCS. Utilization of the two modalities in conjunction or independently should depend on the indicated HM for an optimal approach to detecting chromosomal aberrations.
Keywords: Chromosomal aberration, cytogenetics, fluorescence in situ hybridization, hematological malignancies
Introduction
Hematological malignancies (HM) are a group of diseases characterized by a spectrum of genetic markers which have diagnostic and prognostic implications. Conventional cytogenetic study (CCS) has been the gold standard for more than five decades for detecting genetic alterations that are greater than 10 MB in size (Peterson et al., 2015). CCS has paved the way in identifying specific chromosomal aberrations associated with clinically and morphologically definitive subsets of HMs. In the recent past, fluorescence in situ hybridization (FISH) has become a reliable and rapid complementary test in targeting critical genetic events associated with diagnostics and prognosis in HMs. Although CCS is advantageous in providing a global purview of the chromosome complement, there are many disadvantages. The technique relies upon dividing cells causing high failure rates due to low mitotic index. Even when metaphases are available for analysis, poor morphology of chromosomes hinder identification of aberrations. These factors directly influence its ability in establishing minor clone population during analysis. FISH has addressed these issues by targeting interphase cells in addition to metaphases (Sreekantaiah, 2007). Although complementary FISH testing increases the overall detection of aberrations, its benefit is not uniform across all types of HMs. Recent comparative studies in Myelodysplastic Syndrome (MDS) have showed that FISH does not add value to CCS findings (He et al., 2016) while a similar study contradicted that both modalities are equally important in prognostication of MDS (Kokate et al., 2017). On the other hand, FISH analysis in lymphoid malignancies have resulted in the expansion and identification of distinct subsets of the disease (Sreekantaiah, 2007). Therefore, apt usage of FISH panels in aiding diagnosis or in monitoring follow-up samples of HMs is critical.
The present study was undertaken to compare the diagnostic yield between FISH and CCS in four different hematological malignancies. The hematological diseases considered in this project included Chronic Lymphoid Leukemia (CLL), Acute Lymphoid Leukemia (ALL), Multiple Myeloma (MM) and Myelodysplastic Syndrome. Our findings further inspect recurrence of commonly reported genetic abnormalities detected by FISH in a south Indian population.
Materials and Methods
Patients and sample preparation
The current study included a total of 201 cases consisting of bone marrow and peripheral blood samples queried for several hematological malignancies between October 2014 and June 2017. The study consisted of 93 MDS cases, 42 ALL cases, 40 MM cases and 26 CLL cases. The samples were processed simultaneously for FISH panel and routine cytogenetic assessment. No plasma cell sorting was performed in MM cases. The samples were grouped into three categories based on karyotype and FISH analysis: Group 1 consisted of samples that showed concordant results by FISH and conventional cytogenetic investigation. Group 2 included samples where FISH proved advantageous over karyotype. This also included samples where metaphases were unavailable or insufficient. Finally, samples where karyotype delivered more information such as secondary abnormalities that was not targeted by FISH were classified in Group 3.
Conventional Cytogenetics
Heparinized, blood or whole bone marrow samples were cultured for 24 hours using RPMI-1640 media (Gibco Invitrogen, USA) containing 15% fetal bovine serum (Microphil, USA) and Pen-strep (Gibco Invitrogen, USA). Culture was terminated and processed with hypotonic solution (KCl 0.075 M) and fixed with Cornoy’s Fixative (Methanol: Acetic Acid 3:1). Slides were prepared on grease free slides, checked for metaphases and aged overnight. Following day, G-banding using Trypsin and Geimsa staining was performed. The analysis was performed using Zesis Axio Imager Z2 microscope with Ikaros software (MetaSystems, GmBH, Germany). 20 metaphases were evaluated by two experienced cytogeneticists according to the International System for Human Cytogenomic Nomenclature (ISCN 2016). Clonality was established when more than three metaphases showed the same structural and/or numerical aberrations.
Fluorescent in Situ Hybridization
All FISH assays were carried out in accordance to the manufacturers’ specifications. Fixed cells were dropped onto microscopic slides and incubated at 55°C for up to 5 minutes. 10 µl of the probe mixture was applied to a 22 × 22 mm hybridization area. The marked area was sealed with rubber cement. The sample and probes were co-denatured at 75°C for 5 minutes and allowed to hybridize overnight at 37°C in a hybridization chamber (Statspin, ThermoBrite). Slides were washed in 0.4X SSC at 70°C for 2 minutes followed by 2X SSC/0.5% Tween 20 at room temperature for 30 seconds. Slides were then counterstained and mounted with 10 μl 4’,6-diamidino-2-phenylindole (DAPI II, MetaSystems GmbH, Germany). FISH analyses were independently assessed by two cytogeneticists using Zesis Axio Imager fluorescent microscope with ISIS software (MetaSystems GmbH, Germany). A total of 200 interphase nuclei were scored per probe per slide by two experienced cytogeneticists. All probes were purchased either from MetaSystems, GmbH, Germany or CytoCell, UK. The specific details of probes for the FISH panels are summarized in Table 1.
Table 1.
List of Probes Included in the Present Study
| Disease | Targeted genetic abnormality | FISH Panel Probes | Probe Color | Manufacturer name |
|---|---|---|---|---|
| Acute Lymphoid Leukemia | t(9;22)(q34;q11) | BCR/ABL1 | Green/Red | MetaSystems |
| t(12;21)(p13;q22) | ETV6/RUNX1 | Green/Red | MetaSystems | |
| 11q23 deletion | MLL | Yellow (Green+Red) | CytoCell | |
| Multiple Myeloma | -1/1p32 deletion | CDKN2C | Green | CytoCell |
| +1/1q21 amplification | CKS1B | Red | CytoCell | |
| -13/13q14 deletion | D13S319/13qter | Red/Green | CytoCell | |
| 14q32 rearrangement | IGH | Yellow (Green+Red) | CytoCell | |
| 17p13/D17Z1 deletion | p53/CEP17 | Red/Green | CytoCell | |
| Chronic Lymphoid Leukemia | -11q22/17p13 deletion | ATM/TP53 | Green/Red | CytoCell |
| -13/13q14 deletion | D13S319/ LAMP1 | Red/Blue | CytoCell | |
| -12/12p11-q11 | CEP12 | Green | CytoCell | |
| -6/6q23 deletion | MYB/SEC63 | Red/Green | CytoCell | |
| Myelodysplastic Syndrome | -5/5q31/5q32-33 deletion | EGR1/RPS14 | Red/Green | CytoCell |
| -7/7q22/7q31 deletion | MLL5/MET | Red/Green | CytoCell | |
| -20q12/20q13 deletion | PTPRT/MYBL2 | Red/Green | CytoCell | |
| +8/8p11-q11 | CEP8 | Blue | CytoCell |
Results
In a total of 201 cases of HM, 80 (39.8%) patients were positive for abnormalities by FISH and 36 (17.9%) showed abnormalities by routine cytogenetic analysis as represented in Table 2. The overall concordance between FISH and karyotype was found to be 58.7%. However, FISH could pick up chromosomal aberrations (CA) in 28.2% of the total cases that presented normal karyotype or culture failure. Inversely, karyotype detected chromosomal abnormalities in 12.9% of patients that were not targeted or negative by FISH investigation. Independent analysis of the diseases as shown in Figure 1 revealed that CLL patients benefited the most by FISH when compared to other HM’s. MDS presented highest concordance (>85%) between the two modalities indicating that FISH provided no additional information. Only in ALL, a significant population benefitted from karyotype investigation where 47.6% of patients showed an abnormal karyotype. Overall failure rate for CCS in HMs was 13.9% (28/201). Individual failure rates for CLL, ALL, MM and MDS were found to be 38.4%(10/26), 7.1%(3/42), 17.5%(7/40) and 8.6%(8/93), respectively. FISH provided a result in all 201 samples analyzed.
Table 2.
Abnormal Karyotype and Corresponding FISH Results of the Study Group for All the Hematological Malignancies.
| SN | Karyotype | Fish Panel | ||||
|---|---|---|---|---|---|---|
| Multiple Myeloma | 1p32 (CDKN2C) | 1q21 (CKS1B) | 14q32 (IGH) | 13q14 (D13S319/ LAMP1) | 17p13 (p53) | |
| 1 | 43,-X,-Y,+1,t(1;8)(p14;q24),-2,-4,add(7)(p21), t(13;14)(q10;q10), del(13)(q14;q32), add(16)(p13),+22 [12]/46,XY[8] | 3G | 3-5R | 1Y | 1G1R | 2G2R |
| 2 | 46-48,X,-X,+del(1)(q32),-3,add(4)(q33),-5, del(6) (q15), 10,+add(11)(q23), add(12)(p13)x2, ?del(13) (q32),-15, -17,+18, -20, +4 mar [12]/46,XX[2] | 2G | 3R | 1Y | 1R1G | 1G1R |
| 3 | 54,XY,del(1)(p13p32),del(1)(p13),+1,+3,+5,+7,+8,+9, +10,-13,+21, +21 [16]/46,XY[4] | 2G | 3R | 1Y1G1R | 1R1G | 2G2R |
| 4 | 57,X,-Y,+2,+3,+4,+5,+6,-8, +9,+11, +13,+14, +15, +17,+19,20,+21,+22[5] /46,XY[15] | 2G | 3R | 2Y | 2G2R | 3G3R |
| Chronic Lymphocytic Leukemia | 6q23 (MYB/SEC63) | CEP 12 | 13q14 (D13S319) | 11q22 (ATM) | 17p13 (p53) | |
| 5 | 47,XX,+12[5]/46,XX[25] | 2R2G | 3G | 2R2B | 2R | 2G |
| Acute Lymphocytic Leukemia | t(9;22)(q34;q11) (BCR/ABL1) | t(12;21)(p13;q22) (ETV6/RUNX1) | 11q23 (MLL) | |||
| 6 | 46,XX, t(9;22))(q34;q11) [10]/46,XX[5] | 1G1R2Y | 2G2R | 1Y | ||
| 7 | 46,XY,t(3;12;21)(p13;p13;q22),del(6)(q21),add(12)(p13),del(17)(q22) [12] /46,XY[8] | 2G2R | 2G2R1Y | 1Y | ||
| 8 | 46,XY,t(9;22)(q34;q11)[7]/46,XY,t(2;3)(q31;q27),t(9;22)(q34;q11)[3] | 1G1R2Y | 2G2R | 2Y | ||
| 9 | 46,XX[13]/58,XX,+4,+5,+6,+8,+9,+10, +11,+12,+14, +15,+21,+21[2] | 2G2R | 2R4G | 2Y | ||
| 10 | 46,XY ,?add(9)(p), t(9;22)(q34;q11)[15] | 1G1R2Y | 2G2R | 2Y | ||
| 11 | 52,XY,+X,+6,+14,+17,+21,+21[20] | 1G1R2Y | 2R3G/2R4G | 2Y | ||
| 12 | 46,XX,del(2)(q11),t(10;11)(p13;q23),der(14)t(1;14)(q21;p11)[16]/ 46,XX[4] | 1G1R2Y | 1G1R2Y | 1Y1G1R | ||
| 13 | 47,XX,add(7)(p11),-11,der(12)t(12;?)(p11;?), -20,+21, -22,+3-4 mar [9]/ 46,XX[16] | 1G1R2Y | 1R3G | 2Y | ||
| 14 | 60,XY,+X,dirdup(1)(q21-qter) ,+4,+6, +7, +9, +14, +15,+17, +20, +21, +3 mar[15] | 2G2R | 2R4G | 2Y | ||
| 15 | 45,XY,t(9;22)(q34;q11),-18, der(20)t(18;20) (q11;q13) [20] | 2G1R1F | 2G2R | 2Y | ||
| 16 | 51-60,XX,-Y,add(2 p21),+add(3p21)x2,+del (4q24), +8,+21,+21[15] | 2G2R | 2R3G/2R4G | 2Y | ||
| 17 | 54,XY,+X,+4,+5,+i(7)(q10),+8,+10,+14, +21[18] | 2G2R | 2R3G | 2Y | ||
| 18 | 46,del(1q31),?dup(2q21q31),del(9p13),del(14q) [19]/46,XY[1] | 2G2R | 2G2R | 2Y | ||
| 19 | 44-46,X,-Y,-5,add(7)(p12),add(12),(p11),+1,-2 mar[13] /46,XY[7] | 2G2R | 2G1R | 2Y | ||
| 20 | 46-48,XY,+1,-2mar[12] | 2G2R | 2R1Y | 2Y | ||
| 21 | 46,XY,t(2;5)(q21;p15)[20] | 2G2R | 2G2R | 2Y | ||
| 22 | 46,XY,t(9;15)(q34;q11),?del(13)(q14)[12]/46,XY[8] | 2G2R | 2G2R | 2Y | ||
| 23 | 46, XY t(2;7)(p14;p22) [15] / 46,XY[5] | 2G2R | 2G2R | 2Y | ||
| 24 | 46,XY,i(7)(q10), del(9)(p13)[7]/46,XY[1] | 2G2R | 2G2R | 2Y | ||
| 25 | 46,i(X)(q10),Y[20] | 2G2R | 2G2R | 2Y | ||
| Myelodysplastic Syndrome | -5q31/-5q32-33 (EGR1/RPS14) | -7q22/-7q31 (MLL5/MET) | CEP 8 | -20q12/-20q13 (PTPRT/ MYBL2 | ||
| 26 | 45,XX,-7[1] | 2G2R | 1R1G | 2B | 2G2R | |
| 27 | 45,XX,-7[30] | 2G2R | 1G1R | 2B | 2G2R | |
| 28 | 45,XX,-7,del(20)(q12)[15] | 2G2R | 1G1R | 2B | 1G1R | |
| 29 | 46,XX,del(5)(q21)[10] | 1G2R | 2G2R | 2B | 2G2R | |
| 30 | 60,XY,+X,add(1)(p35),+5,+5,+5,+del(6)(q23),+7,+10, +12,+14,+15,+17,+18,+21,+22/46,XY,del(6)(q23) [3]/46,XY[20] | 3G3R | 3G3R | 2B | 2G2R | |
| 31 | 47,XX,+8[1] | 2G1R | 2G2R | 3B | 2G2R | |
| 32 | 45,XY,t(3;11)(p?;q32),-5[8] | 1G1R | 2G2R | 2B | 2G2R | |
| 33 | 47,XX,dup(5)(q13;q31),+8,del(14)(q23;q32)[10] | 2G1R | 2R2G | 3B | 2G2R | |
| 34 | 47,XY,del(3)(q22),-5,?del (7)(q32q33),+8,+15, ?del(20)(q13)[6] | 2G1R | 1R1G | 3B | 1R1G | |
| 35 | 46,XY,trp(1)(q21;q32),add(3)(q26),add(19)(p13)[15] | 2G2R | 2G2R | 2B | 2G2R | |
| 36 | 46,XY,t(6;9)(p22;q34)[20] | 2G2R | 2G2R | 2B | 2G2R | |
Figure 1.
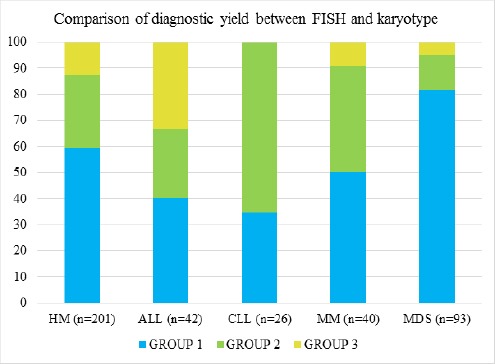
Graph Showing Percentage of Group 1 (concordance between the two modalities), Group 2 (FISH advantageous) and Group 3 (CCS advantageous). X-axis: Sample size (%), Y-axis: Individual HMs.
CLL
Of the 26 samples received for CCS and FISH analysis, only one case (3.8%) was diagnosed positive by karyotype, contrastingly 17 cases (65.4%) were diagnosed positive by FISH. In 16 cases (61.5%) FISH identified an abnormality not detected by CCS, which either yielded a normal karyotype or culture failure. Using our CLL FISH panel on fixed samples, abnormalities were analyzed for each of the probes, with the highest rate observed in deletion of 13q14 (D13S319). As illustrated in Figure 2, 47.1% of the positive cases (8/17) showed loss of the 13q14 (D13S319) segment in a range of 25-80% of interphase nuclei, 41.1% (7/17) depicted trisomy 12 in 45-95% of nuclei and 11.7% (2/17) showed deletions of 17p13 (p53) in 30-65% of cells and 11q22 (ATM) in 20-70% of cells. In only one case, CCS picked up trisomy 12 in 20% of the metaphases analyzed. FISH investigation established higher population of the clone where the aberration was picked up in greater than 60% of interphase nuclei. No abnormality was detected for probe locus 6q23 (MYB).
Figure 2.
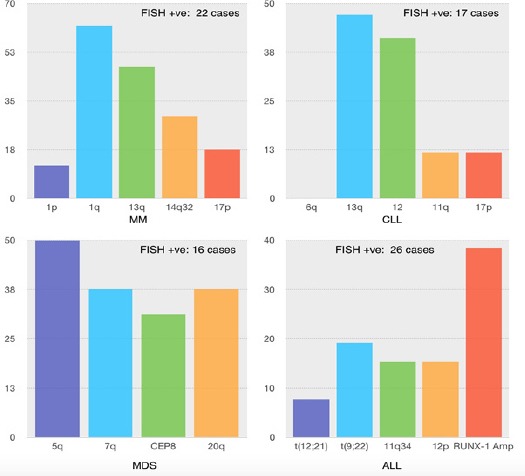
Graphical Representation of CA Picked up by FISH in HM, X-axis: Chromosomal Aberration targeted by specific FISH probe, Y-axis: Incidence in percentage within FISH positive sample group
MM
Analysis of 40 whole bone marrow samples revealed only 4 cases (10.0%) were diagnosed positive by karyotype. On the contrary, 21 cases (52.5%) were diagnosed positive by FISH. In 42.5% of cases, FISH proved advantageous in identifying an abnormality not detected by CCS that exhibited a normal karyotype or culture failure. Using our MM FISH panel, abnormalities were tested for each of the probes, with the most frequent CA being amplification of 1q21 (CKS1B). As illustrated in Figure 2, 61.9% of positive cases (13/21) showed amplification of the 1q21 (CKS1B) segment in a range of 15-55% of interphase nuclei, 11.7% (2/21) displayed deletion of 1p32 (CDKN2C) in 15-55% of nuclei, 47.1% (8/21) had deletion of 13q14 (D13S319) in 15-75% of nuclei, 29.4% (5/21) presented rearrangement of 14q32 (IgH) in 20-55% of cells and 17.6% (3/21) had deletion of 17p13 (p53) in 25-40% of interphase nuclei. CCS could pick up structural and numerical abnormalities in addition to FISH findings in only 10% (4/40) of the total cases. The abnormalities included t(1;8)(p13;q24), add(4)(q?), del(6)(q?), add(7)(p21) and copy number changes of other chromosomes not targeted by FISH.
MDS
Of the 93 samples received for cytogenetic and FISH analysis, 11 cases (11.8%) were diagnosed positive by karyotype. In addition, 16 cases (17.2%) were diagnosed positive by FISH. FISH was observed to be least advantageous in providing additional information compared to other HM’s where CA was detected in less than 15% of cases exhibiting normal karyotype or culture failure. FISH and karyotype showed 90.9% positive concordance and 94.6% negative concordance. Using our MDS FISH panel, abnormalities were verified for each of the probes, and deletion of 5q31 (EGR1/RPS14) was observed to be most predominant. As illustrated in Figure 2, 50% (8/16) of positive cases showed deletion of 5q31 (EGR1/RPS14) segment in a range of 30 - 70% of interphase nuclei, 37.5% (6/16) carried deletion of 7q22 (MLL5/MET) in 20-50% of interphase nuclei and deletion of 20q12 (PTPRT/MYBL2) in 20-60% of nuclei, and 31.2% (5/16) presented trisomy 8 in 15-60% of nuclei. CCS could pick up structural and numerical abnormalities in addition to FISH findings in only 4.3% (4/93) of the total cases. The abnormalities included t(3;11)(p?;q32), dup(1)(q21q32), add(3)(q26), add(19)(p13) and copy number changes of chromosomes not targeted by FISH.
ALL
Of the 42 samples received for cytogenetic and FISH analysis 80.9% (34/42) were pediatric cases. 20 of total cases (47.6%) were diagnosed positive by karyotype. In contrast, 26 of total cases (61.9%) were diagnosed positive by FISH, 23 of which were pediatric. In 15 cases (35.7%), FISH identified an abnormality not detected by CCS yielding a normal karyotype or culture failure. Copy number changes of RUNX1 was observed in 38.4% (10/26) of total cases and 39.1% (9/23) in pediatric cases which was the most common CA in this study. As illustrated in Figure 2, 7.6% (2/26) of the positive cases showed t(12;21)(p13;q22) (ETV6/RUNX1) translocation and 15.3% (5/26) showed deletion of ETV6 gene in a range of 10-95% of interphase nuclei, 19.2% (5/26) presented t(9;22)(q34;q11) (BCR/ABL) translocation in 50-80% of nuclei and 15.4% (4/26) showed rearrangement of 11q23 (MLL) in 40-95% of nuclei analyzed. CCS could pick up additional numerical abnormalities which were not identified by FISH in 33.3% (14/42) such as +4, +5, +6, +8, +10, +14, +15, +17, +20 and structural aberrations such as t(2;5)(q21;p15), t(9;15)(q34;q11), del(13)(q14), t(2;7)(p14;p22), i(7)(q10), del(9p13), del(6)(q21), del(17)(q22), 46,XY,t(2;3)(q31;q27), del(2)(q11), der(14)t(1;14)(q21;p11), add(7)(p11), dirdup(1)(q21-qter), der(20)t(18;20)(q11;q13), dup(2q21q31).
Discussion
In this comparative study, the diagnostic utility of FISH in comparison to CCS was found to be most advantageous in CLL and least advantageous in MDS. The percentages of additional genetic aberrations identified by FISH alone were 65.3% for CLL, 45% for MM, 31% for ALL and 14% for MDS.
According to our study, it is evident that FISH did not add relevant information with respect to chromosomal aberrations in comparison to CCS in MDS. In agreement with our results, CA detection rate of lesser than 15% by FISH has been reported in several studies (Ketterling et al., 2002; Cherry et al., 2003; Costa et al., 2010; Pitchford et al., 2010; Yang et al., 2010). In the current study, MDS had the highest concordance value of more than 90% between the two modalities. FISH testing in MDS could have an advantage when CCS fails or yields chromosomes of poor morphology. A study by Yang et al. (2010) concluded that FISH testing maybe informative in high grade MDS where CCS yields a normal karyotype. We report deletions of 5q31/-5, 7q22/-7, Trisomy 8 and -20q12/-20 in 50%, 37.5%, 31.2% and 37.5% respectively of all abnormal cases of MDS by FISH. Combined positive pick up by CCS and FISH was less than 20% in the current study. This can be attributed to the case selection which included a heterogeneous population of patients at primary diagnosis and post therapy monitoring. CA in CLL are important independent predictors of disease progression and survival and improving the detection rate of these aberrations can help develop a better design for treatment strategies (Wiktor and Van Dyke, 2004). Many recent studies have demonstrated the benefits of using a single mitogen or in combination to augment diagnostic yield by CCS and FISH in detection of CA (Shi et al., 2013; Dubuc et al., 2016; Holmes et al., 2016). Contradictory to the findings in MDS, our study showed that FISH was found to be an invaluable tool for picking up CA in lymphoid malignancies. FISH identified CA where CCS was normal or insufficient in 61.5% (16/26) of CLL, 35.7% (15/42) of ALL and 42.5% (17/40) of MM. In CLL, while a low mitotic index may preclude complete cytogenetic analysis, FISH testing reveals aberrations in non-dividing cells and the identification of a minor cell population or an emerging clone. A Chinese study conducted by Qin et al., (2016) demonstrated that CLL FISH showed higher sensitivity in unveiling chromosomal abnormalities than by CCS. 73.8% of cases demonstrated a CA detected by FISH in comparison to 9.5% by CCS. This corroborates our findings where 65.4% of positive cases were diagnosed by FISH and only 3.8% (1/26) of cases by CCS. CLL exhibited the least concordance between the two modalities. Deletion of 13q14/-13 (D13S319/LAMP1), Trisomy 12, deletion of 11q22 (ATM) and deletion of 17p13 (p53) were found in 47.1%, 41.1%, 11.7% and 11.7% respectively of all CLL positive cases. As represented in Figure 3, hemizygous deletion and monosomic FISH patterns were observed for 13q14. Previous Indian and other Asian studies have also reported deletion of 13q14 as the most prevalent CA identified by FISH (Xu et al., 2008; Amare et al., 2013; Wu et al., 2013; Yi et al., 2017). None of the cases exhibited deletion of 6q23 in our study making it the least prevalent in our population. Similar conclusions have been drawn in other Asian studies (Xu et al., 2008; Amare et al., 2013).
Figure 3.
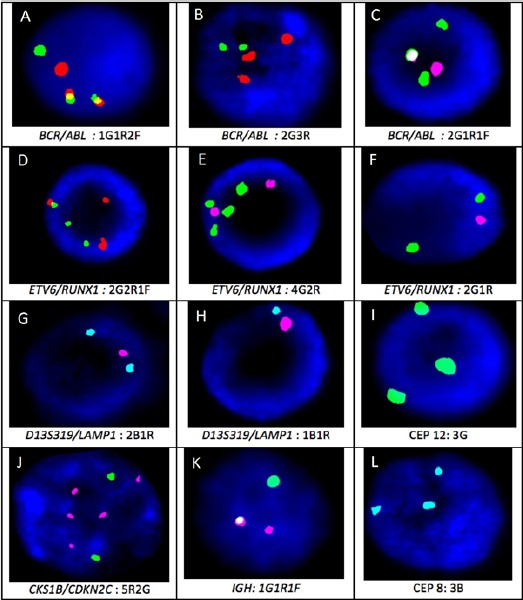
Signal Pattern for Select FISH Probes Included; A: BCR/ABL double fusion (1G1R2Y), B: 3 copies of ABL (3R), C: Single fusion of BCR/ABL with ABL deletion (2G1R1Y), D: Atypical ETV6/RUNX1 fusion indicating 3-way translocation (1Y2G2R), E: RUNX1 amplification (2R4G), F. ETV6 deletion (1R2G), G. Hemizygous deletion of D13S319(2B1R), H. Monosomy 13 (1R1B), I. Trisomy 12 (3G), J. CKS1B amplification (5R), K. IGH break apart (1Y1R1G), L. Trisomy 8 (3B)
We observed that FISH testing also benefitted multiple myeloma significantly. Contrasting to other HMs in which the malignant clone tends to dominate the bone marrow at diagnosis, MM is hindered by low detection of CA because of hypoproliferative nature of plasma cells and normal metaphases that originate from the myeloid cells in the marrow. This limitation in MM diagnostics is countered by implementing plasma cell enrichment by sorting or immunofluorescence assays such as cIg-FISH (Cytoplasmic Immuno Globin) (Shetty et al., 2012; Gole et al., 2014). Although in our present study, CCS and FISH were performed on whole bone marrow samples, CA was identified in 52.5% (21/40) of cases by FISH and 10.0% (4/40) by CCS. In a study, Amare et al (2016) reported a positive pick up rate of 66% by FISH on plasma cell sorted samples. MM exhibited 50% concordance between CC and FISH, with FISH adding value in 45% (18/40) of the cases. Amplification of 1q21, deletion of 1p32, deletion of 13q14, rearrangement of 14q32 and deletion of 17p13 were present in 61.9%, 11.7%, 47.1%, 29.4%, 17.6% of abnormal cases respectively. We observed 1q21 amplification to be the most common CA diagnosed by FISH confirming similar studies (Kwon et al., 2010; Amare et al., 2016). Amplification of 1q21 has been reported to be more prevalent than deletion of 1p32 and has been positively correlated with IGH rearrangement and 13q/13 deletion (Lim et al., 2013). This was evident in our study where amplification of 1q21 was seven times more frequent than deletion of 1p32 (13:2 cases, 1p21:1q32) concurrent with IGH rearrangement in 30.7% and -13q/-13 in 38.4% of 1q21 positive cases. As reported by many studies, -13/-13q14 is present in about 40% to 50% of cases and thus is one of the most frequent abnormalities in MM (Liebisch and Dohner, 2006; Zang et al., 2015). But our study revealed -13/-13q14 to be the second most common CA with a percentage of 47. Among all prognostic markers, deletion of TP53/monosomy 17 is considered worst prognosis and a late event in the evolution of MM (Cremer et al., 2005). Our study reports only 3 patients with TP53 deletion of which two patients also presented with 1q21 amplification and one patient presented with 13q14 deletion.
In ALL, non-random chromosomal abnormalities have important biological, diagnostic, and prognostic significance. In this study, CA was identified in 61.9% (26/42) of cases by FISH and 47.6% (20/42) by CCS with a concordance of 40.3% between the modalities. CCS was advantageous in 33.3% (14/42) of the ALL cases which is the highest across all the HMs included in this study. CCS was able to pick up numerical aberrations such as aneusomies of +4, +5, +6, +8, +10, +14, +15, +17, +20 and structural aberrations such as t(2;5)(q21;p15), t(9;15)(q34;q11), del(13)(q14), t(2;7)(p14;p22), i(7)q, del(9p13), del(6)(q21), del(17)(q22), 46,XY,t(2;3)(q31;q27), del(2)(q11), der(14)t(1;14)(q21;p11), add(7)(p11), dirdup(1)(q21-qter), der(20)t(18;20)(q11;q13), dup(2q21q31). Increased copy number changes of RUNX-1 was the most common aberration diagnosed by FISH representing 38% of the cases considered. Similar findings have reported increased copy numbers of RUNX-1 as the most frequent CA diagnosed by FISH (Udaykumar et al., 2007; Haltrichl et al., 2008). t(12;21), t(9;22) and 11q23 MLL break apart was found in 7.6%, 19.2% and 15.4% of samples respectively. Deletion of 12p13 (ETV6) was found in 15.4% of the total FISH positive cases and copy number changes in other chromosomes (trisomy 9, 11, 22) targeted by FISH was found in 15.4%. These results were similar to that reported by Mazloumi et al (2012) where the most common translocation was t(9;22)(q34;q11) in 12.7% of patients. As represented in Figure 3, varied FISH patterns were observed for BCR/ABL1 representing dual fusion, single fusion and trisomy of chromosome 9. Consistent with our results, previous studies also conclude that CCS is still a relevant and important test in the diagnostics of ALL (Freidman and Weinstein et al., 2000; Harrison, 2001; Soszynska et al., 2008).
As summarised in Figure 1, it is evident that each haematological malignancy presents chromosomal aberrations which can be detected effectively either by CCS or FISH or in tandem. Although, interphase FISH was found to be a reliable and targeted technique for detecting cryptic abnormalities in whole bone marrow samples of HMs, we observed that CCS is sufficient or adds value in diagnostics of certain haematological malignancies. Presenting a high concordance value greater than 90% in MDS, FISH testing is limited to specific scenarios where CCS fails to furnish a conclusive report or presents with a normal karyotype with low mitotic index. On the contrary, the characteristic complexity of genetics in ALL benefitted most by CCS testing which detected additional abnormalities not focussed in FISH panel, thus recommending combined application of these modalities.
A widespread combination of cytogenetic markers specific to disease profile is necessary to augment risk stratification and monitoring of hematological malignancies. Karyotyping continues to remain the gold standard in cytogenetic investigation of hematological malignancies owing to its comprehensive purview of the genome and requirement of relatively simple infrastructure. However, by overcoming the limitations of karyotyping, FISH has proven to become an important tool in routine diagnosis. Therefore, a cost-effective approach for genetic studies by I-FISH and G-banding should be considered such that maximum information about the disease may be achieved.
Statement conflict of Interest
The authors whose names are listed certify that they have no affiliations with or involvement in any organization or entity with any financial or non-financial interest in the subject matter or materials discussed in this manuscript.
Funding Statement
No funding.
Acknowledgements
We are grateful to the management of Anand Diagnostic Laboratory for providing the necessary infrastructure. We express our gratitude to Mrs. Shrivalli BS and Ms. Nivedha VK from the department of cytogenetics for their technical assistance.
References
- Amare Kadam PS, Gadage V, Jain H, et al. Clinico-pathological impact of cytogenetic subgroups in B-cell chronic lymphocytic leukemia:experience from India. Indian J Cancer. 2013;50:261–7. doi: 10.4103/0019-509X.118730. [DOI] [PubMed] [Google Scholar]
- Amare Kadam PS, Jain H, Nikalje S, et al. Observation on frequency &clinic-pathological significance of various cytogenetic risk groups in multiple myeloma:an experience from India. Indian J Med Res. 2016;144:536–43. doi: 10.4103/0971-5916.200890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry AM, Brockman SR, Paternoster SF, et al. Comparison of interphase FISH and metaphase cytogenetics to study myelodysplastic syndrome:an eastern cooperative oncology group (ECOG) study. Leuk Res. 2003;27:1085–90. doi: 10.1016/s0145-2126(03)00104-8. [DOI] [PubMed] [Google Scholar]
- Costa D, Valera S, Carrio A, et al. Do we need to do fluorescence in situ hybridization analysis in myelodysplastic syndromes as often as we do? Leuk Res. 2010;34:1437–41. doi: 10.1016/j.leukres.2010.02.019. [DOI] [PubMed] [Google Scholar]
- Cremer FW, Bila J, Buck I, et al. Delineation of distinct subgroups of multiple myeloma and a model for clonal evolution based on interphase cytogenetics. Genes Chromosomes Cancer. 2005;44:194–203. doi: 10.1002/gcc.20231. [DOI] [PubMed] [Google Scholar]
- Dubuc AM, Davids MS, Pulluqi M, et al. FISHing in the dark:How the combination of FISH and conventional karyotyping improves the diagnostic yield in CpG-stimulated chronic lymphocytic leukemia. Am J Clin Pathol. 2016;91:978–83. doi: 10.1002/ajh.24452. [DOI] [PubMed] [Google Scholar]
- Freidmann AM, Weinstein HJ. The role of prognostic features in the treatment of childhood acute lymphoblastic leukemia. Oncologist. 2000;5:321–8. doi: 10.1634/theoncologist.5-4-321. [DOI] [PubMed] [Google Scholar]
- Gole L, Lin A, Chua C, Chng WJ. Modified cIg-FISH protocol for multiple myeloma in routine cytogenetic laboratory practice. Cancer Genet. 2014;207:31–4. doi: 10.1016/j.cancergen.2013.12.001. [DOI] [PubMed] [Google Scholar]
- Haltrich I, Csoka M, Kovacs G, Fekete G. Cytogenetic and FISH findings are complementary in childhood ALL. Magy Onkol. 2008;52:283–91. doi: 10.1556/MOnkol.52.2008.3.6. [DOI] [PubMed] [Google Scholar]
- Harrison CJ. The detection and significance of chromosomal abnormalities in childhood acute lymphoblastic leukemia. Blood Rev. 2001;15:49–59. doi: 10.1054/blre.2001.0150. [DOI] [PubMed] [Google Scholar]
- He R, Wiktor AE, Durnick DK, et al. Bone marrow conventional karyotyping and Fluorescence In Situ Hybridization:Defining an effective utilization strategy for evaluation of myelodysplastic syndromes. Am J Clin Pathol. 2016;146:86–94. doi: 10.1093/ajcp/aqw077. [DOI] [PubMed] [Google Scholar]
- Holmes PJ, Peiper SC, Uppal GK, et al. Efficacy of DSP30-IL2/TPA for detection of cytogenetic abnormalities in chronic lymphocytic leukemia/small lymphocytic lymphoma. Int J Lab Hematol. 2016;38:483–9. doi: 10.1111/ijlh.12513. [DOI] [PubMed] [Google Scholar]
- McGowan-Jordan J, Simons A, Schmid M. An international system for human cytogeneomic nomenclature:ISCN, 2016. Basel, Switzerland: Karger; 2016. [Google Scholar]
- Ketterling RP, Wyatt WA, VanWier SA, et al. Primary myelodysplastic syndrome with normal cytogenetics:utility of 'FISH panel testing'and M-FISH. Leuk Res. 2002;26:235–40. doi: 10.1016/s0145-2126(01)00117-5. [DOI] [PubMed] [Google Scholar]
- Kokate P, Dalvi R, Koppaka N, et al. Prognostic classification of MDS is improved by inclusion of FISH panel testing with conventional cytogenetics. J Cancer Gen. 2017;21:120–7. doi: 10.1016/j.cancergen.2017.05.004. [DOI] [PubMed] [Google Scholar]
- Kwon WK, Lee JY, Mun YC, et al. Clinical utility of FISH analysis in addition to G-banded karyotype in hematologic malignancies and proposal of a practical approach. Korean J Hematol. 2010;45:171–6. doi: 10.5045/kjh.2010.45.3.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebisch P, Dohner H. Cytogenetics and molecular cytogenetics in multiple myeloma. Eur J Cancer. 2006;42:1520–9. doi: 10.1016/j.ejca.2005.12.028. [DOI] [PubMed] [Google Scholar]
- Lim AS, Lim TH, See KH, et al. Cytogenetic and molecular aberrations of multiple myeloma patients:a single-center study in Singapore. Chin Med J (Engl) 2013;126:1872–7. [PubMed] [Google Scholar]
- Mazloumi SH, Madhumathi DS, Appaji L, Prasannakumari Combined study of cytogenetics and fluorescent in situ hybridization (FISH) analysis in childhood acute lymphoblastic leukemia (ALL) in a tertiary cancer centre in South India. Asian Pac J Cancer Prev. 2012;13:3825–7. doi: 10.7314/apjcp.2012.13.8.3825. [DOI] [PubMed] [Google Scholar]
- Peterson JF, Aggarwal N, Smith CA, et al. Integration of microarray analysis into the clinical diagnosis of hematological malignancies:How much can we improve cytogenetic testing? Oncotarget. 2015;6:18845–62. doi: 10.18632/oncotarget.4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitchford CW, Hettinga AC, Reichard KK. Fluorescence in situ hybridization testing for -5/5q, -7/7q, +8, and del(20q) in primary myelodysplastic syndrome correlates with conventional cytogenetics in the setting of an adequate study. Am J Clin Pathol. 2010;133:260–4. doi: 10.1309/AJCPZ4JL5ZMRPFTD. [DOI] [PubMed] [Google Scholar]
- Qin YW, Wang XR, Yang YN, Wang C. Value of a panel Fluorescence In Situ Hybridization in three kinds of hematological malignancies. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2016;24:1289–93. doi: 10.7534/j.issn.1009-2137.2016.05.001. [DOI] [PubMed] [Google Scholar]
- Shetty S, Siady M, Mallempati KC, et al. Utility of column-free cell sorting system for separation of plasma cells in multiple myeloma FISH testing in clinical laboratories. Int J Hematol. 2012;95:274–81. doi: 10.1007/s12185-012-1021-1. [DOI] [PubMed] [Google Scholar]
- Shi M, Cipollini MJ, Crowley-Bish PA, et al. Improved detection rate of cytogenetic abnormalities in chronic lymphocytic leukemia and other mature B-cell neoplasms with use of CpG-oligonucleotide DSP30 and interleukin 2 stimulation. Am J Clin Pathol. 2013;139:662–9. doi: 10.1309/AJCP7G4VMYZJQVFI. [DOI] [PubMed] [Google Scholar]
- Soszynska K, Mucha B, Debski R, et al. The application of conventional cytogenetics, FISH, and RT-PCR to detect genetic changes in 70 children with ALL. Ann Hematol. 2008;87:991–1002. doi: 10.1007/s00277-008-0540-6. [DOI] [PubMed] [Google Scholar]
- Sreekantaiah C. FISH panels for hematologic malignancies. Cytogenet Genome Res. 2007;118:284–96. doi: 10.1159/000108312. [DOI] [PubMed] [Google Scholar]
- Udayakumar AM, Bashir WA, Pathare AV, et al. Cytogenetic profile of childhood acute lymphoblastic leukemia in Oman. Arch Med Res. 2007;38:305–12. doi: 10.1016/j.arcmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Wiktor A, Van Dyke DL. Combined cytogenetic testing and fluorescence in situ hybridization analysis in the study of chronic lymphocytic leukemia and multiple myeloma. Cancer Genet Cytogenet. 2004;153:73–6. doi: 10.1016/j.cancergencyto.2004.03.006. [DOI] [PubMed] [Google Scholar]
- Wu SJ, Lin CT, Huang SY, et al. Chromosomal abnormalities by conventional cytogenetics and interphase fluorescence in situ hybridization in chronic lymphocytic leukemia in Taiwan, an area with low incidence –clinical implication and comparison between the West and the East. Ann Hematol. 2013;92:799–806. doi: 10.1007/s00277-013-1700-x. [DOI] [PubMed] [Google Scholar]
- Xu W, Li JY, Yu H, et al. Fluorescent in situ hybridization with a panel of probes detects molecular cytogenetic abnormalities in patients with chronic lymphocytic leukemia. Zhonghua Yi Xue Za Zhi. 2008;88:2537–40. [PubMed] [Google Scholar]
- Yang W, Stotler B, Sevilla DW, et al. FISH analysis in addition to G-band karyotyping:utility in evaluation of myelodysplastic syndromes? Leuk Res. 2010;34:420–5. doi: 10.1016/j.leukres.2009.09.013. [DOI] [PubMed] [Google Scholar]
- Yi S, Zou D, An G, et al. Intratumoral genetic heterogeneity and number of cytogenetic aberrations provide additional prognostic significance in chronic lymphocytic leukemia. Genet Med. 2017;19:182–91. doi: 10.1038/gim.2016.81. [DOI] [PubMed] [Google Scholar]
- Zang M, Zou D, Yu Z, et al. Detection of recurrent cytogenetic aberrations in multiple myeloma:a comparison between MLPA and iFISH. Oncotarget. 2015;6:34276–87. doi: 10.18632/oncotarget.5371. [DOI] [PMC free article] [PubMed] [Google Scholar]


