Abstract
Members of the 14-3-3 family of proteins function as adapters/modulators that recognize phosphoserine/phosphothreonine-based binding motifs in many intracellular proteins and play fundamental roles in signal transduction pathways of eukaryotic cells. In platelets, 14-3-3 plays a wide range of regulatory roles in phosphorylation-dependent signaling pathways, including G-protein signaling, cAMP signaling, agonist-induced phosphatidylserine exposure, and regulation of mitochondrial function. In particular, 14-3-3 interacts with several phosphoserine-dependent binding sites in the major platelet adhesion receptor, the glycoprotein Ib-IX complex (GPIb-IX), regulating its interaction with von Willebrand factor (VWF) and mediating VWF/GPIb-IX–dependent mechanosignal transduction, leading to platelet activation. The interaction of 14-3-3 with GPIb-IX also plays a critical role in enabling the platelet response to low concentrations of thrombin through cooperative signaling mediated by protease-activated receptors and GPIb-IX. The various functions of 14-3-3 in platelets suggest that it is a possible target for the treatment of thrombosis and inflammation.
Introduction
Budded from nucleated megakaryocytes, nucleusless mammalian platelets share many common cellular mechanisms and molecules with nucleated cells. However, a unique aspect of platelet function is to survey the vessel wall while in blood flow, with the need to adhere quickly at sites of vascular injury and aggregate into thrombi. These functions require mechanisms of adhesion and signaling under elevated shear stress, which differentiate platelet-specific cellular mechanisms from those of other cell types. Whereas the trails of evolution are not clear, some of the proteins expressed in platelets not only play fundamental roles in general eukaryotic cell biology but also become adapted to mediate unique platelet functions. One such example is the 14-3-3 family of proteins.
14-3-3 and 14-3-3 binding proteins
The 14-3-3 family of 27- to 32-kDa acidic proteins is expressed in the cytoplasm of eukaryotic cells.1 Seven 14-3-3 isotypes have been found in mammals (α/β, γ, τ/θ, ε, η, σ, and ζ/δ), all of which can form homodimers and heterodimers.1 All 14-3-3 isotypes have highly conserved sequences across species and share the key feature of phosphorylation-dependent binding to serine/threonine-based peptide motifs, which are found in >200 different intracellular phosphoproteins.2 These motifs are categorized as mode 1 (RSXpSXP), mode 2 (RXY/FXpSXP),3 and mode 3 (pS/TX1-2COOH),4 where X represents an interchangeable amino acid residue and lower-case p indicates phosphorylation.5 However, amino acid sequences in some identified 14-3-3 binding sites, although homologous, do not conform precisely to these modes, and some can be homologous to >1 mode. For example, the C-terminal 14-3-3 binding sites on platelet GPIbα contain the SIRYSGHSL610-CO2H sequence, which conforms to mode 3, but also show homology to modes 1 and 2.6 Additionally, some unphosphorylated peptide sequences were also shown to interact with 14-3-3.7,8 On the basis of the known binding motifs, tools including software,9,10 databases,11 and Web servers,12 such as Scansite,9 ELM,10 and A Nnotation and Integrated Analysis of the 14-3-3 interactome (ANIA),11 have been developed to help predict 14-3-3 binding peptide sequences.
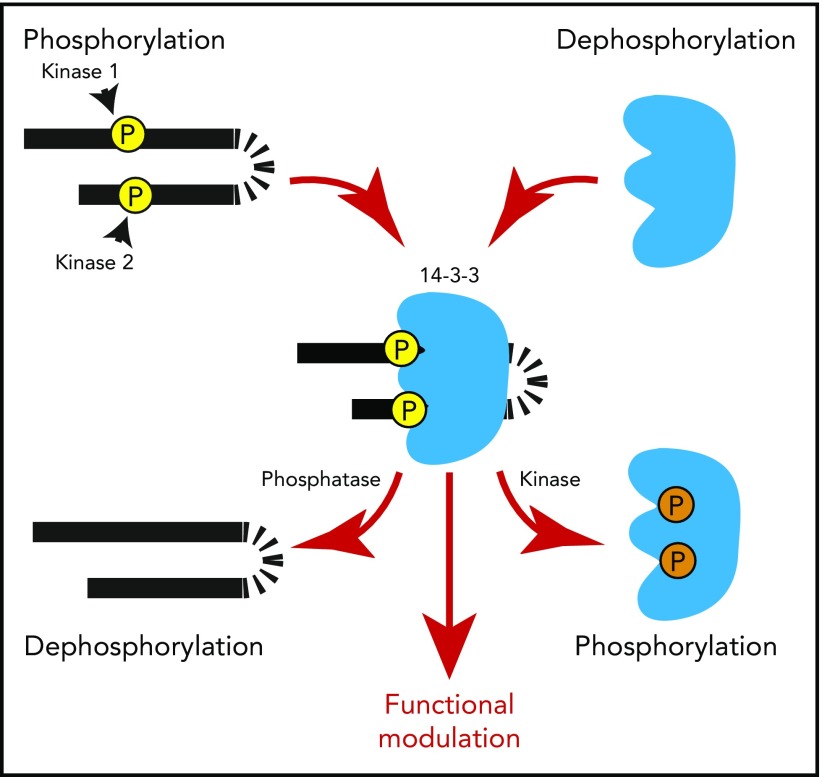
In target proteins, key Ser/Thr residues of the 14-3-3 binding motifs can be phosphorylated by Ser/Thr protein kinases, including AGC kinases (eg, protein kinase A), calcium/calmodulin-dependent kinases, and LIM kinase,13-15 and dephosphorylated by phosphatases (eg, PP2A and PP1),16-18 thereby enhancing or inhibiting, respectively, 14-3-3 binding (Figure 1).19 In some proteins, >1 serine residue can be phosphorylated, with variable effects. Phosphorylation at both Ser residues in the GPIbα cytoplasmic RRPS587ALS590 sequence seems to be required for 14-3-3 binding. In contrast, in the 14-3-3 binding sequence (RRS216RS218FT) of the G-protein regulator RGS18, phosphorylation of Ser218 is important for high-affinity 14-3-3γ binding, but phosphorylation at the neighboring S216 negatively regulates the interaction.20,21 Binding of 14-3-3 to its partners can also be negatively regulated by 14-3-3 phosphorylation at Thr233 and Ser185 (Figure 1).7
Figure 1.
Phosphorylation-regulated binding between 14-3-3 and its target proteins. Each 14-3-3 monomer contains a binding site for a serine/threonine-phosphorylated 14-3-3 binding motif. Upon phosphorylation of target proteins, a 14-3-3 dimer can bind to 2 phosphorylated motifs in tandem in 1 target protein, modulating its conformation/structure. A 14-3-3 dimer can also bind to 2 separate phosphoproteins, acting as an adapter/scaffold for assembly of protein complexes. Phosphorylation of 14-3-3 at Thr233 and Ser185 negatively regulates its ability to interact with target proteins. These features enable 14-3-3 to regulate protein function or transmit signals in a phosphorylation-dependent manner, which can be controlled by a single protein kinase/phosphatase pair or multiple protein kinases/phosphatases.
Members of the 14-3-3 protein family are involved in a variety of phosphorylation-dependent cellular processes, either physiological or pathological. The former include proliferation,22 differentiation,23-25 migration,26-28 cytoskeleton reorganization, apoptosis,29,30 and cell-cycle checkpoint control,31-34 and the latter, cancer progression and metastasis,33,35 although the mechanisms of action are not totally clear. The dimeric nature of 14-3-3 proteins allows association with 2 phosphorylated serine/threonine motifs in the same or in 2 different target proteins (Figure 1).8,36 Thus, 14-3-3 dimers can act as phosphorylation-dependent adaptors/scaffolds to influence the interactions between 2 phosphoproteins,37-39 or they may also modulate the conformation of a single polypeptide chain by binding to 2 phosphorylated sites on the same polypeptide chain.37,38,40 Additionally, it is also possible that 14-3-3 binding inhibits the interaction of its binding partner with other molecules.8,41,42
14-3-3 and 14-3-3 binding proteins in platelet biology
The ζ isoform of 14-3-3 was first identified in human platelets as a 29-kDa species associated with the glycoprotein Ib-IX complex (GPIb-IX) membrane receptor43; β, γ, ε, η,44 and θ,45 but not σ,46 isoforms have also been identified. All are present in the platelet cytoplasm, but there are also reports on 14-3-3ζ presence in and secretion from dense granules.47,48 Platelets express many proteins that interact with 14-3-3, most of which are shared with other cell types. By comparing the platelet proteome reported by Burkhart et al49 with the ANIA database, we identified 71 proteins with experimentally confirmed phosphorylated 14-3-3 binding sites (Table 1). These proteins likely represent only a small fraction of the full spectrum of 14-3-3 binding targets in platelets, because this comparison also identified >1000 proteins that were previously present in the eluates of a high-throughout 14-3-3 affinity capture assay.11 Moreover, ANIA identified another 427 candidates that potentially possess 14-3-3 binding motifs.
Table 1.
Experimentally confirmed platelet-expressed 14-3-3 binding proteins with identified phosphorylated binding sites
| UniProt | Protein ID | Protein name | Binding site | Representative functions in cell |
|---|---|---|---|---|
| P05556 | ITB1 | Integrin β1 | T788 | Forms integrin receptors α2β1 and α5β1 for fibronectin, vitronectin, and collagen binding |
| P00519 | ABL1 | Tyrosine-protein kinase ABL1 | T735 | Cell growth and survival |
| Q9UJU6 | DBNL | Drebrin-like protein | T291, S269 | Endocytosis, cytoskeleton reorganization |
| Q9Y3C5 | RNF11 | RING finger protein 11 | T135 | Protein-protein interactions |
| Q6R327 | RICTR | Rapamycin-insensitive companion of mTOR | T1135 | Cell growth |
| O96013 | PAK4 | Serine/threonine-protein kinase PAK 4 | S99 | Cytoskeleton reorganization |
| Q99683 | M3K5 | MAPK kinase kinase 5 | S966 | Cell differentiation and survival |
| Q92974 | ARHG2 | Rho guanine nucleotide exchange factor 2 | S886, T114 | Multiple cellular processes related to G-protein–coupled receptors |
| P16333 | NCK1 | Cytoplasmic protein NCK1 | S85 | Cytoplasmic adaptor protein, transducing signals from receptor tyrosine kinases |
| P27986 | P85A | PI3K regulatory subunit α | S83 | Cytoplasmic adaptor protein/insulin metabolism |
| Q99959 | PKP2 | Plakophilin-2 | S82 | Protein binding and bridging |
| O00159 | MYO1C | Unconventional myosin-Ic | S736 | Intracellular movements |
| Q8N122 | RPTOR | Regulatory-associated protein of mTOR | S722, S792 | Protein bridging/cell growth, survival, and autophagy |
| P40818 | UBP8 | Ubiquitin carboxyl-terminal hydrolase 8 | S718 | Hydrolysis of ester, thioester, amide, and peptides/regulates protein turnover |
| P19634 | SL9A1 | Sodium/hydrogen exchanger 1 | S703 | pH regulation/intracellular signal transduction |
| Q5PRF9 | SMAG2 | Protein Smaug homolog 2 | S642 | Transcription repression |
| P07359 | GP1BA | GPIbα | S606, S575 | Mediates platelet adhesion and aggregation by binding to VWF A1 domain and other ligands |
| Q9Y4H2 | IRS2 | Insulin receptor substrate 2 | S577, S1148 | Mediate various cellular processes related to insulin |
| Q9NSK0 | KLC4 | Kinesin light chain 4 | S554, S590 | Part of kinesin, a microtubule-associated molecular motor |
| Q07866 | KLC1 | Kinesin light chain 1 | S545 | Part of kinesin, a microtubule-associated molecular motor |
| Q06187 | BTK | Tyrosine-protein kinase BTK | S51, T495 | Protein and metal ion binding/Cell activation and signaling |
| O14974 | MYPT1 | Protein phosphatase 1, regulatory subunit 12A | S472 | Part of protein phosphatase 1C |
| O60825 | F262 | 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 2 | S466, S483 | Synthesis and degradation of fructose 2,6-bisphosphate |
| Q96TC7 | RMD3 | Regulator of microtubule dynamics protein 3 | S46 | Calcium homeostasis |
| Q9BZL4 | PP12C | Protein phosphatase 1 regulatory subunit 12C | S452 | Myosin phosphatase regulator |
| Q86VP3 | PACS2 | Phosphofurin acidic cluster sorting protein 2 | S437 | Controls endoplasmic reticulum–mitochondria communication/protein trafficking/apoptosis |
| Q14432 | PDE3A | cGMP-inhibited 3′,5′-cyclic phosphodiesterase A | S428 | Phosphodiesterase activity/cAMP- and cGMP-mediated signaling |
| Q7KZI7 | MARK2 | Serine/threonine-protein kinase MARK2 | S400, T596 | Protein phosphorylation/microtubule dynamics regulation |
| Q13094 | LCP2 | Lymphocyte cytosolic protein 2 | S376 | Protein kinase activity regulation/platelet activation |
| P15056 | BRAF | Serine/threonine-protein kinase B-raf | S365, S729 | Signal transduction |
| O15530 | PDPK1 | 3-phosphoinositide-dependent protein kinase 1 | S354 | Regulates the phosphorylation and activation of a group of protein kinases/signal transduction |
| Q9Y4H4 | GPSM3 | G-protein-signaling modulator 3 | S35 | Regulates the activation of G(i) α proteins |
| Q8IVT5 | KSR1 | Kinase suppressor of Ras 1 | S309, S404 | Acts as a scaffold protein that promotes phosphorylation of Raf family members and activation of MAPKs |
| Q9Y6R0 | NUMBL | Numb-like protein | S305, S324 | Signal transduction |
| P23528 | COF1 | Cofilin-1 | S3, S24 | Regulates actin cytoskeleton organization |
| Q14247 | SRC8 | Src substrate cortactin | S298 | Regulates actin cytoskeleton organization and cell deformation/intracellular protein transport |
| P48729 | KC1A | Casein kinase I isoform α | S218, S242 | Phosphorylates a large number of proteins and participates in a wide variety of cell signaling pathways |
| Q9NS28 | RGS18 | RGS18 | S218 | Mediates G-protein–coupled receptor signaling pathway |
| P10398 | ARAF | Serine/threonine-protein kinase A-Raf | S214, S582 | Protein phosphorylation and metal ion binding/cell signaling |
| Q8NHG8 | ZNRF2 | E3 ubiquitin-protein ligase ZNRF2 | S19, S82 | Protein ubiquitination and ubiquitin-protein transfer |
| Q53ET0 | CRTC2 | CREB-regulated transcription coactivator 2 | S171 | Glucose homeostasis/cell signaling |
| P42575 | CASP2 | Caspase-2 | S164 | Apoptosis execution |
| Q96F86 | EDC3 | Enhancer of mRNA-decapping protein 3 | S161 | mRNA degradation and decapping |
| Q13363 | CTBP1 | C-terminal-binding protein 1 | S158 | Histone modification/protein binding and phosphorylation |
| Q9GZY8 | MFF | Mitochondrial fission factor | S157 | Mediates mitochondrial and peroxisomal fission |
| Q12802 | AKP13 | A-kinase anchor protein 13 | S1565 | Acts as a scaffold protein/mediates signaling downstream of G-protein–coupled receptors |
| Q15418 | KS6A1 | Ribosomal protein S6 kinase α-1 | S154 | Protein phosphorylation/cell signal transduction and proliferation |
| Q13813 | SPTN1 | Spectrin α chain, non-erythrocytic 1 | S1302 | Cytoskeleton movement |
| Q9UQQ2 | SH2B3 | SH2B adapter protein 3 | S13, S150 | Signal transduction/megakaryocyte development and platelet production |
| P49815 | TSC2 | Tuberin | S1211, S1254, S939 | Regulates protein kinase activity/mediates signal transduction, endocytosis, and cell proliferation |
| Q00536 | CDK16 | Cyclin-dependent kinase 16 | S119 | Protein phosphorylation/mediates vesicle-mediated transport processes and exocytosis |
| O94921 | CDK14 | Cyclin-dependent kinase 14 | S119 | Protein phosphorylation/mediates cell proliferation |
| Q07889 | SOS1 | Son of sevenless homolog 1 | S1134, S1161 | Regulates phosphorylation of MAPK and Ras to Rac signal transduction |
| O43896 | KIF1C | Kinesin-like protein KIF1C | S1092 | Molecular motor for the transport of vesicles |
| Q96PU5 | NED4L | E3 ubiquitin-protein ligase NEDD4-like | S342, S448 | Protein ubiquitination/ion transportation and protein localization |
| Q2PPJ7 | RGPA2 | Ral GTPase-activating protein subunit α-2 | T715 | Subunit of the heterodimeric RalGAP2 complex |
| O43524 | FOXO3 | Forkhead box protein O3 | T32, S253 | Cell response and apoptosis/protein and DNA binding |
| Q8WYL5 | SSH1 | Protein phosphatase Slingshot homolog 1 | S978, S937, S834 | Protein dephosphorylation/actin cytoskeleton organization |
| P49796 | RGS3 | Regulator of G-protein signaling 3 | S943 | Heterotrimeric G-protein signaling suppression |
| P21802 | FGFR2 | Fibroblast growth factor receptor 2 | S782 | Cell proliferation, differentiation, migration, and apoptosis |
| P35222 | CTNB1 | Catenin β-1 | S552 | Regulates Wnt signaling pathway/insulin internalization/protein phosphorylation and ubiquitination/cell adhesion, proliferation, differentiation, and apoptosis |
| Q9UBF8 | PI4KB | Phosphatidylinositol 4-kinase β | S294 | Protein phosphorylation/signal transduction and endocytosis |
| Q9UPA5 | BSN | Protein bassoon | S2851 | Metal ion binding/synapse assembly |
| Q86TI0 | TBCD1 | TBC1 domain family member 1 | S237, T596 | Vesicle and protein trafficking/glucose uptake |
| Q15418 | KS6A1 | Ribosomal protein S6 kinase α-1 | S154 | Protein phosphorylation/signal transduction/cell proliferation, survival, and differentiation |
| Q9H6H4 | REEP4 | Receptor expression-enhancing protein 4 | S152 | Microtubule binding |
| P08069 | IGF1R | Insulin-like growth factor 1 receptor | S1313 | Protein phosphorylation/signal transduction/cell migration and proliferation |
| P43405 | KSYK | Tyrosine-protein kinase SYK | S297 | Signal transduction downstream of transmembrane receptors/mediates platelet adhesion and activation |
| P49757 | NUMB | Protein numb homolog | S276, S295 | Cell junction organization and cell migration |
| Q9UKF7 | PITC1 | Cytoplasmic phosphatidylinositol transfer protein 1 | S274, S299 | Lipid transportation/signal transduction |
| O75791 | GRAP2 | GRB2-related adapter protein 2 | S262 | Interacts with SLP-76 to regulate NF-AT activation/signal transduction |
BTK, Bruton tyrosine kinase; mRNA, messenger RNA; mTOR, mammalian target of rapamycin.
The large number of 14-3-3 binding proteins shared between platelets and other eukaryotic cell types suggests that 14-3-3 is likely to play a general role in serine/threonine phosphorylation-dependent regulation of intracellular signaling pathways. These include G-protein signaling, mitochondrial function,46 regulation of protein kinases, including protein kinase C,44 and melatonin synthesis.37,50-52 However, 14-3-3 also plays critical roles in platelet-specific functions, such as regulation of megakaryocyte proliferation and ploidy53 and GPIb-IX–mediated platelet adhesion and signaling. In the following sections, we summarize some of the major recent advances regarding the role of 14-3-3 in platelets.
14-3-3 in phosphorylation-dependent regulation of G-protein signaling
Members of the 14-3-3 family are important in phosphorylation-dependent regulation of both heterotrimeric G-protein and small GTPase signaling pathways in platelets. Platelet agonists stimulate 14-3-3γ binding to regulator of G-protein signaling 18 (RGS18), a protein that accelerates the hydrolysis of GTP and the conversion of Gα subunits of heterotrimeric G-proteins to their resting form. The binding of 14-3-3γ to RGS18 attenuates the inhibitory effect of RGS18 on G-protein signaling, thereby potentiating platelet activation.20,21 Two 14-3-3 binding sites were reported in RGS18: S49 and S218.21 The RGS18–14-3-3γ interaction is negatively regulated by phosphorylation of RGS18 at S216 by cAMP-dependent (and possibly cyclic guanidine monophosphate–dependent) protein kinase (PKA). During platelet activation, 14-3-3 proteins also bind to Rap1GAP2, a GTPase-activating protein of the small GTPase Rap1. This interaction, possibly involved in the negative regulation of integrin activation, requires phosphorylation of Rap1GAP2 at Ser9.54 In addition, PKA-mediated phosphorylation of the guanine nucleotide exchange factor ARHGEF6 stimulates the association of 14-3-3 with the ARHGEF6/G protein–coupled receptor kinase–interactor 1 complex,55 which may be involved in the negative regulation of Rac1 activity.56 These studies not only suggest a role for 14-3-3 in regulating G-protein signaling in platelets but also suggest a role for cAMP-dependent protein kinases in regulating 14-3-3 binding. Conversely, 14-3-3 may also regulate cAMP signaling by binding to phosphodiesterase 3A during platelet activation.57
14-3-3 in platelet procoagulant activity
Recently, 14-3-3ζ was found to be important in platelet procoagulant activity,46 because membrane exposure of phosphatidylserine (PS) induced by costimulation of thrombin and collagen-related peptide was impaired in 14-3-3ζ–deficient mouse platelets and in platelets treated with an inhibitor of 14-3-3 dimerization. A reduction in thrombin generation was also observed in these experiments. Defective PS exposure in 14-3-3ζ–deficient platelets was associated with elevated mitochondrial respiratory reserve and increased ATP synthesis46 but not with the Bak/Bax-mediated apoptosis pathway, in which 14-3-3 has been known to play a regulatory role.58 GPIb-IX association with other 14-3-3 proteins and agonist-stimulated platelet granule secretion/aggregation are not affected by 14-3-3ζ deficiency. Surprisingly, platelet deficiency of the 14-3-3ζ isoform alone was found to be sufficient to inhibit arterial thrombosis without affecting platelet aggregation or hemostasis.46 Another study suggested that 14-3-3 promotes platelet PS exposure by binding to GPIbα and facilitates the activation of apoptosis signaling pathways during rewarming of cold platelets.59
GPIb-IX, a platelet-specific target of 14-3-3
GPIb-IX is a major receptor for platelet adhesion under conditions of both arterial and venous blood flow.60 It was the first identified platelet receptor interacting with 14-3-361 and unique among 14-3-3 binding proteins for its platelet-specific expression and functions in hemostasis, thrombosis, and inflammation. At the site of vascular injury or inflammation, circulating platelets adhere to the vessel wall and aggregate to form thrombi. Platelet adhesion must be sufficiently fast and strong to overcome the adverse hemodynamic forces of blood flow. This is achieved by a 2-step process where flowing platelets are first captured by the blood vessel wall via the rapid interaction between platelet GPIb-IX and von Willebrand factor (VWF) immobilized on the exposed vascular subendothelial matrix or inflamed/injured endothelium.60,62,63 Subsequently, GPIb-IX induces intracellular signals to activate another platelet adhesion receptor, integrin αIIbβ3 (glycoprotein IIb-IIIa), which mediates stable platelet adhesion and aggregation.63-69
GPIb-IX is composed of disulfide-linked GPIbα and GPIbβ with noncovalently associated GPIX.70-73 The GPIb-IX complex is also noncovalently associated with GPV, a possible negative regulator of GPIb-IX function.74-77 The ligand binding sites of GPIb-IX are located within the N-terminal region of the GPIbα extracellular domain containing 7 tandem leucine rich repeats (LRRs), where the VWF A1 domain binds to a concave structure covering a large area (Figure 2).78-80 The N-terminal region of GPIbα also binds to thrombin,81-83 Mac-1 integrin (αmβ2),84 P-selectin,85 coagulation factors XI86 and XII,87 and high molecular weight kininogen.88 Crystal structures of thrombin in complex with the GPIbα N-terminal domain have shown the possibility of 2 distinctive sites of interaction,89,90 but their respective functional significance remains controversial.91,92 Nonetheless, a region surrounding 3 sulfated tyrosine residues (Tyr276, Tyr278, and Tyr279) was determined to be critical for thrombin binding.91 A heavily glycosylated long stalk (macroglycopeptide) connects the GPIbα N-terminal region to its membrane-spanning region, where GPIbα, GPIbβ, and GPIX are associated.72 The cytoplasmic domain of GPIbα is linked to the actin filament network underlining the cell membrane (membrane skeleton) through filamin A crosslinking. Filamin A binding to the GPIbα cytoplasmic domain has been shown to involve residues Thr536-Phe568,93 Leu556-Val576,94 Leu569-Pro579,95 Val571-Leu589, and possibly also Leu583-Ser591.96,97
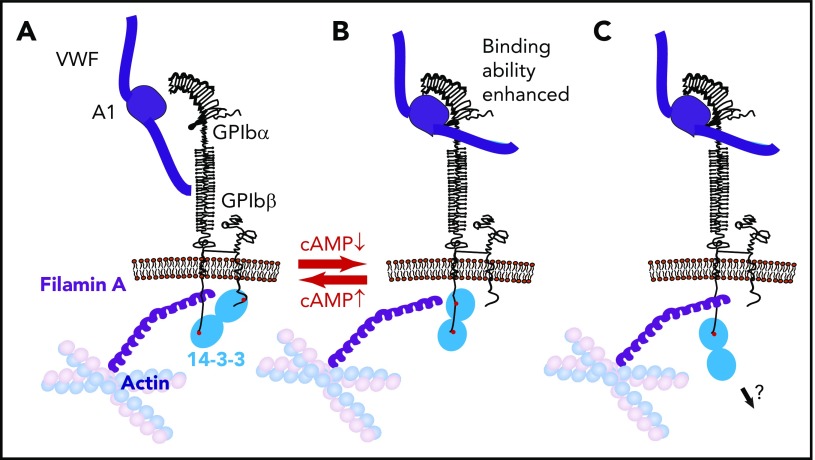
Figure 2.
Three modes of 14-3-3 binding to GPIb and the toggle switch hypothesis on VWF binding to GPIb-IX regulation by cAMP signaling. (A) Dimeric 14-3-3 binds to both GPIbα C-terminus and PKA-phosphorylated GPIbβ. This binding mode is associated with a low GPIb-IX affinity for VWF. (B) Dimeric 14-3-3 binds to the GPIbα C-terminus and to an internal site in GPIbα, which overlaps with the filamin binding site; this mode is associated with GPIbβ dephosphorylation and high GPIb-IX affinity for VWF. In this interaction mode, 14-3-3 may compete with or modulate GPIb-IX interaction with filamin A. (C) 14-3-3 dimer binds to GPIbα C-terminus, potentially linking GPIb-IX to another intracellular protein.
The binding of VWF to GPIb-IX63 is tightly regulated by various mechanisms. It is currently thought that VWF-GPIbα binding does not occur unless VWF is activated, which can be achieved by binding to collagen,98 immobilization on the surface of inflamed endothelial cells,99 pathologically high shear stress,100,101 desialation,102 gain-of-function mutations (mostly in the A1 domain),80,103,104 or deficiency of the VWF cleaving enzyme ADAMTS13.105 ADAMTS13 deficiency causes the persistence of ultralarge VWF multimers in the circulation, which are active in binding to GPIb-IX and mediate platelet adhesion, agglutination, and microvascular thrombosis, thus causing thrombotic thrombocytopenic purpura.106-108 In the laboratory, agents like ristocetin109 and botrocetin110 are used to induce VWF activation. Flow shear force greatly promotes VWF binding to GPIb-IX.111 Shear stress on VWF may stretch the macromolecule and change its conformation around the A1 domain, relieving the autoinhibitory interdomain associations within the A1A2A3 tridomain and between A1 and D’D3, thus enhancing the affinity of the A1 domain for GPIb-IX.112-116 Importantly, GPIb and VWF form a shear-resistant bond, the affinity of which is increased under the dislodging force as exerted by shear (observed as catch bond or flex bond by different groups).117-121
The binding of VWF to GPIbα may also be regulated by mutational changes in GPIb-IX (eg, platelet type VWD)122 and intracellular signaling. PGI2 and PGE1, which elevate intracellular cAMP, reduce VWF binding and inhibit VWF-dependent platelet agglutination,97,123,124 hypothetically by stimulating the phosphorylation of GPIbβ (Ser166) by cAMP-dependent PKA.97,124,125 PKA may potentially regulate GPIb-IX by modulating the association of GPIb-IX with the membrane cytoskeleton, because the inhibitory effect of PGE1 can be reversed by actin depolymerizing agents.97
Association of 14-3-3 with GPIb-IX
All 6 14-3-3 isotypes expressed in platelets can bind GPIb-IX.45 High-affinity binding of 14-3-3 isotypes to GPIb-IX requires the engagement of the GPIbα cytoplasmic domain C-terminal S602IRYSGHpSL610 sequence,45,61,124 in which Ser609 phosphorylation is important. To note, Ser609 is almost 100% constitutively phosphorylated, with only limited and localized dephosphorylation after platelet spreading on VWF,126,127 suggesting that this binding site should mostly remain in a high-affinity state. Even in the Ser609-dephosphorylated state, GPIbα–14-3-3 interaction may still be detectable, although the affinity is reduced.128 Two additional 14-3-3 binding sites in GPIbα were reported at residues Ala551-Arg564,6,129 and Leu580-Ser590.127,130 Both contain phosphoserines (Ser559 and Ser587/Ser590) that are important for the binding of 14-3-3.127,129 The Leu580-Ser590 sequence is close to the C-terminal S602IRYSGHSL610 binding site; however, it remains to be determined whether they are independent tandem binding sequences for a 14-3-3 dimer or whether the entire region can only accommodate the binding of 1 14-3-3 monomer. Nevertheless, it is possible that, under certain conditions, dimers of 14-3-3ζ may simultaneously interact with 2 of these binding sites in GPIbα.
In addition to the 3 binding sites in GPIbα, another 14-3-3 binding site was suggested to exist in the GPIbβ cytoplasmic domain at Arg164-Pro170, in which phosphorylation of Ser166 is required.6,131 Disrupting the GPIbβ binding site does not affect 14-3-3ζ–GPIb interaction,124 suggesting that GPIbβ is not required for 14-3-3 binding to GPIbα. Therefore, the following possible modes of 14-3-3ζ dimer-GPIb interaction are proposed: a 14-3-3 dimer interacts with GPIbα and GPIbβ when GPIbβ is phosphorylated; a 14-3-3 dimer interacts with 2 binding sites in GPIbα; and a 14-3-3 dimer binds to GPIbα and possibly a different protein, such as a signaling molecule (Figure 2).
The role of 14-3-3 in regulating the binding of VWF to GPIb-IX and platelet adhesion
A myristoylated peptide modeled on the 14-3-3 interaction site of GPIbα, MPαC, inhibited the binding of 14-3-3 to GPIb-IX and interfered with VWF binding, ristocetin-induced platelet agglutination, and platelet adhesion to VWF under flow.124 Likewise, another 14-3-3 binding site peptide (557-561) was also reportedly inhibitory.129 The effect of these compounds is consistent with reports demonstrating reduced VWF binding to GPIb-IX mutants lacking the C-terminal 14-3-3 binding site of GPIbα.125,132 Thus, the interaction of GPIbα with 14-3-3 is important in promoting the VWF binding function of GPIb-IX. In contrast, phosphorylation of GPIbβ Ser166 by PKA enabled the binding of 14-3-3 to GPIbβ and seemed to negatively regulate VWF binding to GPIb-IX. A conserved mutation of GPIbβ Ser166 to alanine (but not glycine) was reported to enhance VWF binding.124,125,133 Although controversial, it was hypothesized that when GPIbβ is phosphorylated by PKA, the binding of dimeric 14-3-3 to both GPIbα and GPIbβ (Figure 2A) allows GPIb-IX to stay in a resting conformation with a low affinity for VWF. However, when GPIbβ becomes dephosphorylated, the binding of the 14-3-3 dimer is switched to 2 sites in GPIbα, thereby facilitating VWF binding to GPIb-IX (Figure 2B). In this toggle switch hypothesis, the binding of GPIbα C-terminal sequence to 14-3-3 serves as an anchor point that facilitates a cAMP- and 14-3-3ζ–dependent switch between resting and activated states of GPIb-IX binding to VWF (Figure 2B).124,134 Consistent with this hypothetical model, fusicoccin, a fungal toxin that enhances 14-3-3 binding to GPIbα while negatively affecting 14-3-3 binding to the phosphorylated GPIbβ peptide, stimulates VWF-dependent platelet adhesion, agglutination, and aggregation.135
If the toggle switch model is correct, it raises the question of how 14-3-3 dimers interacting with GPIbα enhance VWF binding. Interestingly, the GPIbα sequence that is important for filamin A binding overlaps with 2 proposed 14-3-3 interaction sites at Ala551-Arg564 and Leu580-Ser590,95 suggesting that the binding of 14-3-3 to these sites likely interferes with the binding of filamin A. Notably, VWF binding to GPIb-IX is enhanced when the filamin A interaction site (together with all 14-3-3 binding sites) is deleted.97 This suggests that the VWF binding function of GPIb-IX is active when filamin binding is abolished, even when 14-3-3 binding is also absent. Furthermore, actin depolymerization also enhanced VWF binding to GPIb-IX as well as shear- or ristocetin-induced platelet agglutination.97,136 Thus, it is hypothesized (but yet to be proven) that the binding of 14-3-3 to both the GPIbα C-terminal domain and the filamin binding domain promotes the VWF binding function by regulating the interaction of filamin with GPIbα (Figure 2A-B).
It is unclear how the filamin-mediated GPIb-IX association with the membrane skeleton regulates VWF binding. If this association, as suggested,97 affects the binding of VWF multimers but not A1 domain fragments, 1 possibility is that cytoskeleton linkage maintains a conformational resting state in the GPIbα ligand binding domain. Alternatively, association with the membrane skeleton may restrict GPIb-IX membrane distribution,137 limiting lateral cis-interactions between GPIb-IX molecules, thus preventing clustered binding between tightly grouped GPIb-IX molecules and A1 domains in VWF multimers. Filamin A–GPIb-IX association is important for adherent platelets to resist shear-induced detachment,95 and GPIb-IX–14-3-3 association was partially reduced, but not abolished, after high shear-induced platelet aggregation,96 suggesting a reduction in affinity after VWF-dependent platelet aggregation. These reports again raise the possibility that filamin A and 14-3-3 may compete with or modulate each other in binding to their overlapping sites, with 14-3-3 occupying 2 sites in the GPIbα C-terminal domain before VWF-GPIbα binding (Figure 2B) but only the C-terminal site after VWF-GPIbα establishes an interaction (Figure 2C).
The role of 14-3-3 proteins in VWF-induced, GPIb-mediated platelet mechanosensing and activation signaling
The binding of VWF to GPIb-IX, independent of other platelet receptors, induces signals leading to the activation of integrin αIIbβ3.66-68,138 Whereas VWF binding to GPIb-IX activates integrin αIIbβ3 even under static conditions,66,68 more robust platelet activation signaling is induced under shear. During platelet adhesion to VWF, shear stress causes an early wave of GPIb-dependent calcium elevations followed by more pronounced integrin-mediated calcium elevations.65,69,139,140 GPIb-IX may thus act as a shear force sensor, converting VWF-mediated mechanical force into robust intracellular chemical signals that lead to integrin activation, integrin-mediated platelet firm adhesion, and signal amplification.141
The binding of VWF to the GPIbα N-terminal ligand binding domain under flow exerts a pulling force on GPIb-IX anchored in the platelet membrane. By using dynamic force spectroscopic techniques and a VWF A1 domain–coated probe, recent studies by 2 different groups suggested that pulling via engaged VWF unfolds and extends a mechanosensitive domain (MSD) in the extracellular membrane-proximal/spanning region of GPIbα (Figure 3Ai-ii).142,143 Unfolding of the MSD was associated with intracellular calcium elevation, suggesting the conversion of pulling force into chemical signaling.142 Pulling on GPIbα by VWF also unfolds the LRR domain near the ligand binding site of GPIbα (Figure 3Aii), which facilitates MSD unfolding.142 Thus, it is proposed that the binding of VWF to GPIbα transmits a pulling force to unfold the LRR domain and MSD and propagates these conformational changes into the membrane, inducing intracellular signaling. MSD unfolding was also suggested to be important in triggering in vivo platelet clearance.144
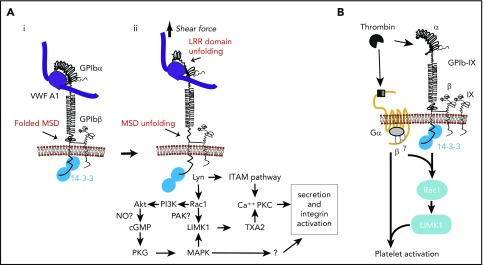
Figure 3.
The role of 14-3-3 in GPIb-IX signaling. (A) VWF binding and shear force–induced, GPIb-mediated mechanosensing and 14-3-3ζ–dependent GPIb-IX signaling pathways. (i) VWF binding to GPIb-IX in a conformation with folded MSD and LRRs. (ii) Pulling force generated by shear stress induces unfolding of LRR and MSD, converting mechanical signal into GPIb conformational changes in the membrane-proximal/spanning domain, which induces a 14-3-3–dependent signaling cascade leading to granule secretion and integrin activation. (B) Cooperative signaling between GPIb-IX and protease-activated receptors (PARs) dependent on 14-3-3 (adapted from Estevez et al146). Thrombin cleavage of PAR1 or PAR4 and binding to GPIb-IX induces 14-3-3–dependent cooperative signaling between GPIb-IX and PARs, enabling platelet response to low thrombin concentrations. cGMP, cyclic guanidine monophosphate; ITAM, immunoreceptor tyrosine-based activation motif; LIMK1, LIM kinase 1; NO, nitric oxide; PI3K, phosphatidylinositol 3-kinase; PKC, protein kinase C; PKG, cGMP-dependent protein kinase; TXA2, thromboxane A2.
14-3-3 proteins are important in GPIb-IX signaling and mechanosensing. Deletion of the C-terminal 14-3-3 binding site in GPIbα inhibited VWF-induced integrin activation in a CHO cell line expressing human GPIb-IX and αIIbβ3, as well as integrin-dependent spreading of these cells on VWF.66 Decreased cell spreading on VWF was also demonstrated when the 14-3-3 binding site between Leu580-Ser590 of GPIbα was deleted or Ser590 was mutated to Ala127 and when GPIb-IX/αIIbβ3-expressing cells were transfected with a small 14-3-3 fragment containing the GPIb binding site.66 Recent work demonstrated that MPαC strongly inhibited intraplatelet Ca2+ fluxes induced by the pulling of GPIbα with recombinant VWF A1 domain.142 Because the binding of A1 domain to GPIbα, unlike that of VWF multimers, is not inhibited by the 14-3-3/filamin-dependent regulatory mechanism described in the previous section, this study has provided strong evidence that 14-3-3 directly participates in the mechanosignaling of GPIb-IX.
The role of 14-3-3 proteins in thrombin-induced platelet activation
Thrombin-induced platelet activation requires thrombin-mediated cleavage of the N-terminal regions of PAR1 (humans) and PAR4 (humans and mice) to expose new N-terminal sequences that act as tethered ligands for the same receptors.145 In mice, PAR3 facilitates thrombin cleavage of PAR4.145 Tethered ligand binding activates receptor-coupled G-proteins Gαq, Gα12/13, and Gαi (possibly indirectly), leading to integrin activation. However, PARs cannot fully activate platelets at low thrombin concentrations. GPIb-IX, a high-affinity thrombin receptor,81-83 is also required.146-148 This is relevant because thrombin is present locally at low concentrations after laser-induced experimental arterial injury and yet is critical for thrombosis,146 consistent with a reportedly VWF-independent role of GPIb-IX in arterial thrombosis.149 How GPIb-IX functions in thrombin-induced platelet activation is still debated, because it has been proposed either as a passive dock presenting thrombin to PARs150 or a proper receptor signaling independently of PARs.151,152 A recent study indicated that neither may be true and provided evidence that GPIb-IX and PARs signal cooperatively and in a mutually dependent fashion to induce platelet activation in response to low-dose thrombin (Figure 3B).146
GPIb-IX–mediated signaling and platelet activation in response to low-dose thrombin requires the binding of 14-3-3 to the GPIbα C-terminal domain.146 Low-dose thrombin-induced Ca2+ signals and platelet activation146 were both impaired in CHO cells expressing a truncation mutant of GPIbα lacking the 14-3-3ζ binding region, as well as in platelets or CHO cells treated with MPαC, which blocks the interaction of 14-3-3 with GPIbα. Furthermore, the role of 14-3-3 in low-dose thrombin-induced platelet activation is independent of thrombin binding to GPIbα, which is not affected by inhibiting GPIbα–14-3-3 interaction.146 Instead, 14-3-3 seems to mediate a thrombin-induced, GPIb-IX–specific signaling pathway leading to LIMK1 activation through Lyn and Rac1 (Figure 3B).146 Thus, a Lyn/Rac1/LIMK1 pathway is important for cooperative signaling of GPIb-IX and PARs in response to low-dose thrombin.
14-3-3–dependent GPIb-IX signaling pathways
GPIb-IX–dependent platelet activation signals induced by VWF or low-dose thrombin both require the binding of 14-3-3 to the cytoplasmic domain of GPIbα. They also share downstream signaling molecules and pathways, including Rac1,146,153 Lyn,68,154,155 PI3K, and Akt156-158; the cyclic guanidine monophosphate–dependent protein kinase pathway67; and mitogen-activated protein kinases p38 and ERK159,160 and LIMK1.146,161 In particular, the stimulatory role of LIMK1 in platelet activation is selective for the GPIb-IX signaling pathway. Therefore, it is likely that VWF and thrombin share the same 14-3-3–dependent GPIb-IX signaling pathway, as illustrated in Figure 3Aii and reviewed elsewhere.141
It remains unclear how 14-3-3 mediates the rather complex GPIb-IX signal transduction pathway. The ability of 14-3-3 to function as a scaffold makes it tempting to hypothesize that 14-3-3 links GPIb-IX to a key signaling molecule (Figure 2C). There was a report that 14-3-3 is involved in the complex that forms between GPIb-IX and PI3K.162 However, a subsequent report suggested that PI3K directly interacts with GPIb-IX.162 There have been reports that the Src family kinases (c-Src and Lyn) and PI3K are coimmunoprecipitated with GPIb-IX.163 However, the role of 14-3-3 in these associations is not definitively established.
The therapeutic potential of 14-3-3 inhibitors in platelets
The involvement of 14-3-3 in multiple biological processes has inspired the development of inhibitors and modulators of 14-3-3 proteins as new drugs, including peptides and small-molecule compounds that either destabilize 14-3-3 dimerization or inhibit or stabilize the interaction of 14-3-3 with its targets. In platelets, GPIb-IX contributes to thrombosis, particularly in stenotic arteries and arterioles.164 The importance of 14-3-3 in GPIb-IX–dependent platelet adhesion and signaling under high shear stress and in thrombin-induced platelet activation makes it an excellent target for drug development. MPαC, which selectively interferes with the binding of 14-3-3 to GPIbα, is a potent inhibitor of platelet adhesion to VWF, signaling under shear force, and platelet activation induced by low-dose thrombin in vitro and reduces arterial thrombosis in vivo with minor bleeding consequences.124 MPαC also inhibited microvascular thrombosis and mortality in endotoxemic mice, suggesting a potential use for treating systemic inflammation.165 RB-011, a 14-3-3 dimer destabilizer, inhibited the procoagulant activity of platelets similar to 14-3-3ζ deficiency.46 These data indicate that 14-3-3 is an attractive target for the development of new antithrombotic/anti-inflammatory drugs. However, the many fundamental roles that 14-3-3 plays in eukaryotic cell biology raise the possibility for nonspecific effects, warranting caution in stages of development involving humans.
Conclusions
The past 2 decades have witnessed the discovery of 14-3-3 proteins and their important roles in eukaryotic cell biology. In platelets, 14-3-3 is not only involved in phosphorylation-dependent signaling common to eukaryotic cells, but also critical for specific platelet adhesion and activation signaling pathways, particularly GPIb-IX signaling. The most recent advances include the discovery that 14-3-3 is a central player in GPIb-IX–mediated mechanosensing,142 cooperative signaling between PARs and GPIb-IX in response to low-dose thrombin,146 and platelet procoagulant activity.46 These advances provide the rationale for considering 14-3-3 as a new target for antiplatelet drug development. Future studies will test the concept that inhibiting 14-3-3 could improve our ability to prevent and treat thrombosis in severely stenotic arteries with extremely high shear stress, in the microvasculature during episodes of thrombotic thrombocytopenic purpura, and during vascular inflammation.
Acknowledgments
The authors acknowledge Cheng Zhu, Woodruff School of Mechanical Engineering and Petit Institute for Bioengineering and Biosciences, Georgia Institute of Technology, Atlanta, GA, for participation in the writing of this review.
This work was supported by National Institute of Health, National Heart, Lung, and Blood Institute grants HL-062350 (X.D.), HL-080264 (X.D.), HL-125356 (X.D.), HL132019 (principal investigator: Cheng Zhu), HL-117722 (Z.M.R.), and HL-135294 (Z.M.R.).
Authorship
Contribution: All authors wrote and revised the manuscript; and Y.C. and X.D. drew the figures.
Conflict-of-interest disclosure: The University of Illinois at Chicago holds patents relevant to the topic (X.D.). The remaining authors declare no competing financial interests.
Correspondence: Xiaoping Du, Department of Pharmacology, University of Illinois at Chicago, 835 South Wolcott Ave, Room E403, M/C 868, Chicago, IL 60612; e-mail: xdu@uic.edu.
References
- 1.Fu H, Subramanian RR, Masters SC. 14-3-3 proteins: structure, function, and regulation. Annu Rev Pharmacol Toxicol. 2000;40:617-647. [DOI] [PubMed] [Google Scholar]
- 2.Johnson C, Crowther S, Stafford MJ, Campbell DG, Toth R, MacKintosh C. Bioinformatic and experimental survey of 14-3-3-binding sites. Biochem J. 2010;427(1):69-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yaffe MB, Rittinger K, Volinia S, et al. . The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91(7):961-971. [DOI] [PubMed] [Google Scholar]
- 4.Coblitz B, Wu M, Shikano S, Li M. C-terminal binding: an expanded repertoire and function of 14-3-3 proteins. FEBS Lett. 2006;580(6):1531-1535. [DOI] [PubMed] [Google Scholar]
- 5.Tzivion G, Shen YH, Zhu J. 14-3-3 proteins; bringing new definitions to scaffolding. Oncogene. 2001;20(44):6331-6338. [DOI] [PubMed] [Google Scholar]
- 6.Andrews RK, Harris SJ, McNally T, Berndt MC. Binding of purified 14-3-3 zeta signaling protein to discrete amino acid sequences within the cytoplasmic domain of the platelet membrane glycoprotein Ib-IX-V complex. Biochemistry. 1998;37(2):638-647. [DOI] [PubMed] [Google Scholar]
- 7.Aitken A. 14-3-3 proteins: a historic overview. Semin Cancer Biol. 2006;16(3):162-172. [DOI] [PubMed] [Google Scholar]
- 8.Obsil T, Obsilova V. Structural basis of 14-3-3 protein functions. Semin Cell Dev Biol. 2011;22(7):663-672. [DOI] [PubMed] [Google Scholar]
- 9.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Res. 2003;31(13):3635-3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Puntervoll P, Linding R, Gemünd C, et al. . ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31(13):3625-3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tinti M, Madeira F, Murugesan G, Hoxhaj G, Toth R, Mackintosh C. ANIA: ANnotation and Integrated Analysis of the 14-3-3 interactome. Database (Oxford). 2014;2014:bat085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madeira F, Tinti M, Murugesan G, et al. . 14-3-3-Pred: improved methods to predict 14-3-3-binding phosphopeptides. Bioinformatics. 2015;31(14):2276-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aitken A. Functional specificity in 14-3-3 isoform interactions through dimer formation and phosphorylation. Chromosome location of mammalian isoforms and variants. Plant Mol Biol. 2002;50(6):993-1010. [DOI] [PubMed] [Google Scholar]
- 14.van Hemert MJ, Steensma HY, van Heusden GP. 14-3-3 proteins: key regulators of cell division, signalling and apoptosis. BioEssays. 2001;23(10):936-946. [DOI] [PubMed] [Google Scholar]
- 15.Gohla A, Bokoch GM. 14-3-3 regulates actin dynamics by stabilizing phosphorylated cofilin. Curr Biol. 2002;12(19):1704-1710. [DOI] [PubMed] [Google Scholar]
- 16.Chiang CW, Kanies C, Kim KW, et al. . Protein phosphatase 2A dephosphorylation of phosphoserine 112 plays the gatekeeper role for BAD-mediated apoptosis. Mol Cell Biol. 2003;23(18):6350-6362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ory S, Zhou M, Conrads TP, Veenstra TD, Morrison DK. Protein phosphatase 2A positively regulates Ras signaling by dephosphorylating KSR1 and Raf-1 on critical 14-3-3 binding sites. Curr Biol. 2003;13(16):1356-1364. [DOI] [PubMed] [Google Scholar]
- 18.Margolis SS, Walsh S, Weiser DC, Yoshida M, Shenolikar S, Kornbluth S. PP1 control of M phase entry exerted through 14-3-3-regulated Cdc25 dephosphorylation. EMBO J. 2003;22(21):5734-5745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dougherty MK, Morrison DK. Unlocking the code of 14-3-3. J Cell Sci. 2004;117(Pt 10):1875-1884. [DOI] [PubMed] [Google Scholar]
- 20.Gegenbauer K, Nagy Z, Smolenski A. Cyclic nucleotide dependent dephosphorylation of regulator of G-protein signaling 18 in human platelets. PLoS One. 2013;8(11):e80251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gegenbauer K, Elia G, Blanco-Fernandez A, Smolenski A. Regulator of G-protein signaling 18 integrates activating and inhibitory signaling in platelets. Blood. 2012;119(16):3799-3807. [DOI] [PubMed] [Google Scholar]
- 22.Rushworth LK, Hindley AD, O’Neill E, Kolch W. Regulation and role of Raf-1/B-Raf heterodimerization. Mol Cell Biol. 2006;26(6):2262-2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Toyo-oka K, Wachi T, Hunt RF, et al. . 14-3-3ε and ζ regulate neurogenesis and differentiation of neuronal progenitor cells in the developing brain. J Neurosci. 2014;34(36):12168-12181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lim GE, Johnson JD. 14-3-3ζ: a numbers game in adipocyte function? Adipocyte. 2015;5(2):232-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lim GE, Albrecht T, Piske M, et al. . 14-3-3ζ coordinates adipogenesis of visceral fat. Nat Commun. 2015;6:7671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen M, Liu T, Xu L, et al. . Direct interaction of 14-3-3ζ with ezrin promotes cell migration by regulating the formation of membrane ruffle. J Mol Biol. 2014;426(18):3118-3133. [DOI] [PubMed] [Google Scholar]
- 27.Kim HS, Ullevig SL, Nguyen HN, Vanegas D, Asmis R. Redox regulation of 14-3-3ζ controls monocyte migration. Arterioscler Thromb Vasc Biol. 2014;34(7):1514-1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toyo-oka K, Shionoya A, Gambello MJ, et al. . 14-3-3epsilon is important for neuronal migration by binding to NUDEL: a molecular explanation for Miller-Dieker syndrome. Nat Genet. 2003;34(3):274-285. [DOI] [PubMed] [Google Scholar]
- 29.Porter GW, Khuri FR, Fu H. Dynamic 14-3-3/client protein interactions integrate survival and apoptotic pathways. Semin Cancer Biol. 2006;16(3):193-202. [DOI] [PubMed] [Google Scholar]
- 30.Niemantsverdriet M, Wagner K, Visser M, Backendorf C. Cellular functions of 14-3-3 zeta in apoptosis and cell adhesion emphasize its oncogenic character. Oncogene. 2008;27(9):1315-1319. [DOI] [PubMed] [Google Scholar]
- 31.Jones DH, Martin H, Madrazo J, et al. . Expression and structural analysis of 14-3-3 proteins. J Mol Biol. 1995;245(4):375-384. [DOI] [PubMed] [Google Scholar]
- 32.Jones DH, Ley S, Aitken A. Isoforms of 14-3-3 protein can form homo- and heterodimers in vivo and in vitro: implications for function as adapter proteins. FEBS Lett. 1995;368(1):55-58. [DOI] [PubMed] [Google Scholar]
- 33.Freeman AK, Morrison DK. 14-3-3 proteins: diverse functions in cell proliferation and cancer progression. Semin Cell Dev Biol. 2011;22(7):681-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gardino AK, Yaffe MB. 14-3-3 proteins as signaling integration points for cell cycle control and apoptosis. Semin Cell Dev Biol. 2011;22(7):688-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao J, Meyerkord CL, Du Y, Khuri FR, Fu H. 14-3-3 proteins as potential therapeutic targets. Semin Cell Dev Biol. 2011;22(7):705-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen YH, Godlewski J, Bronisz A, et al. . Significance of 14-3-3 self-dimerization for phosphorylation-dependent target binding. Mol Biol Cell. 2003;14(11):4721-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Obsil T, Ghirlando R, Klein DC, Ganguly S, Dyda F. Crystal structure of the 14-3-3zeta:serotonin N-acetyltransferase complex. A role for scaffolding in enzyme regulation. Cell. 2001;105(2):257-267. [DOI] [PubMed] [Google Scholar]
- 38.Ottmann C, Marco S, Jaspert N, et al. . Structure of a 14-3-3 coordinated hexamer of the plant plasma membrane H+ -ATPase by combining X-ray crystallography and electron cryomicroscopy. Mol Cell. 2007;25(3):427-440. [DOI] [PubMed] [Google Scholar]
- 39.Rajagopalan S, Jaulent AM, Wells M, Veprintsev DB, Fersht AR. 14-3-3 activation of DNA binding of p53 by enhancing its association into tetramers. Nucleic Acids Res. 2008;36(18):5983-5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mackintosh C. Dynamic interactions between 14-3-3 proteins and phosphoproteins regulate diverse cellular processes. Biochem J. 2004;381(Pt 2):329-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Benzing T, Yaffe MB, Arnould T, et al. . 14-3-3 interacts with regulator of G protein signaling proteins and modulates their activity. J Biol Chem. 2000;275(36):28167-28172. [DOI] [PubMed] [Google Scholar]
- 42.Abramow-Newerly M, Ming H, Chidiac P. Modulation of subfamily B/R4 RGS protein function by 14-3-3 proteins. Cell Signal. 2006;18(12):2209-2222. [DOI] [PubMed] [Google Scholar]
- 43.Du X, Harris SJ, Tetaz TJ, Ginsberg MH, Berndt MC. Association of a phospholipase A2 (14-3-3 protein) with the platelet glycoprotein Ib-IX complex. J Biol Chem. 1994;269(28):18287-18290. [PubMed] [Google Scholar]
- 44.Wheeler-Jones CP, Learmonth MP, Martin H, Aitken A. Identification of 14-3-3 proteins in human platelets: effects of synthetic peptides on protein kinase C activation. Biochem J. 1996;315(Pt 1):41-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mangin PH, Receveur N, Wurtz V, David T, Gachet C, Lanza F. Identification of five novel 14-3-3 isoforms interacting with the GPIb-IX complex in platelets. J Thromb Haemost. 2009;7(9):1550-1555. [DOI] [PubMed] [Google Scholar]
- 46.Schoenwaelder SM, Darbousset R, Cranmer SL, et al. . 14-3-3ζ regulates the mitochondrial respiratory reserve linked to platelet phosphatidylserine exposure and procoagulant function. Nat Commun. 2016;7:12862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hernandez-Ruiz L, Valverde F, Jimenez-Nuñez MD, et al. . Organellar proteomics of human platelet dense granules reveals that 14-3-3zeta is a granule protein related to atherosclerosis. J Proteome Res. 2007;6(11):4449-4457. [DOI] [PubMed] [Google Scholar]
- 48.Coppinger JA, Cagney G, Toomey S, et al. . Characterization of the proteins released from activated platelets leads to localization of novel platelet proteins in human atherosclerotic lesions. Blood. 2004;103(6):2096-2104. [DOI] [PubMed] [Google Scholar]
- 49.Burkhart JM, Vaudel M, Gambaryan S, et al. . The first comprehensive and quantitative analysis of human platelet protein composition allows the comparative analysis of structural and functional pathways. Blood. 2012;120(15):e73-e82. [DOI] [PubMed] [Google Scholar]
- 50.Yu D, dos Santos CO, Zhao G, et al. . miR-451 protects against erythroid oxidant stress by repressing 14-3-3zeta. Genes Dev. 2010;24(15):1620-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Maronde E, Saade A, Ackermann K, et al. . Dynamics in enzymatic protein complexes offer a novel principle for the regulation of melatonin synthesis in the human pineal gland. J Pineal Res. 2011;51(1):145-155. [DOI] [PubMed] [Google Scholar]
- 52.Pagan C, Goubran-Botros H, Delorme R, et al. . Disruption of melatonin synthesis is associated with impaired 14-3-3 and miR-451 levels in patients with autism spectrum disorders. Sci Rep. 2017;7(1):2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kanaji T, Russell S, Cunningham J, Izuhara K, Fox JE, Ware J. Megakaryocyte proliferation and ploidy regulated by the cytoplasmic tail of glycoprotein Ibalpha. Blood. 2004;104(10):3161-3168. [DOI] [PubMed] [Google Scholar]
- 54.Hoffmeister M, Riha P, Neumüller O, Danielewski O, Schultess J, Smolenski AP. Cyclic nucleotide-dependent protein kinases inhibit binding of 14-3-3 to the GTPase-activating protein Rap1GAP2 in platelets. J Biol Chem. 2008;283(4):2297-2306. [DOI] [PubMed] [Google Scholar]
- 55.Nagy Z, Wynne K, von Kriegsheim A, Gambaryan S, Smolenski A. Cyclic nucleotide-dependent protein kinases target ARHGAP17 and ARHGEF6 complexes in platelets. J Biol Chem. 2015;290(50):29974-29983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chahdi A, Sorokin A. Protein kinase A-dependent phosphorylation modulates beta1Pix guanine nucleotide exchange factor activity through 14-3-3beta binding. Mol Cell Biol. 2008;28(5):1679-1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hunter RW, Mackintosh C, Hers I. Protein kinase C-mediated phosphorylation and activation of PDE3A regulate cAMP levels in human platelets. J Biol Chem. 2009;284(18):12339-12348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schoenwaelder SM, Yuan Y, Josefsson EC, et al. . Two distinct pathways regulate platelet phosphatidylserine exposure and procoagulant function. Blood. 2009;114(3):663-666. [DOI] [PubMed] [Google Scholar]
- 59.van der Wal DE, Du VX, Lo KS, Rasmussen JT, Verhoef S, Akkerman JW. Platelet apoptosis by cold-induced glycoprotein Ibα clustering. J Thromb Haemost. 2010;8(11):2554-2562. [DOI] [PubMed] [Google Scholar]
- 60.André P, Denis CV, Ware J, et al. . Platelets adhere to and translocate on von Willebrand factor presented by endothelium in stimulated veins. Blood. 2000;96(10):3322-3328. [PubMed] [Google Scholar]
- 61.Du X, Fox JE, Pei S. Identification of a binding sequence for the 14-3-3 protein within the cytoplasmic domain of the adhesion receptor, platelet glycoprotein Ib alpha. J Biol Chem. 1996;271(13):7362-7367. [DOI] [PubMed] [Google Scholar]
- 62.Litvinov RI, Shuman H, Bennett JS, Weisel JW. Binding strength and activation state of single fibrinogen-integrin pairs on living cells. Proc Natl Acad Sci USA. 2002;99(11):7426-7431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Savage B, Saldívar E, Ruggeri ZM. Initiation of platelet adhesion by arrest onto fibrinogen or translocation on von Willebrand factor. Cell. 1996;84(2):289-297. [DOI] [PubMed] [Google Scholar]
- 64.Savage B, Almus-Jacobs F, Ruggeri ZM. Specific synergy of multiple substrate-receptor interactions in platelet thrombus formation under flow. Cell. 1998;94(5):657-666. [DOI] [PubMed] [Google Scholar]
- 65.Mazzucato M, Pradella P, Cozzi MR, De Marco L, Ruggeri ZM. Sequential cytoplasmic calcium signals in a 2-stage platelet activation process induced by the glycoprotein Ibalpha mechanoreceptor. Blood. 2002;100(8):2793-2800. [DOI] [PubMed] [Google Scholar]
- 66.Gu M, Xi X, Englund GD, Berndt MC, Du X. Analysis of the roles of 14-3-3 in the platelet glycoprotein Ib-IX-mediated activation of integrin alpha(IIb)beta(3) using a reconstituted mammalian cell expression model. J Cell Biol. 1999;147(5):1085-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li Z, Xi X, Gu M, et al. . A stimulatory role for cGMP-dependent protein kinase in platelet activation. Cell. 2003;112(1):77-86. [DOI] [PubMed] [Google Scholar]
- 68.Kasirer-Friede A, Cozzi MR, Mazzucato M, De Marco L, Ruggeri ZM, Shattil SJ. Signaling through GP Ib-IX-V activates alpha IIb beta 3 independently of other receptors. Blood. 2004;103(9):3403-3411. [DOI] [PubMed] [Google Scholar]
- 69.Nesbitt WS, Kulkarni S, Giuliano S, et al. . Distinct glycoprotein Ib/V/IX and integrin alpha IIbbeta 3-dependent calcium signals cooperatively regulate platelet adhesion under flow. J Biol Chem. 2002;277(4):2965-2972. [DOI] [PubMed] [Google Scholar]
- 70.Du X, Beutler L, Ruan C, Castaldi PA, Berndt MC. Glycoprotein Ib and glycoprotein IX are fully complexed in the intact platelet membrane. Blood. 1987;69(5):1524-1527. [PubMed] [Google Scholar]
- 71.Li R, Emsley J. The organizing principle of the platelet glycoprotein Ib-IX-V complex. J Thromb Haemost. 2013;11(4):605-614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.López JA. The platelet glycoprotein Ib-IX complex. Blood Coagul Fibrinolysis. 1994;5(1):97-119. [PubMed] [Google Scholar]
- 73.López JA, Leung B, Reynolds CC, Li CQ, Fox JE. Efficient plasma membrane expression of a functional platelet glycoprotein Ib-IX complex requires the presence of its three subunits. J Biol Chem. 1992;267(18):12851-12859. [PubMed] [Google Scholar]
- 74.Ramakrishnan V, Reeves PS, DeGuzman F, et al. . Increased thrombin responsiveness in platelets from mice lacking glycoprotein V. Proc Natl Acad Sci USA. 1999;96(23):13336-13341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Roth GJ, Yagi M, Bastian LS. The platelet glycoprotein Ib-V-IX system: regulation of gene expression. Stem Cells. 1996;14(suppl 1):188-193. [DOI] [PubMed] [Google Scholar]
- 76.Andrews RK, Berndt MC. Bernard-Soulier syndrome: an update. Semin Thromb Hemost. 2013;39(6):656-662. [DOI] [PubMed] [Google Scholar]
- 77.Kahn ML, Diacovo TG, Bainton DF, Lanza F, Trejo J, Coughlin SR. Glycoprotein V-deficient platelets have undiminished thrombin responsiveness and do not exhibit a Bernard-Soulier phenotype. Blood. 1999;94(12):4112-4121. [PubMed] [Google Scholar]
- 78.Mohri H, Fujimura Y, Shima M, et al. . Structure of the von Willebrand factor domain interacting with glycoprotein Ib. J Biol Chem. 1988;263(34):17901-17904. [PubMed] [Google Scholar]
- 79.Huizinga EG, Tsuji S, Romijn RA, et al. . Structures of glycoprotein Ibalpha and its complex with von Willebrand factor A1 domain. Science. 2002;297(5584):1176-1179. [DOI] [PubMed] [Google Scholar]
- 80.Dumas JJ, Kumar R, McDonagh T, et al. . Crystal structure of the wild-type von Willebrand factor A1-glycoprotein Ibalpha complex reveals conformation differences with a complex bearing von Willebrand disease mutations. J Biol Chem. 2004;279(22):23327-23334. [DOI] [PubMed] [Google Scholar]
- 81.Okumura T, Hasitz M, Jamieson GA. Platelet glycocalicin. Interaction with thrombin and role as thrombin receptor of the platelet surface. J Biol Chem. 1978;253(10):3435-3443. [PubMed] [Google Scholar]
- 82.Harmon JT, Jamieson GA. The glycocalicin portion of platelet glycoprotein Ib expresses both high and moderate affinity receptor sites for thrombin. A soluble radioreceptor assay for the interaction of thrombin with platelets. J Biol Chem. 1986;261(28):13224-13229. [PubMed] [Google Scholar]
- 83.De Marco L, Mazzucato M, Masotti A, Fenton JW II, Ruggeri ZM. Function of glycoprotein Ib alpha in platelet activation induced by alpha-thrombin. J Biol Chem. 1991;266(35):23776-23783. [PubMed] [Google Scholar]
- 84.Simon DI, Chen Z, Xu H, et al. . Platelet glycoprotein ibalpha is a counterreceptor for the leukocyte integrin Mac-1 (CD11b/CD18). J Exp Med. 2000;192(2):193-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Romo GM, Dong JF, Schade AJ, et al. . The glycoprotein Ib-IX-V complex is a platelet counterreceptor for P-selectin. J Exp Med. 1999;190(6):803-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Baglia FA, Shrimpton CN, Emsley J, et al. . Factor XI interacts with the leucine-rich repeats of glycoprotein Ibalpha on the activated platelet. J Biol Chem. 2004;279(47):49323-49329. [DOI] [PubMed] [Google Scholar]
- 87.Bradford HN, Pixley RA, Colman RW. Human factor XII binding to the glycoprotein Ib-IX-V complex inhibits thrombin-induced platelet aggregation. J Biol Chem. 2000;275(30):22756-22763. [DOI] [PubMed] [Google Scholar]
- 88.Joseph K, Nakazawa Y, Bahou WF, Ghebrehiwet B, Kaplan AP. Platelet glycoprotein Ib: a zinc-dependent binding protein for the heavy chain of high-molecular-weight kininogen. Mol Med. 1999;5(8):555-563. [PMC free article] [PubMed] [Google Scholar]
- 89.Celikel R, McClintock RA, Roberts JR, et al. . Modulation of alpha-thrombin function by distinct interactions with platelet glycoprotein Ibalpha. Science. 2003;301(5630):218-221. [DOI] [PubMed] [Google Scholar]
- 90.Dumas JJ, Kumar R, Seehra J, Somers WS, Mosyak L. Crystal structure of the GpIbalpha-thrombin complex essential for platelet aggregation. Science. 2003;301(5630):222-226. [DOI] [PubMed] [Google Scholar]
- 91.Zarpellon A, Celikel R, Roberts JR, et al. . Binding of alpha-thrombin to surface-anchored platelet glycoprotein Ib(alpha) sulfotyrosines through a two-site mechanism involving exosite I. Proc Natl Acad Sci USA. 2011;108(21):8628-8633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lechtenberg BC, Freund SM, Huntington JA. GpIbα interacts exclusively with exosite II of thrombin. J Mol Biol. 2014;426(4):881-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Andrews RK, Fox JE. Identification of a region in the cytoplasmic domain of the platelet membrane glycoprotein Ib-IX complex that binds to purified actin-binding protein. J Biol Chem. 1992;267(26):18605-18611. [PubMed] [Google Scholar]
- 94.Nakamura F, Pudas R, Heikkinen O, et al. . The structure of the GPIb-filamin A complex. Blood. 2006;107(5):1925-1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Williamson D, Pikovski I, Cranmer SL, et al. . Interaction between platelet glycoprotein Ibalpha and filamin-1 is essential for glycoprotein Ib/IX receptor anchorage at high shear. J Biol Chem. 2002;277(3):2151-2159. [DOI] [PubMed] [Google Scholar]
- 96.Feng S, Reséndiz JC, Lu X, Kroll MH. Filamin A binding to the cytoplasmic tail of glycoprotein Ibalpha regulates von Willebrand factor-induced platelet activation. Blood. 2003;102(6):2122-2129. [DOI] [PubMed] [Google Scholar]
- 97.Englund GD, Bodnar RJ, Li Z, Ruggeri ZM, Du X. Regulation of von Willebrand factor binding to the platelet glycoprotein Ib-IX by a membrane skeleton-dependent inside-out signal. J Biol Chem. 2001;276(20):16952-16959. [DOI] [PubMed] [Google Scholar]
- 98.Springer TA. von Willebrand factor, Jedi knight of the bloodstream. Blood. 2014;124(9):1412-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Padilla A, Moake JL, Bernardo A, et al. . P-selectin anchors newly released ultralarge von Willebrand factor multimers to the endothelial cell surface. Blood. 2004;103(6):2150-2156. [DOI] [PubMed] [Google Scholar]
- 100.Sing CE, Alexander-Katz A. Elongational flow induces the unfolding of von Willebrand factor at physiological flow rates. Biophys J. 2010;98(9):L35-L37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Colace TV, Diamond SL. Direct observation of von Willebrand factor elongation and fiber formation on collagen during acute whole blood exposure to pathological flow. Arterioscler Thromb Vasc Biol. 2013;33(1):105-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.De Marco L, Girolami A, Russell S, Ruggeri ZM. Interaction of asialo von Willebrand factor with glycoprotein Ib induces fibrinogen binding to the glycoprotein IIb/IIIa complex and mediates platelet aggregation. J Clin Invest. 1985;75(4):1198-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sadler JE. New concepts in von Willebrand disease. Annu Rev Med. 2005;56:173-191. [DOI] [PubMed] [Google Scholar]
- 104.Celikel R, Ruggeri ZM, Varughese KI. von Willebrand factor conformation and adhesive function is modulated by an internalized water molecule. Nat Struct Biol. 2000;7(10):881-884. [DOI] [PubMed] [Google Scholar]
- 105.Levy GG, Nichols WC, Lian EC, et al. . Mutations in a member of the ADAMTS gene family cause thrombotic thrombocytopenic purpura. Nature. 2001;413(6855):488-494. [DOI] [PubMed] [Google Scholar]
- 106.Arya M, Anvari B, Romo GM, et al. . Ultralarge multimers of von Willebrand factor form spontaneous high-strength bonds with the platelet glycoprotein Ib-IX complex: studies using optical tweezers. Blood. 2002;99(11):3971-3977. [DOI] [PubMed] [Google Scholar]
- 107.Zheng XL. ADAMTS13 and von Willebrand factor in thrombotic thrombocytopenic purpura. Annu Rev Med. 2015;66:211-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Farrell DH, Thiagarajan P, Chung DW, Davie EW. Role of fibrinogen alpha and gamma chain sites in platelet aggregation. Proc Natl Acad Sci USA. 1992;89(22):10729-10732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hoylaerts MF, Nuyts K, Peerlinck K, Deckmyn H, Vermylen J. Promotion of binding of von Willebrand factor to platelet glycoprotein Ib by dimers of ristocetin. Biochem J. 1995;306(Pt 2):453-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fukuda K, Doggett TA, Bankston LA, Cruz MA, Diacovo TG, Liddington RC. Structural basis of von Willebrand factor activation by the snake toxin botrocetin. Structure. 2002;10(7):943-950. [DOI] [PubMed] [Google Scholar]
- 111.Wijeratne SS, Botello E, Yeh HC, et al. . Mechanical activation of a multimeric adhesive protein through domain conformational change. Phys Rev Lett. 2013;110(10):108102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Barg A, Ossig R, Goerge T, et al. . Soluble plasma-derived von Willebrand factor assembles to a haemostatically active filamentous network. Thromb Haemost. 2007;97(4):514-526. [PubMed] [Google Scholar]
- 113.Schneider SW, Nuschele S, Wixforth A, et al. . Shear-induced unfolding triggers adhesion of von Willebrand factor fibers. Proc Natl Acad Sci USA. 2007;104(19):7899-7903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Martin C, Morales LD, Cruz MA. Purified A2 domain of von Willebrand factor binds to the active conformation of von Willebrand factor and blocks the interaction with platelet glycoprotein Ibalpha. J Thromb Haemost. 2007;5(7):1363-1370. [DOI] [PubMed] [Google Scholar]
- 115.Ulrichts H, Udvardy M, Lenting PJ, et al. . Shielding of the A1 domain by the D’D3 domains of von Willebrand factor modulates its interaction with platelet glycoprotein Ib-IX-V. J Biol Chem. 2006;281(8):4699-4707. [DOI] [PubMed] [Google Scholar]
- 116.Auton M, Sowa KE, Behymer M, Cruz MA. N-terminal flanking region of A1 domain in von Willebrand factor stabilizes structure of A1A2A3 complex and modulates platelet activation under shear stress. J Biol Chem. 2012;287(18):14579-14585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yago T, Lou J, Wu T, et al. . Platelet glycoprotein Ibalpha forms catch bonds with human WT vWF but not with type 2B von Willebrand disease vWF. J Clin Invest. 2008;118(9):3195-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ju L, Dong JF, Cruz MA, Zhu C. The N-terminal flanking region of the A1 domain regulates the force-dependent binding of von Willebrand factor to platelet glycoprotein Ibα. J Biol Chem. 2013;288(45):32289-32301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dembo M, Torney DC, Saxman K, Hammer D. The reaction-limited kinetics of membrane-to-surface adhesion and detachment. Proc R Soc Lond B Biol Sci. 1988;234(1274):55-83. [DOI] [PubMed] [Google Scholar]
- 120.Kim J, Zhang CZ, Zhang X, Springer TA. A mechanically stabilized receptor-ligand flex-bond important in the vasculature. Nature. 2010;466(7309):992-995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ju L, Chen Y, Zhou F, Lu H, Cruz MA, Zhu C. Von Willebrand factor-A1 domain binds platelet glycoprotein Ibα in multiple states with distinctive force-dependent dissociation kinetics. Thromb Res. 2015;136(3):606-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dayananda KM, Singh I, Mondal N, Neelamegham S. von Willebrand factor self-association on platelet GpIbalpha under hydrodynamic shear: effect on shear-induced platelet activation. Blood. 2010;116(19):3990-3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Coller BS. Effects of tertiary amine local anesthetics on von Willebrand factor-dependent platelet function: alteration of membrane reactivity and degradation of GPIb by a calcium-dependent protease(s). Blood. 1982;60(3):731-743. [PubMed] [Google Scholar]
- 124.Dai K, Bodnar R, Berndt MC, Du X. A critical role for 14-3-3zeta protein in regulating the VWF binding function of platelet glycoprotein Ib-IX and its therapeutic implications. Blood. 2005;106(6):1975-1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bodnar RJ, Xi X, Li Z, Berndt MC, Du X. Regulation of glycoprotein Ib-IX-von Willebrand factor interaction by cAMP-dependent protein kinase-mediated phosphorylation at Ser 166 of glycoprotein Ib(beta). J Biol Chem. 2002;277(49):47080-47087. [DOI] [PubMed] [Google Scholar]
- 126.Bodnar RJ, Gu M, Li Z, Englund GD, Du X. The cytoplasmic domain of the platelet glycoprotein Ibalpha is phosphorylated at serine 609. J Biol Chem. 1999;274(47):33474-33479. [DOI] [PubMed] [Google Scholar]
- 127.Mangin P, David T, Lavaud V, et al. . Identification of a novel 14-3-3zeta binding site within the cytoplasmic tail of platelet glycoprotein Ibalpha. Blood. 2004;104(2):420-427. [DOI] [PubMed] [Google Scholar]
- 128.Gu M, Du X. A novel ligand-binding site in the zeta-form 14-3-3 protein recognizing the platelet glycoprotein Ibalpha and distinct from the c-Raf-binding site. J Biol Chem. 1998;273(50):33465-33471. [DOI] [PubMed] [Google Scholar]
- 129.Yuan Y, Zhang W, Yan R, et al. . Identification of a novel 14-3-3zeta binding site within the cytoplasmic domain of platelet glycoprotein Ibalpha that plays a key role in regulating the von Willebrand factor binding function of glycoprotein Ib-IX. Circ Res. 2009;105(12):1177-1185. [DOI] [PubMed] [Google Scholar]
- 130.Feng S, Christodoulides N, Reséndiz JC, Berndt MC, Kroll MH. Cytoplasmic domains of GpIbalpha and GpIbbeta regulate 14-3-3zeta binding to GpIb/IX/V. Blood. 2000;95(2):551-557. [PubMed] [Google Scholar]
- 131.Calverley DC, Kavanagh TJ, Roth GJ. Human signaling protein 14-3-3zeta interacts with platelet glycoprotein Ib subunits Ibalpha and Ibbeta. Blood. 1998;91(4):1295-1303. [PubMed] [Google Scholar]
- 132.Dong JF, Li CQ, Sae-Tung G, Hyun W, Afshar-Kharghan V, López JA. The cytoplasmic domain of glycoprotein (GP) Ibalpha constrains the lateral diffusion of the GP Ib-IX complex and modulates von Willebrand factor binding. Biochemistry. 1997;36(41):12421-12427. [DOI] [PubMed] [Google Scholar]
- 133.Perrault C, Mangin P, Santer M, et al. . Role of the intracellular domains of GPIb in controlling the adhesive properties of the platelet GPIb/V/IX complex. Blood. 2003;101(9):3477-3484. [DOI] [PubMed] [Google Scholar]
- 134.Du X. Signaling and regulation of the platelet glycoprotein Ib-IX-V complex. Curr Opin Hematol. 2007;14(3):262-269. [DOI] [PubMed] [Google Scholar]
- 135.Camoni L, Di Lucente C, Visconti S, Aducci P. The phytotoxin fusicoccin promotes platelet aggregation via 14-3-3-glycoprotein Ib-IX-V interaction. Biochem J. 2011;436(2):429-436. [DOI] [PubMed] [Google Scholar]
- 136.Mistry N, Cranmer SL, Yuan Y, et al. . Cytoskeletal regulation of the platelet glycoprotein Ib/V/IX-von willebrand factor interaction. Blood. 2000;96(10):3480-3489. [PubMed] [Google Scholar]
- 137.Shimaoka M, Xiao T, Liu JH, et al. . Structures of the alpha L I domain and its complex with ICAM-1 reveal a shape-shifting pathway for integrin regulation. Cell. 2003;112(1):99-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Ju L, Chen Y, Li K, et al. . Dual Biomembrane Force Probe enables single-cell mechanical analysis of signal crosstalk between multiple molecular species. Sci Rep. 2017;7(1):14185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yap CL, Hughan SC, Cranmer SL, et al. . Synergistic adhesive interactions and signaling mechanisms operating between platelet glycoprotein Ib/IX and integrin alpha IIbbeta 3. Studies in human platelets ans transfected Chinese hamster ovary cells. J Biol Chem. 2000;275(52):41377-41388. [DOI] [PubMed] [Google Scholar]
- 140.Nesbitt WS, Giuliano S, Kulkarni S, Dopheide SM, Harper IS, Jackson SP. Intercellular calcium communication regulates platelet aggregation and thrombus growth. J Cell Biol. 2003;160(7):1151-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Estevez B, Du X. New concepts and mechanisms of platelet activation signaling. Physiology (Bethesda). 2017;32(2):162-177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ju L, Chen Y, Xue L, Du X, Zhu C. Cooperative unfolding of distinctive mechanoreceptor domains transduces force into signals. Elife. 2016;5:e15447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang W, Deng W, Zhou L, et al. . Identification of a juxtamembrane mechanosensitive domain in the platelet mechanosensor glycoprotein Ib-IX complex. Blood. 2015;125(3):562-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Deng W, Xu Y, Chen W, et al. . Platelet clearance via shear-induced unfolding of a membrane mechanoreceptor. Nat Commun. 2016;7:12863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Coughlin SR. Thrombin signalling and protease-activated receptors. Nature. 2000;407(6801):258-264. [DOI] [PubMed] [Google Scholar]
- 146.Estevez B, Kim K, Delaney MK, et al. . Signaling-mediated cooperativity between glycoprotein Ib-IX and protease-activated receptors in thrombin-induced platelet activation. Blood. 2016;127(5):626-636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Jamieson GA, Okumura T, Hasitz M. Structure and function of platelet glycocalicin. Thromb Haemost. 1980;42(5):1673-1678. [PubMed] [Google Scholar]
- 148.McKeown LP, Williams SB, Hansmann KE, Krutzsch H, Gralnick HR. Glycoprotein Ib alpha peptides inhibit thrombin and SFLLRN-induced platelet aggregation. J Lab Clin Med. 1996;128(5):492-495. [DOI] [PubMed] [Google Scholar]
- 149.Bergmeier W, Piffath CL, Goerge T, et al. . The role of platelet adhesion receptor GPIbalpha far exceeds that of its main ligand, von Willebrand factor, in arterial thrombosis. Proc Natl Acad Sci USA. 2006;103(45):16900-16905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.De Candia E, Hall SW, Rutella S, Landolfi R, Andrews RK, De Cristofaro R. Binding of thrombin to glycoprotein Ib accelerates the hydrolysis of Par-1 on intact platelets. J Biol Chem. 2001;276(7):4692-4698. [DOI] [PubMed] [Google Scholar]
- 151.Ramakrishnan V, DeGuzman F, Bao M, Hall SW, Leung LL, Phillips DR. A thrombin receptor function for platelet glycoprotein Ib-IX unmasked by cleavage of glycoprotein V. Proc Natl Acad Sci USA. 2001;98(4):1823-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Adam F, Guillin MC, Jandrot-Perrus M. Glycoprotein Ib-mediated platelet activation. A signalling pathway triggered by thrombin. Eur J Biochem. 2003;270(14):2959-2970. [DOI] [PubMed] [Google Scholar]
- 153.Delaney MK, Liu J, Zheng Y, Berndt MC, Du X. The role of Rac1 in glycoprotein Ib-IX-mediated signal transduction and integrin activation. Arterioscler Thromb Vasc Biol. 2012;32(11):2761-2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yin H, Liu J, Li Z, Berndt MC, Lowell CA, Du X. Src family tyrosine kinase Lyn mediates VWF/GPIb-IX-induced platelet activation via the cGMP signaling pathway. Blood. 2008;112(4):1139-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Li Z, Zhang G, Liu J, et al. . An important role of the SRC family kinase Lyn in stimulating platelet granule secretion. J Biol Chem. 2010;285(17):12559-12570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Yap CL, Anderson KE, Hughan SC, Dopheide SM, Salem HH, Jackson SP. Essential role for phosphoinositide 3-kinase in shear-dependent signaling between platelet glycoprotein Ib/V/IX and integrin alpha(IIb)beta(3). Blood. 2002;99(1):151-158. [DOI] [PubMed] [Google Scholar]
- 157.Hodivala-Dilke KM, McHugh KP, Tsakiris DA, et al. . Beta3-integrin-deficient mice are a model for Glanzmann thrombasthenia showing placental defects and reduced survival. J Clin Invest. 1999;103(2):229-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Stojanovic A, Marjanovic JA, Brovkovych VM, et al. . A phosphoinositide 3-kinase-AKT-nitric oxide-cGMP signaling pathway in stimulating platelet secretion and aggregation. J Biol Chem. 2006;281(24):16333-16339. [DOI] [PubMed] [Google Scholar]
- 159.Li Z, Zhang G, Feil R, Han J, Du X. Sequential activation of p38 and ERK pathways by cGMP-dependent protein kinase leading to activation of the platelet integrin alphaIIb beta3. Blood. 2006;107(3):965-972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Li Z, Xi X, Du X. A mitogen-activated protein kinase-dependent signaling pathway in the activation of platelet integrin alpha IIbbeta3. J Biol Chem. 2001;276(45):42226-42232. [DOI] [PubMed] [Google Scholar]
- 161.Estevez B, Stojanovic-Terpo A, Delaney MK, et al. . LIM kinase-1 selectively promotes glycoprotein Ib-IX-mediated TXA2 synthesis, platelet activation, and thrombosis. Blood. 2013;121(22):4586-4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Mu FT, Andrews RK, Arthur JF, et al. . A functional 14-3-3zeta-independent association of PI3-kinase with glycoprotein Ib alpha, the major ligand-binding subunit of the platelet glycoprotein Ib-IX-V complex. Blood. 2008;111(9):4580-4587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Wu Y, Asazuma N, Satoh K, et al. . Interaction between von Willebrand factor and glycoprotein Ib activates Src kinase in human platelets: role of phosphoinositide 3-kinase. Blood. 2003;101(9):3469-3476. [DOI] [PubMed] [Google Scholar]
- 164.Le Behot A, Gauberti M, Martinez De Lizarrondo S, et al. . GpIbα-VWF blockade restores vessel patency by dissolving platelet aggregates formed under very high shear rate in mice. Blood. 2014;123(21):3354-3363. [DOI] [PubMed] [Google Scholar]
- 165.Yin H, Stojanovic-Terpo A, Xu W, et al. . Role for platelet glycoprotein Ib-IX and effects of its inhibition in endotoxemia-induced thrombosis, thrombocytopenia, and mortality. Arterioscler Thromb Vasc Biol. 2013;33(11):2529-2537. [DOI] [PMC free article] [PubMed] [Google Scholar]