Abstract
Background:
Breast cancer is the most common cancer among women worldwide. Tamoxifen (TAM), a selective estrogen receptor modulator, is widely used in its treatment. TAM is metabolized by cytochrome P450 (CYP450) enzymes, including CYP2D6, CYP3A5 and CYP2C19, whose genetic variations may have clinicopathological importance. However, reports on the association of various P450 polymorphisms with certain cancers are contradictory.
Methods:
We here investigated whether the prevalence of the four most common polymorphism in the CYP2D6*4 (G1934A), CYP2D6*10 (C188T), CYP3A5*3 and CYP2C19*2 alleles has any link with breast cancer using genomic DNA and polymerase chain reaction restriction fragment length polymorphism (PCR-RFLP) analysis.
Results:
Prevalences of CYP2D6*4, CYP2D6*10 and CYP2C19*2 genotypes were differed significantly (P = 0.01 and P = 0.004) between breast cancer patients and controls. The CYP3A5*3 genotype did not demonstrate statistically significant variation.
Conclusion:
Polymorphisms in CYP2 appear to be associated with breast cancer risk. Our data taken together with other reports indicates that drug resistance gene polymorphisms might be indicators of response to tamoxifen therapy in breast cancer cases.
Keywords: Cytochrome P450, polymorphism, breast cancer, Tamoxifen
Introduction
The Cytochrome P450 is the major enzyme involved in drug metabolism. CYP enzymes are responsible for the metabolism of most clinically used drugs. Individual variability in drug efficacy and toxicity resulting in different clinical responses is common in therapeutic areas, including breast cancer. It is an important problem in clinical practice because it can lead to therapeutic failure and adverse effects. Polymorphisms in the genes encoding enzymes responsible for the metabolism of drugs and other xenobiotics and the functional significance of these polymorphisms are critical for predicting clinical outcomes (Ruiter, et al 2010).
Tamoxifen, selective estrogen receptor modulators (SERM) is the most commonly prescribed drug for the treatment and prevention of recurrence for patients with estrogen and/or progesterone receptor positive disease. The biotransformation of tamoxifen is mediated by cytochrome P450 enzymes, amongst which CYP3A4, CYP2B6, CYP2C9, CYP2C19 and CYP2D6 are presumed to be the most important isoenzymes. Two major antiestrogen metabolites, 4-hydroxy-N-desmethyltamoxifen (endoxifen) and 4-hydroxytamoxifen are 30-100 times more potent than itself (Lim et al., 2006). Endoxifen, the greatest potent anti-estrogen, is converted from tamoxifen by sequential biotranformation involving CYP3A4/5 mediated N-demethylation of tamoxifen to form N-desmethyltamoxifen (NDM) and CYP2D6 which is rate-limiting enzyme catalyzed 4-hydroxylation of NDM to form endoxifen (Desta et al., 2004; Hoskins et al., 2009). Major genetic polymorphisms affecting tamoxifen-metabolizing enzyme activity of potential clinical relevance are those related to the CYP450 enzymes: CYP2D6, CYP3A5 and CYP2C19. To date, more than 300 variants of the CYP2D6 gene have been identified CYP2D6 * 4 and CYP2D6 * 10 is the most common mutants in Caucasians and in Asians (Sosa Macias, et al 2013).
The aim of our study is to investigate the prevalence of the most common allelic variants of CYP2D6*4, CYP2D6*10, CYP3A5*3 and CYP2C19*2 for tamoxifen resistance. Previous studies have carried out on each genotype analysis. Therefore we focused to study about each genotype and also the combination of each genotype for progression and prognosis of breast cancer.
Materials and Methods
Study Population
The study population comprised 110 breast cancer (BC) patients and 100 controls. Patients were consecutively recruited from the MNJ cancer hospital, Hyderabad, Telangana, India, between January 2014 and October 2016. All patients were diagnosed with breast cancer. Senior pathologists confirmed all diagnoses. The controls were recruited from general population from Hyderabad during the same time period. Each control was matched to each case by age and sex. All subjects were given a questionnaire to investigate the demographic characteristics, family history of cancer. The clinical characteristics were collected from medical records, including tumor differentiation, tumor size and hormonal and chemotherapy. This study was approved by the hospital Ethnical Committee, Hyderabad, and informed consent was obtained from all participants. . Patients were recruited following certain inclusion and exclusion criteria, which were determined before the beginning of the study.
Human Genomic DNA Isolation
Approximately 4ml of blood was collected from all participants, mixed with EDTA and kept at -20°C Genomic DNA was extracted from the blood and isolated using epicenter DNA isolation kit. Isolated DNA was stored in Tris-EDTA buffer, pH 8.0. DNA samples were stored at -20°C till analysis. Their concentrations were determined by a spectrophotometer (Nanodrop Thermo 2000c). The genomic polymorphism was identified by polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) technique.
Polymerase chain reaction
The primers were synthesized at a commercial facility in Hyderabad (Bioserve, Hyderabad, India). The primers used for amplification of CYP2D6*4, CYP2D6*10, CYP3A5*3 and CYP2C19*2 genes were listed in table -1. PCR was carried out using a PCR kit (Bioserve) in a total volume of 50 μl. The PCR mixture contained 2.5 μl of 25 mM MgCl2, 10 mM dNTP mixture, 160 pmol of each primer (forward and reverse primers), 0.2 μl of Taq (5 U/μl) and a DNA template. The reaction volume was made up to 50 μl with sterile water. The PCR reaction was carried out in a CFX 96 thermocycler (Bio-Rad, Hercules, CA, USA) using the following optimal conditions. Initial denaturation was carried out at 95°C for 5 min, followed by 35 cycles of denaturation at 94°C for 35 sec, annealing at 58°C for 35 sec and extension at 72°C for 40 sec. After completion of 35 cycles, a final extension step was carried out at 72°C for 5 min. Amplification products corresponding to 354bp, respectively, were visualized after electrophoresis in an ethidium-bromide-stained 2% agarose gel. The amplified PCR products were performed RFLP using BstN1, (Fermentas) restriction enzyme for 37°C overnight PCR products subjected to enzyme digestion was visualized on 3% agarose gel stained with ethidium bromide (Figures 1, 2, 3 and 4).
Table 1.
Details of the Primers Used for the Sequences Amplified by PCR, Along with the Target Band Size and Annealing Temperatures are Given in Table Below
| Genes and Primer sequences | Annealing Temp. | Amplicon size |
|---|---|---|
| CYP2D6*4: F: 5’- GCCTTCGCCAACCACTCCG-3’ | 58°C | 355 bp |
| CYP2D6*4: R: 5’- AAATCCTGCTCTTCCGAGGC-3’ | ||
| CYP2D6*10: F: 5’- TCAACACAGCAGGTTCA -3’ | 58°C | 362 bp |
| CYP2D6*10: R: 5’- CTGTGGTTTCACCCACC -3’ | ||
| CYP3A5*3:F: 5’- ATGGAGAGTGGCATAGGAGATA-3’ | 58°C | 130 bp |
| CYP3A5*3:R: 5’- TGTGGTCCAAACAGGGAAGAAATA -3’ | ||
| CYP2C19*2:F: 5’- CAGAGCTTGGCATATTGTATC- -3’ | 58°C | 312 bp |
| CYP2C19*2:R: 5’- GTAAACACACAAAACTAGTCAATG -3’ |
Figure. 1.
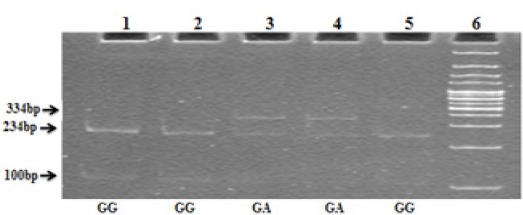
CYP2D6*4 RFLP Agarose Gel Picture; Lane 6, 100bp ladder; Lane 1, 2, 5, wild type 234, 100bp; Lane 3 and 4, Heterozygous mutant 334, 234, and 100 bp
Figure 2.
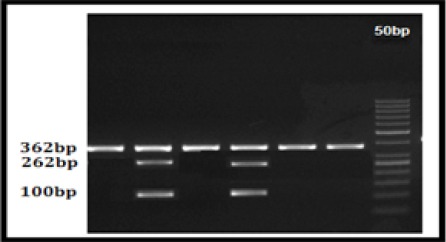
CYP2D6*10 RFLP Agarose Gel Picture; Lane 0, 50bp Ladder; Lane 2 and 4, 362, 262 and 100bp bp heterozygous mutant (CT) (Abnormal); Lane 1, 3, 4 and 5 362 bp Wild type (Normal)
Figure 3.
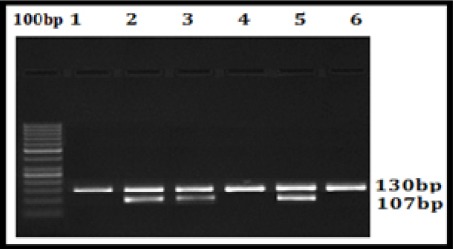
CYP3A5*3 RFLP Agarose Gel Picture; Lane 0, 100bp Ladder; Lane 1, 4 and 6, 130bp Wild type (Normal); Lane 2, 3 and 5, 130bp, 107bp and 23bap heterozygous mutant (Abnormal)
Figure 4.
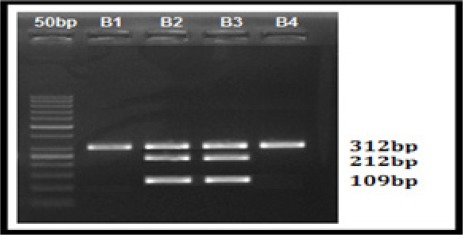
CYP2C19*2 RFLP Agarose Gel Picture; Lane 0, 50bp Ladder; Lane 1 and 3, 312, 212 and 109bp bp heterozygous mutant (Abnormal); Lane 1 and 4 312bp Wild type (Normal)
Statistical analysis
The frequency of genotypes was determined by calculating their percent in the control and cancer groups. Next, the frequency of alleles was obtained in both groups according to the Hardy–Weinberg equilibrium. The fisher’s exact test (2×2 only) was performed by using MedCalc software for Windows (version 7.4.1.0; Mariakerke, Belgium) to examine the association of mir polymorphisms between cases and controls and different clinical and pathological parameters. The difference between the groups was considered significant if the p value was 0.05.
Results
Clinical Characteristics of the study population
In the present study 110 blood samples from breast cancer patients and 100 blood samples from healthy controls were used. General characteristics of the breast cancer patients and controls are given in table 1. All 128 subjects were histopathalogical confirmed breast cancer cases and 98 were healthy controls with age and sex matched. Ages ranged from 20-70 years for cases and 22-70 years for controls. Breast cancer patients were divided into 5 groups according to age at diagnosis; these were 21-30, 31-40, 41-50, 51-60 and 61-70 years. Incidence of breast cancer cases and a control group was higher in the age groups 41-50 (40% (44 out of 110) years when compared to other age groups. Of the patients, majority of sporadic breast cancer cases were higher in postmenopausal 62% (n= 69) and 41% (n= 38) were premenopausal. The frequency of ER+/PR+ and Her2 positive tumor types were high (52% (56 out of 110) and 71% (79 out of 110)), when compared to other types. Of the cases, 22% (n= 23) were Grade I, 48% (n= 53) were Grade II and 30% (n=34) were Grade 3. While 26% (n= 29) of the patients were triple negative ER, PR, HER2 (-ve) breast cancer. Of those observed, 57% (n= 63) had received FAC/tamoxifen, 31% (n= 36) had received only FAC and 12% (n= 11) were unknown cases (Table.2).
CYP2D6*4 (G1934A) Genotype Frequencies
The overall genotype and allele frequencies of CYPs in breast cancer cases and health controls are shown in Table 2 . CYP2D6 gene polymorphisms were classified by metabolizer groups. The frequencies of the EM, IM, and PM genotypes of CYP2D6*4 (G1864A) were 79%, 15%, and 6% in the patients, and 96%, 4%, and 0%, respectively, in the healthy control group. We found significant difference in the frequencies of genotypes (OR: 4.3; 95% CI: 1.4230 to 13.5257, P = 0.01) between Breast Cancer Patients and Controls (Table 3). Significant difference was found between CYP2D6 IM metabolizer group and age group between 51-60 (p= 0.001), and also ER/PR, HER2 grade and therapy were compared with metabolizer groups of the patients. No statistically significant change was observed across the groups (p= 0.57, p= 0.22, p= 0=0.27, p= 0.44) (Table 4).
Table 2.
Pathological Characteristics of Breast Cancer Patients
| Patients Characteristics | Cases n=110(%) |
|---|---|
| Age of Diagnosis | |
| 21-30 | 10 (9%) |
| 31-40 | 26 (24%) |
| 41-50 | 44 (40%) |
| 51-60 | 22 (20%) |
| 61-70 | 8 (7%) |
| Menopausal Status | |
| Post Menopausal | 69 (62%) |
| Pre Menopausal | 41 (38%) |
| Age at Menarche | |
| 11-12 | 32 (29%) |
| 13-14 | 60 (54%) |
| 15-16 | 18 (17%) |
| Hormone Receptor Status | |
| ER+ /PR+ | 56 (52%) |
| ER+/PR- | 8 (7%) |
| ER-/PR+ | 4 (3%) |
| ER-/PR- | 42 (38%) |
| Her-2 Status | |
| Positive | 79 (71%) |
| Negative | 31 (28%) |
| Triple Negative | 29 (26%) |
| Histological Grade | |
| Grade I | 23 (22%) |
| Grade II | 53 (48%) |
| Grade III | 34 (30%) |
| Chemotherapy and Hormonal Therapy | |
| FAC/Tamoxifen | 63 (57%) |
| FAC/Non Tamoxifen | 36 (31%) |
| Unknown | 11 (12%) |
Table 3.
Genotype and Allele Frequencies of CYP2D6*4, CYP2D6*10, CYP3A5*3 & CYP2C19*2 Gene Polymorphism in the South Indian Women with Breast Cancer
| CYP2D6*4 | Cases (110) | Controls (100) | Odds Ratio | 95% CI | P- Value |
|---|---|---|---|---|---|
| GG | 87 (79%) | 96 (96%) | 0.156 | 0.0524 to 0.4738 | P = 0.0010 |
| GA | 17 (15%) | 4 (4%) | 4.3871 | 1.4230 to 13.5257 | 0.0101 |
| AA | 6 (6%) | 0 (00%) | 12.5 | 0.6952 to 224.8466 | 0.0866 |
| G Allele | 191 (86.8%) | 196 (89.0) | 0.1344 | 0.0464 to 0.3896 | 0.0002 |
| A Allele | 29 (13.18) | 4 (2%) | |||
| CYP2D6*10 | Cases (110) | Controls | Odds Ratio | 95% CI | P- Value |
| CC | 83 (75%) | 92 (92%) | 0.2673 | 0.1151 to 0.6210 | 0.0022 |
| CT | 24 (23%) | 7 (7%) | 3.7076 | 1.5202 to 9.0425 | 0.004 |
| TT | 3 (2%) | 1 (1%) | 2.6075 | 0.2666 to 25.4977 | 0.4101 |
| C Allele | 190 (86.3%) | 192 (96%) | 0.2969 | 0.1373 to 0.6421 | 0.002 |
| T Allele | 30 (13.6) | 9 (4.5%) | |||
| CYP3A5*3 | Cases (110) | Controls | Odds Ratio | 95% CI | P- Value |
| AA | 97 (88%) | 95 (95%) | 0.3927 | 0.1348 to 1.1444 | 0.0867 |
| AG | 11 (10%) | 5 (5%) | 2.1111 | 0.7070 to 6.3041 | 0.1807 |
| GG | 2 (2%) | 0 (00%) | 4.6313 | 0.2197 to 97.6476 | 0.3244 |
| A Allele | 205 (93.1%) | 195 (97.5%) | 0.3504 | 0.1250 to 0.9825 | 0.0462 |
| G Allele | 15 (6.8) | 5 (2.5%) | |||
| CYP2C19*2 | Cases (110) | Controls | Odds Ratio | 95% CI | P- Value |
| GG | 93 (85%) | 97 (97%) | 0.1692 | 0.0480 to 0.5964 | 0.0057 |
| GA | 13 (12%) | 3 (3%) | 4.3333 | 1.1969 to 15.6881 | 0.0255 |
| AA | 4 (3%) | 0 | 8.493 | 0.4515 to 159.7675 | 0.1531 |
| G Allele | 199 (90.4%) | 197 (98.5%) | 0.1443 | 0.0424 to 0.4916 | 0.002 |
| A Allele | 21 (9.5%) | 3 (1.5%) | |||
Table 4.
Correlation of Breast Cancer Patients Demographic Factors and CYP2D6*4 Genotype Frequencies
| Characteristics | Cases (n=110) | EM (n=87) | IM (n=17) | PM (n=6) | P- Value |
|---|---|---|---|---|---|
| Age of Diagnosis | |||||
| 21-30 | 10 (9%) | 9 (90%) | 1 (10%) | 0 (0%) | |
| 31-40 | 26 (24%) | 23 (89 %) | 2 (8 %) | 1 (3%) | |
| 41-50 | 44 (40%) | 36 (82%) | 6 (13%) | 2 (5%) | |
| 51-60 | 22 (20%) | 11 (50%) | 8 (36%) | 3 (14%) | 0.001 |
| 61-70 | 8 (7%) | 8 (100%) | 0 (00%) | 0 (00%) | |
| Menopausal Status | |||||
| Post Menopausal | 65 (60%) | 50 (77%) | 12 (19%) | 3 (4%) | 0.31 |
| Premenopausal | 45 (40%) | 37 (83%) | 5 (11%) | 3 (6%) | |
| Hormone Receptor status | |||||
| ER /PR Positive | 56 (66%) | 43 (76%) | 11 (20%) | 2 (4%) | 0.57 |
| ER/PR Negative | 42 (25%) | 32 (76%) | 6 (14%) | 4 (10%) | |
| HER2 neu Positive | 79 (25%) | 64 (82%) | 10 (12%) | 5 (6%) | 0.22 |
| HER2 neu Negative | 31 (25%) | 23 (74%) | 7 (22%) | 1 (3%) | |
| Triple Negative | 29 (26%) | 16 (55%) | 9 (31%) | 4 (14%) | |
| Histological Grade | |||||
| Grade I | 23 (41%) | 21 (92%) | 2 (8%) | 0 (0%) | 0.27 |
| Grade II | 53 (41%) | 40 (68%) | 10 (30%) | 3 (2%) | |
| Grade III | 34 (18%) | 26 (65%) | 5 (22%) | 3 (13%) | |
| Therapy | |||||
| FAC/Tamoxifen | 63 (57%) | 47 (75%) | 11 (17%) | 5 (8%) | 0.04 |
| FAC | 36 (31%) | 30 (80%) | 5 (14%) | 1 (6%) | |
| Unknown | 11 (12%) | 10 (91%) | 1 (9%) | 0 (0%) |
CYP2D6*10 (C188T) Genotype Frequencies
The frequencies of the CC, CT, and TT genotypes of CYP2D6*10 (C100T) were 75%, 23%, and 2% in the patients, and 92%, 7%, and 1%, respectively, in the healthy control group (Table 2). The CT genotype was significantly different between breast cancer patients and control subjects. Frequency of CT genotype was significantly associated with breast cancer (OR: 3.7; 95% CI: 1.5202 to 9.0425, P = 0.004) (Table 3). Significant difference was found between CYP2D6*10 CT genotype group and tamoxifen received group and grade II patients (p= 0.002 and p= 0.02), ER/PR and HER2 were compared with CYP2D6*10 genotypes of the patients. No statistically significant change was observed across the groups (p= 0.98 and p= 0.98) (Table 5).
Table 5.
Correlation of Breast Cancer Patients Demographic Factors and CYP2D6*10 Genotype Frequencies
| Characteristics | Cases (n=110) | EM (n=83) | IM (n=24) | PM (n=3) | P- Value |
|---|---|---|---|---|---|
| Age of Diagnosis | |||||
| 21-30 | 10 (9%) | 10 (100%) | 0 (00%) | 0 (00%) | |
| 31-40 | 26 (24%) | 23 (89 %) | 3 (8 %) | 0 (0%) | 0.02 |
| 41-50 | 44 (40%) | 27 (82%) | 14 (13%) | 3 (5%) | |
| 51-60 | 22 (20%) | 15 (50%) | 7 (36%) | 0 (00%) | |
| 61-70 | 8 (7%) | 8 (100%) | 0 (00%) | 0 (00%) | |
| Menopausal Status | |||||
| Post Menopausal | 65 (60%) | 47 (77%) | 15 (19%) | 3 (4%) | 0.6 |
| Premenopausal | 45 (40%) | 36 (83%) | 9 (11%) | 0 (0%) | |
| Hormone Receptor status | |||||
| ER /PR Positive | 56 (66%) | 41 (76%) | 14 (20%) | 1 (4%) | 0.98 |
| ER/PR Negative | 42 (25%) | 29 (76%) | 10 (14%) | 3 (10%) | |
| HER2 neu Positive | 79 (25%) | 59 (75%) | 17 (21%) | 3 (4%) | 0.98 |
| HER2 neu Negative | 31 (25%) | 24 (77%) | 7 (22%) | 0 (00%) | |
| Triple Negative | 29 (26%) | 16 (55%) | 12 (41%) | 1 (3%) | |
| Histological Grade | |||||
| Grade I | 23 (41%) | 23 (100%) | 0 (00%) | 0 (00%) | |
| Grade II | 53 (41%) | 36 (68%) | 16 (30%) | 1 (2%) | 0.04 |
| Grade III | 34 (18%) | 24 (70%) | 8 (24%) | 2 (6%) | |
| Therapy | |||||
| FAC/Tamoxifen | 63 (57%) | 40 (63%) | 14 (22%) | 3 (5%) | 0.002 |
| FAC | 36 (31%) | 28 (77%) | 8 (23%) | 0 (0%) | |
| Unknown | 11 (12%) | 9 (82%) | 2 (18%) | 0 (0%) |
CYP3A5*3 (A6986G) Genotype Frequencies
The frequencies of the AA, AG, and GG genotypes of CYP3A5*3 (A6986G) were 88%, 10%, and 2% in the patients, and 95%, 11%, and 2%, respectively, in the healthy control group (Table 2). The AG genotype was significantly different between breast cancer patients and control subjects. We show that no difference between the patients and controls for A6986G polymorphism (OR: 2.1; 95% CI: 0.7070 to 6.3041, P = 0.18) (Table 3). In the present study we found that ER/PR, HER2, grade and therapy were compared with metabolizer groups of the patients. No statistically significant change was observed across the groups (p= 0.56, p= 0.98 p= 0.11 and 0.09) (Table 6).
Table 6.
Correlation of Breast Cancer Patients Demographic Factors and CYP3A5*3 Genotype Frequencies
| Characteristics | Cases (n=110) | EM (n=97) | IM (n=11) | PM (n=2) | P-Value |
|---|---|---|---|---|---|
| Age of Diagnosis | |||||
| 21-30 | 10 (9%) | 10 (100%) | 0 (00%) | 0 (00%) | |
| 31-40 | 26 (24%) | 26 (100 %) | 0 (00 %) | 0 (3%) | |
| 41-50 | 44 (40%) | 34 (77%) | 8 (19%) | 2 (4%) | |
| 51-60 | 22 (20%) | 19 (87%) | 3 (13%) | 0 (00%) | |
| 61-70 | 8 (7%) | 8 (100%) | 0 (00%) | 0 (00%) | 0.02 |
| Menopausal Status | |||||
| Post Menopausal | 65 (60%) | 56 (77%) | 7 (19%) | 2 (4%) | 0.7 |
| Premenopausal | 45 (40%) | 41 (83%) | 4 (11%) | 0 (0%) | |
| Hormone Receptor status | |||||
| ER /PR Positive | 56 (66%) | 41 (76%) | 5 (20%) | 0 (4%) | 0.56 |
| ER/PR Negative | 42 (25%) | 34 (76%) | 6 (14%) | 2 (10%) | |
| HER2 neu Positive | 79 (25%) | 59 (75%) | 8 (21%) | 3 (4%) | 0.98 |
| HER2 neu Negative | 31 (25%) | 24 (77%) | 3 (23%) | 0 (00%) | |
| Triple Negative | 29 (26%) | 20 (68%) | 8 (28%) | 1 (4%) | |
| Histological Grade | |||||
| Grade I | 23 (41%) | 23 (100%) | 0 (00%) | 0 (00%) | |
| Grade II | 53 (41%) | 45 (85%) | 8 (15%) | 0 (00%) | 0.11 |
| Grade III | 34 (18%) | 29 (85%) | 3 (91%) | 2 (6%) | |
| Therapy | |||||
| FAC/Tamoxifen | 63 (57%) | 52 (83%) | 9 (14%) | 2 (3%) | 0.09 |
| FAC | 36 (31%) | 34 (95%) | 2 (5%) | 0 (0%) | |
| Unknown | 11 (12%) | 11 (100%) | 0 (00%) | 0 (0%) |
CYP2C19*2 (G681A) Genotype Frequencies
The frequencies of the GG, GA, and AA genotypes of CYP2C19*2 (G681A) were 88%, 10%, and 2% in the patients, and 95%, 5%, and 1%, respectively, in the healthy control group (Table 2).. The GA genotype was significantly different between breast cancer patients and control subjects. Frequency of GA genotype was significantly associated with breast cancer (OR: 3.7; 95% CI: 1.5202 to 9.0425, P = 0.004) (Table 3).. Significant difference was found between CYP2C19*2 GA genotype group and tamoxifen received group (p= 0.03), ER/PR, HER and grade were compared with CYP2C19*2 genotypes of the patients. No statistically significant change was observed across the groups (p= 0.54, p= 0.97 and 0.11) (Table 7).
Table 7.
Correlation of Breast Cancer Patients Demographic Factors and CYP2C19*2 Genotype Frequencies
| Characteristics | Cases (n=110) | EM (n=93) | IM (n=13) | PM (n=4) | P- Value |
|---|---|---|---|---|---|
| Age of Diagnosis | |||||
| 21-30 | 10 (9%) | 10 (100%) | 0 (00%) | 0 (00%) | |
| 31-40 | 26 (24%) | 24 (100 %) | 2 (00 %) | 0 (3%) | |
| 41-50 | 44 (40%) | 22 (77%) | 8 (19%) | 4 (4%) | 0.008 |
| 51-60 | 22 (20%) | 19 (87%) | 3 (13%) | 0 (00%) | |
| 61-70 | 8 (7%) | 8 (100%) | 0 (00%) | 0 (00%) | |
| Menopausal Status | 0.36 | ||||
| Post Menopausal | 65 (60%) | 52 (77%) | 9 (19%) | 4 (4%) | |
| Premenopausal | 45 (40%) | 41 (83%) | 4 (11%) | 0 (0%) | |
| Hormone Receptor status | |||||
| ER /PR Positive | 56 (66%) | 48 (76%) | 6 (20%) | 2 (4%) | 0.37 |
| ER/PR Negative | 42 (25%) | 33 (76%) | 7 (14%) | 2 (10%) | |
| HER2 neu Positive | 79 (25%) | 65 (75%) | 10 (21%) | 4 (4%) | 0.6 |
| HER2 neu Negative | 31 (25%) | 28 (77%) | 3 (23%) | 0 (00%) | |
| Triple Negative | 29 (26%) | 20 (68%) | 8 (28%) | 1 (4%) | |
| Histological Grade | |||||
| Grade-I | 23 (41%) | 23 (100%) | 0 (00%) | 0 (00%) | 0.14 |
| Grade II | 53 (41%) | 45 (85%) | 8 (15%) | 3 (00%) | |
| Grade III | 34 (18%) | 29 (85%) | 5 (91%) | 1 (6%) | |
| Therapy | |||||
| FAC/Tamoxifen | 63 (57%) | 48 (83%) | 11 (14%) | 4 (3%) | 0.03 |
| FAC | 36 (31%) | 34 (95%) | 2 (5%) | 0 (0%) | |
| Unknown | 11 (12%) | 11 (100%) | 0 (00%) | 0 (0%) |
Distribution genotype frequencies of tamoxifen treated cases
The CYP2D6*4 EM, IM and PM in the tamoxifen treated patients were 57.6% and 38% and 4% respectively and for the CYP2D6*10 EM, IM and PM group were 74%, 13% and 0% respectively and for the CYP3A5*3 EM, IM and PM group were 93%, 7% and 0% and for the CYP2C19*2 EM, IM and PM group were 86%, 14% and 0% (Table 8). The frequency of intermediate metabolizes were high in CYP2D6 gene when compared to CYP3A5 and CYP2C19.
Table 8.
Distribution of Tamoxifen Treated Cases
| Genotypes | CYP2D6*4 | CYP2D6*10 | CYP3A5*3 | CYP2C19*2 |
|---|---|---|---|---|
| (n=42) | (n=42) | (n=42) | (n=42) | |
| EM | 24 (57.6%) | 31 (74%) | 39 (93%) | 36 (86%) |
| IM | 16 (38%) | 13 (31%) | 3 (7%) | 6 (14%) |
| PM | 2 (4%) | 0 (00%) | 0 (00%) | 0 (00%) |
Discussion
Tamoxifen is a selective estrogen receptor (ER) modulator and is mainly indicated for the treatment of breast cancer in postmenopausal women. Tamoxifen is a standard treatment to prevent recurrences in women suffering from breast cancer, the drug is still not effective for 20 to 30% of patients receiving it. The important role of cytochrome P450 (CYP) proteins in the metabolism of tamoxifen has led to speculation that polymorphisms in CYP2D6, CYP3A5 and CYP2C19 may contribute to the observed variations in treatment efficacy. The objectives of the present study were to study the pharmacogenetics of CYP3A5, CYP2C9, CYP2C19 and CYP2D6 polymorphisms in south Indian women with breast cancer.
CYP2D6 is responsible for the metabolism of tamoxifen to create its active metabolite, endoxifen. CYP2D6*4 and CYP2D6*10 polymorphisms plays an important role in the breast cancer etiology and might help in planning hormonal therapy where tamoxifen is used. In the present study CYP2D6*4 and CYP2D6*10 intermediate and poor metabolizers were significantly associated with south Indian women with breast cancer. Reduced CYP2D6 activity is associated with poor treatment outcomes, in terms of increased risk of recurrence and shorter recurrence free survival, in breast cancer patients on adjuvant tamoxifen therapy. Kiyotani K, Mushiroda, et al 2010, reported patients with poor metabolizers leads to poor survival. The frequency of intermediate metabolizes of CYP2D6*4 and CYP2D6*10 alleles were high in postmenopausal women treated with tamoxifen. This suggests that identifying such patients before the start of treatment may be useful in optimizing therapy with tamoxifen.
Apart from CYP2D6, other cytochrome P450 enzymes (CYP3A5 and CYP2C19) also contribute to the overall metabolism of tamoxifen and its metabolites, albeit to different extents CYP3A5 catalyses the N-demethylation of tamoxifen to NDM or 4-OHT to endoxifen. In the present study CYP3A5*3 was not statistically significant. Our results are consistent with previous studies. Jin et al 2005 and Tucker et al, 2005 did not find any significant association between CYP3A5*3 (6986A>G; rs776746) and the plasma concentrations of tamoxifen and its metabolites. It is likely that the functional impact of CYP3A5*3 (6986A>G; rs776746) on the metabolism of tamoxifen is nullified. CYP2C19*2 was statistically significant (p=0.02). Previous studies have also found the presence of CYP2C19*2 to be associated with elevated efficacy of tamoxifen treatment, this study agrees with the findings of other research (Rikje Ruiter, et al 2009). The role of CYP3A5 and CYP2C19 seem to be minor.
In the present study the association between genotypes and clinicopathological parameters of breast cancer was also analyzed in our study. It was noted that there was significant association between intermediate metabolizers of CYP2D6 and CYP2C19 and tamoxifen therapy received and grade II patients. It was observed that there was no significant association between CYP2D6 CYP3A5 and CYP2C19 polymorphisms and age, ER/PR, Her2 neu and garde. 15 percent of all breast cancers are triple negative, tamoxifen is not currently used to treat triple negative breast cancer (TNBC). In our study, 26% cases are triple negative breast cancer, the CYP2D6*4, CYP2D6*10, CYP3A5*3 and CYP2C19*2 genes intermediate metabolizer group was showed high frequency but statistically not significant.
References
- Damodaran SE, Pradhan SC, Umamaheswaran G, et al. Genetic polymorphisms of CYP2D6 increases the risk for recurrence of breast cancer in patients receiving tamoxifen as an adjuvant therapy. Cancer Chemother Pharmacol. 2012;70:75–81. doi: 10.1007/s00280-012-1891-1. [DOI] [PubMed] [Google Scholar]
- Desta Z, Ward BA, Soukhova NV, et al. Comprehensive evaluation of tamoxifen sequential biotransformation by the human cytochrome P450 system in vitro:prominent roles for CYP3A and CYP2D6. J Pharmacol Exp Ther. 2004;310:1062–75. doi: 10.1124/jpet.104.065607. [DOI] [PubMed] [Google Scholar]
- Hoskins JM, Carey LA, McLeod HL, et al. CYP2D6 and tamoxifen:DNA matters in breast cancer. Nat Rev Cancer. 2009;9:576–86. doi: 10.1038/nrc2683. [DOI] [PubMed] [Google Scholar]
- Irvin WJ, Walko CM, Weck KE, et al. Genotype-guided tamoxifen dosing increases active metabolite exposure in women with reduced CYP2D6 metabolism:a multicenter study. J Clin Oncol. 2011;29:3232–9. doi: 10.1200/JCO.2010.31.4427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T, Hirano H, Onishi Y, et al. Functional evaluation of ABCB1 (P-glycoprotein) polymorphisms:high-speed screening and structure-activity relationship analyses. Drug Metab Pharmacokinet. 2004;19:1–14. doi: 10.2133/dmpk.19.1. [DOI] [PubMed] [Google Scholar]
- Jin Y, Desta Z, Stearns V, et al. CYP2D6 genotype, antidepressant use, and tamoxifen metabolism during adjuvant breast cancer treatment. J Natl Cancer Inst. 2005;5(97):30–9. doi: 10.1093/jnci/dji005. [DOI] [PubMed] [Google Scholar]
- Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7:573–584. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- Kiyotani K, Mushiroda T, Imamura CK, et al. Significant effect of polymorphisms in CYP2D6 and ABCC2 on clinical outcomes of adjuvant tamoxifen therapy for breast cancer patients. J Clin Oncol. 2010;28:1287–93. doi: 10.1200/JCO.2009.25.7246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Feng L, Xu Y, et al. The association of CYP2D6 *10 polymorphism with breast cancer risk and clinico-pathologic characteristics in Chinese women. Acta Oncol. 2006;45:597–601. doi: 10.1080/02841860600660803. [DOI] [PubMed] [Google Scholar]
- Lim JS, Chen XA, Singh O, et al. Impact of CYP2D6, CYP3A5, CYP2C9 and CYP2C19 polymorphisms on tamoxifen pharmacokinetics in Asian breast cancer patients. Br J Clin Pharmacol. 2011;71:737–50. doi: 10.1111/j.1365-2125.2011.03905.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim YC, Li L, Desta Z, et al. Endoxifen, a secondary metabolite of tamoxifen, and 4-OH-tamoxifen induce similar changes in global gene expression patterns in MCF-7 breast cancer cells. J Pharmacol Exp Ther. 2006;318:503–12. doi: 10.1124/jpet.105.100511. [DOI] [PubMed] [Google Scholar]
- Park HS, Choi J-Y, Lee M-J, et al. Association between genetic polymorphisms of CYP2D6 and outcomes in breast cancer patients with Tamoxifen treatment. J Korean Med Sci. 2011;26:1007–13. doi: 10.3346/jkms.2011.26.8.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberti A, La Sala D, Cinti C, et al. Multiple genetic and epigenetic interacting mechanisms contribute to clonally selection of drug-resistant tumors:current views and new therapeutic prospective. J Cell Physiol. 2006;207:571–58. doi: 10.1002/jcp.20515. [DOI] [PubMed] [Google Scholar]
- Ruiter R, Bijl MJ, van Schaik RH, et al. CYP2C19*2 polymorphism is associated with increased survival in breast cancer patients using tamoxifen. Pharmacogenomics. 2010;11:1367–75. doi: 10.2217/pgs.10.112. [DOI] [PubMed] [Google Scholar]
- Sosa-Macías M, LLerena A. Cytochrome P450 genetic polymorphisms of Mexican indigenous populations. Drug Metabol Drug Interact. 2013;28:193–208. doi: 10.1515/dmdi-2013-0037. [DOI] [PubMed] [Google Scholar]
- Surekha D, Sailaja K, Nageswara Rao D, et al. CYP2D6*4 polymorphisms and breast cancer risk. Biol Med. 2010;2:49–55. [Google Scholar]
- Tucker AN, Tkaczuk KA, Lewis LM, et al. Polymorphisms in cytochrome P4503A5 (CYP3A5) may be associated with race and tumor characteristics, but not metabolism and side effects of tamoxifen in breast cancer patients. Cancer Lett. 2005;10:61–72. doi: 10.1016/j.canlet.2004.08.027. [DOI] [PubMed] [Google Scholar]


