ABSTRACT
Rhizospheric bacteria play important roles in plant tolerance and activation of heavy metals. Understanding the bacterial rhizobiome of hyperaccumulators may contribute to the development of optimized phytoextraction for metal-polluted soils. We used 16S rRNA gene amplicon sequencing to investigate the rhizospheric bacterial communities of the cadmium (Cd) hyperaccumulating ecotype (HE) Sedum alfredii in comparison to its nonhyperaccumulating ecotype (NHE). Both planting of two ecotypes of S. alfredii and elevated Cd levels significantly decreased bacterial alpha-diversity and altered bacterial community structure in soils. The HE rhizosphere harbored a unique bacterial community differing from those in its bulk soil and NHE counterparts. Several key taxa from Actinobacteria, Bacteroidetes, and TM7 were especially abundant in HE rhizospheres under high Cd stress. The actinobacterial genus Streptomyces was responsible for the majority of the divergence of bacterial community composition between the HE rhizosphere and other soil samples. In the HE rhizosphere, the abundance of Streptomyces was 3.31- to 16.45-fold higher than that in other samples under high Cd stress. These results suggested that both the presence of the hyperaccumulator S. alfredii and Cd exposure select for a specialized rhizosphere bacterial community during phytoextraction of Cd-contaminated soils and that key taxa, such as the species affiliated with the genus Streptomyces, may play an important role in metal hyperaccumulation.
IMPORTANCE Sedum alfredii is a well-known Cd hyperaccumulator native to China. Its potential for extracting Cd relies not only on its powerful uptake, translocation, and tolerance for Cd but also on processes underground (especially rhizosphere microbes) that facilitate root uptake and tolerance of the metal. In this study, a high-throughput sequencing approach was applied to gain insight into the soil-plant-microbe interactions that may influence Cd accumulation in the hyperaccumulator S. alfredii. Here, we report the investigation of rhizosphere bacterial communities of S. alfredii in phytoremediation of different levels of Cd contamination in soils. Moreover, some key taxa in its rhizosphere identified in the study, such as the species affiliated with genus Streptomyces, may shed new light on the involvement of bacteria in phytoextraction of contaminated soils and provide new materials for phytoremediation optimization.
KEYWORDS: cadmium, rhizosphere, hyperaccumulator, bacterial community, Streptomyces
INTRODUCTION
Cadmium (Cd) contamination in soils has become a great concern worldwide because of its high mobility in the environment and toxicity in humans (1). Phytoextraction of heavy metals from contaminated soils is an attractive remediation technology because of advantages such as low cost, sustainability, and eco-friendliness (2). The efficiency of plants at extracting metals from contaminated soils is largely dependent on plant contaminant tolerance and accumulation capacity. Hyperaccumulating plants have potential for phytoextraction of polluted soils; however, further optimization for their application is needed because of their overall low biomass and limited expansion of their root system (3, 4). The bacterial rhizobiome, a set of specialized microbes that colonize the plant rhizosphere, makes an important contribution to hyperaccumulators (5). Metal-resistant bacteria isolated from the rhizosphere of hyperaccumulators have been reported to promote plant growth and bioavailability of heavy metals in soils, leading to significantly improved metal accumulation in plants (6, 7). Although bacterium-assisted phytoextraction may at first seem ideal for remediation of heavy metal-polluted soils, its practical use is generally limited by the complex biochemical environments of contaminated soils (8). A deeper understanding of the complex plant-microbiome environment in plant rhizospheres, particularly in those of hyperaccumulators, may contribute to the development of optimized phytoextraction technology for metal-polluted soils (5).
The rhizosphere is a microenvironment comprising root surfaces and their surrounding soil area (9). It supplies organic compounds from plant roots to microbes, enabling their growth and reproduction (10), and is enriched in microbes that are adapted to the rhizospheric environment (11). The rhizosphere habitat is affected by spatial and temporal variations in soil properties and the physiological state of the plant (12). Heavy metals in soils could alter the structure of soil microbial communities (13). Previous studies found that presence of heavy metals, including Cd, in soils decreased the microbial biomass and resulted in a shift in the microbial community structure (14–16). On the other hand, heavy metals in soils could increase the prevalence of specific heavy metal-resistant bacteria (13). Although a considerable amount of metal-resistant bacteria from the rhizosphere of hyperaccumulators have been isolated using traditional culture-dependent techniques, the microbial assemblages associated with hyperaccumulation remain poorly understood (5, 17). Furthermore, previous research tended to focus on the important effect of a single microbial inoculum on the enhancement of phytoextraction but overlooked the complexity of the plant-soil environment in which a variable indigenous microbial community may outcompete or inhibit phytoremediation-assisting inocula (13, 18).
Sedum alfredii is a well-known Zn/Cd cohyperaccumulator native to China (19). The potential application of this hyperaccumulator plant species to soil remediation has recently attracted increasing interest, particularly in China (20–22). Previous studies showed that metal-tolerant bacteria isolated from the rhizosphere of S. alfredii significantly enhanced Cd/Zn extraction (23, 24). However, little knowledge is available regarding the basic ecology of indigenous soil microbial communities associated with the phytoextraction process in polluted soils by this plant species. Although our recent study indicated that the rhizosphere microcharacteristics, including the bacterial community, facilitate phytoextraction of multiple metals in soil by S. alfredii (25), the relationships between the bacterial rhizobiome and the influencing factors or variables, such as soil type, pollutant species, and pollution levels, are still unknown. Therefore, the present study investigated the characteristics of the rhizosphere bacterial community of the hyperaccumulator S. alfredii under different Cd treatments and the interaction of Cd presence and roots of S. alfredii on the structure of soil bacterial communities, using 16S rRNA gene amplicon sequencing. We aimed to (i) compare the differences in bacterial rhizobiome between the hyperaccumulating ecotype (HE) and the contrasting nonhyperaccumulating ecotype (NHE) of S. alfredii in response to different Cd levels, (ii) evaluate the effect of planting and Cd exposure on the rhizospheric bacterial community diversity and composition of S. alfredii, and (iii) explore the key taxa involved in the phytoextraction process of Cd-contaminated soil by HE S. alfredii.
RESULTS
Plant growth and Cd phytoextraction.
After 150 days of plant growth in Cd-contaminated soils, the two ecotypes of S. alfredii exhibited significant differences in metal tolerance and accumulation. The NHE plants presented typical symptoms of metal poisoning, such as dwarfism and chlorotic leaves (Fig. 1). Compared to the controls (0.47 mg Cd kg−1 soil, Cd 0), the root and shoot biomass of the NHE grown in highly Cd-contaminated soil (22.01 mg Cd kg−1 soil, Cd 20) decreased by 53.66 and 27.54%, respectively. In contrast, no visible growth inhibition or symptoms of phytotoxicity were observed for the HE plants (Fig. 1). There was also a significant increase in HE plant growth when the plants were grown in Cd 5 level soils (6.66 mg Cd kg−1 soil) compared with other treatments. At harvest (Table 1), the Cd concentrations in shoots of HE grown in soils treated with Cd 5 and Cd 20 reached 304.56 ± 15.04 mg kg−1 and 678.53 ± 114.92 mg kg−1, respectively, which were 18.54- and 13.53-fold higher than those in the NHE plants, respectively. The translocation factors of Cd (i.e., measuring the amount of Cd transferred from roots to shoots) in the HE plants were 7.88- and 4.69-fold higher than those for the NHE plants, respectively.
FIG 1.
Growth status and dry biomass (g pot−1) of hyperaccumulator ecotype (HE) and nonhyperaccumulator ecotype (NHE) S. alfredii planted in different Cd-contaminated soils. Cd 0 denotes the control treatment; Cd 5 and Cd 20 denote 5-mg kg−1 and 20-mg kg−1 Cd-spiked treatments.
TABLE 1.
Cd concentrations and accumulation in roots and shoots of S. alfredii and Cd translocation factors under different Cd treatmentsa
| Cd treatment | Cd concn (mg kg−1) |
Cd accumulation (μg pot−1) |
Translocation factorb | |||
|---|---|---|---|---|---|---|
| Shoot | Root | Shoot | Root | |||
| Cd 0 | HE | 4.10 ± 0.31c** | 9.64 ± 0.89e** | 10.21 ± 0.76c** | 3.29 ± 0.30d** | 0.43 ± 0.06cd |
| NHE | 0.55 ± 0.14c | 1.71 ± 0.14f | 1.64 ± 0.80c | 0.71 ± 0.06e | 0.32 ± 0.10d | |
| Cd 5 | HE | 304.56 ± 15.04b** | 34.37 ± 2.33b** | 1,039.16 ± 51.32b** | 14.59 ± 0.99b** | 8.91 ± 1.01a** |
| NHE | 16.43 ± 2.23c | 14.52 ± 1.83d | 44.27 ± 6.02c | 4.94 ± 0.62c | 1.13 ± 0.09bc | |
| Cd 20 | HE | 678.53 ± 114.92a* | 84.13 ± 7.00a** | 1,923.63 ± 325.80a** | 26.84 ± 2.23a** | 8.63 ± 1.03a** |
| NHE | 50.15 ± 9.35c | 27.12 ± 4.13c | 100.33 ± 18.72c | 5.20 ± 0.79c | 1.84 ± 0.07b | |
All data are presented as means ± the standard deviations (n = 3). Different letters within the same column indicate the significance of different Cd treatment at P < 0.05 based on a protected Fisher LSD test. Asterisks indicate significant differences between hyperaccumulating (HE) and nonhyperaccumulating (NHE) ecotypes (*, P < 0.05; **, P < 0.01).
Translocation factors = Cd concentration ratio of plant shoots to roots.
Because of its relatively high capacity for extracting Cd from the soils, planting of HE S. alfredii significantly decreased total Cd levels in its rhizospheric zone (Fig. 2). The Cd contents in soil treated with Cd 0, Cd 5, and Cd 20 decreased to 0.12, 3.59, and 12.89 mg kg−1 dry soil in the rhizosphere zones from initial Cd concentrations of 0.47, 6.66, and 22.01 mg kg−1, respectively. This result indicates that the HE plants removed 74.50, 46.10, and 41.43% of the total Cd in the rhizospheric soils under Cd 0, Cd 5, and Cd 20 treatments, respectively.
FIG 2.
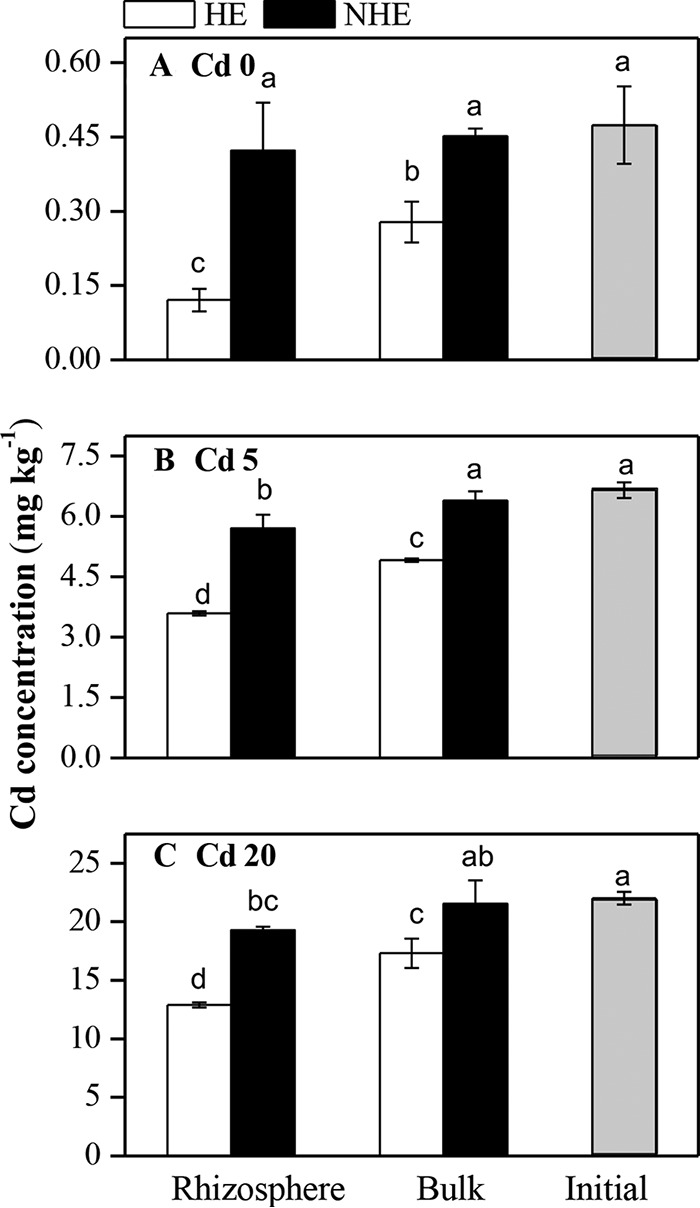
Cd concentrations in the rhizosphere, bulk soils of S. alfredii, unplanted soils, and initial soils under Cd 0 (A), Cd 5 (B), and Cd 20 (C) treatments. Data points represent means ± the standard deviations (n = 3). The various lowercase letters indicate significant differences among all treatments at P < 0.05. Refer to Fig. 1 for treatment notation.
Alpha-diversity of bacterial community.
The high-quality reads produced by using 16S rRNA gene amplicon sequencing were clustered using a >97% sequence identity threshold, which yielded 71,239 bacterial operational taxonomic units (OTUs). A total of 23,437 OTUs were obtained after randomly resampling Illumina 16S sequence reads to the same depth (16,700 sequences per sample). The alpha-diversity of the bacterial community using the observed species and Simpson's index was compared between the rhizosphere and bulk zone for both HE and NHE S. alfredii (Fig. 3). The observed species of bacterial community were significantly influenced by sampled compartments and Cd levels, while Simpson's index was significantly influenced by soil compartments, Cd levels, ecotypes, and their interactions (P < 0.001; see Table S2 in the supplemental material). Both of the alpha-diversity indices exhibited similar decreasing trends in response to the increasing Cd levels in soils regardless of sampling compartments. The metrics in the HE rhizosphere, however, showed the greatest divergences between three different Cd levels, where the highest Cd treatment (Cd 20) resulted in the lowest microbial diversity (Fig. 3). In contrast, variations in alpha-diversity were less pronounced for bacterial communities in the NHE rhizosphere, as well as for those of the bulk and unplanted soils at different Cd levels (Fig. 3).
FIG 3.
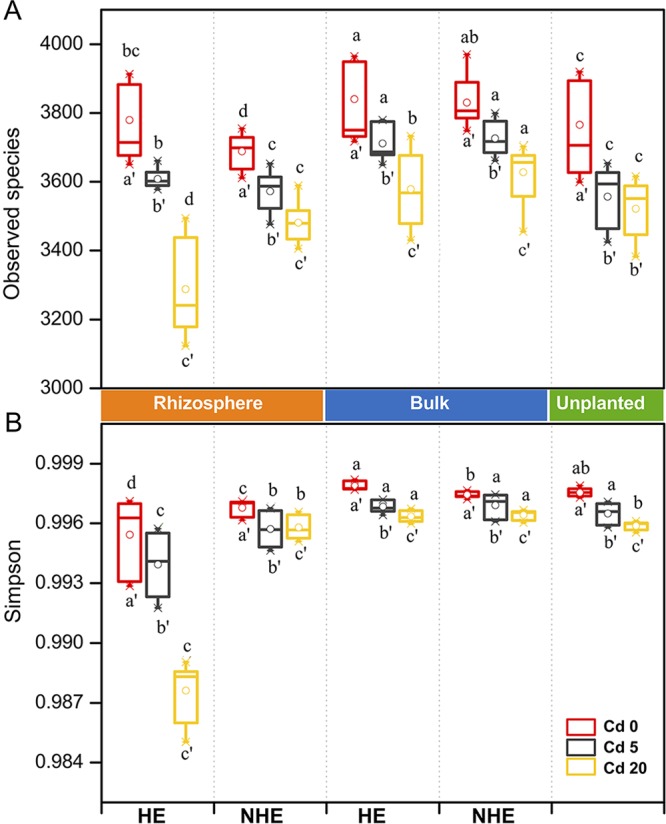
Box plots for alpha-diversity indices, including the number of observed species (A) and the Simpson's index of the bacterial communities (B) in different compartments under different Cd treatments. The lines at the top, bottom, and middle of the box correspond to the 75th, 25th, and 50th percentiles (median), respectively. Whiskers indicate the standard deviations. The white dots indicate the mean values. The various letters above the box indicate significant differences among samples of the same Cd level at P < 0.05. The various letters below the box indicate significant differences among different Cd levels at P < 0.05. Refer to Fig. 1 for treatment notation.
Bacterial community structure.
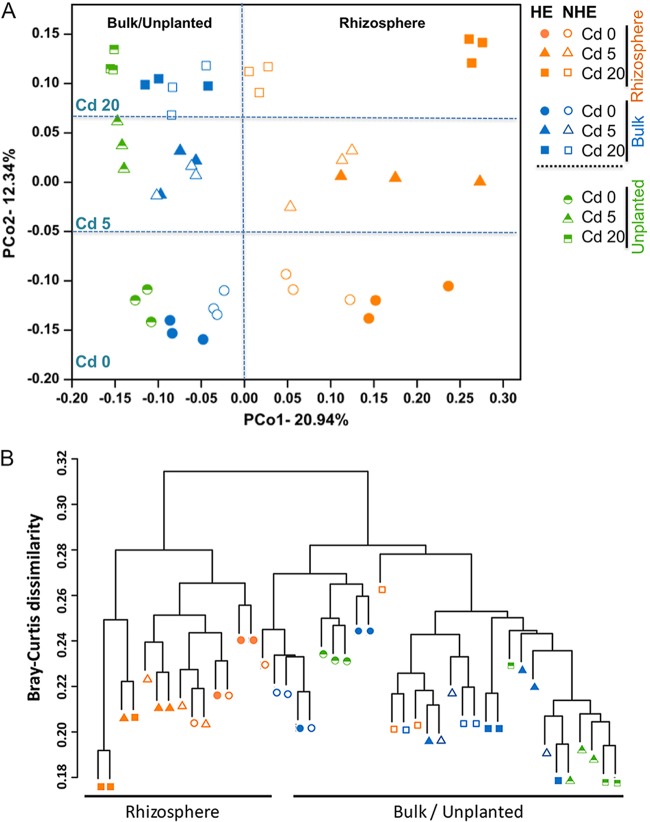
Principal-coordinate analyses (PCoAs) revealed a clear differentiation between different sampling compartment (the rhizosphere, bulk soil, and unplanted soil) (Fig. 4A). Cluster analysis based on the Bray-Curtis dissimilarity showed that bacterial community composition of rhizosphere and bulk/unplanted soils formed distinct clusters for both HE and NHE (with the exception of the community of NHE rhizosphere in Cd 20 level, which clustered with bulk/unplanted soil), confirming that compartment was the major determinant of community structure (Fig. 4B). Permutational multivariate analysis of variance (PERMANOVA) using Bray-Curtis distance corroborates that rhizospheric compartmentalization comprises the largest source of variation (20.58%, P < 0.001; Table 2). Both PCoAs and PERMANOVA described Cd levels as the second largest source of variation (10.82%, P < 0.001; Fig. 4A and Table 2). Nevertheless, a significant variation in the bacterial community composition between the two ecotypes (HE and NHE) was pronounced in the rhizospheric zone, and an increasing variation in the community along the Cd gradient at rhizosphere was clearly observed (Fig. 4A).
FIG 4.
Roots of different S. alfredii ecotypes and Cd concentrations shape bacterial community composition. (A) PCoA plots to visualize the Bray-Curtis distance among the bacterial communities. (B) Bray-Curtis dissimilarity of samples collected from the unplanted soils, rhizosphere, or bulk soils planted with HE and NHE S. alfredii. Refer to Fig. 1 for treatment notation.
TABLE 2.
PERMANOVA results using Bray-Curtis as a distance metric
| Factor | % explained | F | R2 | Pa |
|---|---|---|---|---|
| Compartment | 20.58 | 6.7704 | 0.21 | 0.001*** |
| Cd | 10.82 | 7.1198 | 0.11 | 0.001*** |
| Ecotype | 4.11 | 2.7036 | 0.04 | 0.002** |
| Compartment:Ecotype | 3.45 | 2.2708 | 0.03 | 0.008** |
| Compartment:Cd | 3.60 | 1.1832 | 0.04 | 0.164 |
| Cd:Ecotype | 2.44 | 1.6072 | 0.02 | 0.050* |
| Compartment:Cd:Ecotype | 1.80 | 1.1824 | 0.02 | 0.225 |
| Residuals | 53.20 | 0.53 | ||
| Total | 100.00 | 1.00 |
*, P < 0.05; **, P < 0.01; ***, P < 0.001.
Specific bacterial assemblages in the HE rhizosphere.
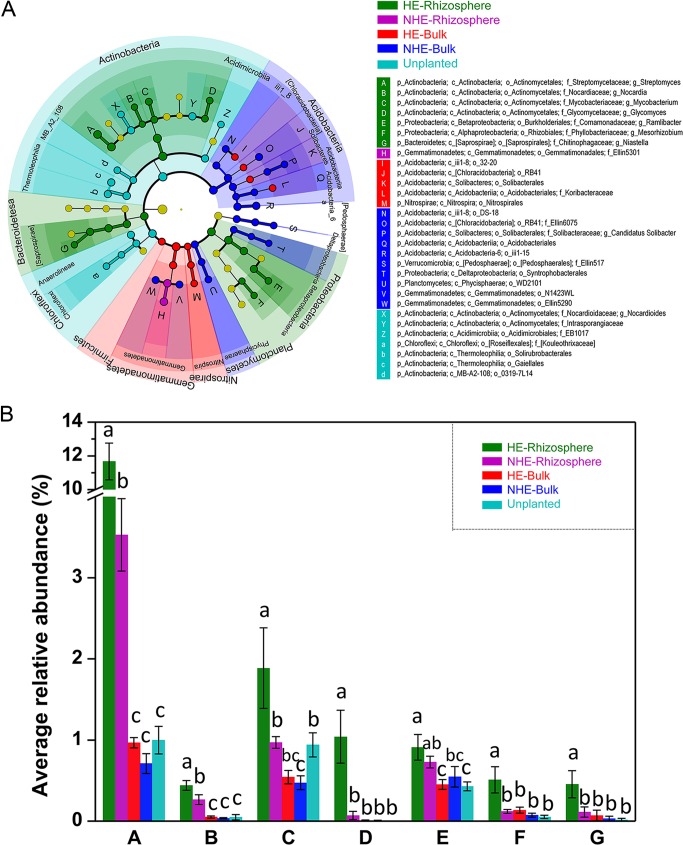
The least discriminant analysis (LDA) effect size taxonomic cladogram was used to analyze the predominant discriminant taxa of the bacterial communities in the HE rhizospheric soil differing from counterparts in the other soil samples exposed to Cd 20 (Fig. 5A). The results showed the specific localization of genera Streptomyces, Nocardia, Mycobacterium, Glycomyces (phylum Actinobacteria), Ramlibacter, Mesorhizobium (phylum Proteobacteria), and Niastella (phylum Bacteroidetes) in the rhizosphere of HE S. alfredii. The average abundances of the seven genera were significantly increased by severalfold in the HE rhizosphere, relative to their counterparts in the bulk/unplanted soils (Fig. 5B). For example, Streptomyces accounted for the largest proportion (11.67%) of the bacterial community in the HE rhizosphere, and its abundance was 16.45- and 3.31-fold higher than that in the bulk/unplanted soils and in the NHE rhizosphere, respectively (Fig. 5B).
FIG 5.
Root-specific taxa from the rhizosphere compartment of HE S. alfredii compared to those from other samples. (A) Least discriminant analysis (LDA) effect size taxonomic cladogram to compare samples collected from the rhizosphere of both S. alfredii ecotypes. The discriminant taxon nodes are colored and branches are shaded according to the highest-ranked group for that taxon. If the taxon did not represent significant differences among sample groups, the corresponding node is yellow. Selected highly abundant taxa are indicated with uppercase or lowercase letters. (B) The average relative abundance of seven taxa (column groups A to G) in HE rhizosphere shown in panel A.
Key taxa associated with Cd-contaminated levels.
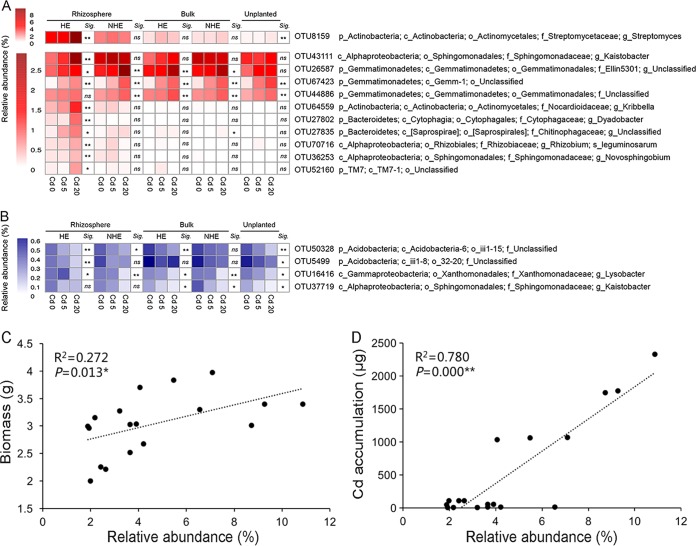
According to the correlation coefficients (see Table S3 in the supplemental material), 15 representative phylotypes (OTUs) correlated with Cd levels were separated into a positively correlated group and a negatively correlated group (Fig. 6). The closely matched type strains provide additional information on taxonomic identification of some OTUs (see Table S4 in the supplemental material). Eleven key phylotypes showed significant positive correlation with Cd levels, among which the phylum Gemmatimonadetes (two members of the genus Gemmatirosa and a member of the genus Gemmatimonas) increased essentially in response to the elevated Cd levels in soils, regardless of plantation (Fig. 6A, Table S4 in the supplemental material). However, most of the phylotypes, including Actinobacteria (a member of the genera Streptomyces and Kribbella), Bacteroidetes (a member of genera Dyadobacter and Niastella), TM7 (a member of the genus Saccharimonas), and class Alphaproteobacteria (a member of the genus Kaistobacter, Rhizobium, and Novosphingobium), were positively correlated with the increasing Cd levels in the HE rhizosphere, and seldom in the other soil samples (Fig. 6A; Table S4 in the supplemental material).
FIG 6.
Relationships between key taxa, soil Cd levels, plant biomass, and Cd accumulation. Heat maps of average relative abundances of key taxa positively (A) and negatively (B) associated with Cd levels. At each compartment, the significance (Sig.) is noted as follows: **, significant Pearson's correlation (P < 0.01); *, significant Pearson's correlation (P < 0.05); and ns, not significant. The taxonomy of the OTUs was identified in the Greengenes database (13_8_release). Refer to Fig. 1 for treatment notation. Also shown are linear regressions between the average relative abundance of OTU8159 and plant biomass (C) and Cd accumulation (D).
A species (OTU8159) affiliated with genus Streptomyces (phylum Actinobacteria) had the highest relative abundance both in genus Streptomyces and in all bacterial OTUs found in the HE rhizosphere and contributed highly (5.90%) to the overall dissimilarity along the Cd gradient (see Table S3 in the supplemental material). OTU8159 was highly similar (99.55%) to Streptomyces lucensis NBRC 13056T and nine other Streptomyces-affiliated species, based on a 16S rRNA gene sequence query against the EzTaxon database. In the rhizosphere of HE S. alfredii, the abundance of OTU8159 increased from 4.81 to 9.62% as soil Cd increased from 0.47 to 22.01 mg kg−1, whereas it remained at 2.44 to 2.76% in the NHE rhizosphere, and even less (0.40 to 1.16%) in the bulk and unplanted soils (Fig. 6A). In addition to being positively correlated with soil Cd concentration, the abundance of OTU8159 was significantly positively correlated with plant biomass (Fig. 6C) and Cd accumulation in plants (Fig. 6D).
DISCUSSION
The hyperaccumulator HE S. alfredii has been reported to not only tolerate high levels of Cd, but also to efficiently take up and accumulate large amounts of Cd in its aboveground parts (26), and thereby could be useful in the phytoremediation of Cd-polluted soils (19). This point of view is supported by the results of this study (Table 1, Fig. 2). This potential of HE S. alfredii for extracting Cd relies not only on its powerful metal uptake, translocation ability, and tolerance for Cd in stem and leaf cells (27, 28), but also on processes underground that facilitate root uptake and tolerance of the metals. A high-throughput sequencing approach was therefore applied to gain insight into the soil-plant-microbe interactions that may influence metal accumulation in the hyperaccumulator S. alfredii.
Bacterial diversity reduced with root presence and Cd exposure.
Plant root-driven changes in the rhizosphere microbial community are well documented (29–31). In this research, plant roots of both HE and NHE S. alfredii tended to have decreased bacterial diversity in their rhizosphere zone (Fig. 3). Reductions in bacterial community diversity in rhizospheric soils of some plant systems have been suggested to result from a combination of reduced taxon richness and evenness (32, 33). Soil is thought to be among the most diverse microbial habitats on Earth because of its microsite niche heterogeneity (34, 35). The impact of the root on soil C availability, pH, water, and atmosphere could overwhelm and homogenize differences among soil microsites and then reduce the microbial community richness in the rhizosphere (32). More importantly, the selectivity of plant roots to surrounding microbes may alter species abundance and reduce the evenness of taxon distribution (enrichment of select members or loss of detectable low-abundance taxa); a decrease in taxon-based diversity indices may consequently result (e.g., the Simpson's index, Fig. 3B).
Bacterial growth rate and biomass production are also influenced by metal ion concentration (26). Bacterial diversity in soils consistently diminished along the Cd gradient regardless of planting HE or NHE S. alfredii (Fig. 3). This pattern suggests that certain sensitive taxa were extinguished by Cd poisoning (such as inhibition of growth and destruction of cell morphology and biochemical processes), which eventually resulted in reduced bacterial diversity in highly Cd-contaminated soils (36, 37).
Unique rhizosphere bacterial community assembled by roots of HE S. alfredii under Cd exposure.
In addition to bacterial diversity, the results suggested the rhizosphere bacterial community structure of HE S. alfredii differed from those of bulk soils and the rhizospheric soils of NHE S. alfredii, especially under high Cd stress (Fig. 4 and 5). Rhizosphere microbial communities can be recruited by the plant host to either supply it with nutrients or to help in the survival under stressful conditions (38). Root-driven changes in the rhizosphere microbial community composition were reportedly a consequence of altered plant root exudates (11, 39–41). For example, an Arabidopsis ABC transporter mutant caused significant changes to the natural microbial community compared to the wild type (40), and these differences in root exudation supported distinct rhizosphere bacterial communities (41). The mechanisms underlying the occurrence of different bacterial communities in the rhizospheric zones of HE and NHE under Cd stress requires further investigation, though differences of the exudate profile between the two S. alfredii ecotypes have been reported (42, 43).
It is well documented that heavy metals present in contaminated soils can alter the structure of soil microbial communities (13). In this study, on the basis of plant presence, the increase of Cd pollution further altered the soil bacterial community structure (Fig. 4), which is in line with results from a previous study focusing on the response of bacterial community to Cd in coastal water microcosms (44). The gradually increasing divergence of bacterial community assemblage along the Cd gradient between HE rhizosphere and other samples indicated the interactions between root system and Cd exposure that occurred during the phytoextraction of Cd pollution by HE S. alfredii. Note that the bacterial community in the NHE rhizosphere exposed to Cd 20 levels clustered with bulk/unplanted samples (Fig. 4B), indicating that the inhibited root growth of NHE under high Cd stress had little effect on rhizosphere bacterial community.
Key taxa in response to Cd pollution and HE plantation.
In general, Cd toxicity acts on a portion of low-resistance microbes, suppressing their growth rate and reducing their abundance (36). As shown in Fig. 6B, the abundances of several bacterial phyla (e.g., Acidobacteria and Proteobacteria) were negatively correlated with elevated Cd concentrations. On the other hand, Cd stress could cause an increase in the prevalence of Cd-resistant bacteria (13). High tolerance to heavy metals of HE resulted in an extensive root system and high root exudates secretion to the soil environment (3, 23), thus changing the ecology in the rhizosphere. Therefore, Cd-induced root responses of plants as an indirect factor also play an important role in shaping the rhizosphere bacterial communities and key taxa composition. The present findings indicate a unique bacterial community in the HE rhizosphere under high Cd stress, comprising several key taxa from Actinobacteria, Bacteroidetes, TM7, and the class Alphaproteobacteria (Fig. 6A), which have been identified as the most dominant phyla in heavy metal-contaminated soils and may provide a stabilizing influence in heavy metal-contaminated soils (45, 46). Actinobacteria was the most important discriminant phylum in HE rhizosphere under high Cd stress (Fig. 5). This bacterial phylum is regarded as an important group of microbes that colonizes plant rhizospheres (47). Many members of this phylum can produce IAA to stimulate plant growth, as well as siderophores to form stable complexes with Cd that protect against phytopathogenic fungi (48). The actinobacterial genus Streptomyces was responsible for the majority of divergence among the rhizospheres of HE S. alfredii and bulk soils, contrasting with the rhizosphere of NHE S. alfredii. Our previous study also found HE harbored abundant Streptomyces in its rhizosphere during the phytoremediation of multimetal-contaminated soil (25). Streptomyces is the largest antibiotic-producing genus in the microbial world (49). More importantly, it has a high tolerance to heavy metals and has a potential use in bioremediation (50). Some bacterial strains belonging to Streptomyces isolated from the rhizosphere and root of Betula pendula L and Alnus glutinosa showed the most effective ability of siderophore synthesis under Cd stress (51). A representative Streptomyces species has been reported to stimulate metal solubility and auxin synthesis by secretion of siderophores (52). Streptomyces sp. F4, isolated from a former uranium mine, was capable of Cd(II) uptake and complexation in culture medium and soil samples (53).
Our results showed that key taxa from the phylum Gemmatimonadetes were positively associated with Cd levels, regardless of planting with or without the seedlings of HE or NHE S. alfredii (Fig. 6A). The stable correlation of these taxa with Cd suggests a possible ecological role of them in response to Cd exposure in soils (44). Even so, they are not likely key players in Cd hyperaccumulation in HE S. alfredii, since they are not specifically localized to the HE rhizosphere. Unlike these taxa affiliated with Gemmatimonadetes, an unclassified species OTU8159, affiliated with the genus Streptomyces, showed its specific colonization in HE rhizosphere in response to the elevated Cd stress (Fig. 6A). It was also found that the abundance of OTU8159 was significantly associated with plant growth (Fig. 6C) and Cd accumulation (Fig. 6D). Although there is no more direct evidence here, this member of Streptomyces from the HE rhizosphere might play vital role in facilitating the phytoextraction of Cd from contaminated soil. Further identification and exploration of this specific bacterial species and other key taxa found in the rhizosphere of HE S. alfredii would shed light on the optimization of its usage on the remediation of Cd-polluted soils.
MATERIALS AND METHODS
Plant material and soil characterization.
Seedlings of the hyperaccumulating ecotype (HE) of S. alfredii were obtained from an ancient Pb/Zn mine site in Quzhou. Nonhyperaccumulating ecotype (NHE) S. alfredii plants were collected from a tea plantation in Hangzhou, Zhejiang Province, China. Seedlings were grown in noncontaminated soil for more than three generations to minimize internal metal content. Healthy and equally sized plant shoots were selected and precultivated hydroponically for 2 weeks in a basic nutrient solution to grow roots (54).
Purplish clayed soil, which is commonly used for growing rice in China, was collected from the surface layer (0–20 cm) of a paddy field located in Shaoxing, Zhejiang province. Soil samples were air dried, ground, and sieved through a 2-mm mesh. Geochemical properties of this soil were analyzed (see Table S1 in the supplemental material). The Cd concentration in the clayed soil was ∼0.47 mg kg−1 soil. Three levels of Cd (0, 5, and 20 mg Cd kg−1 soil) were added as a solution of Cd(NO3)2 in the pretreated soil, and the Cd-spiked soil was aged for 3 months at a moisture content of 60% of water holding capacity prior to its use for pot experiments. At the end of the aging, the corresponding concentrations of Cd in three treatments—(i) low-level control (Cd 0), (ii) intermediate level (Cd 5), and (iii) high level (Cd 20)—were 0.47, 6.66, and 22.01 mg kg−1, respectively.
Pot culture experiment.
Two root bags (polyester mesh; 300 mesh pore size, 2-cm diameter, 10-cm height) were filled with the prepared soil (0.1 kg) and evenly distributed in each microcosm (12-cm height × 14-cm diameter). The remaining soil (0.8 kg) was filled in between the root bags. Two-week-old seedlings of the HE and NHE S. alfredii were transplanted to the root bags. For each Cd level, three microcosms with two plants each were prepared, and the treatments without planting (unplanted) were also repeated three times. The microcosms were watered to maintain soil moisture at approximately 65% of the maximum water-holding capacity. The plants were grown for 150 days in a greenhouse with natural light, average day/night temperatures of 30/24°C, and a day/night humidity of 70/85%.
Soil sampling, plant harvesting, and elemental analyses.
At harvest, rhizosphere soil was collected by gently shaking the entire plant root system; the soil adhering to the root system was placed in 20 ml of phosphate-buffered saline. The soil that was washed from the two plant roots of the same microcosm was mixed and stored as rhizosphere compartment. Approximately 5 g of bulk soil was collected from the middle depth of soil layer between two plants. All samples, including those from the unplanted soils, were stored at −80°C until DNA extraction. Soil properties and elemental concentrations were analyzed for soils of the rhizosphere, bulk zone, and unplanted microcosms at both the beginning and the end of the experiment. To analyze the total Cd concentrations, soil samples (0.1 g) were digested with 5 ml of HNO3, 1 ml of HClO4, and 1 ml of HF at 180°C for 10 h. The plants were separated into roots and shoots, washed thoroughly, and rinsed with distilled water. The plant samples were then oven dried at 65°C, weighed, ground, and passed through a 60 mesh sieve. Plant samples (0.1 g) were digested with 5 ml of HNO3 and 1 ml of H2O2 at 180°C for 8 h for elemental analysis. The concentrations of Cd in the digestive solutions were determined by inductively coupled plasma-mass spectrometry (ICS-MS 7500a; Agilent, USA). Sample replicates, reagent blanks, rice flour (IRMM-804; Sigma, USA), and soil (GBW07429; the National Research Center for Certified Reference Materials of China) standard reference materials were included in each batch of analysis to ensure the quality of analysis. The recovery of standard for each element was 90 to 110%.
All data are reported as means ± the standard deviations, based on three replicates. Significant differences (P ≤ 0.05) among treatments were analyzed using a protected Fisher least significant difference (LSD) test, after a one-way analysis of variance (ANOVA). These statistical analyses were conducted using SPSS 18.0.
DNA extraction, bacterial 16S amplification, and MiSeq sequencing.
Soil genomic DNA was extracted by the PowerSoil DNA isolation kit (MOBIO, USA). Photospectrometric quantification of DNA extracts was completed using a NanoDrop ND 2000 (PEQLAB Biotechnology GmbH, Germany) and stored at −80°C prior to amplification. The V3-V4 regions of the bacterial 16S rRNA genes were amplified using the primer set 341F (5′-CCTACGGGNGGCWGCAG-3′) and 805R (5′-GACTACHVGGGTATCTAATCC-3′). An aliquot of 10 ng of purified DNA template from each sample was amplified with a 20-μl PCR system under the following conditions: initial denaturation at 95°C for 3 min, followed by 27 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 45 s, with a final extension at 72°C for 10 min. An equimolar amount of PCR product for each sample was combined into one pooled sample and sequenced using a MiSeq platform (Illumina, USA).
16S amplicon processing and analysis.
All sequences of 16S rRNA were demultiplexed with the QIIME pipeline (v1.8.0) (55), and the paired reads were joined with FLASH using the default settings (56). The sequences were assigned to each sample according to the barcodes and were quality controlled using the split_libraries.py script. The remaining sequences were chimera checked using USEARCH (57) and clustered into OTUs using the pick_de_novo_otus.py script with the UCLUST method at a threshold of 97% similarity, and singletons were discarded. The representative sequences of OTUs were taxonomically assigned against the Greengenes database (13_8_release) and aligned by PyNAST against a template alignment of the Greengenes core set (58). Next, archaeal and chloroplast sequences were removed. To reduce the influence of sequencing depth on treatment effects, samples were then randomly sampled again to the same sequence depth, based on the least number of sequences (16,700 sequences per sample).
Alpha-diversity and beta-diversity estimates were calculated using QIIME with multiple indices (number of observed species, Simpson's index) and the Bray-Curtis distance between samples. The overall structure of bacterial communities was ordinated by a PCoA based on Bray-Curtis distance that was carried out using the “pcoa” function of the “ape” package in R (v3.3.2; R Foundation for Statistical Computing). Cluster analysis of samples was performed based on calculated Bray-Curtis dissimilarities between the samples. In addition, PERMANOVA was carried out using adonis function of Vegan to measure effect size and significances on beta-diversity. Pearson's correlations between variables were analyzed using SPSS 18.0. The significantly discriminant taxa in each treatment were determined using the LDA, which employs the factorial Kruskal-Wallis rank sum test (α = 0.05). This analysis identified taxa with significantly different relative abundances between groups (using one-against-all comparisons) (59). Significant taxa were used to generate taxonomic cladograms illustrating differences between the compartments (rhizosphere, bulk soil, and unplanted soil) of two ecotypes under Cd 20 treatment. To find representative taxa associated with the Cd levels, a similarity percentage (SIMPER) analysis was applied to screen OTUs primarily responsible for the overall dissimilarity (contribution more than 0.2% dissimilarity each) in the bacterial communities along the Cd gradient using PAST. The OTUs with abundance significantly correlated with the Cd level (P < 0.05) were selected, and their sequences were aligned in the EzTaxon database (60) to find the closest-matched type strains for taxonomic identification. Heat maps of average relative abundances of the key phylotypes were prepared in the R environment using the “pheatmap” package.
Accession number(s).
The sequences generated in the present study were deposited in the Sequence Read Archive of DDBJ (http://www.ddbj.nig.ac.jp) and are available under accession number DRA005760.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by projects from the National Natural Science Foundation of China (41401366 and 31370040), the National Key Research and Development Program of China (2017YFD0801303 and 2016YFD0800805), the Zhejiang Provincial Major Research and Development Program of China (2015C03020), Foundation for the Author of National Excellent Doctoral Dissertation of People's Republic of China (201469), and the Fundamental Research Funds for the Central Universities (2018FZA6007).
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02797-17.
REFERENCES
- 1.Bertin G, Averbeck D. 2006. Cadmium: cellular effects, modifications of biomolecules, modulation of DNA repair and genotoxic consequences (a review). Biochimie 88:1549–1559. doi: 10.1016/j.biochi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Luo CL, Shen ZG, Li XD. 2005. Enhanced phytoextraction of Cu, Pb, Zn, and Cd with EDTA and EDDS. Chemosphere 59:1–11. doi: 10.1016/j.chemosphere.2004.09.100. [DOI] [PubMed] [Google Scholar]
- 3.Chen B, Zhang Y, Rafiq MT, Khan KY, Pan F, Yang X, Feng Y. 2014. Improvement of cadmium uptake and accumulation in Sedum alfredii by endophytic bacteria Sphingomonas SaMR12: effects on plant growth and root exudates. Chemosphere 117:367–373. doi: 10.1016/j.chemosphere.2014.07.078. [DOI] [PubMed] [Google Scholar]
- 4.Guerinot ML, Salt DE. 2001. Fortified foods and phytoremediation: two sides of the same coin. Plant Physiol 125:164–167. doi: 10.1104/pp.125.1.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Visioli G, D'Egidio S, Sanangelantoni AM. 2015. The bacterial rhizobiome of hyperaccumulators: future perspectives based on omics analysis and advanced microscopy. Front Plant Sci 5:752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Muehe EM, Weigold P, Adaktylou IJ, Planer-Friedrich B, Kraemer U, Kappler A, Behrens S. 2015. Rhizosphere microbial community composition affects cadmium and zinc uptake by the metal-hyperaccumulating plant Arabidopsis halleri. Appl Environ Microbiol 81:2173–2181. doi: 10.1128/AEM.03359-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sessitsch A, Kuffner M, Kidd P, Vangronsveld J, Wenzel WW, Fallmann K, Puschenreiter M. 2013. The role of plant-associated bacteria in the mobilization and phytoextraction of trace elements in contaminated soils. Soil Biol Biochem 60:182–194. doi: 10.1016/j.soilbio.2013.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhuang X, Chen J, Shim H, Bai Z. 2007. New advances in plant growth-promoting rhizobacteria for bioremediation. Environ Int 33:406–413. doi: 10.1016/j.envint.2006.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Yergeau E, Sanschagrin S, Maynard C, St-Arnaud M, Greer CW. 2014. Microbial expression profiles in the rhizosphere of willows depend on soil contamination. ISME J 8:344–358. doi: 10.1038/ismej.2013.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venturi V, Keel C. 2016. Signaling in the rhizosphere. Trends Plant Sci 21:187–198. doi: 10.1016/j.tplants.2016.01.005. [DOI] [PubMed] [Google Scholar]
- 11.Haichar FeZ, Marol C, Berge O, Rangel-Castro JI, Prosser JI, Balesdent J, Heulin T, Achouak W. 2008. Plant host habitat and root exudates shape soil bacterial community structure. ISME J 2:1221–1230. doi: 10.1038/ismej.2008.80. [DOI] [PubMed] [Google Scholar]
- 12.Philippot L, Raaijmakers JM, Lemanceau P, van der Putten WH. 2013. Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol 11:789–799. doi: 10.1038/nrmicro3109. [DOI] [PubMed] [Google Scholar]
- 13.Wood JL, Zhang C, Mathews ER, Tang C, Franks AE. 2016. Microbial community dynamics in the rhizosphere of a cadmium hyper-accumulator. Sci Rep 6:36067. doi: 10.1038/srep36067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J, Zheng YM, Liu YR, Ma YB, Hu HW, He JZ. 2014. Initial copper stress strengthens the resistance of soil microorganisms to a subsequent copper stress. Microb Ecol 67:931–941. doi: 10.1007/s00248-014-0391-8. [DOI] [PubMed] [Google Scholar]
- 15.Gao Y, Zhou P, Mao L, Zhi YE, Shi WJ. 2010. Assessment of effects of heavy metals combined pollution on soil enzyme activities and microbial community structure: modified ecological dose-response model and PCR-RAPD. Environ Earth Sci 60:603–612. doi: 10.1007/s12665-009-0200-8. [DOI] [Google Scholar]
- 16.Hoostal MJ, Bidart-Bouzat MG, Bouzat JL. 2008. Local adaptation of microbial communities to heavy metal stress in polluted sediments of Lake Erie. FEMS Microbiol Ecol 65:156–168. doi: 10.1111/j.1574-6941.2008.00522.x. [DOI] [PubMed] [Google Scholar]
- 17.Long XX, Zhang YG, Jun D, Zhou Q. 2009. Zinc, cadmium and lead accumulation and characteristics of rhizosphere microbial population associated with hyperaccumulator Sedum alfredii Hance under natural conditions. Bull Environ Contam 82:460–467. doi: 10.1007/s00128-009-9660-5. [DOI] [PubMed] [Google Scholar]
- 18.Thies JE, Singleton PW, Benbohlool B. 1991. Influence of the size of indigenous rhizobial populations on establishment and symbiotic performance of introduced rhizobia on field-grown legumes. Appl Environ Microbiol 57:19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang XE, Long XX, Ye HB, He ZL, Calvert DV, Stoffella PJ. 2004. Cadmium tolerance and hyperaccumulation in a new Zn-hyperaccumulating plant species (Sedum alfredii Hance). Plant Soil 259:181–189. doi: 10.1023/B:PLSO.0000020956.24027.f2. [DOI] [Google Scholar]
- 20.Xiao W, Wang H, Li T, Zhu Z, Zhang J, He Z, Yang X. 2013. Bioremediation of Cd and carbendazim co-contaminated soil by Cd-hyperaccumulator Sedum alfredii associated with carbendazim-degrading bacterial strains. Environ Sci Pollut Res 20:380–389. doi: 10.1007/s11356-012-0902-4. [DOI] [PubMed] [Google Scholar]
- 21.Zhu ZQ, Yang XE, Wang K, Huang HG, Zhang X, Fang H, Li TQ, Alva AK, He ZL. 2012. Bioremediation of Cd-DDT co-contaminated soil using the Cd-hyperaccumulator Sedum alfredii and DDT-degrading microbes. J Hazard Mater 235:144–151. doi: 10.1016/j.jhazmat.2012.07.033. [DOI] [PubMed] [Google Scholar]
- 22.Lu M, Zhang ZZ. 2014. Phytoremediation of soil co-contaminated with heavy metals and deca-BDE by co-planting of Sedum alfredii with tall fescue associated with Bacillus cereus JP12. Plant Soil 382:89–102. doi: 10.1007/s11104-014-2147-0. [DOI] [Google Scholar]
- 23.Li WC, Ye ZH, Wong MH. 2007. Effects of bacteria on enhanced metal uptake of the Cd/Zn-hyperaccumulating plant, Sedum alfredii. J Exp Bot 58:4173–4182. doi: 10.1093/jxb/erm274. [DOI] [PubMed] [Google Scholar]
- 24.Pan F, Meng Q, Luo S, Shen J, Chen B, Khan KY, Japenga J, Ma X, Yang X, Feng Y. 2017. Enhanced Cd extraction of oilseed rape (Brassica napus) by plant growth-promoting bacteria isolated from Cd hyperaccumulator Sedum alfredii Hance. Int J Phytoremediat 19:281–289. doi: 10.1080/15226514.2016.1225280. [DOI] [PubMed] [Google Scholar]
- 25.Hou DD, Wang K, Liu T, Wang HX, Lin Z, Qian J, Lu LL, Tian SK. 2017. Unique rhizosphere micro-characteristics facilitate phytoextraction of multiple metals in soil by the hyperaccumulating plant Sedum alfredii. Environ Sci Technol 51:5675–5684. doi: 10.1021/acs.est.6b06531. [DOI] [PubMed] [Google Scholar]
- 26.Deng DM, Shu WS, Zhang J, Zou HL, Lin Z, Ye ZH, Wong MH. 2007. Zinc and cadmium accumulation and tolerance in populations of Sedum alfredii. Environ Pollut 147:381–386. doi: 10.1016/j.envpol.2006.05.024. [DOI] [PubMed] [Google Scholar]
- 27.Tian S, Lu L, Labavitch J, Yang X, He Z, Hu H, Sarangi R, Newville M, Commisso J, Brown P. 2011. Cellular sequestration of cadmium in the hyperaccumulator plant species Sedum alfredii. Plant Physiol 157:1914–1925. doi: 10.1104/pp.111.183947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu L, Tian S, Yang X, Wang X, Brown P, Li T, He Z. 2008. Enhanced root-to-shoot translocation of cadmium in the hyperaccumulating ecotype of Sedum alfredii. J Exp Bot 59:3203–3213. doi: 10.1093/jxb/ern174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bulgarelli D, Rott M, Schlaeppi K, van Themaat EVL, Ahmadinejad N, Assenza F, Rauf P, Huettel B, Reinhardt R, Schmelzer E, Peplies J, Gloeckner FO, Amann R, Eickhorst T, Schulze-Lefert P. 2012. Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488:91–95. doi: 10.1038/nature11336. [DOI] [PubMed] [Google Scholar]
- 30.Chaparro JM, Badri DV, Vivanco JM. 2014. Rhizosphere microbiome assemblage is affected by plant development. ISME J 8:790–803. doi: 10.1038/ismej.2013.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edwards J, Johnson C, Santos-Medellin C, Lurie E, Podishetty NK, Bhatnagar S, Eisen JA, Sundaresan V. 2015. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc Natl Acad Sci U S A 112:E911–E920. doi: 10.1073/pnas.1414592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shi S, Nuccio E, Herman DJ, Rijkers R, Estera K, Li J, da Rocha UN, He Z, Pett-Ridge J, Brodie EL, Zhou J, Firestone M. 2015. Successional trajectories of rhizosphere bacterial communities over consecutive seasons. mBio 6:e00746-15. doi: 10.1128/mBio.00746-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li T, Xu Z, Han X, Yang X, Sparks DL. 2012. Characterization of dissolved organic matter in the rhizosphere of hyperaccumulator Sedum alfredii and its effect on the mobility of zinc. Chemosphere 88:570–576. doi: 10.1016/j.chemosphere.2012.03.031. [DOI] [PubMed] [Google Scholar]
- 34.Fierer N, Lennon JT. 2011. The generation and maintenance of diversity in microbial communities. Am J Bot 98:439–448. doi: 10.3732/ajb.1000498. [DOI] [PubMed] [Google Scholar]
- 35.Leibold MA, McPeek MA. 2006. Coexistence of the niche and neutral perspectives in community ecology. Ecology 87:1399–1410. doi: 10.1890/0012-9658(2006)87[1399:COTNAN]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 36.Pishchik VN, Vorob'ev NI, Provorov NA, Khomyakov YV. 2016. Mechanisms of plant and microbial adaptation to heavy metals in plant-microbial systems. Microbiology 85:257–271. doi: 10.1134/S0026261716030097. [DOI] [Google Scholar]
- 37.Roane TM, Pepper IL. 1999. Microbial responses to environmentally toxic cadmium. Microb Ecol 38:358–364. doi: 10.1007/s002489901001. [DOI] [PubMed] [Google Scholar]
- 38.Lopes LD, Silva M, Andreote FD. 2016. Bacterial abilities and adaptation toward the rhizosphere colonization. Front Microbiol 7:1341. doi: 10.3389/fmicb.2016.01341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaparro JM, Badri DV, Bakker MG, Sugiyama A, Manter DK, Vivanco JM. 2013. Root exudation of phytochemicals in arabidopsis follows specific patterns that are developmentally programmed and correlate with soil microbial functions. PLoS One 8:e55731. doi: 10.1371/journal.pone.0055731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Badri DV, Quintana N, El Kassis EG, Kim HK, Choi YH, Sugiyama A, Verpoorte R, Martinoia E, Manter DK, Vivanco JM. 2009. An ABC transporter mutation alters root exudation of phytochemicals that provoke an overhaul of natural soil microbiota. Plant Physiol 151:2006–2017. doi: 10.1104/pp.109.147462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Micallef SA, Shiaris MP, Colon-Carmona A. 2009. Influence of Arabidopsis thaliana accessions on rhizobacterial communities and natural variation in root exudates. J Exp Bot 60:1729–1742. doi: 10.1093/jxb/erp053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li WC, Ye ZH, Wong MH. 2010. Metal mobilization and production of short-chain organic acids by rhizosphere bacteria associated with a Cd/Zn hyperaccumulating plant, Sedum alfredii. Plant Soil 326:453–467. doi: 10.1007/s11104-009-0025-y. [DOI] [Google Scholar]
- 43.Luo Q, Sun L, Hu X, Zhou R. 2014. The variation of root exudates from the hyperaccumulator Sedum alfredii under cadmium stress: metabonomics analysis. PLoS One 9:e115581. doi: 10.1371/journal.pone.0115581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang K, Zhang D, Xiong J, Chen X, Zheng J, Hu C, Yang Y, Zhu J. 2015. Response of bacterioplankton communities to cadmium exposure in coastal water microcosms with high temporal variability. Appl Environ Microbiol 81:231–240. doi: 10.1128/AEM.02562-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ellis RJ, Morgan P, Weightman AJ, Fry JC. 2003. Cultivation-dependent and -independent approaches for determining bacterial diversity in heavy-metal-contaminated soil. Appl Environ Microbiol 69:3223–3230. doi: 10.1128/AEM.69.6.3223-3230.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Idris R, Trifonova R, Puschenreiter M, Wenzel WW, Sessitsch A. 2004. Bacterial communities associated with flowering plants of the Ni hyperaccumulator Thlaspi goesingense. Appl Environ Microbiol 70:2667–2677. doi: 10.1128/AEM.70.5.2667-2677.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schrey SD, Tarkka MT. 2008. Friends and foes: streptomycetes as modulators of plant disease and symbiosis. Antonie Van Leeuwenhoek 94:11–19. doi: 10.1007/s10482-008-9241-3. [DOI] [PubMed] [Google Scholar]
- 48.Rajkumar M, Ae N, Prasad MNV, Freitas H. 2010. Potential of siderophore-producing bacteria for improving heavy metal phytoextraction. Trends Biotechnol 28:142–149. doi: 10.1016/j.tibtech.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 49.Watve MG, Tickoo R, Jog MM, Bhole BD. 2001. How many antibiotics are produced by the genus Streptomyces? Arch Microbiol 176:386–390. doi: 10.1007/s002030100345. [DOI] [PubMed] [Google Scholar]
- 50.Alvarez A, Saez JM, Davila Costa JS, Colin VL, Fuentes MS, Cuozzo SA, Benimeli CS, Polti MA, Amoroso MJ. 2017. Actinobacteria: current research and perspectives for bioremediation of pesticides and heavy metals. Chemosphere 166:41–62. doi: 10.1016/j.chemosphere.2016.09.070. [DOI] [PubMed] [Google Scholar]
- 51.Zloch M, Thiem D, Gadzala-Kopciuch R, Hrynkiewicz K. 2016. Synthesis of siderophores by plant-associated metallotolerant bacteria under exposure to Cd2+. Chemosphere 156:312–325. doi: 10.1016/j.chemosphere.2016.04.130. [DOI] [PubMed] [Google Scholar]
- 52.Dimkpa CO, Svatos A, Dabrowska P, Schmidt A, Boland W, Kothe E. 2008. Involvement of siderophores in the reduction of metal-induced inhibition of auxin synthesis in Streptomyces spp. Chemosphere 74:19–25. doi: 10.1016/j.chemosphere.2008.09.079. [DOI] [PubMed] [Google Scholar]
- 53.Sineriz ML, Kothe E, Abate CM. 2009. Cadmium biosorption by Streptomyces sp. F4 isolated from former uranium mine. J Basic Microbiol 49(Suppl 1):S55–S62. doi: 10.1002/jobm.200700376. [DOI] [PubMed] [Google Scholar]
- 54.Lu L, Tian S, Zhang J, Yang X, Labavitch JM, Webb SM, Latimer M, Brown PH. 2013. Efficient xylem transport and phloem remobilization of Zn in the hyperaccumulator plant species Sedum alfredii. New Phytol 198:721–731. doi: 10.1111/nph.12168. [DOI] [PubMed] [Google Scholar]
- 55.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Tumbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Magoc T, Salzberg SL. 2011. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27:2957–2963. doi: 10.1093/bioinformatics/btr507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Caporaso JG, Bittinger K, Bushman FD, DeSantis TZ, Andersen GL, Knight R. 2010. PyNAST: a flexible tool for aligning sequences to a template alignment. Bioinformatics 26:266–267. doi: 10.1093/bioinformatics/btp636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, Huttenhower C. 2011. Metagenomic biomarker discovery and explanation. Genome Biol 12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon YS, Lee JH, Yi H, Won S, Chun J. 2012. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721. doi: 10.1099/ijs.0.038075-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.