ABSTRACT
Lettuce has been implicated in human norovirus (HuNoV) outbreaks. The virus is stable on the leaf surface for at least 2 weeks; however, the dynamics of virus internalization have not been fully investigated. The purpose of this study was to assess the internalization and distribution of HuNoV and two surrogate viruses, porcine sapovirus (SaV) and Tulane virus (TV), in lettuce and spinach. Viral inoculations through the roots of seedlings and the petiole of leaves from mature plants were performed, and the viruses were tracked on days 1 and 6 post-root inoculation and at 16 h and 72 h post-petiole inoculation. Confocal microscopy was used to visualize root-internalized HuNoV. In both lettuce and spinach, (i) HuNoV was internalized into the roots and leaves at similar RNA titers, whereas surrogate viruses were more restricted to the roots, (ii) all three viruses were stable inside the roots and leaves for at least 6 days, and (iii) HuNoV disseminated similarly inside the central veins and leaf lamina, whereas surrogate viruses were more restricted to the central veins. Infectious TV, but not SaV, was detectable in all tissues, suggesting that TV has greater stability than SaV. HuNoV was visualized inside the roots' vascular bundle and the leaf mesophyll of both plants. In conclusion, using surrogate viruses may underestimate the level of HuNoV internalization into edible leaves. The internalization of HuNoV through roots and cut leaves and the dissemination into various spinach and lettuce tissues raise concerns of internal contamination through irrigation and/or wash water.
IMPORTANCE Human noroviruses are the leading cause of foodborne outbreaks, with lettuce being implicated in the majority of outbreaks. The virus causes acute gastroenteritis in all age groups, with more severe symptoms in children, the elderly, and immunocompromised patients, contributing to over 200,000 deaths worldwide annually. The majority of deaths due to HuNoV occur in the developing world, where limited sanitation exists along with poor wastewater treatment facilities, resulting in the contamination of water resources that are often used for irrigation. Our study confirms the ability of lettuce and spinach to internalize HuNoV from contaminated water through the roots into the edible leaves. Since these leafy greens are consumed with minimal processing that targets only surface pathogens, the internalized HuNoV presents an added risk to consumers. Thus, preventive measures should be in place to limit the contamination of irrigation water. In addition, better processing technologies are needed to inactivate internalized viral pathogens.
KEYWORDS: human norovirus, porcine sapovirus, Tulane virus, lettuce, spinach, internalization, confocal microscopy, tissue distribution, water contamination, leafy greens
INTRODUCTION
Human norovirus (HuNoV) is the most common cause of viral gastroenteritis outbreaks globally in people of all ages. Gastrointestinal symptoms (diarrhea, vomiting, stomach pain, and nausea) of HuNoV infections are self-limiting in healthy adults but can be severe and prolonged in young children, the elderly, and immunocompromised patients (1). The global economic burden of HuNoV infections has been estimated to be ∼$64 billion in direct (health care) and indirect (loss of productivity) costs (2). In the United States, HuNoV is the leading cause of foodborne illness (58%), causing an estimated 71,000 hospitalizations and 570 to 800 deaths annually and over $700 million in health care costs (3). However, in developing countries, it is estimated that up to 200,000 deaths occur annually in children under 5 years of age due to HuNoV infections (4). In spite of the HuNoV global disease burden, research on disease transmission and effective control strategies has been historically hampered due to the lack of a routine cell culture system, although promising advances have been reported in 2016 for culturing the virus in human intestinal stem cell-derived enteroids (5). Much of our understanding of HuNoV in the environment has been based on human volunteer studies, the use of culturable surrogate caliciviruses, such as feline calicivirus (FCV), murine norovirus (MNV), porcine sapovirus (SaV), and nonhuman primate Tulane virus (TV), and various molecular tools to estimate virus infectivity, inactivation, and persistence in the environment.
Human noroviruses are small (28 to 35 nm in diameter) nonenveloped single-stranded RNA viruses that belong to the Caliciviridae family. Human norovirus has been referred to as the perfect pathogen because of its (i) low infectious dose (estimated at 18 to 1,000 viral particles in one study [6] and at 1,320 genomic equivalents [GE] in another [7]), (ii) environmental stability (remains infectious at freezing temperature, above 60°C, and on surfaces for 2 weeks), (iii) resistance to common disinfectants (1, 6, 8), (iv) no long-term protective immunity (9), (v) high genetic diversity leading to the emergence of new pandemic strains with distinct antigenicity every 2 to 4 years, and (vi) multiple modes of transmission, including direct person-to-person transmission or indirect transmission via fecal or vomit contamination of food, water, surfaces, and the environment (10, 11). In the absence of approved antivirals or vaccines against HuNoV, the virus continues to be a significant burden globally.
Leafy greens are frequently associated with HuNoV outbreaks and are globally recognized to be a high priority in terms of the microbial safety of fresh produce (12). Reported HuNoV outbreaks in leafy greens have been mainly linked to the genogroup II genotype 4 (GII.4) strains (13–16). Contamination of leafy greens with HuNoVs can occur at any stage along the farm-to-fork chain through a number of sources, including feces-contaminated water used for irrigation or processing, improperly treated sewage sludge used for fertilization, and asymptomatically HuNoV-infected food harvesters or food handlers who do not follow proper hygiene practices (12, 17). In the United States between 2009 and 2012, 70% of HuNoV outbreaks occurring in restaurants were due to improper hygienic practices of food workers, and leafy greens contributed to about 30% of the outbreaks when a specific food was implicated (1). In Canada, 28.2% of the leafy greens (mostly imported from the United States but packaged in Canada) sampled from supermarkets were found to be contaminated with HuNoV (18). Since leafy greens are consumed raw or minimally processed, the elimination of HuNoV prior to retail is vital to protect consumers.
On the farm, because HuNoV is known to survive in water for long periods, contaminated irrigation water can be a source of HuNoV on leafy greens (12, 19). In one study, HuNoV remained infectious for >60 days in water, as assessed by challenging human volunteers, while other studies reported the detection of HuNoV RNA for >2 years in water (reviewed in reference 20). HuNoV contaminating leafy green surfaces has been found to be stable for at least 2 weeks (21). This is a critical duration of time because mature leafy greens are marketable within 2 weeks. Multiple factors have been found to enhance the virus survival on the leaf surface, including the presence of phytopathogens, physical damage, and/or the natural phyllosphere bacteria (21–23). Furthermore, although multiple studies investigated the possibility of HuNoV internalization to the edible leaves through the roots, the studies either were based solely on the use of surrogate viruses or reported conflicting results under similar settings or for the same produce. One research team has used HuNoV in their studies and found significant internalization of the virus in lettuce and green onion under hydroponic growth conditions (24, 25), while another team failed to detect any internalization of HuNoV in lettuce under hydroponic or soil growth conditions (26). This discrepancy, in addition to the fact that different surrogate viruses have been found to exhibit various degrees of similarity to HuNoV on leafy greens (21, 27), necessitates a better assessment of the internalization dynamics of HuNoV in leafy greens, especially lettuce. Therefore, the objectives of this study were to (i) investigate whether lettuce and spinach can internalize HuNoV into the edible leaves, (ii) simultaneously assess the behavior of surrogate viruses TV and SaV in comparison with that of HuNoV, and (iii) determine whether HuNoV particles can be visualized inside lettuce and spinach tissues. We selected SaV and TV in this study because, like HuNoV, both viruses cause gastroenteritis in their corresponding host species (28, 29). Also, like HuNoV, TV binds to carbohydrates of histo-blood group antigens (HBGAs) (30). Although SaVs do not bind to HBGA (31) (rather, they bind to sialic acids on gangliosides and on glycoproteins), they are important to study on their own, as they serve as surrogates for human sapoviruses, which are also important foodborne pathogens (32, 33).
RESULTS
Transport of HuNoV, SaV, and TV through lettuce and spinach roots.
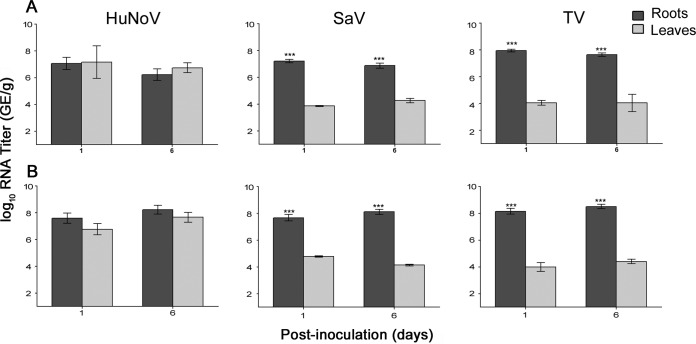
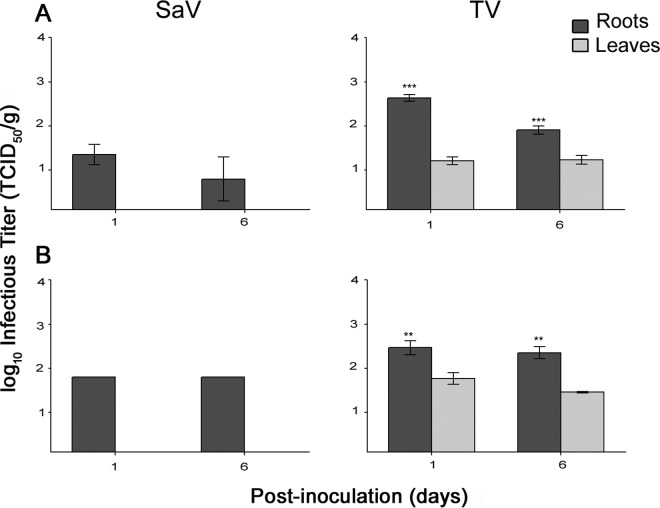
Analysis of reverse transcription-quantitative PCR (RT-qPCR) data showed that HuNoV was internalized through the roots of lettuce and spinach plants and was detected in their leaves for at least 6 days postinoculation (Fig. 1, left graphs). Both surrogate viruses, SaV and TV, were internalized into the leaves of lettuce and spinach (Fig. 1, middle and right graphs for SaV and TV, respectively). On days 1 and 6, there were no significant differences between HuNoV RNA titers in leaves and those in the roots in both lettuce and spinach plants. In contrast, on days 1 and 6, the viral RNA titers for both TV and SaV were significantly higher in the roots of lettuce and spinach than in their corresponding leaves. In addition, there were no significant changes in SaV, TV, or HuNoV RNA titers in roots or leaves from day 1 to day 6. Infectious SaV was detected inside lettuce and spinach root samples only, whereas infectious TV was detected in both the leaves and roots of both plants (Fig. 2). Again, infectious TV titers in both lettuce and spinach roots were significantly higher than those in leaves.
FIG 1.
RNA titers of HuNoV, SaV, and TV internalized through the roots into the leaves of lettuce (A) and spinach (B) (asterisks indicate significant differences in RNA titers between roots and leaves; see Materials and Methods). The assay detection limit was 2.69 log10 GE/g.
FIG 2.
Infectivity (in 50% tissue culture infective dose per gram) of SaV and TV internalized through the roots into the leaves of lettuce (A) and spinach (B) (asterisks indicate significant differences in RNA titers between roots and leaves; see Materials and Methods). The assay detection limit was 0.3 log10 GE/g.
Transport of HuNoV, SaV, and TV through the petiole of lettuce and spinach leaves.
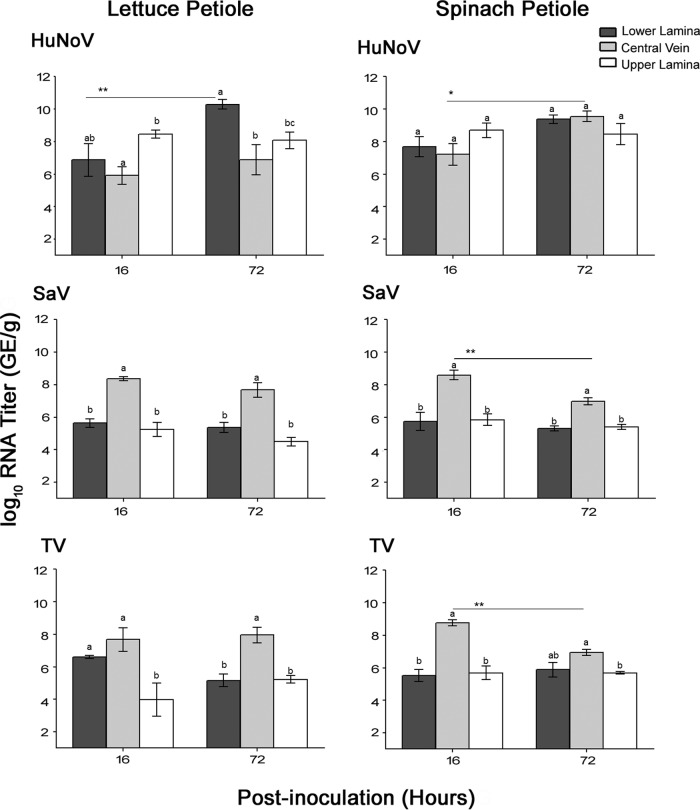
Analysis of RT-qPCR data showed that HuNoV was detected inside the central vein and the upper and lower leaf lamina in both lettuce and spinach plants at 16 h and 72 h postinoculation (Fig. 3, top graphs). The RNA titers of HuNoV increased in the lower lamina of lettuce and in the central veins of spinach at 72 h. In contrast, both SaV and TV showed significantly higher RNA titers in the central veins, especially in comparison to the upper lamina in both lettuce and spinach plants at 16 h and 72 h (Fig. 3, middle and bottom graphs). In lettuce, the RNA titers of SaV and TV in the central veins did not change significantly over the 72-h period, suggesting viral persistence in the central veins. In contrast, in spinach, the RNA titers of SaV and TV decreased significantly in the central veins at the end of 72 h, without any significant changes to the viral RNA titers in the upper or lower leaf lamina, suggesting that these viruses die off in the central veins rather than transfer to the nearby tissues. Accordingly, TV and SaV may be more restricted than HuNoV to the central veins.
FIG 3.
RNA titers (in genomic equivalents per gram) of HuNoV, SaV, and TV internalized through the petiole of lettuce and spinach plants into their central veins and leaf lamina (lower and upper portions). Asterisks indicate significant differences in RNA titers between time points (see Materials and Methods), and different letters indicate significant differences among tissues. The assay detection limit was 2.69 log10 GE/g.
Infectious SaV was detected inside the central veins of lettuce and spinach samples only at 16 h (1.80 ± 0.28 and 1.75 ± 0.30 50% tissue culture infective dose [TCID50]/g, respectively). In contrast, infectious TV was detected in all tissues of lettuce and plants over the 72-h period. In lettuce at 72 h, TV showed significantly higher infectivity titers in central veins (1.8 ± 0.42 TCID50/g) than in the lower lamina (1.45 ± 0.1 TCID50/g) but not the upper lamina (1.62 0.25 TCID50/g). In spinach at 72 h, TV showed significantly higher infectivity titers in the central veins (1.9 ± 0.26 TCID50/g) than in both the upper (1.37 ± 0.12 TCID50/g) and lower (1.37 ± 0.22 TCID50/g) lamina.
Visualization of HuNoV inside lettuce and spinach tissues.
In lettuce, using confocal microscopy, HuNoV viral particles were detected inside the roots' vascular bundles and in the tissue surrounding them (Fig. 4F). In the leaves, the virus was observed inside the leaf mesophyll between the secondary vascular bundles (Fig. 4C). A similar trend was also observed in spinach leaves and roots (Fig. 4I and L). Vacuum-infiltrated lettuce plants showed the virus to be mostly on the edges of the leaves (Fig. 4B), and the virus was not conclusively detectable in the vascular bundles of roots (Fig. 4E), suggesting inefficient infiltration of the virus under vacuum to the internal tissues of lettuce leaves and roots. A similar trend was also observed for vacuum-infiltrated spinach leaves (Fig. 4H); however, HuNoV particles were detected in the vascular bundles of the roots (Fig. 4K). Negative-control plants (non-HuNoV inoculated) showed no nonspecific binding of the primary or secondary antibodies to any of the leaf or root tissues (Fig. 4A, D, G, and J).
FIG 4.
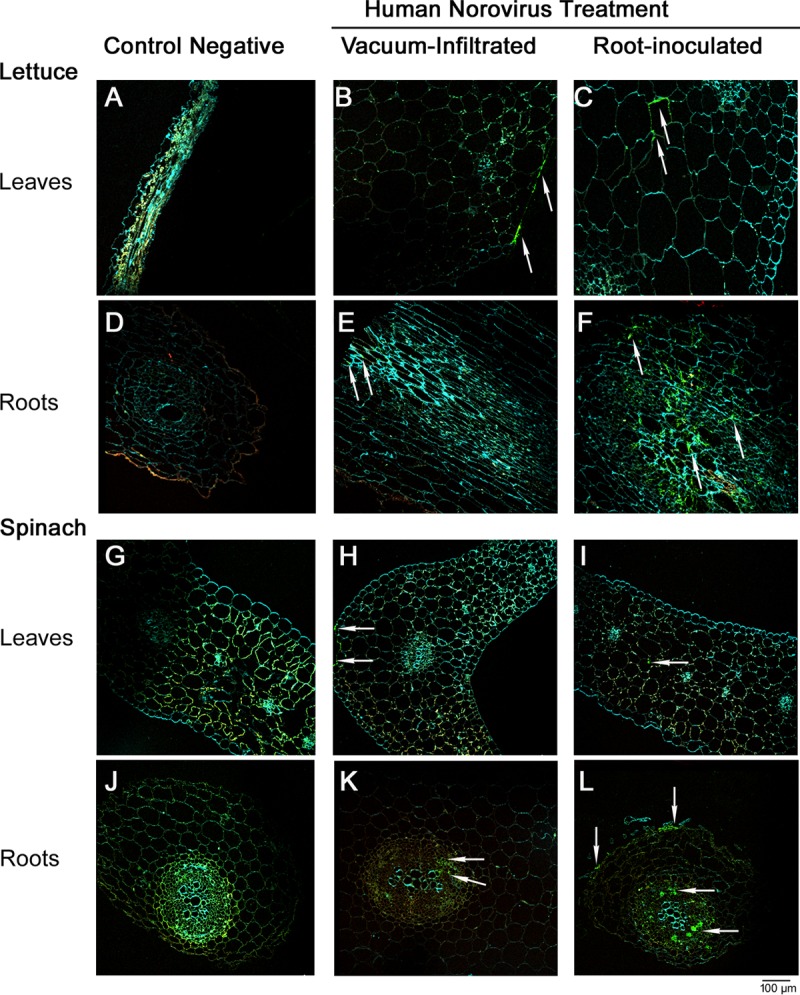
(B, C, E, and F) Confocal microscopy images showing HuNoV inside leaves of lettuce plants that were vacuum infiltrated (B) or root inoculated with the virus (C) and their corresponding roots (E and F). (H, I, K, and L) HuNoV inside leaves of spinach plants that were vacuum infiltrated (H) or root inoculated with the virus (I) and their corresponding roots (K and L). (A, D, G, and J) Negative-control lettuce leaves (A) and roots (D) and spinach leaves (G) and roots (J). Examples of immunofluorescence associated with HuNoV detection are indicated by arrows.
DISCUSSION
Irrigation water can be a source of human viral pathogens on produce (34–36). The presence of HuNoV in river water used for irrigation has been demonstrated in numerous studies worldwide (reviewed in reference 36). Because the majority of deaths associated with human norovirus worldwide occur in developing countries, where there is limited or even no treatment of wastewater before the waste is discharged into river water, and because HuNoV is shed in high titers (∼1011 GE/ml of stool) in stools for a long time (7), the contamination of produce with HuNoV via water constitutes a serious risk, given the long survival time of the virus in water (reviewed in reference 20).
Previous studies have investigated the potential surface or internal contamination of leafy greens with HuNoV due to irrigation water; however, the majority of studies utilized one or two of the HuNoV surrogate viruses instead of the pathogen itself. The only study that compared the survival of surrogate viruses (MNV, SaV, and TV) to HuNoV on the surface of leafy greens (lettuce and spinach) under preharvest conditions found that these viruses did not mimic HuNoV, which remained stable, whereas the surrogates were inactivated over time (21). In addition, the use of HuNoV surrogate viruses to study internalization in leafy greens revealed that MNV and TV differed significantly in the degree of internalization inside leafy greens (25, 37, 38). Therefore, because the potential viral internalization differs between surrogate viruses and between different plants, it is important to assess HuNoV internalization in leafy greens using the viral pathogen itself.
Furthermore, studies from two laboratories using HuNoV (GII.4) reported conflicting results regarding the internalization of the virus in lettuce. One study reported no root internalization of HuNoV in lettuce seedlings (10 days of age) or in commercial lettuce grown in soil or under hydroponic culture conditions (26). In the same study, the surrogate virus canine calicivirus (CaCV) did not mimic HuNoV behavior and was shown to be internalized infrequently through the roots into the leaves of lettuce seedlings (26). In contrast, another laboratory reported the internalization of HuNoV through the roots into the leaves of lettuce seedlings (3 weeks of age) and in 2-month-old green onions (24, 25). The authors maintained their plants under laboratory conditions of 40% relative humidity, which may have enhanced the passive internalization of the virus, driven by increased transpiration rate of the plants under low relative humidity, as was shown for MNV in lettuce (39).
In our study, we used the same genotype of HuNoV (GII.4) as in the two previous studies and found that internalization of HuNoV occurred in both lettuce and spinach seedlings (5 weeks of age) that were incubated in a biosecurity chamber under conditions optimized for plant growth with 60 to 70% relative humidity. The discrepancy with the results of the first study may be due to the use of very young lettuce seedlings (10 days), which would have little root biomass, and due to the lower viral inocula used (1.7 × 105 or 106 GE/ml in 1, 3, or 50 ml of feed water, i.e., a maximum of 5 × 107 GE/plant) than the inoculum in ours (5 × 108 in 3 ml of feed water, i.e., 1.3 × 109 GE/plant) and the second study (6.7 × 106 GE/ml in 800 ml of feed water, i.e., ∼5 × 109 GE/plant). In fact, during our preliminary analyses, inoculation of lettuce and spinach (5 weeks of age) roots with 5 × 107 GE/ml in 3 ml feed water (absorbed completely in 3 days) resulted in no internalization into the leaves of lettuce seedlings (0/6), while only one spinach seedling showed internalization into the leaves (1/6), despite the virus being detected inside the roots of both plants at 5 ± 0.6 log10 GE/g for lettuce and at 6 ± 0.2 log10 GE/g for spinach (data not shown). This is consistent with the results from a previous study reporting that MNV at low viral inoculum (5 × 105 GE/ml in 50 ml feed water, i.e., ∼2 × 107 GE/plant) showed no internalization of infectious MNV in the leaves of lettuce plants (3 weeks of age), while at a higher inoculum (5 × 108 GE/ml in 50 ml feed water, i.e., ∼2 × 1010 GE/plant), internalization of infectious MNV was successful (39). Therefore, we increased the HuNoV inoculum level to ∼108 GE/ml to enhance the detection and visualization of internalized HuNoV. Although the RNA titer of the HuNoV inoculum seems high, it is less than that of SaV and TV (∼1010 GE/ml, equivalent to ∼106 TCID50/ml) and like these viruses may not translate to similar infectivity titers due to the inherent presence of defective and damaged particles that do not have infectivity.
Internalization may also depend on the virus and the plant type, as TV at a low inoculum (104 GE/ml in 400 ml, i.e., 4 × 106 GE/plant) or a high inoculum (106 GE/ml in 400 ml, i.e., 4 × 108 in total feed water) was not detected in the leaves of green onions despite being detected in their roots (24). Furthermore, although washing the roots during the experimental setup may have caused minimal root damage and enhanced virus uptake, internalization of viruses in our study was not solely based on passive root uptake, as was evidenced by the different patterns of internalization for HuNoV and its surrogates SaV and TV. Taken together, we confirmed that HuNoV can be internalized not only in lettuce but also in spinach plants through their roots; however, the level of internalization is dependent on the magnitude of the initial contamination.
The dynamics of viruses in roots in comparison to leaves were not always addressed in previous studies investigating viral internalization in leafy greens under hydroponic growth conditions (26, 37–39). In studies where internalized viruses were quantified in the roots, a gradient effect (i.e., higher viral titers in roots versus leaves) was reported for SaV in lettuce and for bacteriophage F2 in corn and beans (40, 41), whereas similar viral titers in roots versus leaves were reported for MNV in microgreens, for HuNoV, MNV, and TV in lettuce, and for HuNoV in green onions (24, 25, 37). The studies of Yang et al. in 2018 (24) and DiCaprio et al. in 2012 (25) and 2015 (37) are consistent with our findings for HuNoV in lettuce and spinach roots versus leaves but are in contrast with our finding that, like SaV, TV is more restricted to the roots. The difference observed in TV distribution in lettuce may be due to different volumes of feed water (3 ml versus 800 ml) and/or aeration versus static status of the feed water used. Continuous circulation of the feed water may have increased the chances of passive viral uptake by the roots until saturating the leaves. However, large volumes of feed water (12 liters) and aeration were used in the corn and bean study, which showed gradient distribution of bacteriophage F2 in these plants. It may be then that plant age affected the TV distribution pattern, as we used lettuce at 5 weeks of age, whereas DiCaprio et al. (25) used lettuce at 3 weeks of age.
To further assess the differences observed in our root internalization results between the HuNoV and its surrogates, we subjected leaves taken from mature plants to virus inoculation at the base (lettuce) or the petiole of the leaves (spinach). Our results confirmed that HuNoV behaved differently from SaV and TV. HuNoV was able to unload to leaf lamina and exhibited a tendency to accumulate in both spinach and lettuce leaves, whereas both surrogate viruses were more restricted to central veins and exhibited die-off at 72 h in spinach. This die-off is likely due to the degradation of capsid and genomic RNA for SaV and TV in spinach central veins, as RNA titers decreased significantly at 72 h. Proteomic analysis has shown that some of the proteins in the xylem sap (∼12%) have proteolytic functions involved in the defense against pathogen infections (42). These proteases may differ between lettuce and spinach xylem sap, to which different viruses (SaV and TV) may show different susceptibilities. HuNoV has been previously shown to have higher stability than SaV and TV on lettuce and spinach leaves (21), which may explain its accumulation in certain tissues of lettuce and spinach. These differences in the behavior of surrogate viruses inside leaves of different plants highlight the importance of not extrapolating results from one plant to another, especially when the results are based on surrogate viruses. Therefore, the surrogate viruses SaV and TV showed gradient distribution from the roots to the leaves in our root experiment, in part due to their limited ability to unload to the leaf lamina, which may have blocked further uptake from the roots to the leaves. In contrast, HuNoV showed less restriction in their transfer between central veins to the leaf lamina, which may have promoted viral uptake to the leaves, resulting in levels similar to those in the roots.
Understanding viral distribution within the different plant tissues has implications for sampling and interpretations of results. For example, Urbanucci et al. took the leaf samples from commercial lettuce heads at the base of the leaf but failed to detect HuNoV, although CaCV was detectable (26). Similarly, Wei et al. took 50 mg of leaf samples (unknown location on the leaves) from 3-week-old lettuce but found infrequent internalization in spite of using high MNV inocula (108 GE/ml in 50 ml of feed water for 24 h) (39). In another study, positive internalization in hydroponic static culture was found when whole green onion (10 and 60 days old) and spinach (10 days old), i.e., including roots, were processed for MNV and hepatitis A virus (38). In summary, the use of surrogate viruses alone may underestimate the magnitude of edible leaf contamination due to their different translocation pattern compared with that of HuNoV. It is important to understand how and where HuNoV is internalized inside leafy greens, not only to address technical issues of sampling and result interpretations but also to devise better quantitative risk analysis models and processing technologies that take into consideration initial viral contamination levels, plant type, and the levels of internalized viruses from roots to edible tissues.
Confocal microscopy showed HuNoV inside the vascular bundle in roots of both lettuce and spinach, confirming that the virus is transported through the vasculature into the leaves. The plant vasculature is made up of xylem and phloem. Xylem carries water and micronutrients into the phloem, while phloem unidirectionally transports nutrients to various organs of the plant. Therefore, HuNoV can be transported via water inside xylem vessels, which is consistent with studies finding the surrogate viruses SaV (40) and CaCV in lettuce xylem exudates (26). Although, based on RNA data, HuNoV was internalized similarly in roots and leaves, our observation of ample HuNoV signal in roots compared with less frequent detection of the virus in the leaves is in part due to the large surface area of the leaves versus that of the roots and the technical difficulty in embedding large sections of leaves. This leads to only one small section (∼2 by 2 mm) from each leaf being embedded in one capsule, from which semithin sections of 1 μm are made, making the detection of the virus in the leaves more labor-intensive than in the roots. Nevertheless, we were able to detect HuNoV inside the leaves of both plants. Knowing that surrogate viruses in our root experiments were detected at lower titers than HuNoV in the leaves, we attempted to detect SaV by embedding the leaves in paraffin blocks, which can be cut into thicker sections (5 μm); however, we failed to detect the virus, and paraffin embedding resulted in poor structural integrity of the leaves. Finally, the detection of HuNoV inside leaf tissues by polyclonal antibody suggests that the antigenicity of the virus is preserved inside the leaves. This, in addition to the RNA extraction and detection of the viral RNA following RNase treatment, suggests that internalized HuNoVs in our study were intact and potentially infectious. Future studies will allow the confirmation of HuNoV infectivity by using the newly developed HuNoV cell culture system using human intestinal stem cell-derived enteroids (5).
In conclusion, we confirmed and visualized the internalization of HuNoV in both lettuce and spinach from the roots to the leaves. The viral particles were stable in both tissues at similar titers over the 6-day study period. Also, the virus was readily transported from the central veins to the leaf lamina, with the ability to accumulate inside leaves. Neither TV nor SaV entirely mimicked the HuNoV internalization pattern. Both showed more restriction than HuNoV to the roots and the central veins of leaves. TV may be a better surrogate than SaV to estimate HuNoV infectivity, as infectious TV, but not SaV, was detectable inside the leaves of both lettuce and spinach. Internalized HuNoV in both plants is potentially infectious, as was evident by the detection of viral RNA after RNase treatment and the detection of viral antigens by polyclonal antibodies. Our study emphasizes the need for better preventive and postharvest control measures to limit HuNoV contamination of leafy greens and to protect public health.
MATERIALS AND METHODS
Preparation of viruses.
Cell culture propagation of SaV and TV was performed as described previously (27) using the following cell lines: an LLC-porcine kidney cell line (LLC-PK1, ATCC CL-101) and an LLC-monkey kidney cell line (LLC-MK2, ATCC CCL-7), respectively. The HuNoV used in this study was HS194 strain (GenBank accession no. GU325839), belonging to genogroup II genotype 4 (GII.4) (43), and its preparation was described in our previous work (21). Virus titers were assayed using reverse transcription-quantitative PCR (RT-qPCR) and infectivity assays for surrogate viruses (as described below). The titer of HuNoV RNA used in subsequent experiments was 8.6 ± 0.1 log10 genomic equivalent (GE)/ml. The infectivity titers of SaV and TV used were 5.5 ± 0.2 and 6.0 ± 0.3 log10 50% tissue culture infectious dose (TCID50)/ml, respectively, corresponding to viral RNA titers of 10.3 ± 0.2 and 9.8 ± 0.1 log10 GE/ml, respectively.
Plant cultivation.
Seeds of romaine lettuce cultivar ‘Tall Guzmaine Elite’ (Siegers Seed Co., Holland, MI) and baby's leaf hybrid spinach (Burpee, Warminster, PA) were grown in 200-cell trays containing Fafard super-fine germinating mix (Conrad Fafard, Agawam, MA) under greenhouse conditions of ∼23°C daytime/15°C nighttime, with a 12-h photoperiod (40). At 2 weeks of age, spinach and lettuce seedlings were transferred to 15-cm-diameter pots containing sterile soil (Wooster sandy loam) and were fertilized biweekly using Osmocote slow-release fertilizer. The plants were watered twice a day using an overhead irrigation sprinkler hose until they were used in subsequent experiments.
Inoculation of plants with viruses.
For the inoculation of plants, the fecal suspension of HuNoV and the cell lysates of SaV and TV were buffer exchanged to sterile water (to mimic contamination through irrigation water) using Amicon 100K Ultra-15 centrifugal devices (Millipore, Billerica, MA) at 4,000 × g for 0.5 h at 4°C, as described previously (27). Viruses were suspended in sterile water and immediately used in plant inoculations.
Viral inoculation through the roots.
To study HuNoV transport from contaminated roots to leaves, lettuce and spinach seedlings at 5 weeks of age were used, and the soil was removed from the roots by gentle washing with sterile water. Direct inoculation of the virus through the roots was followed to assess internalization, as it mimics contamination through a hydroponic culture and was used in order to compare our results to those of the previous two studies that reported conflicting internalization results for HuNoV in hydroponically grown lettuce. Roots were submerged in a 3-ml viral solution supplemented with a 1% antibiotic-antimycotic cocktail (Invitrogen, Carlsbad, CA, USA) in glass tubes. Seedling leaves were protected from cross-contamination by covering the tube openings with a Parafilm wrap. All treated seedlings, along with mock-treated (sterile water) control seedlings, were incubated inside biosafety level 2 plant growth chambers under controlled conditions (12-h photoperiod, 20°C daytime/15°C nighttime temperatures, and ∼60 to 70% relative humidity). Between days 1 and 3, all plants absorbed the viral solution or sterile water; then, plants were transferred into 15-ml Falcon tubes filled with sterile water supplemented with a 1% antibiotic-antimycotic cocktail. Water was replenished as needed. Sampling of leaves and roots was performed on postinoculation days (PID) 1 and 6. Samples were processed as described below.
Viral inoculation through the petiole.
To study HuNoV dissemination inside leaves, mature lettuce and spinach at 8 and 6 weeks of age, respectively, were submerged in water to cut their leaves without blocking the vascular system, as described previously for the vascular transport of exogenous proteins (44). Leaves were cut at the base, and immediately, the petiole/base (at ∼1 cm from cut edge) was immersed into viral solutions inside Whirl-Pak bags and incubated in the growth chamber for 16 h and 72 h at 60 to 70% relative humidity. Samples were processed as described below.
Recovery efficiency of infiltrated HuNoV, SaV, and TV.
This experiment was conducted to assess the recovery efficiency of internalized viruses in lettuce and spinach. Leaves of mature lettuce and spinach plants (weeks 8 and 6, respectively) were infiltrated with viruses by gently pressing a 1-ml needless syringe filled with 200 μl of SaV (1010 GE/ml), TV (1010 GE/ml), and HuNoV (108 GE/ml) onto the abaxial side. For spinach, whole leaves (of approximate surface area similar to a 3-cm diameter circle) were used, since a 200-μl dose diffused through the entire leaf area. The infiltration area in lettuce leaves was limited to a marked circle of ∼3 cm in diameter. One set of infiltrated leaves was detached from the plants and processed for viral quantification (as described below), while the second set was immediately immersed at the petiole/base in sterile water supplemented with a 1% antibiotic-antimycotic cocktail inside Whirl-Pak bags and incubated in the biosafety level 2 plant growth chambers for 16 h. Leaves were then processed as described below. The recovery efficiency was calculated for each virus by subtracting the viral titers obtained at 16 h from those at 0 h. For HuNoV, SaV, and TV, there were no significant differences in recovery loss among the viruses and between lettuce and spinach at 16 h (∼0.5, 0.3, and 0.4 log10 GE/g of lettuce tissues versus 0.2, 0.1, and 0.1 log10 GE/g of spinach tissues, respectively).
Processing of plant tissue for virus detection.
At each harvesting time point, samples (all leaves/plant or all roots/plant) were weighed and subjected to a chlorine wash (1,000 ppm chlorine at room temperature [RT] for 5 min, followed by rinsing in sterile water and washing in 0.25 M sodium thiosulfate to neutralize residual chlorine) as described previously to inactivate any viruses on the surface (25). The samples were allowed to dry for 10 min. For the petiole experiment, plant leaves were further sectioned into three parts, the central veins (including petiole in the case of spinach) and leaf lamina that was further cut into upper and lower halves. All samples were further cut into small pieces (0.5 cm by 0.5 cm) and suspended in elution buffer containing minimal essential medium (MEM) with a 1% antibiotic-antimycotic cocktail (Invitrogen, Carlsbad, CA, USA) and 2% heat-inactivated (60°C for 1 h) fetal bovine serum (FBS), as described previously (40). Homogenization was performed on ice using a Polytron homogenizer (Cole-Parmer Instruments, USA) at maximum speed for 1 min. The samples were centrifuged twice at 3,724 × g for 10 min to remove plant debris. The supernatants were used for viral RNA and infectivity assays as described below.
Infectivity assays.
Viruses were titrated for TCID50 using their respective cell lines cultured in 96-well plates, as described previously (27). Briefly, 1- to 2-day-old confluent cell monolayers in 96-well plates were infected in quadruplet with serially diluted (1:4 or 1:10 dilution in the respective cell culture media supplemented with a 1% antibiotic-antimycotic cocktail) samples and incubated at 37°C. The plates were stained using an immunohistochemistry protocol described previously for SaV (45). Hyperimmune serum against TV was generated using guinea pigs according to an approved institutional animal care and use committee (IACUC) protocol (46). The wells with infected cells were scored as positive, and the viral titers were calculated using the Reed-Muench equation for the calculation of TCID50 (47). The infectivity assay's detection limit was 0.3 log10 TCID50/ml.
RNase treatment, RNA extraction, and quantification by RT-qPCR.
A 200-μl aliquot from each sample was treated with RNase (0.5 μg/μl; Invitrogen) for 1 h at 37°C to inactivate free RNA, including naked viral RNA, and then immediately subjected to RNA extraction using RNeasy minikit (Qiagen, Valencia, CA, USA). A sterile water sample was extracted with every run to serve as an RNA extraction control in addition to virus negative-control plant samples. The RNA was eluted in 50 μl of nuclease-free water and stored at −20°C. One-step TaqMan virus-specific RT-qPCR was used to estimate the viral RNA titers in plant samples, as described previously for HuNoV, SaV, and TV (28, 45, 48). Each sample was tested in duplicate. An internal RNA control was spiked into randomly selected samples to check for RT-PCR inhibitors, as described previously (45). None of the RNA samples showed any PCR inhibitors. The detection limit of the RT-qPCR for HuNoV, SaV, and TV was 10 GE per 20-μl reaction mixture, as reported in our previous work (45), which is equivalent to 2.69 log10 GE/g.
HuNoV visualization in plant tissues.
HuNoV inoculation of lettuce and spinach seedlings was performed through the roots as described above. The plants were incubated in the growth chamber for 16 h. Negative-control plants were treated similarly but without the addition of HuNoV. Small pieces (5 by 5 mm) of leaves and roots from each plant were vacuum infiltrated with HuNoV for 2 h, potentially serving as a positive control.
Fixation and embedding.
At 16 h postinoculation, plant roots and leaves were cut into small pieces (∼2 mm by 2 mm) and immediately immersed in fixative solution (4% formaldehyde, 2% glutaraldehyde in 1× PBS). The root sections were taken just below the stem, whereas random pieces were taken from the leaves. The tissues were allowed to transpire the fixative solution under vacuum infiltration, which was maintained for ∼2 h until all the plant pieces sunk to the bottom of the tubes. The plant tissues were then incubated for 16 h at room temperature with gentle rocking. The protocol of Bell et al. (49) was followed, with minor modifications. Briefly, the sections were washed in 1× PBS for 10 min before dehydration in a graded ethanol series (50% [vol/vol], 70% [vol/vol], 90% [vol/vol] and 100% [vol/vol] twice, each for 1 h). The tissues were infiltrated in a medium-grade LR (London Resin) white (Electron Microscopy Sciences, Hatfield, PA) at 1:1, 1:2, and 1:3 ratios of 100% ethanol to resin for 1 h each before 1-h changes in 100% LR. All steps were performed at room temperature. The tissue sections were then transferred to gelatin capsules (Electron Microscopy Sciences) and allowed to polymerize at 50°C for 24 h. Ten capsules (each capsule containing one tissue section) were made for each tissue type (leaves or roots) per treatment (negative control, vacuum infiltrated, or root inoculated) per plant (lettuce or spinach).
Immunolocalization of HuNoV.
Tissue sections (1 μm) were cut using a glass knife on a Leica ultramicrotome (Leica Microsystems). Serial sections were screened from three random capsules per tissue type per treatment per plant. Sections were affixed to poly-l-lysine (Invitrogen) slides (n = 6/slide) and stained using the protocol of Bell et al. (49). Briefly, the sections were incubated in blocking solution (3% [wt/vol] bovine serum albumin in 1× PBS) for 10 min at RT. Following washing with 1× PBS 3 times for 1 min, the sections were incubated with purified antibody against HuNoV (anti-HS66 VLP guinea pig serum) and then Alexa Fluor 488 conjugated goat anti-guinea pig IgG antibodies (Invitrogen). All antibodies were diluted in 1% bovine serum albumin and 0.02% (vol/vol) Tween in 1× PBS and incubated at 37°C. Following each step, the sections were washed 5 times for 1 min each with PBS–0.05% Tween 20 (PBS-T) under shaking conditions. The sections were then stained with 10 mM calcofluor (Invitrogen) for 2 min, followed by rinsing with 1× PBS. Calcofluor was used to stain the cellulose. The sections were air-dried before mounting with the antifade agent Fluoromount-G (Southern Biotech, Birmingham, AL) under a coverslip, which was sealed with nail varnish. Slides were first screened with a fluorescence microscope under UV excitation until a positive signal was observed for HuNoV. These sections were then imaged using confocal microscopy.
Confocal microscopy (Leica TCS-SP5; Leica Microsystems) was used to visualize the Alexa 488 green fluorescence specific for HuNoV, 405-nm excitation for calcofluor, and the plant chloroplast red autofluorescence using the 488-nm wavelength of the argon excitation laser. Negative-control plants (non-HuNoV inoculated) were processed like HuNoV-inoculated plants but with/or without primary and/or secondary antibodies to assess nonspecific binding of these antibodies to the plant tissues. The optimum dilutions for primary and secondary antibodies for both lettuce and spinach tissues were 1:100 and 1:500, respectively.
Statistical analysis.
GraphPad Prism version 5 (GraphPad Software, USA) was used for statistical analyses. The entire data set for viruses was log10 transformed. Significant differences in mean infectivity and RNA titers were determined by using two-way analysis of variance (ANOVA) (followed by Bonferroni posttests). The factors analyzed included time and plant type. Each greenhouse experiment was repeated twice with triplicate plants per time point per inoculation route per plant type. Differences in means were considered significant when the P value was <0.05 and are denoted in the figures by asterisks as follows: *, P < 0.05; **, P < 0.01; and ***, P < 0.001. Data are expressed as the mean ± standard error (SE).
ACKNOWLEDGMENTS
We thank Tibor Farkas for providing Tulane virus, Andrea Kaszas at the Molecular and Cellular Imaging Center (OARDC, The Ohio State University [OSU]) for her technical help with tissue sectioning and confocal microscopy image acquisition, and Sarah Tegtmeier for her initial help in testing different embedding media for immunohistochemistry detection of SaV in lettuce. We also thank Sally Miller for allowing us to use the greenhouse and growth chambers at the plant pathology department, Lee Wilson for coordinating the use of the growth chamber, and Bob James for watering the plants at the greenhouse. Finally, we thank Susan Sommer Wagner, Megan Strother, and Juliet Hanson for their help in generating TV antisera and caring for the guinea pigs and Pat Boley for facilitating the research at Qiuhong Wang's lab.
The USDA National Institute of Food and Agriculture (NIFA) provided funding to Qiuhong Wang under grant 2014-67017-21704. Salaries and research support were provided by state and federal funds provided to the Ohio Agricultural Research and Development Center (OARDC), The Ohio State University.
REFERENCES
- 1.Hall AJ, Wikswo ME, Pringle K, Gould LH, Parashar UD. 2014. Vital signs: foodborne norovirus outbreaks–United States, 2009–2012. MMWR Morb Mortal Wkly Rep 63:491–495. [PMC free article] [PubMed] [Google Scholar]
- 2.Bartsch SM, Lopman BA, Ozawa S, Hall AJ, Lee BY. 2016. Global economic burden of norovirus gastroenteritis. PLoS One 11:e0151219. doi: 10.1371/journal.pone.0151219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hall AJ, Lopman BA, Payne DC, Patel MM, Gastanaduy PA, Vinje J, Parashar UD. 2013. Norovirus disease in the United States. Emerg Infect Dis 19:1198–1205. doi: 10.3201/eid1908.130465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Patel MM, Hall AJ, Vinje J, Parashar UD. 2009. Noroviruses: a comprehensive review. J Clin Virol 44:1–8. doi: 10.1016/j.jcv.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 5.Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng XL, Qu L, Kou B, Opekun AR, Burrin D, Graham DY, Ramani S, Atmar RL, Estes MK. 2016. Replication of human noroviruses in stem cell-derived human enteroids. Science 353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teunis PF, Moe CL, Liu P, Miller SE, Lindesmith L, Baric RS, Le Pendu J, Calderon RL. 2008. Norwalk virus: how infectious is it? J Med Virol 80:1468–1476. doi: 10.1002/jmv.21237. [DOI] [PubMed] [Google Scholar]
- 7.Atmar RL, Opekun AR, Gilger MA, Estes MK, Crawford SE, Neill FH, Ramani S, Hill H, Ferreira J, Graham DY. 2014. Determination of the 50% human infectious dose for Norwalk virus. J Infect Dis 209:1016–1022. doi: 10.1093/infdis/jit620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopman B, Gastanaduy P, Park GW, Hall AJ, Parashar UD, Vinje J. 2012. Environmental transmission of norovirus gastroenteritis. Curr Opin Virol 2:96–102. doi: 10.1016/j.coviro.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 9.Parrino TA, Schreiber DS, Trier JS, Kapikian AZ, Blacklow NR. 1977. Clinical immunity in acute gastroenteritis caused by Norwalk agent. N Engl J Med 297:86–89. doi: 10.1056/NEJM197707142970204. [DOI] [PubMed] [Google Scholar]
- 10.de Graaf M, Villabruna N, Koopmans MPG. 2017. Capturing norovirus transmission. Curr Opin Virol 22:64–70. doi: 10.1016/j.coviro.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 11.Karst SM, Baric RS. 2015. What is the reservoir of emergent human norovirus strains? J Virol 89:5756–5759. doi: 10.1128/JVI.03063-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gil MI, Selma MV, Suslow T, Jacxsens L, Uyttendaele M, Allende A. 2015. Pre- and postharvest preventive measures and intervention strategies to control microbial food safety hazards of fresh leafy vegetables. Crit Rev Food Sci Nutr 55:453–468. doi: 10.1080/10408398.2012.657808. [DOI] [PubMed] [Google Scholar]
- 13.Ethelberg S, Lisby M, Bottiger B, Schultz AC, Villif A, Jensen T, Olsen KE, Scheutz F, Kjelso C, Muller L. 2010. Outbreaks of gastroenteritis linked to lettuce, Denmark, January 2010. Euro Surveill 15:e19484 https://www.eurosurveillance.org/content/10.2807/ese.15.06.19484-en. [PubMed] [Google Scholar]
- 14.Makary P, Maunula L, Niskanen T, Kuusi M, Virtanen M, Pajunen S, Ollgren J, Minh NNT. 2009. Multiple norovirus outbreaks among workplace canteen users in Finland, July 2006. Epidemiol Infect 137:402–407. doi: 10.1017/S0950268808000605. [DOI] [PubMed] [Google Scholar]
- 15.Oogane T, Hirata A, Funatogawa K, Kobayashi K, Sato T, Kimura H. 2008. Food poisoning outbreak caused by norovirus GII/4 in school lunch, Tochigi prefecture, Japan. Jpn J Infect Dis 61:423–424. [PubMed] [Google Scholar]
- 16.Schmid D, Stuger HP, Lederer I, Pichler AM, Kainz-Arnfelser G, Schreier E, Allerberger F. 2007. A foodborne norovirus outbreak due to manually prepared salad, Austria 2006. Infection 35:232–239. doi: 10.1007/s15010-007-6327-1. [DOI] [PubMed] [Google Scholar]
- 17.Mercanoglu Taban B, Halkman AK. 2011. Do leafy green vegetables and their ready-to-eat [RTE] salads carry a risk of foodborne pathogens? Anaerobe 17:286–287. doi: 10.1016/j.anaerobe.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Baert L, Mattison K, Loisy-Hamon F, Harlow J, Martyres A, Lebeau B, Stals A, Van Coillie E, Herman L, Uyttendaele M. 2011. Review: norovirus prevalence in Belgian, Canadian and French fresh produce: a threat to human health? Int J Food Microbiol 151:261–269. doi: 10.1016/j.ijfoodmicro.2011.09.013. [DOI] [PubMed] [Google Scholar]
- 19.Steele M, Odumeru J. 2004. Irrigation water as source of foodborne pathogens on fruit and vegetables. J Food Prot 67:2839–2849. doi: 10.4315/0362-028X-67.12.2839. [DOI] [PubMed] [Google Scholar]
- 20.Cook N, Knight A, Richards GP. 2016. Persistence and elimination of human norovirus in food and on food contact surfaces: a critical review. J Food Prot 79:1273–1294. doi: 10.4315/0362-028X.JFP-15-570. [DOI] [PubMed] [Google Scholar]
- 21.Esseili MA, Gao X, Tegtmeier S, Saif LJ, Wang Q. 2015. Abiotic stress and phyllosphere bacteria influence the survival of human norovirus and its surrogates on preharvest leafy greens. 82:352–363. Appl Environ Microbiol doi: 10.1128/AEM.02763-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esseili MA, Chin A, Saif L, Miller SA, Qu F, Lewis Ivey ML, Wang Q. 2015. Postharvest survival of porcine sapovirus, a human norovirus surrogate, on phytopathogen-infected leafy greens. J Food Prot 78:1472–1480. doi: 10.4315/0362-028X.JFP-14-518. [DOI] [PubMed] [Google Scholar]
- 23.Esseili MA, Wang Q, Saif LJ. 2012. Binding of human GII.4 norovirus virus-like particles to carbohydrates of romaine lettuce leaf cell wall materials. Appl Environ Microbiol 78:786–794. doi: 10.1128/AEM.07081-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang ZH, Chambers H, DiCaprio E, Gao G, Li JR. 2018. Internalization and dissemination of human norovirus and Tulane virus in fresh produce is plant dependent. Food Microbiol 69:25–32. doi: 10.1016/j.fm.2017.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DiCaprio E, Ma YM, Purgianto A, Hughes J, Li JR. 2012. Internalization and dissemination of human norovirus and animal caliciviruses in hydroponically grown romaine lettuce. Appl Environ Microbiol 78:6143–6152. doi: 10.1128/AEM.01081-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Urbanucci A, Myrmel M, Berg I, von Bonsdorff CH, Maunula L. 2009. Potential internalisation of caliciviruses in lettuce. Int J Food Microbiol 135:175–178. doi: 10.1016/j.ijfoodmicro.2009.07.036. [DOI] [PubMed] [Google Scholar]
- 27.Esseili MA, Saif LJ, Farkas T, Wang Q. 2015. Feline calicivirus, murine norovirus, porcine sapovirus, and Tulane virus survival on postharvest lettuce. Appl Environ Microbiol 81:5085–5092. doi: 10.1128/AEM.00558-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sestak K, Feely S, Fey B, Dufour J, Hargitt E, Alvarez X, Pahar B, Gregoricus N, Vinje J, Farkas T. 2012. Experimental inoculation of juvenile rhesus macaques with primate enteric caliciviruses. PLoS One 7:e37973. doi: 10.1371/journal.pone.0037973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang QH, Costantini V, Saif LJ. 2007. Porcine enteric caliciviruses: genetic and antigenic relatedness to human caliciviruses, diagnosis and epidemiology. Vaccine 25:5453–5466. doi: 10.1016/j.vaccine.2006.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang D, Huang P, Zou L, Lowary TL, Tan M, Jiang X. 2015. Tulane virus recognizes the A type 3 and B histo-blood group antigens. J Virol 89:1419–1427. doi: 10.1128/JVI.02595-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim DS, Hosmillo M, Alfajaro MM, Kim JY, Park JG, Son KY, Ryu EH, Sorgeloos F, Kwon HJ, Park SJ, Lee WS, Cho D, Kwon J, Choi JS, Kang MI, Goodfellow I, Cho KO. 2014. Both α2,3- and α2,6-linked sialic acids on O-linked glycoproteins act as functional receptors for porcine sapovirus. PLoS Pathog 10:e1004172. doi: 10.1371/journal.ppat.1004172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Svraka S, Vennema H, van der Veer B, Hedlund KO, Thorhagen M, Siebenga J, Duizer E, Koopmans M. 2010. Epidemiology and genotype analysis of emerging sapovirus-associated infections across Europe. J Clin Microbiol 48:2191–2198. doi: 10.1128/JCM.02427-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oka T, Wang Q, Katayama K, Saif LJ. 2015. Comprehensive review of human sapoviruses. Clin Microbiol Rev 28:32–53. doi: 10.1128/CMR.00011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El-Senousy WM, Costafreda MI, Pinto RM, Bosch A. 2013. Method validation for norovirus detection in naturally contaminated irrigation water and fresh produce. Int J Food Microbiol 167:74–79. doi: 10.1016/j.ijfoodmicro.2013.06.023. [DOI] [PubMed] [Google Scholar]
- 35.Masclaux FG, Hotz P, Friedli D, Savova-Bianchi D, Oppliger A. 2013. High occurrence of hepatitis E virus in samples from wastewater treatment plants in Switzerland and comparison with other enteric viruses. Water Res 47:5101–5109. doi: 10.1016/j.watres.2013.05.050. [DOI] [PubMed] [Google Scholar]
- 36.Mathijs E, Stals A, Baert L, Botteldoorn N, Denayer S, Mauroy A, Scipioni A, Daube G, Dierick K, Herman L, Van Coillie E, Uyttendaele M, Thiry E. 2012. A review of known and hypothetical transmission routes for noroviruses. Food Environ Virol 4:131–152. doi: 10.1007/s12560-012-9091-z. [DOI] [PubMed] [Google Scholar]
- 37.DiCaprio E, Culbertson D, Li JR. 2015. Evidence of the internalization of animal caliciviruses via the roots of growing strawberry plants and dissemination to the fruit. Appl Environ Microbiol 81:2727–2734. doi: 10.1128/AEM.03867-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hirneisen KA, Kniel KE. 2013. Comparative uptake of enteric viruses into spinach and green onions. Food Environ Virol 5:24–34. doi: 10.1007/s12560-012-9093-x. [DOI] [PubMed] [Google Scholar]
- 39.Wei J, Jin Y, Sims T, Kniel KE. 2011. Internalization of murine norovirus 1 by Lactuca sativa during irrigation. Appl Environ Microbiol 77:2508–2512. doi: 10.1128/AEM.02701-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Esseili MA, Wang Q, Zhang Z, Saif LJ. 2012. Internalization of sapovirus, a surrogate for norovirus, in romaine lettuce and the effect of lettuce latex on virus infectivity. Appl Environ Microbiol 78:6271–6279. doi: 10.1128/AEM.01295-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ward RL, Mahler RJ. 1982. Uptake of bacteriophage f2 through plant roots. Appl Environ Microbiol 43:1098–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rodríguez-Celma J, Ceballos-Laita L, Grusak MA, Abadia J, Lopez-Millan AF. 2016. Plant fluid proteomics: delving into the xylem sap, phloem sap and apoplastic fluid proteomes. Biochim Biophys Acta 1864:991–1002. doi: 10.1016/j.bbapap.2016.03.014. [DOI] [PubMed] [Google Scholar]
- 43.Takanashi S, Wang Q, Chen N, Shen Q, Jung K, Zhang Z, Yokoyama M, Lindesmith LC, Baric RS, Saif LJ. 2011. Characterization of emerging GII.g/GII.12 noroviruses from a gastroenteritis outbreak in the United States in 2010. J Clin Microbiol 49:3234–3244. doi: 10.1128/JCM.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Niu C, Smith N, Garteiser P, Towner R, Verchot J. 2011. Comparative analysis of protein transport in the N. benthamiana vasculature reveals different destinations. Plant Signal Behav 6:1793–1808. doi: 10.4161/psb.6.11.17896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Q, Zhang Z, Saif LJ. 2012. Stability of and attachment to lettuce by a culturable porcine sapovirus surrogate for human caliciviruses. Appl Environ Microbiol 78:3932–3940. doi: 10.1128/AEM.06600-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cheetham S, Souza M, Meulia T, Grimes S, Han MG, Saif LJ. 2006. Pathogenesis of a genogroup II human norovirus in gnotobiotic pigs. J Virol 80:10372–10381. doi: 10.1128/JVI.00809-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am J Hyg 27:493–497. [Google Scholar]
- 48.Kitajima M, Oka T, Takagi H, Tohya Y, Katayama H, Takeda N, Katayama K. 2010. Development and application of a broadly reactive real-time reverse transcription-PCR assay for detection of murine noroviruses. J Virol Methods 169:269–273. doi: 10.1016/j.jviromet.2010.07.018. [DOI] [PubMed] [Google Scholar]
- 49.Bell K, Mitchell S, Paultre D, Posch M, Oparka K. 2013. Correlative imaging of fluorescent proteins in resin-embedded plant material. Plant Physiol 161:1595–1603. doi: 10.1104/pp.112.212365. [DOI] [PMC free article] [PubMed] [Google Scholar]