Abstract
We previously reported the sequential recovery of daptomycin-nonsusceptible MRSA clinical isolates with an L431F substitution in the MprF protein. The aim of the present study is to determine the effect of this mutation by replacing the mprF gene on the chromosome of a daptomycin-susceptible progenitor strain, CGK5, to obtain CGK5mut having the L431F MprF mutation. Compared to CGK5, the daptomycin and vancomycin MICs of CGK5mut increased from 0.5 to 3 μg/ml and from 1.5 to 3 μg/ml, respectively; however, its oxacillin MIC decreased from 128 to 1 μg/ml in medium without added 2% NaCl. The expression levels of vraSR and several other cell-wall synthesis-related genes were significantly increased in CGK5mut, and the mutant also had significantly reduced negative cell membrane charge, thicker cell wall, and longer doubling time. These features were abolished in the reverse mutant carrying F431L MprF, confirming the pleiotropic effects of the L431F MprF mutation. We believe that this is the first work that shows a single MprF missense mutation can lead to not only changes in the cell membrane but also increased expression of vraSR and subsequently increased resistance to daptomycin and vancomycin while simultaneously conferring increased susceptibility to oxacillin in an isogenic MRSA strain.
Keywords: MRSA, evolution, drug resistance, daptomycin, vancomycin, oxacillin
Introduction
Daptomycin, a cyclic lipopeptide antibiotic, is one of the last-line agents for the treatment of certain severe multidrug-resistant Staphylococcus aureus infections, including those caused by methicillin-resistant S. aureus (MRSA) (Enoch et al., 2007). Daptomycin functions by inserting itself into the bacterial cell membrane in a calcium-dependent manner to cause membrane depolarization, leading to cell death (Enoch et al., 2007; Baltz, 2009). Daptomycin-nonsusceptible (DAP-NS) MRSA isolates, although still uncommon, have emerged during daptomycin treatment of patients (Lee et al., 2010; Boyle-Vavra et al., 2011). DAP-NS MRSA mutants have also been generated in the laboratory by serial passage of isolates in sublethal concentrations of daptomycin (Enoch et al., 2007; Camargo et al., 2008; Mishra et al., 2009, 2012; Rubio et al., 2012).
The exact mechanisms giving rise to daptomycin-non-susceptibility in S. aureus are not fully elucidated but appear to involve diverse genetic events and several genetic loci, including mprF, yycG (walK), vraSR, tagA, and dltABCD (Friedman et al., 2006; Baltz, 2009; Bertsche et al., 2011; Song et al., 2013). These loci are all part of the cell wall stimulon in S. aureus and include genes encoding proteins involved in the production of membrane phospholipids. The mprF (multiple peptide resistance factor) gene seems to be especially critical, as mprF mutations are the most frequently reported genetic lesions in DAP-NS MRSA isolates (Friedman et al., 2006; Lee et al., 2010; Boyle-Vavra et al., 2011; Mehta et al., 2012a). MprF is a bi-functional membrane protein with lysylphosphatidylglycerol (LPG) synthase and flippase activities (Peschel et al., 2001; Ernst et al., 2009; Ernst and Peschel, 2011). Different point mutations in mprF have been associated with elevated LPG synthesis. This results in increased amounts of LPG relative to phosphatidylglycerol (PG) on the outer leaflet of the cytoplasmic membrane and an accompanying reduction in cell membrane negative charge (Baltz, 2009; Rubio et al., 2012).
A feature that has been seen in both clinical and laboratory-generated DAP-NS MRSA isolates is a concomitant vancomycin intermediate or heterogeneous intermediate resistance (VISA or hVISA) phenotype (Camargo et al., 2008; Mishra et al., 2009); VISA has moderate resistance to vancomycin and hVISA has varying sub-population of cells resistant to vancomycin, thus exhibit mixed susceptibility to vancomycin. Mutations in the vraSR, graSR, or walKR (yycGF) two-component systems have been associated with the VISA/hVISA phenotype (Howden et al., 2010). Among these 3 two-component systems, the VraSR system is particularly important in maintaining cell wall integrity. It serves as a sentinel in response to cell wall damage by positively regulating a unique set of genes involved in cell wall synthesis, resulting in the generation of a resistant phenotype (Kuroda et al., 2003; Gardete et al., 2006; McAleese et al., 2006). Another unusual feature is the so-called “seesaw” effect on β-lactam susceptibility, wherein DAP-NS isolates exhibit reduced β-lactam MICs (Mishra et al., 2009; Lee et al., 2010; Yang et al., 2010; Mehta et al., 2012a). However, these phenomena are not observed in all DAP-NS MRSA isolates.
Our previous study of eight sequential clinical MRSA isolates from a patient with persistent bacteremia revealed an L431F amino acid substitution in the MprF protein of DAP-NS isolates (Lee et al., 2010). Since this mutation had not been reported previously, we undertook the present study to determine the effect of this mutation on the cellular response to daptomycin. To eliminate the possibility of unknown genetic changes that might have occurred in paired clinical strains, we used a base-substitution method to replace a single nucleotide (from CTT to TTT) within the chromosomal mprF gene of the daptomycin-susceptible (DAP-S) progenitor of the DAP-NS strains. To confirm the results obtained by this single amino acid exchange (L431F) in MprF, we also constructed a reverse mutant carrying F431L MprF as well as a silent EcoRV site. Our study demonstrated that this single amino acid change (L431F) confers increased resistance to both daptomycin and vancomycin, with a concurrent decrease in oxacillin MIC. The phenotype and genetic factors associated with these changes were investigated.
Materials and Methods
Bacterial Strains
The bacterial strains, plasmids, and primers used are listed in Tables 1, 2. Unless stated otherwise, Luria-Bertani (LB) broth and plates were used for growth of Escherichia coli and S. aureus at 37°C. E. coli strain XL10-Gold (Stratagene, La Jolla, CA, United States) and GeneHogs® (Invitrogen, Carlsbad, CA, United States) were used for cloning. S. aureus cells were transformed by electroporation, as described previously (Schenk and Laddaga, 1992). Ampicillin (100 μg/ml), chloramphenicol (5 μg/ml), erythromycin (5 μg/ml), spectinomycin (100 μg/ml), and tetracycline (5 μg/ml) were used for plasmid selection in E. coli and S. aureus.
Table 1.
Bacterial strains and plasmids used in this study.
| Strain/plasmid | Description | Source or Reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| XL10-Gold | Ultra-competent cell for site-directed mutagenesis | Stratagene |
| Genehogs | Electrocompetent cells | Invitrogen |
| S. aureus | ||
| RN4220 | Restriction-deficient derivative of 8325-4 | Novick, 1991 |
| RN6911 | RN6390B agr::tetM (agr-null) | Novick et al., 1993 |
| Z172 | Clinical VISA isolate with spc gene | Chen et al., 2013 |
| CGK5 | Daptomycin-susceptible MRSA | Lee et al., 2010 |
| CGK5mut | MprF L431F derivative of CGK5 | This study |
| CGK5mutR | Reversed derivative of CGK5mut with MprF containing F431L and a new EcoRV site | This study |
| CGK6 | Daptomycin-non-susceptible MRSA | Lee et al., 2010 |
| E. faecium | ||
| 2V076 | Clinical isolate with aadE-sat4-aphA-3 gene cluster | This study |
| Plasmids | ||
| pMAD | pE194ts derivative for gene replacement in Gram-positive bacteria | Arnaud et al., 2004 |
| pMAD-SAT4-tetM | Modified pMAD with sat4 and tetM markers | This study |
| pMprFmut | The mprF fragment amplified from CGK6 and cloned into pMAD-SAT4-tetM for allelic exchange in CGK5 | This study |
| pMprF5 | The mprF fragment amplified from CGK5 and cloned into pMAD-SAT4-tetM for site-directed mutagenesis | This study |
| pMprFmutR | A silent EcoRV site was introduced into the middle of the mprF fragment amplified from pMprF5 and cloned into pMAD-SAT4-tetM for allelic exchange in CGK5mut | This study |
| pluxT2 | pSK5630 derivative containing luxABCDE with T2 terminator | Chen et al., 2014 |
| pluxT2-SPC | pluxT2 with spc marker | This study |
tsStands for thermosensitive.
Table 2.
Primers used in this study.
| Primers | Nucleotide sequence (5′–3′)a |
|---|---|
| Cloning of new pMAD | |
| bgaBnew-F | CGGGATCCAGGAATTCGCTCCCGGGCATGCCATGGGTCTAGTTAATGTGTAACGTAACA |
| bgaBnew-R | ACGCGTCGACGTAAGGCCTTCACTAAACCTTCCCGGCTTC |
| sat4-F | P-AGAGAGGCGGGAACAGTG |
| sat4-R | ACGCGTCGACTGCAGGCCTTCAGATCTAAGACGAACTCCAATTCACT |
| tetM-F | P-GGAGATTCCTTTACAAATATG |
| tetM-R | ACGCGTCGACGTAAGGCCTATAACAACATAAAACGCACTA |
| Cloning of mprF gene for allelic exchange | |
| mprF-F | CGGGATCCTAGAATTGATGTGAAAAAATGA |
| mprF-R | TCCCCCGGGCGCATCAGGCATAACTGTATA |
| Site-directed mutagenesis | |
| mprF(EcoRV)-F | P-ATCATTGCTAAAATTCCATCATTGTC |
| mprF(EcoRV)-R | ATCCTTTTGATAAGACATTAAAA |
| Real-time qPCR | |
| mprF-QF | TCATTATTGCTGCATTATCTGGA |
| mprF-QR | TTTTCCTCAGGGACACCTAAAG |
| vraSR-QF | GCCAGATTCAGGTACACG |
| vraSR-QR | TCTGAGTCGTCGCTTC |
| fmtA-QF | AAAACATCTAAGCCTATCCCATTG |
| fmtA-QR | TTTGAATCGCTTTAACTGCTTGAT |
| murZ-QF | AAAATAAGAGGTGGACGCACA |
| murZ-QR | ACTGTTTTTCGCGCCACT |
| pbp2-QF | TCGGTGCAATTGGTAAGAACT |
| pbp2-QR | TTAATGTTGAGGCACCTTCAGA |
| sgtB-QF | TAGCGACAGAGATGTGC |
| sgtB-QR | TTGTGACATAGCCTGTTG |
| tagA-QF | AATAAATCAAGCGAGCTATATTGTTG |
| tagA-QR | ACGATGCGAAGCTTTGACTAC |
| gyrB-QF | CGTTAATTGAAGCAGGCTATGTG |
| gyrB-QR | TGGTGTTGGATTCAATTCAGATT |
| Cloning of promoter-reporter | |
| spc-F | P-AAAGTTCTCGTTCGGAGG |
| spc-R | TCCCCCGGGAAAGTAAGCACCTGTTATTGC |
| PmprF-F | CGGGATCCGAAAATAAAAACAAGTGGTAT |
| PmprF-R | ACGCGTCGACTTAACTTCCTGATTCATTT |
| PvraSR-F | CGGGATCCCGTTTATCTCATCAAATG |
| PvraSR-R | ACGCGTCGACTAGTTCATAACTATCACCTTT |
aRestriction enzyme sites are underlined. P represents phosphorylation. Boldface type indicates a mutant nucleotide.
Construction of MprF Mutant Derivatives
To add new selection markers and cloning sites, the bgaB cassette located between the HindIII sites was removed from pMAD by digesting with HindIII, and religated. A modified bgaB, with extra restriction sites, was cloned by PCR (using primers bgaBnew-F and bgaBnew-R) from the original pMAD into the pMAD lacking the bgaB cassette to generate new pMAD. Two selection markers, as sat4 and tetM cassettes, were incorporated by cloning PCR fragments of the sat4 and tetM genes from the chromosomal DNA of 2V076 and RN6911, respectively, into the StuI-SalI sites of the new pMAD using primers sat4-F and sat4-R, and tetM-F and tetM-R to produce pMAD-SAT4-tetM, which confers resistance to nourseothricin and tetracycline. To assess the effect of MprF L431F on daptomycin non-susceptibility, an mprF-bearing fragment from CGK6 (the first DAP-NS isolate containing the MprF F431 mutant) was amplified by PCR using the primers mprF-F and mprF-R, and ligated into the BamHI-SmaI sites of pMAD-SAT4-tetM. This recombinant plasmid, pMprFmut, was used as the allelic exchange vector for mutation of the mprF gene in CGK5 to create CGK5mut. To validate the phenotypes in CGK5mut, the mutant strain was reverted to wild type by allelic exchange again. The mprF fragment was amplified from CGK5 and then cloned into pMAD-SAT4-tetM. This recombinant plasmid, pMprF5, was used as the template DNA for site-directed mutagenesis to introduce a new EcoRV site1 into the complementation construct (and without altering the coding sequence) to allow it to be differentiated from the CGK5 parental strain. After PCR using the mutant primers mprF(EcoRV)-F and mprF(EcoRV)-R, the mprF reverse mutant was treated with DpnI, ligated into a circle with T4 DNA ligase, and then used as the template DNA. The entire mprF gene was then cloned from the mutated plasmid into the pMAD-SAT4-tetM vector again to eliminate potential mutation of the vector sequence. The constructs were verified by restriction analysis and DNA sequencing. Sequencing was performed at the DNA Sequencing Core Lab of our institutes.
The allele replacement procedure was applied to create a single base replacement in the mprF gene of CGK5, as described previously (Arnaud et al., 2004). Briefly, the pMprFmut plasmid was electroporated into S. aureus strain RN4220, and then electroporated into CGK5. Transformants were selected at 30°C on LB plates containing tetracycline and X-Gal (150 μg/ml). One blue colony was inoculated in Trypticase Soy broth (TSB) containing tetracycline and incubated with shaking for 2 h at 30°C followed by 6 h at 43°C, serial diluted then plated on Trypticase Soy agar (TSA) plates containing tetracycline and X-Gal and incubated at 43°C overnight to obtain light blue colonies caused by a single crossover event. One light blue colony was inoculated in TSB without antibiotic and incubated with shaking at 30°C overnight, then diluted 1:100 and incubated at 43°C for 6 h; serial dilutions were plated on TSA plates in the absence of antibiotics and incubated at 37°C overnight. Several white colonies were selected to verify for tetracycline sensitivity, which indicates loss of the integrated vector resulting from a double crossover event. To confirm the double crossover, PCR amplifications were performed with multiplex primers hybridizing outside and inside of the mprF gene and vector sequences. A colony with a single crossover was used as a negative control. The resulting mutant was verified by DNA sequencing, including the adjacent region of the mprF gene. The complementation construct (CGK5mutR) was constructed by the same procedure.
Antimicrobial Susceptibility Testing
The MICs of daptomycin, oxacillin and vancomycin were determined by Etest® (bioMérieux SA, Marcy l’Étoile, France) following the manufacturer’s instructions and using Mueller Hinton II agar (MHA) (Becton Dickinson, Cockeysville, MD, United States). The daptomycin Etest strips were overlaid with 40 μg/ml of calcium (Package insert) and the MHA contained 2.9–5.9 μg/ml of calcium. The MICs of oxacillin were also determined by broth microdilution (BMD) (CLSI, 2013). The BMD method was performed in Mueller Hinton II broth (MHB) (Becton Dickinson) with and without 2% NaCl from an inoculum of 5 × 105 CFU/ml, and the MIC was read after incubation at 35°C for 24 h. S. aureus ATCC 29213 and Enterococcus faecalis ATCC 29212 were used as quality control organisms for Etest. S. aureus ATCC 29213 and ATCC 43300 were used as quality control organisms for BMD.
Population Analysis Profiles
A population analysis profile (PAP) for vancomycin was performed on CGK5 and CGK5 mutant derivatives following protocols previously described (Hiramatsu et al., 1997; Howden et al., 2006). Briefly, overnight cultures of test isolates were serially diluted in TSB and inoculated onto brain–heart infusion (BHI) agar plates containing 0–8 μg/ml vancomycin. After 48 h incubation at 35°C, colonies were counted and plotted. Mu3 (ATCC 700698) and N315 S. aureus strains were tested in parallel as hVISA positive and negative controls, respectively. The area under the curve (AUC) values of the test strains were compared to that of Mu3.
Cell Wall Thickness
Bacterial cells for transmission electron microscopy were prepared following previously described protocols (Hanaki et al., 1998). Photographic images were obtained at a final magnification of 15,000× using a Hitachi H-7650 microscope (Hitachi High-Technologies Corporation, Tokyo, Japan). Fifty measurements of equatorially cut cells were taken for the calculation of cell wall thickness, and results were expressed as mean ± SD following previously described protocols (Cui et al., 2000).
Growth Rate
Overnight fresh cultures of bacteria were adjusted in 0.85% NaCl to 0.5 McFarland turbidity, then diluted 1:200 in MHB to obtain 5 × 105 CFU/ml starting inoculum. The inoculum was dispensed at 120 μl per well in triplicates into a 100-well plate and incubated at 37°C in Bioscreen C MBR (Oy Growth Curves Ab Ltd, Helsinki, Poland) (Richardson et al., 2008). Triplicate medium-only blank wells were included in each plate. The OD600 of each well was read every 30 min for 24 h. The average OD of the blank wells was subtracted from the average of the triplicate test wells at each time point and plotted. Doubling times were calculated using the exponential growth phase following a previously described protocol (Cui et al., 2003). To verify the OD measurements, the CFU counts were checked by the shaker flask method.
Cytochrome c Binding Assay
Cytochrome c binding assay was performed following an approach similar to that of Mukhopadhyay et al. (2007) with slight modification. Briefly, bacteria grown overnight at 35°C were harvested and washed twice with 20 mM MOPS buffer (pH 7.0) and resuspended in the same buffer to a final OD578 of 7. Cytochrome c (Sigma Chemicals, St. Louis, MO, United States) was prepared in the same buffer. The bacterial suspension was incubated with 1 mg/ml cytochrome c for 10 min, and then centrifuged at 3000 g at 4°C for 10 min. The supernatant containing unbound cytochrome c was collected and measured spectrophotometrically at OD530. The cytochrome c was serially diluted (0.1–0.6 mg/ml) to create a standard curve to measure the cytochrome c concentration of the supernatant.
Western Blot Analysis for PBP2a Detection
Detailed descriptions of mouse monoclonal anti-PBP2a antibody (2F6F) production and detection of PBP2a were presented previously (Chen et al., 2014). However, in that study, PBP2a was detected in the total protein preparation. In the present study, for more precise detection of PBP2a in the membrane, membrane fraction was prepared following previously described protocol with slight modifications (Downer et al., 2002). Briefly, strains were grown in TSB overnight at 37°C with shaking at 200 rpm, then diluted 1/100 in 50 ml fresh medium and further incubated until cells reached an OD600 of 1. Ten milliliter of the bacterial cultures was then centrifuged, and the pellet was resuspended in 1 ml lysis buffer. Cells were disrupted by FastProteinTM Blue Matrix using a FastPrep-24 homogenizer (MP Biomedicals) at 6 M/s for 4 cycles of 20 s with a 2-min ice incubation in between each cycle. Protein samples (1 μg of protein per lane) were separated in 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and then transferred to a polyvinylidene difluoride membrane (Immobilon, Millipore Corp., Bedford, MA, United States). PBP2a was probed with mouse monoclonal anti-PBP2a antibody (2F6F), and bands were visualized with HRP-conjugated secondary antibodies (Abcam Inc., Cambridge, MA, United States) followed by incubation in Western lightning chemiluminescence reagent plus (Perkin Elmer Life Sciences, Boston, MA, United States). Sortase A, a transpeptidase, was used as an internal control, identified by rabbit polyclonal anti-Sortase A primary antibody (Abcam Inc.).
Real-Time Quantitative PCR Analysis for mprF, vraSR, and Cell Wall Synthesis-Related Genes
For RNA isolation, strains were grown overnight with shaking at 37°C in MHB and diluted with 5 ml fresh medium to 1/100. They were then grown at 37°C with shaking at 200 rpm, and samples were collected from four time points (2, 3, 4, 5 h) for analysis. Approximately 2 × 109 cells were harvested for RNA isolation. Total RNA isolation was performed as described previously (Chen et al., 2009). Real-time quantitative PCR analysis was performed using Maxima SYBR Green qPCR Master Mix (Thermo Fisher Scientific, United States) on a Roche LightCycler 480 II Real-Time PCR System (Roche). Results were normalized to the expression of gyrB. The expression levels of mprF, vraSR, and several selected cell wall synthesis-related genes (fmtA, murZ, pbp2, sgtB, tagA) were determined using the primers listed in Table 2. The expression of ldh1, a gene involved in nitric oxide resistance in S. aureus (Richardson et al., 2008), was used as a negative control. The target gene transcripts were quantified by using the basic relative quantification method of the LightCycler 480 Software v1.5.1.62 (Roche). Three independent experiments were performed in duplicate and results are shown as mean ± SD. The mRNA expression levels of genes from CGK5 were defined as 1.
Construction of mprF and vraSR Promoter-Reporter Plasmids
For introducing the pluxT2 reporter into CGK5, the selection marker spc was amplified by PCR from the Z172 strain (Chen et al., 2013), digested with SmaI and then cloned into pluxT2 (Chen et al., 2014) to generate pluxT2-SPC. The mprF and vraSR promoter fragments were PCR-amplified from the chromosome of CGK5 using the primers PmprF-F, PmprF-R and PvraSR-F, PvraSR-R. These amplified promoter fragments were fused upstream of the lux reporter genes of pluxT2-SPC using the BamHI and SalI sites, and the resulting plasmids were used for transformation of the S. aureus strain RN4220 and then of CGK5 and CGK5 mutant derivatives. The resulting promoter-reporter fusion constructs were confirmed by restriction enzyme analysis and DNA sequencing.
Luciferase Reporter Assays
Bioluminescence was measured with a SpectraMax L microplate reader (Molecular Devices, Sunnyvale, CA, United States) to determine the transcriptional level of the mprF and vraSR promoter constructs. Three independent transformants harboring promoter-reporter fusion plasmids were grown overnight with shaking at 37°C in TSB containing spectinomycin for reporter plasmid maintenance, diluted 1:100 in the same medium, and aliquots (200 μl) of the cultures were transferred into 96-well plates in duplicate and incubated at 37°C. Both OD595 and bioluminescence (in relative light units, RLU) were monitored every hour for 7 h.
Statistical Analysis
Assay results are reported as mean ± SD where appropriate. For comparison of differences between CGK5 and CGK5 mutant derivatives, the Student’s t-test was performed using GraphPad Prism 6 software (GraphPad, La Jolla, CA, United States). A P value less than 0.05 was considered statistically significant.
Results
To examine the effects of the MprF L431F amino acid substitution in an isogenic background, we replaced the chromosomal wild-type mprF gene of the DAP-S parental strain CGK5 with the mutant mprF gene from the CGK6 DAP-NS strain, creating the new strain CKG5mut. A single base change (from CTT to TTT) causes the L431F mutation in MprF. Furthermore, we generated a reverse mutation complementation construct from CGK5mut, called CGK5mutR, which carries a wild-type mprF with a new EcoRV restriction site that can be used to differentiate CGK5mutR from CGK5. CGK5mut and CGK5mutR were verified by PCR and sequencing of the entire mprF gene. We then carried out a phenotypic analysis to compare CGK5mut and CGK5mutR against their parental strain CGK5.
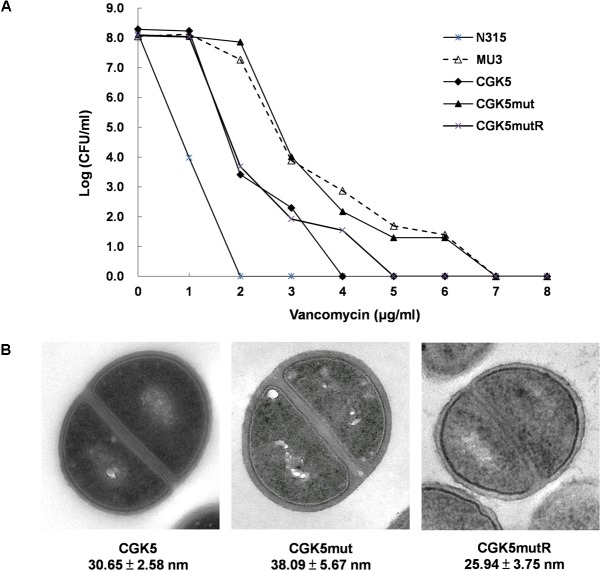
The daptomycin MIC of CGK5mut was 6-fold higher (3 μg/ml) compared to that of the wild-type parental strain CGK5 (0.5 μg/ml), a level of increase similar to what we previously reported for DAP-NS clinical isolates (Lee et al., 2010). To our surprise, the vancomycin MIC of CGK5mut was also increased compared to CGK5 (3 μg/ml vs. 1.5 μg/ml). The reverse complementation construct, CGK5mutR, showed the same level of susceptibility as CGK5. PAP results showed that, of the three strains, only CGK5mut displayed a hVISA phenotype (Figure 1A). The bacterial feature that is often associated with DAP-NS and VISA or hVISA strains is increased cell wall thickness (Camargo et al., 2008; Mehta et al., 2012a). Transmission electron microscopy analysis revealed that CGK5mut has a significantly thicker cell wall (38.09 ± 5.67 nm) than CGK5 (30.65 ± 2.58 nm) and CGK5mutR (25.94 ± 3.75 nm) (p < 0.05) (Figure 1B). It has also been reported that clinical and in vitro-derived S. aureus strains exhibiting increased vancomycin MICs grow slower than their progenitors (Cui et al., 2003; Camargo et al., 2008). This phenomenon has been attributed to increased cell wall synthesis at a biological cost to the resistant strains (Cui et al., 2003). We also found that CGK5mut (doubling time = 240 ± 9.5 min) grows slower than both CGK5 (doubling time = 158 ± 3.5 min) and CGK5mutR (doubling time = 170 ± 4.2 min) in MHB (Figure 2).
FIGURE 1.
Detection of hVISA features. (A) Population analysis profile of vancomycin on CGK5 and its mutant derivatives. Overnight cultures of test isolates were serially diluted in Trypticase Soy broth (TSB) and inoculated onto brain heart infusion (BHI) agar plates containing 0 to 8 μg/ml of vancomycin. After 48 h incubation at 35°C, colonies were counted and the results were plotted on a graph. Mu3 (ATCC 700698) and N315 S. aureus strains were tested in parallel as hVISA positive and negative controls, respectively. (B) Transmission electron microscopy images of CGK5 and its mutant derivatives. Cell wall thickness (in nanometers) was measured at 150,000× magnification. Fifty measurements of equatorially cut cells were taken for calculation of cell wall thickness, and results are expressed as mean ± SD. CGK5, progenitor of CGK5mut; CGK5mut, derivatives of CGK5 with L431F mutation in mprF; CGK5mutR, reverse complementation construct of CGK5mut.
FIGURE 2.
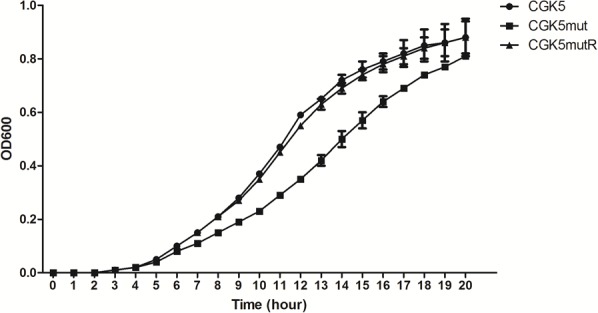
Growth curve of CGK5, CGK5mut, and CGK5mutR in Mueller Hinton II broth (MHB). Overnight fresh cultures of the bacteria were adjusted in 0.85% NaCl to 0.5 McFarland turbidity, then diluted 1:200 in MHB to obtain ∼5 × 105 CFU/ml starting inoculum. The inoculum was dispensed at 120 μl per well in triplicates to a 100-well plate and incubated at 37°C in Bioscreen C MBR. The OD600 of each well was read every 30 min for 24 h. The average OD of the blank wells was subtracted from the average of the triplicate test wells at each time point and plotted.
Altered cell membrane charge has been found in some DAP-NS S. aureus isolates with point mutations in mprF (Peschel et al., 2001; Oku et al., 2004). Cytochrome c is a cationic protein that has been used to estimate the relative bacterial cell surface charge of S. aureus (Mukhopadhyay et al., 2007). We detected a significant increase in unbound cytochrome c for CGK5mut (0.676 ± 0.056 mg/ml) compared to CGK5 (0.393 ± 0.058 mg/ml) and CGK5mutR (0.417 ± 0.02 mg/ml) (p < 0.05) (Figure 3), indicating an increased positive charge density on the cell surface of CGK5mut and suggesting a mechanism by which the L431F mutation of MprF might contribute to daptomycin resistance.
FIGURE 3.
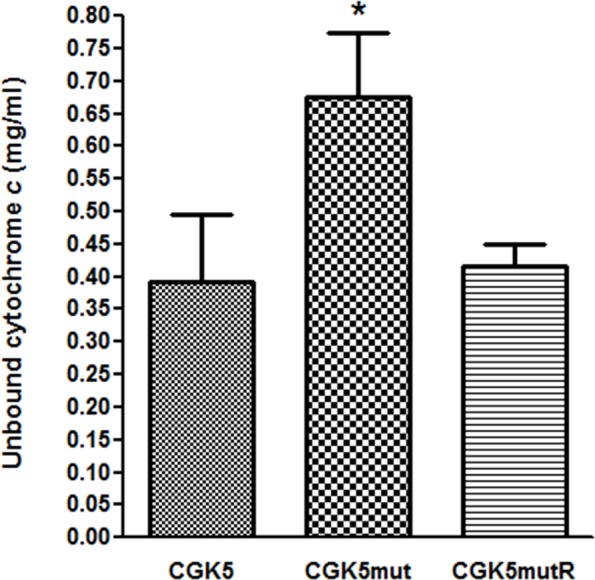
Binding of cationic cytochrome c to CGK5, CGK5mut, and CGK5mutR whole cells. The graph represents the unbound concentrations of cytochrome c after 10 min of incubation with CGK5 and CGK5 mutant derivatives. Results are expressed as mean ± SD from three independent experiments. ∗p < 0.05.
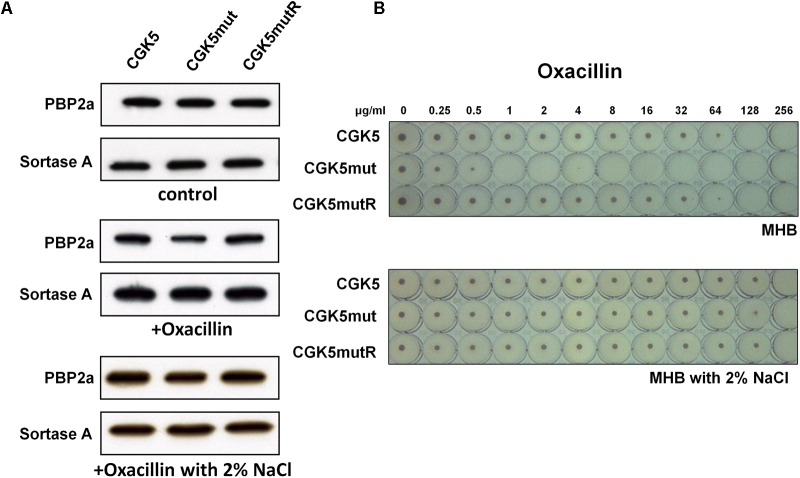
The mechanism of oxacillin resistance in MRSA is thought to be mainly due to the production of penicillin-binding protein 2a (PBP2a, encoded by the mecA gene) with reduced affinity for β-lactams (Hiramatsu et al., 2001). Although first observed between glycopeptide and β-lactams (Sieradzki and Tomasz, 1999), the “seesaw” inverse relationship between daptomycin and β-lactam susceptibility has been reported by several groups (Mishra et al., 2009; Lee et al., 2010; Yang et al., 2010; Mehta et al., 2012a). We determined the level of PBP2a production in CGK5mut and found it to be comparable to that of CGK5 and CGK5mutR (Figure 4A).
FIGURE 4.
PBP2a production and the “seesaw-like” effect. (A) Production of penicillin-binding protein 2a (PBP2a) by CGK5 and CGK5 mutant derivatives was measured by Western blot analysis. Membrane protein was extracted from strains grown in medium alone (control) and in oxacillin (0.25 μg/ml)-containing medium with and without 2% NaCl. PBP2a was probed with mouse monoclonal anti-PBP2a antibody (2F6F), and sortase A was used as an internal control by using rabbit polyclonal anti-Sortase A primary antibody. (B) Oxacillin MICs of CGK5 and CGK5 mutant derivatives were determined by broth microdilution (BMD) in Mueller Hinton II broth (MHB; containing 2% NaCl or not) from an inoculum of 5 × 105 CFU/ml.
However, concurrent to the increased daptomycin MIC, the oxacillin MIC of CGK5mut decreased to 1 μg/ml from 128 μg/ml in CGK5, as determined by the BMD method in medium without 2% NaCl, whereas CGK5mutR showed a similar MIC as CGK5 (Figure 4B). Thus, CGK5mut exhibits the “seesaw-like” effect similar to which we and others have previously observed in DAP-NS in vitro-selected and clinical S. aureus isolates (Lee et al., 2010; Mehta et al., 2012a,b). Interestingly, the oxacillin MICs of CGK5, CGK5mut and CGK5mutR were the same (256 μg/ml) in medium containing 2% NaCl (Figure 4B). Therefore, we further measured the level of PBP2a of these strains grown in oxacillin (0.25 μg/ml) containing medium with and without 2% NaCl. The level of PBP2a of CGK5mut showed a marked reduction after exposure to oxacillin, and 2% NaCl counteracted this effect (Figure 4A).
Mutation of mprF has been shown to impact the expression of mprF itself (Yang et al., 2009b). In addition, VraSR, a two-component regulatory system that plays a major role in cell wall synthesis and is a key player in the cell-wall stress response, has often been implicated in vancomycin and daptomycin resistance (Kuroda et al., 2003; Camargo et al., 2008; Muthaiyan et al., 2008; Mehta et al., 2012a). We therefore compared the expression of mprF and vraSR-related genes in CGK5 and its mutant derivatives by real-time quantitative PCR analysis. We found there was no significant difference in the mprF expression among them (Figure 5). However, significant increases (>4-fold increase, P < 0.01) in the expression of vraSR were seen in CGK5mut compared to CGK5 and CGK5mutR at the 4-h time point. In addition, the expression levels of four examined cell wall synthesis-related genes (fmtA, murZ, pbp2, sgtB) were also significantly increased in CGK5mut (>2∼3-fold increase, P < 0.01) at this time point.
FIGURE 5.
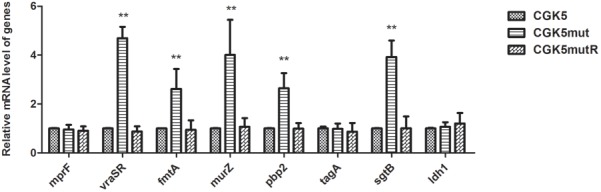
Transcriptional expression of mprF and vraSR and cell wall synthesis-related genes. Expression of mprF and vraSR-related genes in CGK5 and CGK5 mutant derivatives was compared by real-time quantitative PCR analysis. Three independent experiments were performed in duplicate at four time points (2, 3, 4, 5 h) and results at the 4-h time point are shown here as mean ± SD. The expression of ldh1 was used as a negative control. The mRNA expression level of genes from CGK5 was defined as 1. ∗∗p < 0.01 when compared to CGK5.
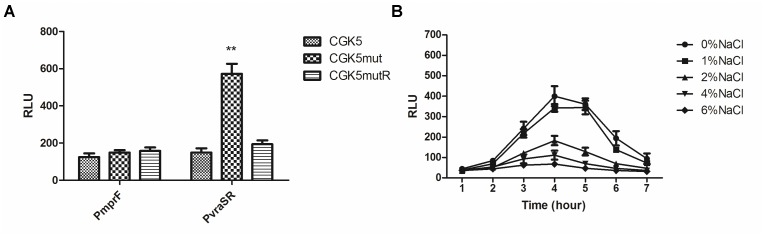
To clarify whether the up-regulation of gene expression in CGK5mut is due to promoter activity, we constructed mprF and vraSR promoter-reporters. The transcriptional activity of the mprF and vraSR promoters was compared in CGK5 and its mutant derivatives by using a bioluminescence assay. mprF promoter activity among the tested strains showed no obvious differences (Figure 6A). However, the vraSR promoter showed significantly higher promoter activity in CGK5mut compared to the other two strains and the maximum discrepancy was at the 4-h time point (Figure 6A), indicating that the up-expression of vraSR in CGK5mut was mediated by its promoter.
FIGURE 6.
Transcriptional activity of promoters of CGK5 and its mutant derivatives. Bioluminescence assay was performed to determine the transcriptional level of the (A) mprF or vraSR promoter in CGK5 and CGK5 mutant derivatives (maximal activity at the 4-h time point is shown) and the effect of (B) salt concentration on vraSR promoter activity in CGK5mut. Data are expressed as relative light units (RLU) from three independent transformants tested in duplicate.
To determine whether the reduced oxacillin MIC of CGK5mut was caused by increased expression of vraSR, and how this might be related to the fact that 2% NaCl could restore the oxacillin MIC of CGK5mut to wild-type levels, we examined whether the high salt concentration could inhibit the expression of vraSR in CGK5mut. A vraSR promoter activity assay showed that NaCl does indeed inhibit vraSR promoter activity (Figure 6B). The effect is dose-dependent and 2% NaCl is the critical concentration for inhibition.
Discussion
Daptomycin is an important last-line agent against serious S. aureus infection, particularly in patients with persistent MRSA bacteremia and endocarditis (Boucher and Sakoulas, 2007; Marco et al., 2008). Daptomycin non-susceptibility in S. aureus, albeit still rare, can emerge during treatment and usually in patients with severe deep-seated infections (Julian et al., 2007; Lee et al., 2010; Boyle-Vavra et al., 2011). The mechanism of daptomycin-non-susceptibility is still not fully understood but has been associated with multiple genetic changes on the bacterial chromosome, with single point mutations in the mprF gene being the most frequently identified alteration (Friedman et al., 2006; Julian et al., 2007; Mehta et al., 2012a; Mishra et al., 2012). Although a few hot spots have been associated with daptomycin resistance, such as L826F, S295L, and S337L, there is considerable variation in the locations of the mutations in the MprF polypeptide structure (Friedman et al., 2006; Julian et al., 2007; Yang et al., 2009a; Lee et al., 2010; Boyle-Vavra et al., 2011; Mehta et al., 2012a; Peleg et al., 2012). A study just published by Yang et al. (2018) investigated the impact of laboratory generated single or dual point mutations within the hot spot loci of mprF and found that extra point mutation in mprF resulted in diminished characteristics associated with DAP-NS, which may explain why no clinically derived DAP-NS strains contained multiple point mutations within the mprF gene.
Our previous study on sequential clinical MRSA isolates from a patient with persistent bacteremia revealed an L431F amino acid substitution in the 3 DAP-NS isolates we characterized (Lee et al., 2010). Since this mutation had not been seen in other DAP-NS isolates reported to date, we carried out the present study to verify its contribution to daptomycin resistance. Because some DAP-NS isolates are also vancomycin non-susceptible, and a few DAP-NS isolates (including our own; Mishra et al., 2009; Lee et al., 2010; Yang et al., 2010; Mehta et al., 2012a) have been reported to exhibit a reduced oxacillin MIC, we also investigated the effects of this L431F MprF change on vancomycin and oxacillin MICs. The approach we employed differed from those of other studies in that we used a single-base-substitution (from CTT to TTT) method to create the L431F mutant (CGK5mut) from CGK5, the wild-type progenitor of our DAP-NS strains. We also generated a reverse mutant from CGK5mut, CGK5mutR, to verify our findings for CGK5mut. This approach ensured that the strains we tested and compared were isogenic, except for the single nucleotide difference at the site corresponding to codon 431 of MprF (and a mutated silent EcoRV site in CGKmutR which has no effect on the sequence of the polypeptide).
The increased daptomycin MIC observed in CGK5mut confirmed that the L431F missense mutation contributes to daptomycin resistance. We also showed that CGK5mut has reduced binding of cytochrome c. Since the L431 amino acid is located in the C-terminal LPG synthase domain of MprF (Ernst et al., 2009), the daptomycin-non-susceptibility in CGK5mut might be an effect of a reduced PG:LPG ratio (Ernst et al., 2009; Yang et al., 2009b). A lower PG:LPG ratio has been shown to reduce the negative charge of the cell membrane (Baltz, 2009; Rubio et al., 2012), thus diminishing the access of calcium-dependent daptomycin to its cellular target.
Another factor that is commonly associated with daptomycin-non-susceptibility is VraSR (Camargo et al., 2008; Mehta et al., 2012a), a two-component system that positively regulates the cell wall biosynthesis pathway and is involved in the cell wall stimulon (Kuroda et al., 2003; Gardete et al., 2006). The VraSR system was originally identified as a key regulator of vancomycin resistance in VISA and hVISA isolates (Kuroda et al., 2000; Cui et al., 2005). We found that the expression levels of vraSR and several vraSR-regulated genes were significantly increased in CGK5mut compared to CGK5 and CGK5mutR. In addition to the increased daptomycin MICs, the vancomycin MIC of CGK5mut was also increased, and population analysis showed that more CGK5mut isolates were able to grow in higher concentrations of vancomycin compared to CGK5 and CGK5mutR. CGK5mut cells also have a thicker cell wall and grow slower than CGK5 and CGK5mutR cells. These cellular features have been associated with both DAP-NS S. aureus and hVISA/VISA (Camargo et al., 2008; Song et al., 2013). The changes in CGK5mut vancomycin MIC and cell wall thickness likely arise from the upregulated vraSR-dependent system. But how the L431F mutant protein brings about the increased expression of vraSR and the cell wall-related genes is unknown. However, CGK5mut had significant increased vraSR promoter activity compared to CGK5 and CGK5mutR using vraSR promoter-reporter assay, indicating that the up-expression of vraSR in CGK5mut was mediated by its promoter. A previous study observed the mutual presence of both mprF point mutations and increased expression levels of vraSR in DAP-NS strains compared to their DAP-susceptible counterparts, and hypothesized that both genes were mechanistically linked to the DAP-NS phenotype (Mehta et al., 2012a). However, our results suggest a causal relationship between mprF point mutation and increased expression of vraSR, which may explain why the daptomycin resistance is often concomitant with vancomycin resistance in clinical isolates. There are a plethora of pathways to heterogeneous and intermediate resistance to vancomycin in S. aureus (Howden et al., 2010). In addition to VraSR, two other two-component systems, GraSR and WalKR, have also been associated with reduced susceptibility of S. aureus to both daptomycin and vancomycin. It has been reported that GraSR also plays an important role in S. aureus resistance to cationic antimicrobial peptides via altered expression of mprF and dltABCD resulting in increased electrostatic repulsion of cationic antimicrobial peptides. GraSR has also been shown to interact with the WalKR system and have significant regulatory overlap (Li et al., 2007; Meehl et al., 2007; Dubrac et al., 2008; Falord et al., 2011). However, we found no significant differences in the expression of mprF and mprF promoter activity among the tested strains. Therefore, it appears unlikely that reduced susceptibility of CGK5mut to both daptomycin and vancomycin are associated with these two two-component systems.
Although expression of PBP2a (encoded by the mecA gene) is thought to play a major role in oxacillin-resistance, several studies have reported that the amount of PBP2a expressed does not correlate with the level of methicillin-resistance (Chambers and Hackbarth, 1987; Murakami and Tomasz, 1989; De Lencastre et al., 1994), and other auxiliary genes have been reported to be essential for the optimal expression of methicillin-resistance (De Lencastre and Tomasz, 1994). When we determined the production level of PBP2a in CGK5mut and found it to be comparable to that of CGK5 and CGK5mutR in the normal condition, we thought its “seesaw-like” effect may be caused by other factors. This indicated that the expression of mecA gene in CGK5mut was not altered by the mprF point mutation. Since the oxacillin MIC of CGK5mut was reduced in medium lacking 2% NaCl, we measured the levels of PBP2a in these strains in the presence of oxacillin with and without 2% NaCl and found that the level of PBP2a production in CGK5mut reduced after exposure to oxacillin and 2% NaCl could abolish this effect. According to the CLSI guideline, 2% NaCl should be added to the medium in determining the oxacillin MIC for staphylococci (CLSI, 2013). Interestingly, our results also showed that the expression of vraSR decreased in the presence of 2% NaCl, with a concurrent restoration of oxacillin resistance in CGK5mut, indicating that the effect of upregulation of vraSR in CGK5mut could be abolished by adding 2% NaCl. Since the “seesaw” effect has been observed in only a few DAP-NS MRSA isolates (Mehta et al., 2012b), the factors associated with this phenomenon warrant further investigation. However, mprF point mutation in CGK5mut only exhibits the “seesaw-like” effect (meaning the seesaw effect was only observed in the absence of NaCl), we speculate that additional mutations at other genes are required to achieve truly “seesaw” effect observed in clinical isolates. In addition, how 2% NaCl counteracts the up-regulation effect of vraSR caused by MprF L431F mutation requires further research.
In fact, Mehta et al. (2012a) previously observed the phenotype in DAP-non-susceptible strains with upregulation of the two-component regulatory system vraSR; however, they did not prove this was caused by the MprF mutation. By contrast, they introduced an overexpression of vraSR in DAP-susceptible strains to show a reduction in oxacillin resistance. In their latest study, they found that introducing an overexpression of vraSR in DAP-susceptible strains triggering MprF mutation and result in impairment of PrsA chaperone functions, both events are required for β-lactam resistance via PBP2a maturation (Renzoni et al., 2017). It has long been recommended that 2% NaCl (or KCl) be added to the culture medium to reliably detect oxacillin resistance in staphylococci (Thornsberry and McDougal, 1983; Huang et al., 1993). When salt is placed in solvent, the solid salt dissolves into its component ions, sodium (Na+), potassium (K+), chloride (Cl-), which are the primary ions of electrolytes in physiology. It is plausible to speculate that these ions maintain the phosphorylation state of VraS in respond to mprF point mutation and/or β-lactam stimulon to precisely regulate its downstream genes for the generation of resistant phenotype.
Conclusion
The present study demonstrated that a single amino acid substitution (L431F) in the MprF protein contributes to both daptomycin and vancomycin resistance as well as increased oxacillin susceptibility in an isogenic MRSA strain. This point mutation in CGK5 also causes up-expression of vraSR. Further studies on the mechanisms contributing to these phenomena may lead to discovery of potential therapeutics against multidrug-resistant staphylococci.
Author Contributions
F-JC contributed to the study design, data analysis, and manuscript preparation. T-LL performed the data analysis and prepared the manuscript. C-HL, Y-CH, I-WH, P-CH, and C-SY performed the work and data analysis.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We wish to thank Ms. Yu-Ching Chen, Hsin-Ying Huang (Research Assistant, Center for Nanomedicine Research), and Yu-Lin Chen (Research Assistant, NIIDV) for their technical assistance. We are also grateful to Prof. Paul Williams (University of Nottingham, United Kingdom) for kindly providing many of important bacterial strains and plasmids used in this study.
Funding. This project was supported by intramural grants from the National Health Research Institutes to the National Institute of Infectious Diseases and Vaccinology (IV-101-SP-10, IV-102-SP-10, and IV-101-SP-11), and grants from the Ministry of Science and Technology in Taiwan (MOST 104-2320-B-400-013-). The transmission electron microscopy analysis was supported by an NHRI intramural grant to the Center for Nanomedicine Research (NM-101-PP-04).
References
- Arnaud M., Chastanet A., Debarbouille M. (2004). New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. 70 6887–6891. 10.1128/AEM.70.11.6887-6891.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baltz R. H. (2009). Daptomycin: mechanisms of action and resistance, and biosynthetic engineering. 13 144–151. 10.1016/j.cbpa.2009.02.031 [DOI] [PubMed] [Google Scholar]
- Bertsche U., Weidenmaier C., Kuehner D., Yang S. J., Baur S., Wanner S., et al. (2011). Correlation of daptomycin resistance in a clinical Staphylococcus aureus strain with increased cell wall teichoic acid production and D-alanylation. 55 3922–3928. 10.1128/AAC.01226-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucher H. W., Sakoulas G. (2007). Perspectives on Daptomycin resistance, with emphasis on resistance in Staphylococcus aureus. 45 601–608. 10.1086/520655 [DOI] [PubMed] [Google Scholar]
- Boyle-Vavra S., Jones M., Gourley B. L., Holmes M., Ruf R., Balsam A. R., et al. (2011). Comparative genome sequencing of an isogenic pair of USA800 clinical methicillin-resistant Staphylococcus aureus isolates obtained before and after daptomycin treatment failure. 55 2018–2025. 10.1128/AAC.01593-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camargo I. L., Neoh H. M., Cui L., Hiramatsu K. (2008). Serial daptomycin selection generates daptomycin-nonsusceptible Staphylococcus aureus strains with a heterogeneous vancomycin-intermediate phenotype. 52 4289–4299. 10.1128/AAC.00417-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers H. F., Hackbarth C. J. (1987). Effect of NaCl and nafcillin on penicillin-binding protein 2a and heterogeneous expression of methicillin resistance in Staphylococcus aureus. 31 1982–1988. 10.1128/AAC.31.12.1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F. J., Lauderdale T. L., Wang L. S., Huang I. W. (2013). Complete genome sequence of Staphylococcus aureus Z172, a vancomycin-intermediate and daptomycin-nonsusceptible methicillin-resistant strain isolated in Taiwan. 1:e01011-13. 10.1128/genomeA.01011-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F. J., Wang C. H., Chen C. Y., Hsu Y. C., Wang K. T. (2014). Role of the mecA gene in oxacillin resistance in a Staphylococcus aureus clinical strain with a pvl-positive ST59 genetic background. 58 1047–1054. 10.1128/AAC.02045-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. C., Tsou L. T., Chen F. J. (2009). Ligand-receptor recognition for activation of quorum sensing in Staphylococcus aureus. 47 572–581. 10.1007/s12275-009-0004-2 [DOI] [PubMed] [Google Scholar]
- CLSI (2013). Performance Standards for Antimicrobial Susceptibility Testing: 23rd Informational Supplement M100-S23. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Cui L., Lian J. Q., Neoh H. M., Reyes E., Hiramatsu K. (2005). DNA microarray-based identification of genes associated with glycopeptide resistance in Staphylococcus aureus. 49 3404–3413. 10.1128/AAC.49.8.3404-3413.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L., Ma X., Sato K., Okuma K., Tenover F. C., Mamizuka E. M., et al. (2003). Cell wall thickening is a common feature of vancomycin resistance in Staphylococcus aureus. 41 5–14. 10.1128/JCM.41.1.5-14.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L., Murakami H., Kuwahara-Arai K., Hanaki H., Hiramatsu K. (2000). Contribution of a thickened cell wall and its glutamine nonamidated component to the vancomycin resistance expressed by Staphylococcus aureus Mu50. 44 2276–2285. 10.1128/AAC.44.9.2276-2285.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lencastre H., De Jonge B. L., Matthews P. R., Tomasz A. (1994). Molecular aspects of methicillin resistance in Staphylococcus aureus. 33 7–24. 10.1093/jac/33.1.7 [DOI] [PubMed] [Google Scholar]
- De Lencastre H., Tomasz A. (1994). Reassessment of the number of auxiliary genes essential for expression of high-level methicillin resistance in Staphylococcus aureus. 38 2590–2598. 10.1128/AAC.38.11.2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downer R., Roche F., Park P. W., Mecham R. P., Foster T. J. (2002). The elastin-binding protein of Staphylococcus aureus (EbpS) is expressed at the cell surface as an integral membrane protein and not as a cell wall-associated protein. 277 243–250. 10.1074/jbc.M107621200 [DOI] [PubMed] [Google Scholar]
- Dubrac S., Bisicchia P., Devine K. M., Msadek T. (2008). A matter of life and death: cell wall homeostasis and the WalKR (YycGF) essential signal transduction pathway. 70 1307–1322. 10.1111/j.1365-2958.2008.06483.x [DOI] [PubMed] [Google Scholar]
- Enoch D. A., Bygott J. M., Daly M. L., Karas J. A. (2007). Daptomycin. 55 205–213. 10.1016/j.jinf.2007.05.180 [DOI] [PubMed] [Google Scholar]
- Ernst C. M., Peschel A. (2011). Broad-spectrum antimicrobial peptide resistance by MprF-mediated aminoacylation and flipping of phospholipids. 80 290–299. 10.1111/j.1365-2958.2011.07576.x [DOI] [PubMed] [Google Scholar]
- Ernst C. M., Staubitz P., Mishra N. N., Yang S. J., Hornig G., Kalbacher H., et al. (2009). The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. 5:e1000660. 10.1371/journal.ppat.1000660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falord M., Mader U., Hiron A., Debarbouille M., Msadek T. (2011). Investigation of the Staphylococcus aureus GraSR regulon reveals novel links to virulence, stress response and cell wall signal transduction pathways. 6:e21323. 10.1371/journal.pone.0021323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman L., Alder J. D., Silverman J. A. (2006). Genetic changes that correlate with reduced susceptibility to daptomycin in Staphylococcus aureus. 50 2137–2145. 10.1128/AAC.00039-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardete S., Wu S. W., Gill S., Tomasz A. (2006). Role of VraSR in antibiotic resistance and antibiotic-induced stress response in Staphylococcus aureus. 50 3424–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanaki H., Kuwahara-Arai K., Boyle-Vavra S., Daum R. S., Labischinski H., Hiramatsu K. (1998). Activated cell-wall synthesis is associated with vancomycin resistance in methicillin-resistant Staphylococcus aureus clinical strains Mu3 and Mu50. 42 199–209. 10.1093/jac/42.2.199 [DOI] [PubMed] [Google Scholar]
- Hiramatsu K., Aritaka N., Hanaki H., Kawasaki S., Hosoda Y., Hori S., et al. (1997). Dissemination in Japanese hospitals of strains of Staphylococcus aureus heterogeneously resistant to vancomycin. 350 1670–1673. 10.1016/S0140-6736(97)07324-8 [DOI] [PubMed] [Google Scholar]
- Hiramatsu K., Cui L., Kuroda M., Ito T. (2001). The emergence and evolution of methicillin-resistant Staphylococcus aureus. 9 486–493. 10.1016/S0966-842X(01)02175-8 [DOI] [PubMed] [Google Scholar]
- Howden B. P., Davies J. K., Johnson P. D., Stinear T. P., Grayson M. L. (2010). Reduced vancomycin susceptibility in Staphylococcus aureus, including vancomycin-intermediate and heterogeneous vancomycin-intermediate strains: resistance mechanisms, laboratory detection, and clinical implications. 23 99–139. 10.1128/CMR.00042-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden B. P., Johnson P. D., Ward P. B., Stinear T. P., Davies J. K. (2006). Isolates with low-level vancomycin resistance associated with persistent methicillin-resistant Staphylococcus aureus bacteremia. 50 3039–3047. 10.1128/AAC.00422-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M. B., Gay T. E., Baker C. N., Banerjee S. N., Tenover F. C. (1993). Two percent sodium chloride is required for susceptibility testing of staphylococci with oxacillin when using agar-based dilution methods. 31 2683–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julian K., Kosowska-Shick K., Whitener C., Roos M., Labischinski H., Rubio A., et al. (2007). Characterization of a daptomycin-nonsusceptible vancomycin-intermediate Staphylococcus aureus strain in a patient with endocarditis. 51 3445–3448. 10.1128/AAC.00559-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M., Kuroda H., Oshima T., Takeuchi F., Mori H., Hiramatsu K. (2003). Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. 49 807–821. 10.1046/j.1365-2958.2003.03599.x [DOI] [PubMed] [Google Scholar]
- Kuroda M., Kuwahara-Arai K., Hiramatsu K. (2000). Identification of the up- and down-regulated genes in vancomycin-resistant Staphylococcus aureus strains Mu3 and Mu50 by cDNA differential hybridization method. 269 485–490. 10.1006/bbrc.2000.2277 [DOI] [PubMed] [Google Scholar]
- Lee C. H., Wang M. C., Huang I. W., Chen F. J., Lauderdale T. L. (2010). Development of daptomycin nonsusceptibility with heterogeneous vancomycin-intermediate resistance and oxacillin susceptibility in methicillin-resistant Staphylococcus aureus during high-dose daptomycin treatment. 54 4038–4040. 10.1128/AAC.00533-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Cha D. J., Lai Y., Villaruz A. E., Sturdevant D. E., Otto M. (2007). The antimicrobial peptide-sensing system aps of Staphylococcus aureus. 66 1136–1147. 10.1111/j.1365-2958.2007.05986.x [DOI] [PubMed] [Google Scholar]
- Marco F., De La Maria C. G., Armero Y., Amat E., Soy D., Moreno A., et al. (2008). Daptomycin is effective in treatment of experimental endocarditis due to methicillin-resistant and glycopeptide-intermediate Staphylococcus aureus. 52 2538–2543. 10.1128/AAC.00510-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McAleese F., Wu S. W., Sieradzki K., Dunman P., Murphy E., Projan S., et al. (2006). Overexpression of genes of the cell wall stimulon in clinical isolates of Staphylococcus aureus exhibiting vancomycin-intermediate- S. aureus-type resistance to vancomycin. 188 1120–1133. 10.1128/JB.188.3.1120-1133.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehl M., Herbert S., Gotz F., Cheung A. (2007). Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. 51 2679–2689. 10.1128/AAC.00209-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S., Cuirolo A. X., Plata K. B., Riosa S., Silverman J. A., Rubio A., et al. (2012a). VraSR two-component regulatory system contributes to mprF-mediated decreased susceptibility to daptomycin in in vivo-selected clinical strains of methicillin-resistant Staphylococcus aureus. 56 92–102. 10.1128/AAC.00432-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta S., Singh C., Plata K. B., Chanda P. K., Paul A., Riosa S., et al. (2012b). beta-lactams increase the antibacterial activity of daptomycin against clinical methicillin-resistant Staphylococcus aureus strains and prevent selection of daptomycin-resistant derivatives. 56 6192–6200. 10.1128/AAC.01525-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra N. N., Rubio A., Nast C. C., Bayer A. S. (2012). Differential adaptations of methicillin-resistant Staphylococcus aureus to serial in vitro passage in daptomycin: evolution of daptomycin resistance and role of membrane carotenoid content and fluidity. 2012:683450. 10.1155/2012/683450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra N. N., Yang S. J., Sawa A., Rubio A., Nast C. C., Yeaman M. R., et al. (2009). Analysis of cell membrane characteristics of in vitro-selected daptomycin-resistant strains of methicillin-resistant Staphylococcus aureus. 53 2312–2318. 10.1128/AAC.01682-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay K., Whitmire W., Xiong Y. Q., Molden J., Jones T., Peschel A., et al. (2007). In vitro susceptibility of Staphylococcus aureus to thrombin-induced platelet microbicidal protein-1 (tPMP-1) is influenced by cell membrane phospholipid composition and asymmetry. 153 1187–1197. 10.1099/mic.0.2006/003111-0 [DOI] [PubMed] [Google Scholar]
- Murakami K., Tomasz A. (1989). Involvement of multiple genetic determinants in high-level methicillin resistance in Staphylococcus aureus. 171 874–879. 10.1128/jb.171.2.874-879.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthaiyan A., Silverman J. A., Jayaswal R. K., Wilkinson B. J. (2008). Transcriptional profiling reveals that daptomycin induces the Staphylococcus aureus cell wall stress stimulon and genes responsive to membrane depolarization. 52 980–990. 10.1128/AAC.01121-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P. (1991). Genetic systems in staphylococci. 204 587–636. 10.1016/0076-6879(91)04029-N [DOI] [PubMed] [Google Scholar]
- Novick R. P., Ross H. F., Projan S. J., Kornblum J., Kreiswirth B., Moghazeh S. (1993). Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. 12 3967–3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku Y., Kurokawa K., Ichihashi N., Sekimizu K. (2004). Characterization of the Staphylococcus aureus mprF gene, involved in lysinylation of phosphatidylglycerol. 150 45–51. 10.1099/mic.0.26706-0 [DOI] [PubMed] [Google Scholar]
- Peleg A. Y., Miyakis S., Ward D. V., Earl A. M., Rubio A., Cameron D. R., et al. (2012). Whole genome characterization of the mechanisms of daptomycin resistance in clinical and laboratory derived isolates of Staphylococcus aureus. 7:e28316. 10.1371/journal.pone.0028316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel A., Jack R. W., Otto M., Collins L. V., Staubitz P., Nicholson G., et al. (2001). Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. 193 1067–1076. 10.1084/jem.193.9.1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renzoni A., Kelley W. L., Rosato R. R., Martinez M. P., Roch M., Fatouraei M., et al. (2017). Molecular bases determining daptomycin resistance-mediated resensitization to beta-lactams (seesaw effect) in methicillin-resistant Staphylococcus aureus. 61:e1634-16. 10.1128/AAC.01634-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson A. R., Libby S. J., Fang F. C. (2008). A nitric oxide-inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity. 319 1672–1676. 10.1126/science.1155207 [DOI] [PubMed] [Google Scholar]
- Rubio A., Moore J., Varoglu M., Conrad M., Chu M., Shaw W., et al. (2012). LC-MS/MS characterization of phospholipid content in daptomycin-susceptible and -resistant isolates of Staphylococcus aureus with mutations in mprF. 29 1–8. 10.3109/09687688.2011.640948 [DOI] [PubMed] [Google Scholar]
- Schenk S., Laddaga R. A. (1992). Improved method for electroporation of Staphylococcus aureus. 73 133–138. 10.1111/j.1574-6968.1992.tb05302.x [DOI] [PubMed] [Google Scholar]
- Sieradzki K., Tomasz A. (1999). Gradual alterations in cell wall structure and metabolism in vancomycin-resistant mutants of Staphylococcus aureus. 181 7566–7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Rubio A., Jayaswal R. K., Silverman J. A., Wilkinson B. J. (2013). Additional routes to Staphylococcus aureus daptomycin resistance as revealed by comparative genome sequencing, transcriptional profiling, and phenotypic studies. 8:e58469. 10.1371/journal.pone.0058469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornsberry C., McDougal L. K. (1983). Successful use of broth microdilution in susceptibility tests for methicillin-resistant (heteroresistant) staphylococci. 18 1084–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. J., Kreiswirth B. N., Sakoulas G., Yeaman M. R., Xiong Y. Q., Sawa A., et al. (2009a). Enhanced expression of dltABCD is associated with the development of daptomycin nonsusceptibility in a clinical endocarditis isolate of Staphylococcus aureus. 200 1916–1920. 10.1086/648473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. J., Mishra N. N., Kang K. M., Lee G. Y., Park J. H., Bayer A. S. (2018). Impact of multiple single-nucleotide polymorphisms within mprF on daptomycin resistance in Staphylococcus aureus. 10.1089/mdr.2017.0156. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. J., Xiong Y. Q., Boyle-Vavra S., Daum R., Jones T., Bayer A. S. (2010). Daptomycin-oxacillin combinations in treatment of experimental endocarditis caused by daptomycin-nonsusceptible strains of methicillin-resistant Staphylococcus aureus with evolving oxacillin susceptibility (the “seesaw effect”). 54 3161–3169. 10.1128/AAC.00487-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S. J., Xiong Y. Q., Dunman P. M., Schrenzel J., Francois P., Peschel A., et al. (2009b). Regulation of mprF in daptomycin-nonsusceptible Staphylococcus aureus strains. 53 2636–2637. 10.1128/AAC.01415-08 [DOI] [PMC free article] [PubMed] [Google Scholar]