ABSTRACT
The inclusion of high-quality hay (HQH), in place of concentrates, shifts dietary carbohydrate intake, and the extent to which these shifts effect epimural microbiota and epithelial gene expression of the rumen has not yet been evaluated. Eight ruminally cannulated nonlactating Holstein cows were used in a replicated 4 by 4 Latin square design with four dietary treatments containing HQH, with either 0% concentrate/100% HQH (100HQH), 25% concentrate/75% HQH (75HQH), or 40% concentrate/60% HQH (60HQH). The fourth group (control [CON]) was fed 60% normal fiber-rich hay and 40% concentrate. The data showed that measures of diversity for the rumen epimural population, specifically the Shannon (P = 0.004) and Simpson (P = 0.003) indices, decreased with increasing levels of HQH in the diet. The feeding of HQH shifted the epimural population from predominantly Firmicutes to Proteobacteria. Phylogenetic analysis revealed that HQH feeding markedly shifted the abundance of Campylobacter spp. from 7.8 up to 33.5% (P < 0.001), with greater ingestion of protein (r = 0.63) and sugars (r = 0.65) in HQH diet being responsible for this shift. The expression of genes targeting intracellular pH regulation, barrier function, and nutrient uptake of rumen epithelium remained stable regardless of the carbohydrate source. In conclusion, the data suggest strong alterations of the ruminal epimural microbiota in response to changes in the nutritive patterns of the diet. Further research is warranted to evaluate the long-term effects of these significant microbial changes on rumen health and food safety aspects in cattle at a transcriptional level.
IMPORTANCE Feeding of forages versus starchy concentrates is a highly debated topic. Hay is believed to be healthier and more ecological sustainable for cattle than are concentrates, although the effects of feeding hay with enhanced sugar and protein content on epimural microbiota and host gene expression have not yet been evaluated. This research provides a report of the role of feeding hay with increased sugar and protein content in place of starchy concentrates in altering epimural microbiota and in generating a host response. Our research shows that the addition of high-quality hay to dairy rations shifted nutrient intake, resulting in strong alterations in the epimural microbiota in cattle. This work provides a background for further long-term research regarding the effects of feeding practices on the host-microbiome interaction and its role in rumen health and food safety in cattle.
KEYWORDS: epimural microbiota, gene expression, hay, protein, epimural
INTRODUCTION
Rumen microbiota have profound influences on host nutrition, immune stimulation, and protection against pathogens. Perturbations in the stability of the gut microbial population dispose the host to inflammation (1), digestive disorders (2, 3), pathogenic invasion (4), disease (5), and food safety issues (6). Understanding gut microbial community structure and population dynamics in both symbiosis and dysbiosis is fundamental in developing strategies to improve gut health and promote the establishment of a beneficial rumen microbial ecosystem (7, 8).
The rumen is a complex ecological system, and classical microbiology has determined the rumen epithelial community (epimural microbiome) to be unique from those associated with rumen digesta and fluid. This research also showed that rumen epimural bacteria play a major role in the rumen for oxygen removal, tissue degradation, and urea recycling (9). The advancement of molecular biology and sequencing technologies has revealed the diversity of the epimural community (2), the variation of this community between individuals (10), the effect of dietary change on the epimural population (2, 11), and the impact this population can have on epithelial gene expression (12). In other gut systems, it has been shown that specific nutrients, whether derived from the host diet (13) or from endogenous host sources (14), are also critically important in shaping the structure of the host-associated microbial communities.
The rumen epithelium is the primary source of nutrient uptake and the first line of defense against enteric pathogens, such as Gram-negative Escherichia coli serotypes and some Campylobacter spp. (15), both of which are enteropathogens related to public health and food safety issues. The rumen microbiome is an herbivore-adapted microbiome, with limited tolerance to energy substrates, such as starches, from concentrates commonly found in modern feeding practice. Feeding large amounts of concentrates leads to rumen dysbiosis, which is associated with pathogen growth and a decrease in diversity, local and systemic inflammation, and epithelial barrier dysfunction (7, 10). Specifically, when diets containing large amounts of dietary starch are fed, increases in the expression of Toll-like receptor 4 (TLR4), a surface receptor which binds toxic lipopolysaccharide (LPS) molecules released from the cell surface of Gram-negative bacteria (GNB) (16, 17) and initiates a localized host response (10). Furthermore, the low ruminal pH associated with high-concentrate feeding has been shown to have an effect on the expression of barrier function-related gene expression, indicating a host response to the rumen environment (10). However, increased dietary starch resulting in increased rumen volatile fatty acid (VFA) production has also been shown to increase VFA transport-related gene expression, resulting in a positive increase in energy uptake for the animal (18). While the fermentation of starch in the rumen in more rapid than the fermentation of forages, previous research from our lab has shown the beneficial effects of feeding high-quality hay (HQH) with an elevated content of sugars and metabolizable energy, which shifts the energy intake away from starch-rich concentrates and toward a healthier forage-based product (19). We found that the addition of such forages provides the rumen microbiota with digestible substrate for VFA production while also providing sufficient structural fiber to promote rumen pH buffering through rumination (19). Furthermore, it was seen that changes in the substrates provided with HQH resulted in changes in the digesta-associated microbiota of the rumen (20). However, the impacts of increased dietary sugar and protein on both the rumen epimural microbiome and epithelial gene expression in diets replacing HQH for high-starch concentrates have not yet been studied.
Therefore, in this study, we evaluated the effects of feeding sugar-rich complex carbohydrates from hay with titrated amounts of starch-rich concentrate on the epimural bacterial community and host gene expression from dry Holstein cows. We hypothesized that by proportionally decreasing the level of concentrate-based starch and increasing HQH in the diet, there would be a linear decrease in Gram-negative epimural genera and TLR4 expression. Furthermore, we hypothesized that the replacement of starch concentrates with HQH would decrease barrier function and pH transporter-related gene expression, but it would not alter the expression of nutrient transport-related genes.
RESULTS
Assessment of rumen epimural population diversity.
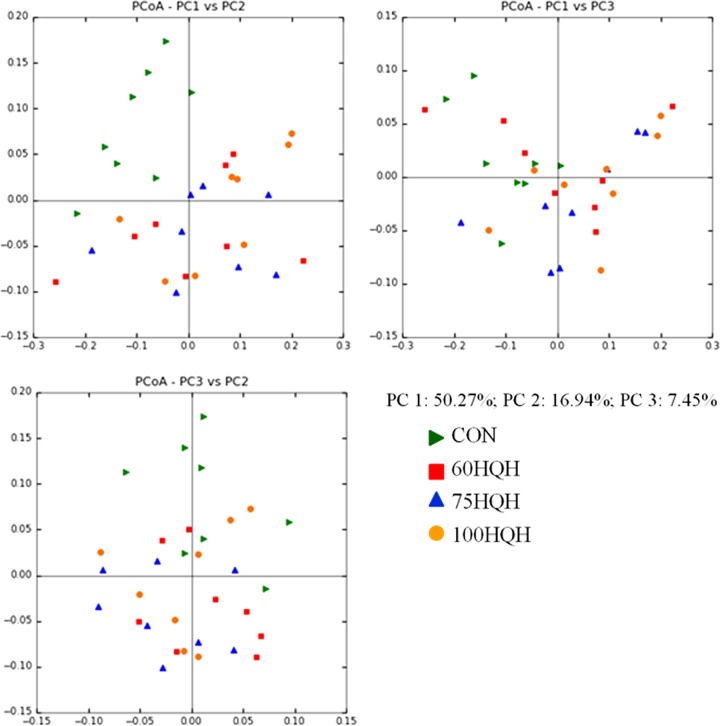
Sequences were quality filtered and then normalized to a depth of 1,217 sequences per sample to account for variation in the number of sequences obtained per sample. Several measures of rumen epimural population richness, abundance, and diversity were measured, and a significant decrease was seen for both the Shannon (P = 0.004) and Simpson (P = 0.003) diversity indices as the percentage of HQH was increased in the diet composition (Table 1). Approximately 67% of the population diversity could be explained with the first and second principal components (PC; Fig. 1). A relationship can be seen for the first and second PC, with the CON group clustering separately in the upper left quadrant of the principal-coordinate analysis (PCoA) weighted Unifrac plot.
TABLE 1.
Alpha-diversity indices for species richness, abundance, and population diversity in the rumen epimural population in diets with increasing levels of high-quality haya
| Index | Results by diet |
SEM | P valueb | |||
|---|---|---|---|---|---|---|
| CON | 60HQH | 75HQH | 100HQH | |||
| Observed OTUs | 729 | 676 | 693 | 640 | 69.3 | 0.75 |
| Chao1 | 993 | 893 | 986 | 897 | 67.6 | 0.48 |
| Simpson index | 0.97 A | 0.92 B | 0.91 B | 0.89 B | 0.016 | 0.003 |
| Shannon index | 6.95 A | 6.18 B | 6.07 B | 5.70 B | 0.268 | 0.004 |
Treatments include control (CON), 60% high-quality hay (60HQH), 75% high-quality hay (75HQH), and 100% high-quality hay (100HQH). Values are the least square means, and means with different letters in a row are significantly different.
P value is an adjusted Tukey HSD value.
FIG 1.
Principal-coordinate analysis (PCoA) of all OTUs grouped by treatment. The first three principal components account for 74.7% of the total sample variance (PC1, 50.3%; PC2, 16.9%; PC3, 7.5%). Green triangles, CON; red squares, 60HQH; blue triangles, 75HQH; orange circles, 100HQH. The control diet tended to cluster apart from the diets containing various levels of HQH, accounting for the variation seen in PC1 and PC2.
Phylogenetic analysis of sequencing data.
Statistical analysis of operational taxonomic units (OTUs) representing more than 0.1% of the sequenced epimural population was performed, and OTUs were ranked based on their relative abundances (see Table S1 in the supplemental material). Of the 126 OTU analyzed, 22 OTU showed an effect of dietary treatment on relative population abundance. This included two different Campylobacter-like OTUs (OTU 1 and 4), which were significantly increased (P < 0.001) as HQH was increased in the diet, and Ruminobacter spp. and Ruminococcus-like spp. (OTUs 31 and 55), which were significantly decreased (P = 0.03 and 0.001, respectively) as HQH was increased.
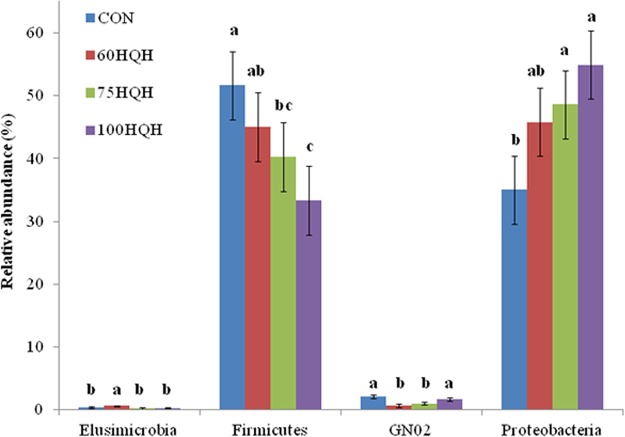
Relative abundances of epimural bacteria at the phylum level show significant effects of dietary treatment on Elusimicrobia, Firmicutes, GN02, and Proteobacteria (Fig. 2). Proteobacteria increased in relative abundance as the percentage of HQH increased in the diet, accounting for more than 54% of the total population in the 100HQH treatment (P = 0.02). This corresponded with the decrease in Firmicutes as the level of HQH increased in the diet (P = 0.04), going from 51.6% relative abundance in the CON group to 33.3% abundance in the 100HQH group.
FIG 2.
Comparison of the distribution of epimural bacteria with significant effects at the phylum level for dairy cows (n = 8) fed a control diet (CON) or 60%, 75%, or 100% high-quality hay (60HQH, 75HQH, or 100HQH, respectively) diets based on 16S rRNA sequences compared to the SILVA version 123 database. Error bars indicate the standard error of the mean. Different lowercase letters above the bars indicate significant differences (P < 0.05). Increases in the Proteobacteria population in the HQH diets correspond largely with decreases in the Firmicutes population.
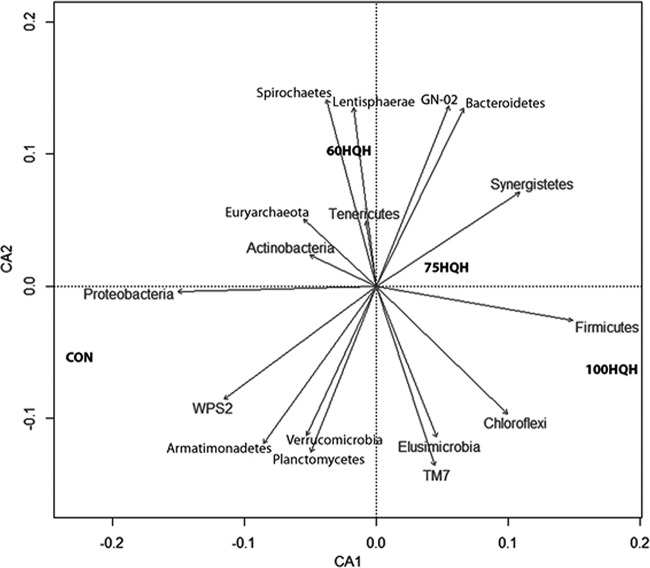
Correspondence analysis of the percent relative abundance of each phylum and the dietary treatment showed a separation of phyla by diet (Fig. 3). Diets containing 60% hay and 40% concentrate (CON and 60HQH) are located on the negative x axis, and diets containing less concentrate (75HQH and 100HQH) are located on the positive x axis. A negative y axis arrow for Firmicutes in the direction of 100HQH and Proteobacteria in the direction of CON corresponds to their decrease in those respective diets. However, only Proteobacteria showed a significant and strong correlation to HQH when analyzed using nonmetric multidimensional scaling (NMDS) vector analysis (r2 = 0.9977, P = 0.04).
FIG 3.
Correspondence analysis (CA) performed in R for epimural bacteria at the phylum level based on the dietary treatments of control diet (CON) or 60%, 75%, or 100% high-quality hay (HQH). NMDS vector analysis showed only Proteobacteria as having a strong significant NMDS vector analysis (r2 = 0.9977, P = 0.04). Results indicate clear separations in the rumen microbiota at the phylum level for the different diets, with each diet falling into a separate quadrant.
Separation and analysis of the sequencing data were also performed at the genus level, with a total of 10 genera being affected by increasing levels of HQH (Table 2). Of these, 5 Gram-negative genera were found, including Anaerovibrio and Selenomonas, which were significantly higher in relative abundances with the 60HQH diet (P = 0.04 and 0.05, respectively). Further Gram-negative genus comparisons showed that Campylobacter spp. increased in relative abundance as the percentage of HQH in the diet increased, and Ruminobacter spp. and Oscillospira spp. decreased significantly as HQH was increased (P = 0.03 and 0.01, respectively). Gram-positive genera showed similarly varied responses to HQH inclusion, including decreases in Carnobacterium spp., as well as increases in Methanosphaera spp. in the CON diet compared to the 100HQH.
TABLE 2.
Percent relative abundance of genera with a significant effect of diets with increasing levels of high-quality haya
| Genusb | Results by diet |
SEM | P valuec | |||
|---|---|---|---|---|---|---|
| CON | 60HQH | 75HQH | 100HQH | |||
| Anaerovibrio | 0.03 B | 0.23 A | 0.16 A | 0.06 B | 0.046 | 0.04 |
| Campylobacter | 7.83 B | 27.06 A | 30.88 A | 33.46 A | 3.717 | <0.0001 |
| Carnobacterium | 0.03 A | 0.00 B | 0.00 B | 0.00 B | 0.008 | 0.05 |
| Methanosphaera | 0.01 B | 0.02 B | 0.02 B | 0.04 A | 0.007 | 0.05 |
| Oscillospira | 0.15 A | 0.09 B | 0.06 B | 0.08 B | 0.018 | 0.01 |
| Ruminobacter | 1.44 A | 0.38 B | 0.29 B | 0.04 B | 0.345 | 0.03 |
| Selenomonas | 0.17 | 1.14 | 0.85 | 0.42 | 0.241 | 0.05 |
| Shuttleworthia | 0.00 B | 0.02 A | 0.00 B | 0.00 B | 0.004 | 0.02 |
| Syntrophomonas | 0.03 | 0.07 | 0.03 | 0.02 | 0.013 | 0.06 |
| Unknown | 55.53 A | 43.26 B | 40.27 B | 37.23 B | 3.339 | 0.002 |
Treatments include control (CON), 60% high-quality hay (60HQH), 75% high-quality hay (75HQH), and 100% high-quality hay (100HQH). Values are the least square means, and means with different letters in a row are significantly different.
All genera in bold type stained Gram negative.
P value is an adjusted Tukey HSD value.
Correlations among genera of the epimural population, rumen pH, dry matter intake, diet composition, and VFA profiles.
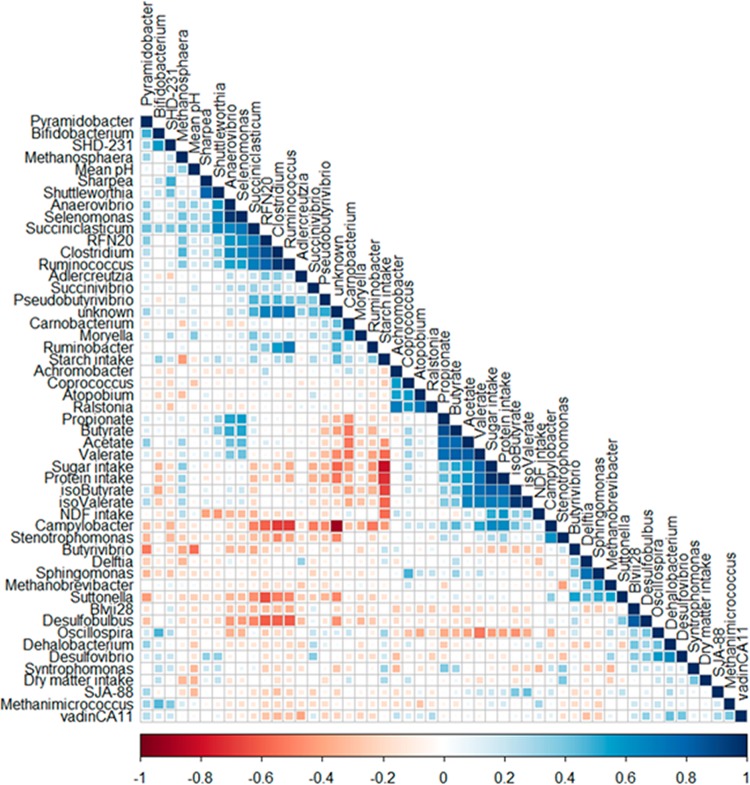
To identify which genera were impacted by the rumen environment, correlation analysis of all genera with dietary parameters, including dry matter intake (DMI), mean ruminal pH (mean pH), starch, sugar, protein, and NDF composition in the diet, as well as VFA composition of the rumen fluid at sampling, consisting of acetate, butyrate, propionate, valerate, isobutyrate, and isovalerate, was performed (17, 19). Unless otherwise indicated, significant correlations are discussed for P value of ≤0.05, whereas highly significant results are indicated for a P value of ≤0.001. The results are summarized in a heatmap, shown in Fig. 4. One distinct cluster of positively correlated epimural bacteria was found, including RFN20 species, Anaerovibrio spp., Selenomonas spp., Succiniclasticum spp., Clostridium spp., Ruminococcus spp., and Shuttleworthia species. Additional positive correlations were found between Ruminobacter spp. and Ruminococcus spp., as well as between Sharpea spp. and SHD-231 species. Succiniclasticum had positive correlations to both genera Sharpea and Methanosphaera. Methanosphaera spp. also showed a negative correlation to starch intake (r = −0.43). Total concentrations of VFA, propionate, butyrate, acetate, and valerate were positively correlated with Anaerovibrio spp. and highly significantly correlated with Selenomonas species. In contrast, the relative abundance of Carnobacterium spp. was negatively correlated with all individual VFAs (0.001 ≤ P ≤ 0.01). Strong negative correlations were found between the Campylobacter and both Ruminococcus and Clostridium genera, as well as between RFN20 and both Sutterella and Desulfobulbus species. Additionally, a strong negative correlation was seen between the genus Oscillospira and rumen concentrations of valerate at time of sampling (P = 0.002). Campylobacter spp. were positively correlated with protein (r = 0.63) and sugar (r = 0.65) intake and negatively correlated with starch intake (r = −0.42).
FIG 4.
Correlation heatmap using hierarchical cluster analysis of 16S rRNA sequences labeled by 97% identity to the genus level with rumen and dietary parameters, including mean pH, dry matter intake, nutrient intake (starch, sugar, protein, neutral detergent fiber [NDF] and volatile fatty acid [VFA]) composition of the rumen fluid at time of sampling, including acetate, propionate, butyrate, valerate, isobutyrate, and isovalerate. Data for dietary composition and VFA profiles of the rumen fluid have been previously published (Kleefisch et al. [19] and Klevenhusen et al. [20]). Scale indicates Pearson correlation coefficient from −1 (red) to 1 (blue). Scale is indicated by color and size of square within the heatmap. Most notably, there is a strong negative correlation between starch and sugar intake based on dietary composition, and this matches a strong negative correlation, e.g., see between Campylobacter populations and Ruminococcus populations.
Rumen gene expression related to diet.
The relative gene expression of 11 genes under four categories was analyzed, including pattern recognition receptor Toll-like receptor 4 (TLR4), barrier function (claudin [CLDN] 1, 4, and 7), pH regulation (anion-exchanger 2 [AE2], sodium-hydrogen exchanger [NHE] 1, 2, and 3), and nutrient transport (mono-carboxylate transporter [MCT] 1 and 4 and sodium-dependent glucose transporter 1 [SGLT1]) (Table 3). Significant changes (P ≤ 0.05) in gene expression based on dietary treatment were not seen. However, NHE3 showed a trend toward increasing (P = 0.07) in 60HQH compared to all other treatments.
TABLE 3.
Relative expression of barrier function, pH regulation, and nutrient transport genes based on increasing levels of high-quality hay in the dieta
| Geneb | Expression (2−ΔΔCq) by diet |
SEM | P valuec | |||
|---|---|---|---|---|---|---|
| CON | 60HQH | 75HQH | 100HQH | |||
| Pattern recognition receptor | ||||||
| TLR4 | 3.18 | 3.02 | 3.00 | 2.94 | 1.357 | 0.99 |
| Barrier function | ||||||
| CLDN1 | 0.78 | 1.15 | 1.17 | 1.38 | 0.200 | 0.14 |
| CLDN4 | 0.89 | 1.19 | 1.03 | 1.03 | 0.093 | 0.13 |
| CLDN7 | 1.29 | 1.23 | 1.15 | 1.37 | 0.349 | 0.97 |
| pH regulation | ||||||
| AE2 | 1.00 | 1.02 | 1.16 | 0.99 | 0.076 | 0.49 |
| NHE1 | 1.10 | 1.06 | 1.00 | 0.93 | 0.051 | 0.22 |
| NHE2 | 0.89 | 1.31 | 1.01 | 1.02 | 0.119 | 0.18 |
| NHE3 | 1.05 | 1.70 | 1.20 | 0.89 | 0.189 | 0.07 |
| Nutrient transporters | ||||||
| MCT1 | 1.08 | 1.32 | 1.02 | 0.89 | 0.201 | 0.47 |
| MCT4 | 0.97 | 1.20 | 1.05 | 0.91 | 0.082 | 0.13 |
| SGLT1 | 0.93 | 0.66 | 0.36 | 0.74 | 0.175 | 0.11 |
Treatments include control (CON), 60% high-quality hay (60HQH), 75% high-quality hay (75HQH), and 100% high-quality hay (100HQH). Values are the least square means.
Gene family symbols: CLDN, claudin; NHE, sodium-hydrogen exchanger; AE, anion-exchanger; MCT, monocarboxylate transporter; SGLT, sodium-dependent glucose transporter.
P value is an adjusted Tukey HSD value.
DISCUSSION
Dietary composition is a predominant factor influencing both the community structure and the metabolic function of the rumen microbiota, including the epimural bacteria (2, 3). However, the influence of changes in nutrient composition with relation to carbohydrate and protein sources on both the epimural community structure and rumen epithelial gene expression remains largely unknown. The epimural community is known to contain LPS-producing GNB genera that are associated with changes in both the TLR and the tight junction structure of the rumen epithelial wall (10, 17). Under conditions of increased starch intake, increases in both the concentration of LPS (21) and the percentage of GNB have been found (22). Therefore, we hypothesized that the incorporation of HQH into a high-energy ration, replacing starch substrate, would reduce the population of Gram-negative LPS-producing epithelial genera and alter rumen gene expression patterns to indicate improved barrier function and nutrient transport.
Incorporation of HQH in the ration was associated with a decrease in diversity which contradicts previous research showing increased diversity within the epimural population in cattle fed a 100% forage diet compared to a concentrate-rich diet (2). However, a decreased diversity was also observed with feeding HQH on the digesta-associated bacteria (20) and might therefore rather depend on dietary sugar content than on starchy concentrate proportion. At the community level, higher diversity and richness tend to be associated with host health and productivity in many systems (23). However, the role of reduced epimural diversity in the causality of rumen inflammation and epithelial diseases, such as rumenitis, remains unclear. The decrease in diversity seen in this study was based largely on the shift from a predominantly Firmicutes-based population to one dominated by Proteobacteria, more specifically a significant increase in the proportion of two Campylobacter-like OTUs (OTUs 1 and 4). A comparison of these 2 OTUs to the NCBI database using the basic local alignment search tool identified OTU 1 as Campylobacter fetus subsp. testudinum (96% identity), and OTU 4 (identified to 95%) as Campylobacter gracilis. While it is known that campylobacters are usually commensal bacteria within the gastrointestinal tract, detection rates above 20% in cattle are uncommon (2, 3, 11). In this study, Campylobacter-like OTUs 1 and 4 accounted for the decrease in diversity seen as HQH levels increased in the diet and showed opportunistic growth under conditions of increased protein and sugar intake (17). This increase in relative abundance combined with the Gram-negative status of Campylobacter spp. is in opposition to our hypothesis that the inclusion of increasing levels of forage to the diet would decrease dysbiosis and the associated GNB genera. However, previous research (2) has noted decreases in Campylobacter populations in animals undergoing ruminal acidosis with the feeding of increased dietary concentrates. This would imply that Campylobacter spp. are sensitive to low ruminal pH and therefore are part of the GNB population associated with increased ruminal LPS due to bacterial cell lysis under low pH. However, the mean daily ruminal pH in this study was 6.5 and seemingly favored the growth of Campylobacter-like OTUs in the HQH diets (19). The lack of significant increase in the relative abundance of Campylobacter spp. in other epimural studies related to increases in energy supplementation would further indicate that their dietary niche is based on protein degradation, likely predominantly from epithelial sloughing, making them a relatively stable population within a moderate rumen pH range (11). The additional provision of protein and nonprotein nitrogen through the HQH in this experiment could have provided an alternative protein source resulting in the increase in population growth. Interestingly, it has also been speculated that the rapid growth of GNB in the rumen also results in increased LPS shedding resulting in a host response (4). However, in this study, the expression of barrier function genes and TLR4 was stable regardless of diet and microbiota. This may indicate that, despite the proliferation of this genus with increased protein and sugars in the HQH diet, it was not recognized as a pathogen by the rumen epithelium in this study. However, sampling in this experiment was performed on day 28, after extended exposure to this increasing population, and it is possible that any LPS released by this community resulted in the establishment of a tolerance state (24). This is supported by other studies which have shown changes to the TLR expression of animals under high-grain feeding programs, with the exception of TLR4 (12, 17). However, due to the limitations of a single sampling time point, as well as the targeting of only transmembrane gene families, an assessment of the overall effect of increased Campylobacter spp. on the rumen health cannot be made. Further studies are required to look at epithelial cell changes at a transcriptional level over both short- and long-term exposures to LPS. Additionally, the extensive presence of Campylobacter spp. in the epimural community in this study brings into question the impact of the rumen epimural bacteria on food safety.
Comparatively, the other GNB genera identified, Anaerovibrio and Selenomonas, were significantly higher in relative abundance in the 60HQH diet and correlated positively with total concentrations of the VFA propionate, butyrate, acetate, and valerate. Correlation analysis of the genera and rumen environmental parameters showed a cluster of positively correlated epimural bacteria, including RFN20 species, Anaerovibrio spp., Selenomonas spp., Succiniclasticum spp., Clostridium spp., Ruminococcus spp., and Shuttleworthia species. The phylogenetic diversity of these clustered genera from within the Firmicutes phylum may indicate cross-feeding or a niche establishment within a biofilm on the rumen papillae.
In contrast to the Campylobacter-like OTUs, the majority of Gram-positive Ruminococcus OTUs decreased with the addition of HQH, which is contradictory to the general status of ruminococci as fiber digesters (25). However, of the several Ruminococcaceae-like OTUs identified as being affected by dietary composition, only one could be identified to 97% as the genus Ruminococcus. The lack of database identification is strong evidence that there are members of this community which are fastidious and remain uncultured and unidentified, possibly with divergent metabolic functions. Interestingly, Oscillospira spp., additional Gram-positive bacteria, were also seen at significantly higher levels with the CON diet than with the HQH diets. While this genus is reported extensively within gastrointestinal ecosystems, including the rumen (3, 26), its presence in the rumen is generally associated with fresh pasture and negatively associated with valerate.
Changes in rumen epithelial expression have been previously documented under rumen environmental stress conditions, such as subacute ruminal acidosis (SARA) (10). However, in the present study, gene expression remained unchanged with changes to nutrient composition. For those genes related to nutrient uptake (MCT1, MCT4, and SGLT1), expression in the rumen is strongly related to both substrate availability and the energy requirements of the host. Since substrate availabilities were similar, in spite of the sources of carbohydrate fed, and the cows were nonlactating, having a lower demand for energy than lactating cows, no changes in nutrient uptake gene expression were expected. For the genes related to pH regulation, alterations in gene expression patterns are to be associated with changes in ruminal pH. The rumen pH remained stable (19) and therefore, changes to the expression of proton transport gene targets were not expected. Gene targets associated with barrier function have also previously been reported to be altered by SARA (10). However, throughout this experiment, pH levels remained in a physiological range (19), and therefore, no significant alteration in barrier function was to be expected. However, as previously mentioned, due to the limitations of analyzing cell surface receptors, it is difficult to interpret if the stability of those genes targeted in this experiment corresponds to stability at the epithelial transcriptional level.
In conclusion, the increase in HQH in the diet shifted the population of rumen epimural bacteria from being predominantly Firmicutes to Proteobacteria, resulting in an overall decrease in epimural diversity. These changes may be explained by the increase in protein and nonprotein nitrogen content of HQH and the proliferation of Campylobacter-like OTUs. The stability of the relative gene expression of nutrient transporter and pH targets under differing nutrient profiles in the rumen could be the result of a number of variables which could not be identified due to the limitation of sampling at a singular time point. Future consideration should be given to the time required for epithelial adaptation to both environmental and microbial changes. This research provides knowledge of the responses of epimural bacterial community to a shift in carbohydrate source and an increase in protein. As well, this study provides evidence that GNB proliferation (e.g., Campylobacter-like OTUs) can occur without initiating a host response, being a potential threat for food safety. Further research is warranted to evaluate both short- and long-term effects of the observed changes of epimural microbiota on the host and food safety aspects in cattle.
MATERIALS AND METHODS
Feeding experiment.
The trial was conducted at the Dairy Research Facilities in Kremesberg of the University of Veterinary Medicine Vienna, Austria. The details of the experimental setup, including diets, feeding, and husbandry conditions, are provided in companion papers (19, 20). In brief, eight ruminally cannulated (100-mm inner diameter [i.d.]; Bar Diamond, Parma, ID) nonlactating and nonpregnant Holstein cows (initial mean ± standard deviation [SD] body weight [BW], 851 ± 75 kg) were used for the trial to enable sampling and biopsying of rumen papillae. The cows were housed on a concrete floor covered with rubber mats and deep-litter lying areas covered and arranged in a double 4 by 4 Latin square design, balanced for carryover effects. The experiment was conducted in four consecutive runs, each run lasting 25 days. Three of the four dietary treatment groups contained the sugar- and protein-rich hay (called high-quality hay [HQH]), with either 0% concentrate and 100% HQH (100HQH), 25HQH concentrate and 75% HQH (75HQH), or 40% concentrate and 60% HQH (60HQH). The fourth group (CON), a control group, was fed 60% fiber-rich hay and 40% concentrate, which is a typical feeding for dairy cows in Austria and several other countries in the European Union (EU). Chewing activity, nutrient intake, and digestion profile, as well as rumen fermentation parameters and rumen microbial populations of the digesta, have been previously published (19, 20). The normal fiber-rich hay (second-cut meadow hay at the beginning of blooming; approximately 40% Dactylis glomerata, 30% other grasses, including Festuca pratensis, Alopecurus pratensis, and Arrhenatherum elatius, 20% clover, and 10% herbs) contained, per kilogram of dry matter (DM), 113 g sugars, 577 g neutral detergent fiber (NDF), 350 g acid detergent fiber (ADF), 35.3 g acid detergent lignin (ADL), and 113 g CP, whereas HQH (Lolium perenne as main grass, mixed equal proportions of 1st and 2nd cut, beginning of ear emergence) contained, per kg (DM), 187 g sugars, 463 g NDF, 235 g ADF, 14.8 g ADL, and 235 g CP. The concentrate contained, on average, per kilogram (DM), 41 g sugars, 200 g NDF, 94 g ADF, and 450 g starch. The details of the chemical composition and energy content of ingredients and the four diets are shown in a study by Kleefisch et al. (19). The animal experimental protocol of this research was discussed and approved by the institutional ethics committee of the University of Veterinary Medicine Vienna in accordance with Good Scientific Practice guidelines and the national authority according to paragraph 26 of the Law for Animal Experiments, Tierversuchsgesetz–TVG 2012 (GZ 68.205/0111-WF/II/3b/2014) (44).
Quantification of volatile fatty acids in the rumen.
The percentages of individual VFA (acetate, propionate, isobutyrate, butyrate, isovalerate, and valerate) based on total VFA concentrations in the rumen, were determined by gas chromatography, as described by Pourazad et al. (45). Briefly, thawed samples were centrifuged at 20,000 × g for 25 min at 4°C (Avanti 30; Beckmann, Krefeld, Germany) to remove solid materials. Supernatant (0.6 ml) was transferred into a fresh tube with 0.2 ml of distilled water, 0.2 ml of HCl (1.8 mol liter−1), and 0.2 ml of internal standard (4-methylvalerian acid) and then centrifuged at 20,000 × g for 25 min at 4°C to remove precipitated substrates. The supernatant was again removed and then analyzed for VFA concentrations using gas chromatography (GC; Fisons GC model 8060 MS DPFC, catalog no. 950713; Rodena, Italy). The GC was equipped with a flame-ionization detector and a 30 m by 0.530 mm by 0.53 μm capillary column (Trace TR-Wax; Thermo Fisher Scientific, Waltham, MA). The injector temperature was 170°C, while the detector temperature was 190°C, with helium as a carrier gas (flow rate, 6 ml · min−1). Chromatograms were generated and evaluated using the Stratos software (Stratos version 4.5.0.0; Polymer Laboratories, Church Stretton, Shropshire, UK). The data were previously published by Klevenhusen et al. (20). The percentage of each VFA was calculated as the average value between liquid and solid fractions based on the total VFA concentration within an individual sample (Table S2).
Epithelial sampling.
Rumen papilla samples were taken on the last day of each experimental run at 10:00 a.m., immediately prior to feeding, according to the procedure suggested by Wetzels et al. (11). First, rumen digesta was evacuated via the cannula, and rumen contents were kept at body temperature during the sampling procedure via water baths. The ventral rumen wall was manually inverted through the cannula and washed with phosphate-buffered saline (PBS), and then rumen papillae were cut with aseptic scissors and tweezers and put directly into liquid nitrogen. The shock-frozen papilla samples were put in 2 cryotubes for both DNA and RNA analysis and stored at −80°C until analysis. After sampling, the rumen content was put back into the rumen.
Microbial composition analysis.
To preserve anaerobic cultures, rumen papillae were thawed on ice until they became pliant and were cut into small pieces, and a 250-mg homogenized subsample was used for genomic DNA isolation using the PowerSoil DNA isolation kit (Mo Bio Laboratories, Inc., Carlsbad, CA). Samples were mixed with sodium dodecyl sulfate-containing buffer C1 and heated at 70°C for 10 min to ensure proper lysis of the bacteria (27). Samples were then bead-beaten to dissociate microbes from feed particles and to disrupt the bacterial cells (28). This was followed by chemical removal of cell debris and PCR inhibitors and column-based isolation of total genomic DNA, according to the manufacturer's instructions. The isolated DNA concentration was determined using a Qubit 2.0 fluorometer (Life Technologies, Carlsbad, CA, USA) using the Qubit double-stranded DNA (dsDNA) HS assay kit (Life Technologies). One 20-μl aliquot of each sample for a total of 32 genomic DNA samples was sent for amplicon sequencing using a MiSeq Illumina sequencing platform and paired-end technology (Microsynth AG, Balgach, Switzerland). Sequencing targeted the V3-V5 hypervariable region of the 16S rRNA gene using the 357F (5′-CCTACGGGAGGCAGCAG-3′) and 926R (5′-CCGTCAATTCMTTTRAGT-3′) primer set (8) to generate an approximate amplicon size of ∼570 bp. The 16S rRNA gene PCRs, library preparation, and sequencing were performed by Microsynth. Libraries were constructed by ligating sequencing adapters and indices onto purified PCR products using the Nextera XT sample preparation kit (Illumina), according to the recommendations of the manufacturer. Equimolar amounts of each of the libraries were pooled and submitted for sequencing on an Illumina MiSeq personal sequencer using a 300-bp-read-length paired-end protocol. After sequencing, the corresponding overlapping paired-end reads were stitched by Microsynth, resulting in a total of 394,197 unfiltered reads, with an average of 537 nucleotides in length and a mean of 8,714 sequences per sample. Sequence quality control and analyses were performed using the QIIME pipeline (29).
Sequences were first quality filtered according to previously published recommendations (30), screened for chimeras using the gold.fa database and USEARCH (version 8.1), filtered, and then picked using UCLUST (31). Finally, samples were aligned and clustered to define operational taxonomic units (OTUs) using PyNAST (29) and the SILVA database as a reference template (version 123) (32). The degree of similarity between sequences was defined at 97% to obtain OTU identity at the species level. Any OTUs which clustered with <10 reads were manually removed to ensure that unique OTUs were not overestimated. A total of 316,884 sequences clustered into 1,490 OTU, of which 25 OTU were unassigned at the kingdom level and therefore removed from the analysis. Due to the large number of OTUs, all OTUs with a relative abundance greater than 0.1% (126 OTU) were taken for statistical analysis. Samples were rarefied based on the minimum number of sequences found in a sample (1,217 sequences) prior to diversity analysis.
RNA isolation and cDNA synthesis.
Frozen rumen epithelium samples for RNA analysis were stored on ice and weighed, and approximately 20 mg was combined with lysis buffer (RNeasy Mini QIAcube kit; Qiagen, Hilden, Germany), according to the manufacturer's protocols, and autoclaved ceramic beads (0.6 g, 1.4 mm diameter; VWR) added. Samples were homogenized at a 6.5 ms−1 for 30 s using the FastPrep-24 (MP Biomedicals, Santa Ana, CA, USA). The remainder of the RNA extraction protocol was completed using the automated QIAcube robotic workstation (Qiagen). After extraction, all samples were treated with the Turbo DNA kit (Ambion) to remove genomic DNA, and the quantification of RNA was determined with the Qubit 2.0 fluorometer (Life Technologies, Carlsbad, CA, USA), applying the Qubit RNA assay kit (Life Technologies). The quality of extracted RNA was evaluated using the Agilent Bioanalyzer 2100 (Agilent RNA 6000 Nano assay; Agilent, Santa Clara, CA, USA), with an average RNA integrity number (RIN) of 6.0, with the majority of samples having a RIN value between 7 and 9; however, 5 samples measured later had a RIN value range between 4.4 and 5.7. cDNA was synthesized using 2 μg RNA combined with a total of 20 μl of master mix (high-capacity cDNA reverse transcription [RT] kit; Applied Biosystems) composed of 4 μl of 10× reaction buffer, 1.6 μl of 100 mM 25× dinucleoside triphosphate (dNTP) mix, 4 μl of 10× random primers, 2 μl reverse transcriptase, 10 μl DNase/RNase-free water (diethyl pyrocarbonate [DEPC] treated; G-Biosciences, St. Louis, MO, USA), and the addition of 1 μl of RNase inhibitor (Biozym, Hessisch Oldendorf, Germany). The 2-step PCR was performed by a Mastercycler nexus (Eppendorf, Hamburg, Germany) using the following temperature program: incubation at 25°C for 10 min, reverse transcription at 37°C for 120 min, and a final heating step at 85° for 5 min. Reverse transcription controls were completed as a control for residual DNA contamination.
Real-time PCR.
Primers utilized for rumen epithelium quantitative reverse transcription-PCR (qRT-PCR) are listed with efficiencies in Table 4. Target genes for a pattern recognition receptor (Toll-like receptor isoform 4 [TLR4]), barrier function (claudin [CLDN]1, 4, and 7), cell transport (monocarboxylate cotransporter [MCT] isoforms 1 and 4), cellular pH regulation (anion exchanger isoform 2 [AE2], sodium/proton exchanger [NHE] isoforms 1, 2, and 3, and sodium/potassium ATPase [ATP1A1]), sodium-glucose transport (sodium-glucose transporter 1 [SGLT1]), and housekeeping genes (glyceraldehyde-3-phosphate dehydrogenase [GAPDH], beta actin [ACTB], hypoxanthine phosphoribosyltransferase 1 [HPRT1], ornithine decarboxylase antizyme 1 [OAZ1], and tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta [YWHAZ]) were analyzed. These primers were verified with PrimerBLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and tested for efficiencies and specificity using melting curves (R. M. Petri, S. U. Wetzels, B. U. Metzler-Zebeli, and Q. Zebeli, unpublished data), except SGLT1, which was previously published by Metzler-Zebeli et al. (33; also see references 34–36). Each 20-μl reaction mixture consisted of 50 ng in 1 μl of sample cDNA, 10 μl 1× Fast Plus Eva Green master mix with low ROX (Biotium, Hayward, CA, USA), 1 μl each of 200 nM forward and reverse primers, and 10 μl DNase/RNase-free water in a 96-well plate (Agilent Technologies). All reactions were performed on a real-time PCR MX3000P (Agilent Technologies) thermocycler using the following conditions: 95°C for 5 min, followed by 60°C for 30 s and 72°C for 30 s for 40 cycles. The presence of a single PCR product was verified with an additional dissociation stage. All reactions were run in duplicate, and replicates were done when greater than 0.5 cycles were between replicates. Analysis of 5 housekeeping genes was performed and the 3 best-fitting housekeeping genes were selected using NormFinder (37) and BestKeeper (38). Relative quantities of the target genes were normalized using the geometric mean of the genes OAZ1 and YWHAZ based on their comprehensive gene stability. The mRNA abundance of target genes was calculated using the quantification cycle (Cq) method, as previously described (39). Briefly, samples from all cows were analyzed on the same plate for a single gene. For each sample, the Cq value for the gene was calculated and subtracted from the geometric mean of the 3 best-fitting housekeeping genes to provide the ΔCq value. The average ΔCq value of all samples was used as a reference value to calculate the ΔΔCq. The relative expression values were calculated by using the formula relative expression = 2−ΔΔCq (Table 3).
TABLE 4.
Primers for Bos taurus genes and annealing temperatures for real-time PCR analysis
| Official gene symbol | Common gene symbola | Primer direction | Oligonucleotide sequence (5′ to 3′) | Annealing temp (°C) | Efficiency (%) | Reference |
|---|---|---|---|---|---|---|
| CLDN1 | Forward | CACAGCATGGTATGGCAATAGAA | 60 | 99.016 | New design | |
| Reverse | CAGCAGCCCAGCCAATGA | |||||
| CLDN4 | Forward | GTGTTTGGCGTGCTGTTGTC | 60 | 98.695 | New design | |
| Reverse | GGCCTTGGAGCTCTCATCAT | |||||
| CLDN7 | Forward | CAGATTTCTATAACCCATTGGTCC | 60 | 92.536 | 2 | |
| Reverse | GTATCCAGCTTTGCTCTCACTCC | |||||
| SLC16A1 | MCT1 | Forward | CATCATGTTGGCTGTCATGTATGG | 60 | 90.468 | 27 |
| Reverse | TCCTGCACAGTGTTACAGAAGGA | |||||
| SLC16A3 | MCT4 | Forward | CTCACCACAGGGGTCCTTAC | 60 | 103.345 | 27 |
| Reverse | AAGTAGCGGTTGAGCATGATGA | |||||
| SLC4A2 | AE2 | Forward | CGGCTGACCTGGACCTCAT | 60 | 106.331 | New design |
| Reverse | CGTTCTTCCGCACCAAGTGT | |||||
| SLC9A1 | NHE1 | Forward | GAAAGACAAGCTCAACCGGTTT | 60 | 92.1 | 4 |
| Reverse | GGAGCGCTCACCGGCTAT | |||||
| SLC9A2 | NHE2 | Forward | TTGTGCGATGACCATGAATAAGT | 60 | 92.064 | 4 |
| Reverse | TGATGGTCGTGTAGGATTTCTGA | |||||
| SLC9A3 | NHE3 | Forward | AGCCTTCGTGCTCCTGACA | 60 | 97.121 | 4 |
| Reverse | TGACCCCTATGGCCCTGTAC | |||||
| ATP1A1 | Forward | GACTCATCTCCATGATTGACC | 60 | 90.637 | 27 | |
| Reverse | TCCGGTGACCATGATGACCT | |||||
| TLR4 | Forward | GGTTTCCACAAAAGCCGTAA | 60 | 102.589 | New design | |
| Reverse | AGGACGATGAAGATGATGCC | |||||
| GAPDH | Forward | TGGAAAGGCCATCACCATCT | 60 | 90.947 | 5 | |
| Reverse | CCCACTTGATGTTGGCAG | |||||
| ACTB | Forward | CGTGAGAAGATGACCCAGATCA | 60 | 90.17 | 5 | |
| Reverse | TCACCGGAGTCCATCACGAT | |||||
| HPRT1 | Forward | TTGTATACCCAATCATTATGCTGAG | 60 | 95.515 | New design | |
| Reverse | ACCCATCTCCTTCATCACATCT | |||||
| OAZ1 | Forward | CACAAGAACCGTGATGATCGA | 60 | 108.157 | New design | |
| Reverse | TCTCACAATCTCAAAGCCCAAA | |||||
| YWHAZ | Forward | TGAAAGGAGACTACTACCGCTACTTG | 60 | 93.768 | New design | |
| Reverse | GCTGTGACTGGTCCACAATCC | |||||
| SLC5A1 | SGLT1 | Forward | ACATCGCCTACCCGACCTT | 60 | 96.638 | 27 |
| Reverse | CAGCATGACCGACAGCATCA |
Given when the common symbol is different than the official gene symbol.
Statistical analysis and visualization.
Microbiota OTU data, phylum and genus relative abundances, as well as normalized epithelial gene expression data were analyzed using the MIXED procedure of SAS 9.4 (40). Terms in the model included fixed effects of diet and square, and cow within square as a random effect. Group means were calculated using the LSMEANS option, and data are expressed with standard error of the mean and Tukey's honestly significant difference (HSD)-adjusted P values. Microbiota, gene expression data, and rumen environmental parameters of nutrient intake and DMI, and mean VFA concentrations in the ventral rumen liquid were analyzed for correlation using the CORR procedure of SAS. Correlation visualization heatmap and nonmetric multidimensional scaling (NMDS) analysis were performed with the corrplot (41), MASS (42), and ggplot2 packages (43) in R Studio (version 1.0.136). Significance was declared at a P value of <0.05, while tendencies are discussed at a P value of <0.10.
Accession number(s).
The sequences determined in this study have been deposited in the European Nucleotide Archive (ENA) under the study accession numbers ERX2119048 to ERX2119079.
Supplementary Material
ACKNOWLEDGMENTS
We thank the staff of the research station “Kremesberg” at the Vetmeduni Vienna for their assistance throughout the animal trial, including Iris Kröger for her assistance in running the animal trial. ARGE Heumilch is acknowledged for supplying the hay for this experiment. We also recognize the contribution of A. Sener for her significant work in the laboratory.
Contributions to this article include conception and design of the project by F.K. and Q.Z., project supervision by Q.Z., experiment execution and data acquisition from M.T.K. and R.M.P., data analysis and interpretation by R.M.P., and manuscript editing by M.T.K., F.K., B.U.M.-Z., and Q.Z. This paper was drafted and written by R.M.P.
We declare no conflicts of interest.
Funding was provided by the Austrian Ministry for Agriculture, Forestry, Environment and Water Management (grant BMLFUW100928), as well as by the University of Veterinary Medicine Vienna Postdoc Programme.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.00384-18.
REFERENCES
- 1.Chow J, Lee SM, Shen Y, Khosravi A, Mazmanian SK. 2010. Host-bacterial symbiosis in health and disease. Adv Immunol 107:243–274. doi: 10.1016/B978-0-12-381300-8.00008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Petri RM, Schwaiger T, Penner GB, Beauchemin KA, Forster RJ, McKinnon JJ, McAllister TA. 2013. Changes in the rumen epimural bacterial diversity of beef cattle as affected by diet and induced ruminal acidosis. Appl Environ Microbiol 79:3744–3755. doi: 10.1128/AEM.03983-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petri RM, Schwaiger T, Penner GB, Beauchemin KA, Forster RJ, McKinnon JJ, McAllister TA. 2013. Characterization of the core rumen microbiome in cattle during transition from forage to concentrate as well as during and after an acidotic challenge. PLoS One 8:e83424. doi: 10.1371/journal.pone.0083424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khafipour E, Li S, Plaizier JC. 2009. Alfalfa pellet-induced subacute ruminal acidosis in dairy cows increases bacterial endotoxin in the rumen without causing inflammation. J Dairy Sci 92:1712–1724. doi: 10.3168/jds.2008-1656. [DOI] [PubMed] [Google Scholar]
- 5.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. 2006. Microbial ecology: human gut microbes associated with obesity. Nature 444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 6.Han Z, Willer T, Li L, Pielsticker C, Rychlik I, Velge P, Kaspers B, Rautenschlein S. 2017. Influence of the gut microbiota composition on Campylobacter jejuni colonization in chickens. Infect Immun 85:e00380-. doi: 10.1128/IAI.00380-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zebeli Q, Metzler-Zebeli BU. 2012. Interplay between rumen digestive disorders and diet-induced inflammation in dairy cattle. Res Vet Sci 93:1099–1108. doi: 10.1016/j.rvsc.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 8.Weimer PJ. 2015. Redundancy, resilience, and host specificity of the ruminal microbiota: implications for engineering improved ruminal fermentations. Front Microbiol 6:296. doi: 10.3389/fmicb.2015.00296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng KJ, Bailey CB, Hironaka R, Costerton JW. 1979. A technique for depletion of bacteria adherent to the epithelium of the bovine rumen. Can J Anim Sci 59:207–209. doi: 10.4141/cjas79-025. [DOI] [Google Scholar]
- 10.McCann JC, Luan S, Cardoso FC, Derakhshani H, Khafipour E, Loor JJ. 2016. Induction of subacute ruminal acidosis affects the ruminal microbiome and epithelium. Front Microbiol 7:701. doi: 10.3389/fmicb.2016.00701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wetzels SU, Mann E, Pourazad P, Qumar M, Pinior B, Metzler-Zebeli BU, Wagner M, Schmitz-Esser S, Zebeli Q. 2017. Epimural bacterial community structure in the rumen of Holstein cows with different responses to a long-term subacute ruminal acidosis diet challenge. J Dairy Sci 100:1829–1844. doi: 10.3168/jds.2016-11620. [DOI] [PubMed] [Google Scholar]
- 12.Shen H, Chen Z, Shen Z, Lu Z. 2017. Maintaining stability of the rumen ecosystem is associated with changes of microbial composition and epithelial TLR signaling. Microbiologyopen 6:e436. doi: 10.1002/mbo3.436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ng KM, Ferreyra JA, Higginbottom SK, Lynch JB, Kashyap PC, Gopinath S, Naidu N, Choudhury B, Weimer BC, Monack DM, Sonnenburg JL. 2013. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502:96–99. doi: 10.1038/nature12503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tong M, McHardy I, Ruegger P, Goudarzi M, Kashyap PC, Haritunians T, Li X, Graeber TG, Schwager E, Huttenhower C, Fornace AJ Jr, Sonnenburg JL, McGovern DP, Borneman J, Braun J. 2014. Reprograming of gut microbiome energy metabolism by the FUT2 Crohn's disease risk polymorphism. ISME J 8:2193–2206. doi: 10.1038/ismej.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alugongo GM, Xiao J, Wu Z, Li S, Wang Y, Cao Z. 2017. Utilization of yeast of Saccharomyces cerevisiae origin in artificially raised calves. J Anim Sci Biotechnol 8:34–46. doi: 10.1186/s40104-017-0165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hooper LV, Littman DR, Macpherson AJ. 2012. Interactions between the microbiota and the immune system. Science 336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu JH, Bian GR, Zhu WY, Mao SY. 2015. High-grain feeding causes strong shifts in ruminal epithelial bacterial community and expression of Toll-like receptor genes in goats. Front Microbiol 6:167. doi: 10.3389/fmicb.2015.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Steele MA, Dionissopoulos L, AlZahal O, Doelman J, McBride BW. 2012. Rumen epithelial adaptation to ruminal acidosis in lactating cattle involves the coordinated expression of insulin-like growth factor-binding proteins and a cholesterolgenic enzyme. J Dairy Sci 95:318–327. doi: 10.3168/jds.2011-4465. [DOI] [PubMed] [Google Scholar]
- 19.Kleefisch MT, Zebeli Q, Humer E, Kröger I, Ertl P, Klevenhusen F. 2017. Effects of the replacement of concentrate and fibre-rich hay by high-quality hay on chewing, rumination and nutrient digestibility in non-lactating Holstein cows. Arch Anim Nutr 71:21–36. doi: 10.1080/1745039X.2016.1253227. [DOI] [PubMed] [Google Scholar]
- 20.Klevenhusen F, Petri RM, Kleefisch MT, Khiaosa-ard R, Metzler-Zebeli BU, Zebeli Q. 2017. Changes in fibre-adherent and fluid-associated microbial communities and fermentation profiles in the rumen of cattle fed diets differing in hay quality and concentrate amount. FEMS Microbiol Ecol doi: 10.1093/femsec/fix100. [DOI] [PubMed] [Google Scholar]
- 21.Gozho GN, Krause DO, Plaizier JC. 2007. Ruminal lipopolysaccharide concentration and inflammatory response during grain-induced subacute ruminal acidosis in dairy cows. J Dairy Sci 90:856–866. doi: 10.3168/jds.S0022-0302(07)71569-2. [DOI] [PubMed] [Google Scholar]
- 22.Nagaraja TG, Titgemeyer EC. 2007. Ruminal acidosis in beef cattle: the current microbiological and nutritional outlook. J Dairy Sci 90:E17–E38. doi: 10.3168/jds.2006-478. [DOI] [PubMed] [Google Scholar]
- 23.Shabat SK, Sasson G, Doron-Faigenboim A, Durman T, Yaacoby S, Miller ME, White BA, Shterzer N, Mizrahi I. 2016. Specific microbiome-dependent mechanisms underlie the energy harvest efficiency of ruminants. ISME J 10:2958–2972. doi: 10.1038/ismej.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Medvedev AE, Lentschat A, Wahl LM, Golenbock DT, Vogel SN. 2002. Dysregulation of LPS-induced Toll-like receptor 4-MyD88 complex formation and IL-1 receptor-associated kinase 1 activation in endotoxin-tolerant cells. J Immunol 169:5209–5216. doi: 10.4049/jimmunol.169.9.5209. [DOI] [PubMed] [Google Scholar]
- 25.Dassa B, Borovok I, Ruimy-Israeli V, Lamed R, Flint HJ, Duncan SH, Henrissat B, Coutinho P, Morrison M, Mosoni P, Yeoman CJ, White BA, Bayer EA. 2014. Rumen cellulosomics: divergent fiber-degrading strategies revealed by comparative genome-wide analysis of six ruminococcal strains. PLoS One 9:e99221. doi: 10.1371/journal.pone.0099221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mackie RI, Aminov RI, Hu W, Klieve AV, Ouwerkerk D, Sundset MA, Kamagata Y. 2003. Ecology of uncultivated Oscillospira species in the rumen of cattle, sheep, and reindeer as assessed by microscopy and molecular approaches. Appl Environ Microbiol 69:6808–6815. doi: 10.1128/AEM.69.11.6808-6815.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Metzler-Zebeli BU, Schmitz-Esser S, Klevenhusen F, Podstatzky-Lichtenstein L, Wagner M, Zebeli Q. 2013. Grain-rich diets differently alter ruminal and colonic abundance of microbial populations and lipopolysaccharide in goats. Anaerobe 20:65–73. doi: 10.1016/j.anaerobe.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 28.Kong Y, Teather R, Forster R. 2010. Composition, spatial distribution, and diversity of the bacterial communities in the rumen of cows fed different forages. FEMS Microbiol Ecol 74:612–622. doi: 10.1111/j.1574-6941.2010.00977.x. [DOI] [PubMed] [Google Scholar]
- 29.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. 2013. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 32.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO. 2012. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Metzler-Zebeli BU, Hollmann M, Sabitzer S, Podstatzky-Lichtenstein L, Klein D, Zebeli Q. 2013. Epithelial response to high-grain diets involves alteration in nutrient transporters and Na/K-ATPase mRNA expression in rumen and colon of goats. J Anim Sci 91:4256–4266. doi: 10.2527/jas.2012-5570. [DOI] [PubMed] [Google Scholar]
- 34.Stumpff F, Georgi MI, Mundhenk L, Rabbani I, Fromm M, Martens H, Günzel D. 2011. Sheep rumen and omasum primary cultures and source epithelia: barrier function aligns with expression of tight junction proteins. J Exp Biol 214:2871–2882. doi: 10.1242/jeb.055582. [DOI] [PubMed] [Google Scholar]
- 35.Oba M, Mewis JL, Zhining Z. 2015. Effects of ruminal doses of sucrose, lactose, and corn starch on ruminal fermentation and expression of genes in ruminal epithelial cells. J Dairy Sci 98:586–594. doi: 10.3168/jds.2014-8697. [DOI] [PubMed] [Google Scholar]
- 36.Steele MA, Croom J, Kahler M, AlZahal O, Hook SE, Plaizier K, McBride BW. Bovine rumen epithelium undergoes rapid structural adaptations during grain-induced subacute ruminal acidosis. Am J Physiol Regul Integr Comp Physiol 300:R1515–R1523. doi: 10.1152/ajpregu.00120.2010. [DOI] [PubMed] [Google Scholar]
- 37.Andersen CL, Jensen JL, Ørntoft TF. 2004. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res 64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- 38.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. 2004. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol Lett 26:509–515. doi: 10.1023/B:BILE.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
- 39.Gui H, Shen Z. 2016. Concentrate diet modulation of ruminal genes involved in cell proliferation and apoptosis is related to combined effects of short-chain fatty acid and pH in rumen of goats. J Dairy Sci 99:6627–6638. doi: 10.3168/jds.2015-10446. [DOI] [PubMed] [Google Scholar]
- 40.SAS Institute. 2004. SAS/STAT users guide. Release 9.1. SAS Institute, Cary, NC. [Google Scholar]
- 41.Wei T, Simko V. 2016. Corrplot: visualization of a correlation matrix. R package version 0.77. https://CRAN.R-project.org/package=corrplot.
- 42.Venables WN, Ripley BD. 2002. Modern applied statistics with S. 4th ed Springer, New York, NY. [Google Scholar]
- 43.Wickham H. 2009. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
- 44.Binder R. 2014. Das österreichische Tierschutzrecht: Tierschutzgesetz und Tierversuchsgesetz 2012 mit ausführliche Kommentierung. Edïtion Juridica Manz, Vienna, Austria. [Google Scholar]
- 45.Pourazad P, Khiaosa-Ard R, Qumar M, Wetzels SU, Klevenhusen F, Metzler-Zebeli BU, Zebeli Q. 2016. Transient feeding of a concentrate-rich diet increases the severity of subacute ruminal acidosis in dairy cattle. J Anim Sci 94:726–738. doi: 10.2527/jas.2015-9605. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.