Abstract
Tests for isolation by distance (IBD) are the most commonly used method of assessing spatial genetic structure. Many studies have exclusively used mitochondrial DNA (mtDNA) sequences to test for IBD, but this marker is often in conflict with multilocus markers. Here, we report a review of the literature on IBD, with the aims of determining (a) whether significant IBD is primarily a result of lumping spatially discrete populations, and (b) whether microsatellite datasets are more likely to detect IBD when mtDNA does not. We also provide empirical data from four species in which mtDNA failed to detect IBD by comparing these with microsatellite and SNP data. Our results confirm that IBD is mostly found when distinct regional populations are pooled, and this trend disappears when each is analysed separately. Discrepancies between markers were found in almost half of the studies reviewed, and microsatellites were more likely to detect IBD when mtDNA did not. Our empirical data rejected the lack of IBD in the four species studied, and support for IBD was particularly strong for the SNP data. We conclude that mtDNA sequence data are often not suitable to test for IBD, and can be misleading about species’ true dispersal potential. The observed failure of mtDNA to reliably detect IBD, in addition to being a single-locus marker, is likely a result of a selection-driven reduction in genetic diversity obscuring spatial genetic differentiation.
Introduction
Species with wide distributions are rarely completely panmictic1, even in potentially open systems such as the world’s oceans2,3. When individual dispersal distances are smaller than a species’ range, genetic drift acting on neutral genetic markers will over time result in a positive correlation between genetic differentiation between locations and the geographic distance separating them, a pattern known as “isolation by distance” (IBD)4,5. IBD is currently the most frequently tested hypothesis in studies of spatial genetic structure, and is considered to provide a suitable means of indirectly estimating a species’ dispersal ability6,7. The method is also considered important in the management of exploited species. For example, in high-dispersal marine species in which the existence of clear stock boundaries is unlikely, tests for IBD can help to define regional geographic units that respond to exploitation relatively independently8.
Despite their popularity, tests for IBD are often criticised. One recent debate concerns the use of Mantel tests in assessing relationships between genetic and spatial data9. It has been suggested that the power of other approaches (e.g. linear correlation, regression and canonical analysis) is far greater than that of Mantel tests in detecting a relationship when one is present10. Another problem, and one that affects IBD per se rather than merely the way in which it is identified, concerns its use under inappropriate conditions. Most prevalently, IBD can be strongly affected by the existence of distinct regional genetic clusters11. For example, secondary contact between populations that have been isolated from each other in glacial refugia12 or the existence of sister lineages in adjacent biogeographical regions13, will give the impression of limited dispersal ability even when each population is genetically homogeneous within its range. The fact that some regional genetic clusters display physiological adaptations to the region they occupy, and some are morphologically distinguishable from each other14,15 suggests that significant IBD may often be the result of inappropriately lumping distinct cryptic species. When these regional units are analysed individually16–20 or when stratified Mantel tests are applied and locations of populations are permuted within clusters21, the pattern of IBD is often no longer found, and the species’ true dispersal potential is revealed. Nonetheless, due to their simplicity and the fact that geographic distance can serve as a proxy for numerous other factors that limit gene flow between locations, tests for IBD will likely continue to be used as a standard method of assessing a species’ dispersal ability. As such, they represent a simple baseline method against which to test more complex landscape genetic approaches22.
One aspect of IBD that has received little prior attention is the question which genetic markers are most suitable to detect it, and under which conditions. Numerous studies have dealt with the fact that the power of different genetic markers to detect genetic structure can differ considerably, and may result in conflicting findings23,24, but the implications of this on tests for IBD remain poorly explored. With the increased application of next-generation sequencing approaches in species-level studies, the information content of the markers that can be used for IBD has increased by several orders of magnitude. It is thus timely to determine how traditional markers such as single-locus mtDNA sequence data or a limited number of microsatellite loci perform relative to the much larger SNP data sets.
In the present paper, we searched the literature to get an overview of the performance of mtDNA compared to other markers. An ad-hoc Google Scholar search using the search terms DNA barcoding, isolation by distance and mitochondria returned 17 800 hits. Although not all these studies would have conducted tests for IBD using this single locus, the result suggests that the use of mtDNA in tests for IBD is very common, and, despite the existence of more powerful alternatives, remains a standard method to assess species’ dispersal potential. We then compared microsatellite and selectively neutral single nucleotide polymorphism (SNP) datasets generated using double-digest restriction site associated DNA sequencing (ddRADseq, i.e. a reduced representation genotyping method employing high-throughput sequencing25) with mtDNA data in four cases where no IBD was found with the latter marker. The data sets used in this study originate from widespread marine species with high dispersal ability (long-lived planktonic larvae and/or active adult dispersal) along continuous coastlines. The finding that the alternative markers consistently found strong evidence for IBD when none was found with mtDNA when accounting for the existence of spatially distinct population clusters suggests that the exclusive use of the latter can result in questionable conclusions about species’ dispersal ability.
Materials and Methods
Literature review
To gain an overview about the performance of mtDNA in identifying IBD, we searched for (a) studies that used this marker to test for IBD along the South African coastline (where the ranges of the evolutionary lineages of high-dispersal coastal species tend to be strongly linked to the region’s temperature-defined marine biogeographic provinces13,26), and along the coast of temperate southern Australia. The aim was to determine whether significant IBD is mostly the result of inappropriately pooling distinct regional genetic clusters (sensu Meirmans et al.11). We only included species with high dispersal potential (i.e. those with active adult dispersal and/or planktonic larvae) because in low-dispersal species (i.e. whose offspring remains in the parent habitat) genetic structure is often not clearly linked with contemporary biogeography13,27. We also searched for (b) studies that compared IBD between mtDNA and microsatellites. For (b), various combinations of the following search terms were used in Google Scholar to identify suitable papers: isolation by distance (or isolation by geographic distance), IBD, mitochondria (or mtDNA), discrepancy, contrasting, conflicting, microsatellite. A table was then constructed for each, and general trends discussed.
Acquisition and generation of genetic data
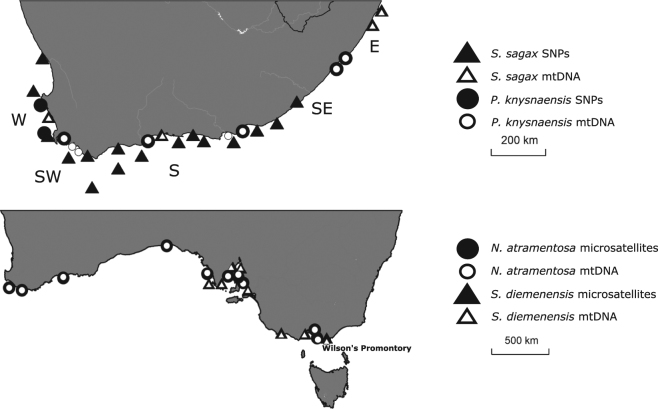
We compared IBD in four marine species that occur along continuous marine regions in temperate southern Australia and South Africa. These include two Australian gastropods (the snail Nerita atramentosa and the limpet Siphonaria diemenensis), and two South African teleosts (the goby Psammogobius knysnaensis and the sardine Sardinops sagax). These species were selected because (a) comprehensive range-wide genetic data sets were available and (b) none exhibits mtDNA-based genetic structure, even though co-distributed species with similar dispersal potential may comprise multiple regional evolutionary lineages26,28. The mtDNA data were obtained from the following studies: N. atramentosa and S. diemenensis26; P. knysnaensis29; S. sagax: B. Chiazzari, unpubl. data. Data from other markers included microsatellites for the Australian gastropods (N. atramentosa30; S. diemenensis31), and SNP data for the South African teleosts. The latter were generated for the present study. Briefly, DNA was extracted from fin clip or muscle tissue using the cetyltrimethylammonium bromide (CTAB) method32. Double-digest restriction site associated DNA sequencing (ddRADseq) was performed at the Molecular Ecology Laboratory at Flinders University following the protocol of ddRADseq.25 with restriction enzymes SbfI and MseI and modifications as described in Brauer et al.33. Libraries were sequenced on an Illumina HiSeq 2000 platform at the McGill University and Genome Québec Innovation Centre. BayeScan v2.034 was then used to identify and exclude loci potentially under selection on the basis of having significantly elevated allele frequency differences between populations. We specified default settings and a false discovery rate of 5%. In order to create data sets comprising selectively neutral loci, we excluded 203 outlier SNPs identified with BayeScan from the total sardine data set (out of a total of 11649 SNPs) and 572 outlier SNPs from the goby data set (out of a total of 8543 SNPs), and used only the remaining loci to test for IBD. Sample sites for all four species are shown in Fig. 1 (see Supplementary Table S1 for details concerning sampling sites and number of samples). Ethics clearance for the sample acquisition of the two teleosts was granted by the ethics committee of the University of Johannesburg. All experiments were performed in accordance with relevant named guidelines and regulations.
Figure 1.
Sampling sites in South Africa (top) and temperate southern Australia (bottom). In both figures, small white symbols indicate mtDNA data and large black symbols represent either microsatellites or SNPs from ddRADseq. Marine bioregions in South Africa are indicated as W (west coast), SW (south-west coast), S (south coast), SE (south-east coast) and E (east coast); in Australia, distinct bioregions are separated by a biogeographic barrier at Wilson’s Promontory.
Isolation by geographical distance
Genetic distance matrices were created in Arlequin v3.5.2.235 that comprised F-statistics36 between pairs of sites, using ΦST for mtDNA data and FST36 for microsatellite and SNP data. The most suitable genetic distance model for each mtDNA data set was determined using the Bayesian Inference Criterion37 in MEGA v.7.0.2638 and were the Kimura 2-Parameter model39 for Sardinops sagax and Psammogobius knysnaensis, the Tamura-Nei model40 for Nerita atramentosa, and the Tamura 3-Parameter model41 for Siphonaria diemenensis. Geographical distances between pairs of sites were measured as the shortest along-coast distances using the path tool in Google Earth Pro v 7.3.0.3832.
Relationships between genetic and geographic distance matrices were calculated using (a) a Mantel test9 and, to account for possible type II error10 (b) simple linear regression. The Mantel tests were performed with the R package adegenet 1.4–042. This program tests for significant IBD by comparing the observed correlation with a histogram of simulated correlation categories and their frequency under the assumption of no IBD. We selected Spearman rank correlations43 and 999 matrix permutations. The program also creates scatterplots of geographic vs. genetic distance, that depict the density between points. Densities between points in a scatterplot can be used to determine whether the data originated from a single genetic cline or from two or more distinct regional clusters for each of which the test for IBD should be performed separately11. To confirm that the neutral SNP data did not comprise multiple genetic clusters, the k-means clustering algorithm in adegenet was used to identify the number of clusters that maximise between-cluster variation. Simple linear regression to corroborate the results of the Mantel tests was performed in SigmaStat 1.0 (Systat Software, San Jose, CA).
Data availability
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.
Results
Tests for IBD: literature review of patterns in two coastal regions
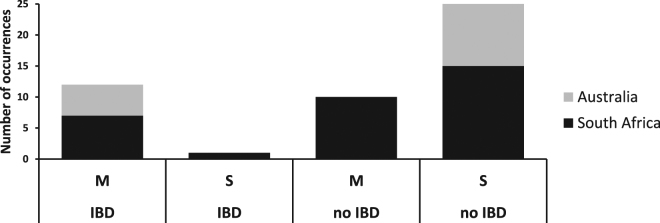
A synthesis of studies on the South African and southern Australian coastlines indicated that significant IBD was found much more frequently when these tests were performed on data sets that originated from multiple (M) biogeographical regions than when tests were performed within single regions (S) (Fig. 2, Supplementary Table S2). This trend was particularly clear in Australia, even though the fact that most studies employed microsatellites would intuitively suggest that the chances of finding significant IBD within provinces would be greater. A single study reported significant IBD within a single province (Parechinus angulosus, based on mtDNA data), but this result was considered unreliable because of a small sample size20.
Figure 2.
Number of instances of significant IBD and no IBD being found in South African (black) and temperate southern Australian (grey) coastal marine species when samples were collected across multiple marine biogeographical provinces (M) or within single provinces (S).
Particularly clear examples of significant IBD that was no longer found when data from individual biogeographical regions were analysed separately include Acanthochiton garnoti, Cyclograpsus puncatus, Oxystele tigrina, O. variegata20 and Tetraclita serrata44 in South Africa, as well as Donax deltoides45 and Haliotis rubra46 in Australia.
Test for IBD: literature review of marker types
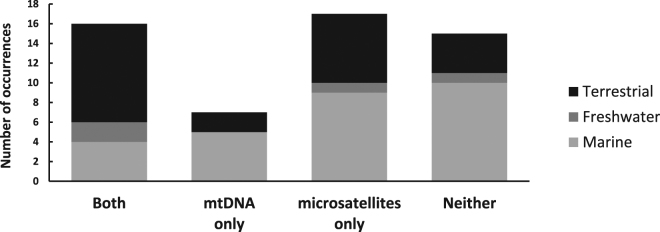
A comparison of IBD between mtDNA and microsatellites in studies that used both markers revealed that marker-specific discrepancies are common, and were found in almost half of the studies reviewed (Fig. 3, Supplementary Table S3). Microsatellites were more likely to identify significant IBD when mtDNA did not than the inverse, and there was no clear indication that this trend is habitat-specific.
Figure 3.
Number of studies that used both mtDNA and microsatellites to test for IBD in which significant IBD was found for both, either or neither marker. For each category, the habitat through which a particular taxon is most likely to disperse is indicated.
Tests for Isolation by Distance: this study
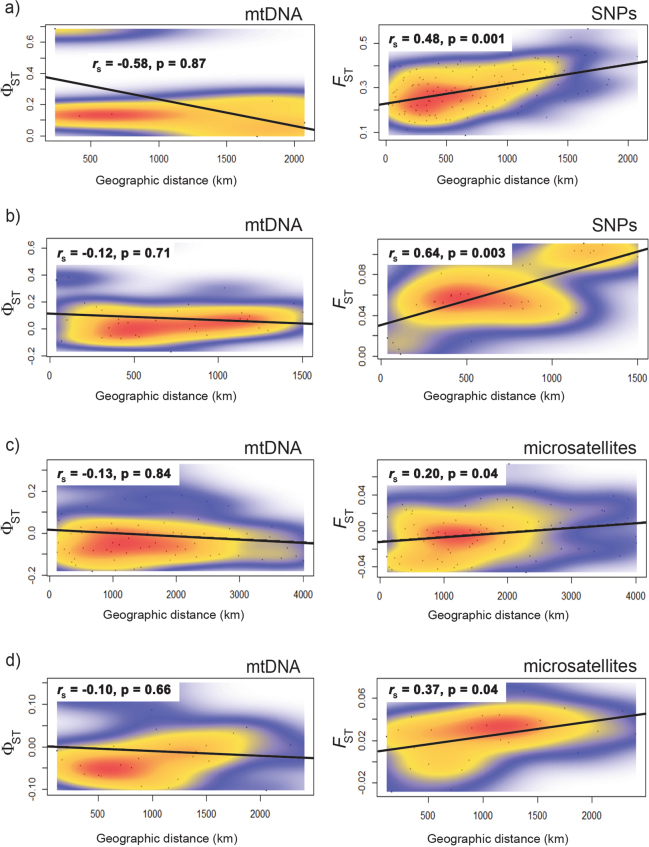
Mantel tests performed on geographic distance matrices and matrices comprising pairwise F-statistics between sites identified no IBD for any of the mtDNA data sets (Fig. 4, left panels); all correlations were negative, which indicates more differentiation within sites than between pairs of sites. In contrast, all correlations were positive for the nuclear markers (right panels) and Mantel tests were significant. Scatter plots with information on point density showed no strong evidence for the existence of multiple regional clusters for any species or marker, and k-means clustering confirmed that both SNP data sets comprised single genetic clusters (Supplementary Fig. S1), information that was already available for the other data sets26,29.
Figure 4.
Plots of geographic distances vs. F-statistics for the following species (plots on the left show mtDNA data, those on the right SNP or microsatellite data): (a) Sardinops sagax; (b) Psammogobius knysnaensis; (c) Nerita atramentosa; (d) Siphonaria diemenensis. The density of data points is indicated by colours.
The Mantel test results were confirmed by linear regression analysis, with the exception of the microsatellite data of N. atramentosa, where significant IBD was no longer found (Supplementary Table S4). The statistical power (i.e. the probability of correctly rejecting the null hypothesis of no IBD when IBD is present) of all non-significant tests was below the desired power of 0.847, indicating that these data sets were not sufficiently informative to identify IBD, and failure to identify it thus needs to be interpreted with caution. The power of the tests performed on the two SNP data sets was highest (both 1.0, i.e. the maximum possible).
As IBD analyses can be affected by the number of sites22, we reanalysed the sardine SNP data set (which included many sites for which no COI data were available, Supplementary Table S1) by including only four sites that were represented in both data sets. Both the Mantel test and the linear regression analyses remained signficant (Mantel rs = 0.83, P = 0.035; Linear regression R2 = 0.8, F = 32.0, P < 0.001), and the power of the latter was only slightly below that of the complete data set (0.97).
Discussion
Tests for isolation by distance that employed mtDNA sequence data became popular in the 1990s, when this became the marker of choice because of its faster mutation rate and smaller genome size compared to allozymes48,49. It has been used rather uncritically ever since, and it is still not fully understood why it is often in conflict with nuclear markers23, although the smaller effective population size50, which results in a higher rate of lineage sorting51, is often cited. It is likely that the increasing popularity of DNA barcoding will result in an increase in the number of studies that will exclusively use mtDNA sequences to test for IBD, as it is now possible to readily apply this approach on entire communities (e.g.52–54). For that reason, a critical assessment of the power of mtDNA in detecting IBD is timely.
mtDNA vs. microsatellites
Our review of studies that used mtDNA together with microsatellites indicated that the two markers were often in conflict, with microsatellites more likely to identify IBD when mtDNA did not. We are aware of no studies that employed power analyses to test for the probability of type II error, but linear regression analyses of our empirical data (Supplementary Table S4) indicate that the power of the mtDNA data sets normally used to test for IBD is often far below the acceptable threshold of 0.847, and non-significant results questionable. It is noteworthy that several additional studies we reviewed reported IBD for only one of these markers even though both were used in other analyses55–58, perhaps because the other was in conflict. Intuitively, one would expect a comprehensive data set of hypervariable microsatellites to be more suitable to detect IBD, not only because of their high mutation rate59,60 but also because different combinations of alleles arise in each generation and are sorted over geographical space61, making microsatellites useful to study spatial patterns at geographic scales as small as tens of meters62,63. As such, it is possible that some of the studies in which significant IBD was found with mtDNA data, but not microsatellites, may be a result of the latter employing microsatellite loci with comparatively little variation and/or a small number of loci, in addition to potential technical artefacts such as large allele dropout and null alleles. Any general conclusions about the usefulness of microsatellites to detect IBD is thus hampered by the fact that, unlike mtDNA sequences, the microsatellite data sets used in different studies are not directly comparable. However, a detailed assessment of the factors responsible for this discrepancy is beyond the scope of the present meta-analysis. Our results for the Australian gastropods, which employed highly informative microsatellite data sets suitable to detect self-recruitment30,31, and for which the same sites and individuals were used as for the corresponding mtDNA data, represent an important contribution towards facilitating more direct comparisons between the two marker types. In the light of these findings, it is clear that reliable results at a minimum require congruence between multiple marker types.
Pooling of distinct regional lineages
A review of studies on marine animals that tested for IBD further indicated that in many cases where a significant positive correlation was found with mtDNA data, it could potentially be attributed to the pooling of spatially discrete evolutionary lineages. There was no convincing evidence for significant IBD within single marine biogeographic provinces for any of the markers employed. Temperature is arguably the most important environmental predictor of coastal biogeographic patterns64, and in South Africa, many species are subdivided into regional evolutionary lineages whose ranges coincide with temperature-defined biogeography13. Physiological data indicate that sister lineages in adjacent marine regions have different thermal tolerance ranges14,15,65. Because of this, the pooling of data from multiple lineages does not provide information on species’ dispersal potential per se, because it ignores the fact that dispersal of regional evolutionary lineages is constrained by their inability to establish themselves in the habitat of their sister lineage rather than the inability to reach that habitat. The situation is similar in temperate Australia, where species that are present on either side of the marine biogeographic barrier at Wilson’s Promontory (Fig. 1) often comprise distinct southern and eastern lineages that do not readily establish themselves in each other’s habitat66,67 and that should be treated as distinct when testing for IBD. Numerous additional examples of distinct evolutionary lineages occurring along continuous coastlines that are separated by biogeographical transition zones have been reported elsewhere, including California and Florida27, Chile68,69 and various sites along the north-eastern Atlantic and the Mediterranean70,71. The above considerations likely apply to all of them.
A process-driven explanation for the poor performance of mtDNA
Mitochondrial DNA sequence data originate from a single locus and, unlike in microsatellite data, there is thus no differentiation in the frequency of alleles from different loci from one generation to the next that can generate spatial genetic structure. Nonetheless, the high mutation rate of mtDNA compared to nuclear DNA72 should theoretically make it suitable to study IBD because of high levels of genetic diversity at the intrapopulation level. When dispersal is limited, the frequency of new mutations that have arisen at a particular location is expected to decrease away from that location. A number of recent studies have rejected the idea that the high mutation rate per se results in high genetic variation at or below the species level, and have indicated that it is in fact much lower than expected under the neutral model of evolution73. Instead, the maximum genetic diversity (MGD) hypothesis, which assumes a saturation plateau beyond which genetic diversity no longer increases74,75 may describe mtDNA evolution more adequately. At the population level, the low level of mtDNA diversity is a result of strong variation-reducing selection that may include strong purifying selection73,76 or selective sweeps driven by physiological constraints within environmentally homogeneous regions77. Interestingly, this selection not only affects non-synonymous sites but also third character codon positions where mutations do not result in amino acid changes78–80. Although there is no consensus, explanations such as linkage of sites affected by synonymous mutations to regions under intense selection81–84 or codon biases during translation78,85,86 have been invoked. The consequence of such processes (which do not merely drive selection at the population level but even at the individual or organelle level73), is that mutations and reversions in mitochondrial genes at the population level are typically limited to relatively few sites, and homoplasies are common76,87. In the context of IBD, it is possible that the same mutations arising independently across the physiologically constrained range of a population may obscure already limited neutral genetic differentiation driven by geographic distance. It has been suggested that the combination of variation-reducing selection at the population or species level, and adaptive selection at the between-species level, makes mtDNA particularly powerful for the molecular differentiation of species because it increases the ‘barcoding gap’76,87. However, the very departures from the expectations of selective neutrality that are so useful for DNA barcoding may be partly responsible for making this marker unsuitable to detect IBD.
Relevance of the study findings to non-marine taxa
Many marine species have very large census population sizes88. This suggests that for selectively neutral loci, marine populations should also have very high genetic diversity, and this has been confirmed using nuclear DNA89–92. However, as explained in the previous paragraph, the assumption of selective neutrality does not apply to mtDNA. A meta-analysis of >1600 animal species91 showed that within-species (or population level) mtDNA diversity is very similar across animal phyla, irrespective whether they have large or small census population sizes, or whether they are aquatic or terrestrial. An explanation for the low genetic diversity of large populations is provided by Gillespie’s77 ‘genetic draft’ hypothesis, which states that beneficial mutations are more likely to arise in large populations, and these result in recurrent selective sweeps that reduce genetic diversity at the mitogenome level. Low variation in animal mtDNA, irrespective of census population size, range or habitat type has been confirmed by numerous phylogeographic and DNA barcoding studies93–97. These considerations indicate that there is little reason to assume that the failure of mtDNA to reliably detect IBD is limited to marine species, and this is confirmed by the fact that of the taxa used for Fig. 3, the proportion of studies that found discrepancies between mtDNA and microsatellites were very similar for different habitat types (39% of terrestrial taxa and 46% of aquatic taxa). As both aquatic and terrestrial species tend to be subdivided into distinct, geographically isolated evolutionary lineages98 that may be under divergent selection, the artefact of identifying IBD when distinct populations are pooled shown in Fig. 2 also applies to non-marine animal taxa.
Application of neutral SNP data
In addition to the issues in identifying IBD that are related to marker type and evolutionary history, the power to detect IBD is also related to the number of populations included in the analysis7,22. Together, these factors indicate that a sampling design hampered by insufficient financial commitment, capacity and sampling effort may result in spurious results that can be misleading about a species’ realized dispersal. Future research needs to focus on the amount of data required to reliably identify IBD when it is present. Our novel SNP data indicate that high throughput sequencing approaches may be suitable to resolve the problem of discrepancies between mtDNA and nuclear markers evident in numerous previous studies. The presence of significant positive relationships between genetic and geographic distance in species where no such relationship was evident for mtDNA data was particularly clear for the neutral SNP data, where the power to correctly detect IBD was maximal and the probability of a type II error was zero. However, given the costs that are presently involved in generating and analysing genome-wide SNP data, this type of marker could be seen as a standard against which less informative but more affordable genetic markers could be measured. We conclude that mtDNA data can be problematic when testing for IBD, as they often do not provide reliable information on a species’ true dispersal potential.
Electronic supplementary material
Acknowledgements
This study was funded by the National Research Foundation (CSUR Grant No. 87702 to P.R.T.), the University of Johannesburg (URC/FRC grant to P.R.T) and the Australian Research Council (FT130101068 and DP110101275 to L.B.B.). T.R.G. and A.E.-K. acknowledge the University of Johannesburg for Global Excellence and Stature (GES) fellowships for doctoral and postdoctoral study, respectively. We are grateful to two anonymous reviewers whose comments improved the quality of this manuscript.
Author Contributions
P.R.T. and L.B.B. designed the study; C.v.d.L., P.R.T., S.v.d.H., B.C. and J.S.-C. collected the samples; P.R.T. and T.R.G. conducted the literature review; P.R.T., J.S.-C., T.R.G. and A.E.-K. analysed the data; P.R.T., L.B.B., T.R.G. and S.v.d.H. wrote the paper, with input from C.v.d.L., J.S.-C., B.v.V. and B.C.
Competing Interests
The authors declare no competing interests.
Footnotes
Peter R. Teske and Tirupathi Rao Golla contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-25138-9.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Rousset F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics. 1997;145:1219. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wirth T, Bernatchez L. Genetic evidence against panmixia in the European eel. Nature. 2001;409:1037. doi: 10.1038/35059079. [DOI] [PubMed] [Google Scholar]
- 3.Gaither MR, et al. Depth as a driver of evolution in the deep sea: Insights from grenadiers (Gadiformes: Macrouridae) of the genus Coryphaenoides. Mol. Phylogenet. Evol. 2016;104:73–82. doi: 10.1016/j.ympev.2016.07.027. [DOI] [PubMed] [Google Scholar]
- 4.Wright S. Isolation by distance. Genetics. 1943;28:114–138. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Slatkin M. Isolation by distance in equilibrium and non-equilibrium populations. Evolution. 1993;47:264–279. doi: 10.1111/j.1558-5646.1993.tb01215.x. [DOI] [PubMed] [Google Scholar]
- 6.Bradbury IR, Bentzen P. Non-linear genetic isolation by distance: implications for dispersal estimation in anadromous and marine fish populations. Mar. Ecol. Prog. Ser. 2007;340:245–257. doi: 10.3354/meps340245. [DOI] [Google Scholar]
- 7.Baco AR, et al. A synthesis of genetic connectivity in deep-sea fauna and implications for marine reserve design. Mol. Ecol. 2016;25:3276–3298. doi: 10.1111/mec.13689. [DOI] [PubMed] [Google Scholar]
- 8.Cunningham KMM, Canino MFF, Spies IBB, Hauser L. Genetic isolation by distance and localized fjord population structure in Pacific cod (Gadus macrocephalus): limited effective dispersal in the northeastern Pacific Ocean. Can. J. Fish. Aquat. Sci. 2009;66:153–166. doi: 10.1139/F08-199. [DOI] [Google Scholar]
- 9.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- 10.Legendre P, Fortin M-J. Comparison of the Mantel test and alternative approaches for detecting complex multivariate relationships in the spatial analysis of genetic data. Mol. Ecol. Resour. 2010;10:831–844. doi: 10.1111/j.1755-0998.2010.02866.x. [DOI] [PubMed] [Google Scholar]
- 11.Meirmans PG. The trouble with isolation by distance. Mol. Ecol. 2012;21:2839–2846. doi: 10.1111/j.1365-294X.2012.05578.x. [DOI] [PubMed] [Google Scholar]
- 12.Taberlet P, Fumagalli L, Wust‐saucy A, Cosson J. Comparative phylogeography and postglacial colonization routes in Europe. Mol. Ecol. 1998;7:453–464. doi: 10.1046/j.1365-294x.1998.00289.x. [DOI] [PubMed] [Google Scholar]
- 13.Teske PR, von der Heyden S, McQuaid CD, Barker NP. A review of marine phylogeography in southernAfrica. South Afr. J. Sci. 2011;107:1–11. [Google Scholar]
- 14.Teske PR, et al. Oceanic dispersal barriers, adaptation and larval retention: an interdisciplinary assessment of potential factors maintaining a phylogeographic break between sister lineages of an African prawn. BMC Evol. Biol. 2008;8:341–341. doi: 10.1186/1471-2148-8-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Papadopoulos I, Teske PR. Larval development reflects biogeography in two formerly synonymised southern African coastal crabs. Afr. J. Aquat. Sci. 2014;39:347–350. doi: 10.2989/16085914.2014.938600. [DOI] [Google Scholar]
- 16.Kuchta SR, Tan A. Isolation by distance and post-glacial range expansion in the rough-skinned newt. Taricha granulosa. Mol. Ecol. 2005;14:225–244. doi: 10.1111/j.1365-294X.2004.02388.x. [DOI] [PubMed] [Google Scholar]
- 17.Marmi J, et al. Mitochondrial DNA reveals a strong phylogeographic structure in the badger across Eurasia. Mol. Ecol. 2006;15:1007–1020. doi: 10.1111/j.1365-294X.2006.02747.x. [DOI] [PubMed] [Google Scholar]
- 18.Lukoschek V, Waycott M, Keosh JS. Relative information content of polymorphic microsatellites and mitochondrial DNA for inferring dispersal and population genetic structure in the olive sea snake, Aipysurus laevis. Mol. Ecol. 2008;17:3062–3077. doi: 10.1111/j.1365-294X.2008.03815.x. [DOI] [PubMed] [Google Scholar]
- 19.Durand DJ, Blel H, Shen KN, Guinand G. Population genetic structure of Mugil cephalus in the Mediterranean and Black Seas: a single mitochondrial clade and many nuclear barriers. Mar. Ecol. Prog. Ser. 2013;474:243–261. doi: 10.3354/meps10080. [DOI] [Google Scholar]
- 20.Wright D, Bishop JM, Matthee CA, von der Heyden S. Genetic isolation by distance reveals restricted dispersal across a range of life histories: implications for biodiversity conservation planning across highly variable marine environments. Divers. Distrib. 2015;21:698–710. doi: 10.1111/ddi.12302. [DOI] [Google Scholar]
- 21.Oksanen J, et al. The vegan package. Community Ecol. Package. 2007;10:631–637. [Google Scholar]
- 22.Jenkins DG, et al. A meta-analysis of isolation by distance: relic or reference standard for landscape genetics? Ecography. 2010;33:315–320. [Google Scholar]
- 23.Toews DPL, Brelsford A. The biogeography of mitochondrial and nuclear discordance in animals. Mol. Ecol. 2012;21:3907–3930. doi: 10.1111/j.1365-294X.2012.05664.x. [DOI] [PubMed] [Google Scholar]
- 24.Teske PR. Connectivity in solitary ascidians: Is a 24-h propagule duration sufficient to maintain large-scale genetic homogeneity? Mar. Biol. 2014;161:2681–2687. doi: 10.1007/s00227-014-2522-7. [DOI] [Google Scholar]
- 25.Peterson, B. K., Weber, J. N., Kay, E. H., Fisher, H. S. & Hoekstra, H. E. Double digest RADseq: An inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PLoS One 7, e37135 (2012). [DOI] [PMC free article] [PubMed]
- 26.Teske PR, Sandoval-Castillo J, Waters J, Beheregaray LB. An overview of Australia’s temperate marine phylogeography, with new evidence from high-dispersal gastropods. J. Biogeogr. 2017;44:217–229. doi: 10.1111/jbi.12783. [DOI] [Google Scholar]
- 27.Pelc RA, Warner RR, Gaines SD. Geographical patterns of genetic structure in marine species with contrasting life histories. J. Biogeogr. 2009;36:1881–1890. doi: 10.1111/j.1365-2699.2009.02138.x. [DOI] [Google Scholar]
- 28.Teske PR, et al. Climate-driven genetic divergence of limpets with different life histories across a southeast African marine biogeographic disjunction: Different processes, same outcome. Mol. Ecol. 2011;20:5025–5041. doi: 10.1111/j.1365-294X.2011.05307.x. [DOI] [PubMed] [Google Scholar]
- 29.Drost, E., Golla, T. R., von der Heyden, S. & Teske, P. R. No divergent evolution, despite restricted connectivity, between Atlantic and Indian Ocean goby populations. Mar. Biodivers. 46, 465–471 (2016).
- 30.Teske P, Sandoval-Castillo J, Sasaki M, Beheregaray L. Invasion success of a habitat-forming marine invertebrate is limited by lower-than-expected dispersal ability. Mar. Ecol. Prog. Ser. 2015;536:221–227. doi: 10.3354/meps11463. [DOI] [Google Scholar]
- 31.Teske, P. R., Sandoval-Castillo, J., Waters, J. & Beheregaray, L. B. An overview of temperate Australian marine phylogeography, with new evidence from high-dispersal gastropods. J. Biogeogr. 44, 217–229 (2016).
- 32.Doyle, J. CTAB total DNA isolation. In Molecular Techniques in Taxonomy 283–293 (1991).
- 33.Brauer CJ, Hammer MP, Beheregaray LB. Riverscape genomics of a threatened fish across a hydroclimatically heterogeneous river basin. Mol. Ecol. 2016;25:5093–5113. doi: 10.1111/mec.13830. [DOI] [PubMed] [Google Scholar]
- 34.Foll M, Gaggiotti O. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: A Bayesian perspective. Genetics. 2008;180:977–993. doi: 10.1534/genetics.108.092221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 36.Wright S. The interpretation of population structure by F-statistics with special regard to systems of mating. Evolution. 1965;19:395–420. doi: 10.1111/j.1558-5646.1965.tb01731.x. [DOI] [Google Scholar]
- 37.Schwarz, Gideon. Estimating the dimension of a model. Ann Stat. 6, 461–464 (1978).
- 38.Kumar S, Stecher G, Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120 (1980). [DOI] [PubMed]
- 40.Tamura, K. & Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 10, 512–526 (1993). [DOI] [PubMed]
- 41.Tamura K. Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol. Biol. Evol. 1992;9:678–687. doi: 10.1093/oxfordjournals.molbev.a040752. [DOI] [PubMed] [Google Scholar]
- 42.Jombart T. Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics. 2008;24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- 43.Spearman C. The proof and measurement of association between two things. Am. J. Psychol. 1904;15:72–101. doi: 10.2307/1412159. [DOI] [PubMed] [Google Scholar]
- 44.Reynolds, T. V., Matthee, C. A. & von der Heyden, S. The influence of pleistocene climatic changes and ocean currents on the phylogeography of the southern African barnacle, Tetraclita serrata (Thoracica; Cirripedia). PLoS One 9, e102115 (2014). [DOI] [PMC free article] [PubMed]
- 45.Miller AD, Versace VL, Matthews TG, Montgomery S, Bowie KC. Ocean currents influence the genetic structure of an intertidal mollusc in southeastern Australia - implications for predicting the movement of passive dispersers across a marine biogeographic barrier. Ecol. Evol. 2013;3:1248–1261. doi: 10.1002/ece3.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miller AD, et al. Contrasting patterns of population connectivity between regions in a commercially important mollusc Haliotis rubra: integrating population genetics, genomics and marine LiDAR data. Mol. Ecol. 2016;25:3845–3864. doi: 10.1111/mec.13734. [DOI] [PubMed] [Google Scholar]
- 47.Cohen, J. Statistical power analysis for the behavioral sciences. Hilsdale. NJ Lawrence Earlbaum Assoc. 2 (1988).
- 48.Hurst CD, Skibinski DOF. Comparison of allozyme and mitochondrial DNA spatial differentiation in the limpet Patella vulgata. Mar. Biol. 1995;122:257–263. [Google Scholar]
- 49.Chenoweth SF, Hughes JM, Keenan CP, Lavery S. When oceans meet: a teleost shows secondary intergradation at an Indian–Pacific interface. Proc. R. Soc. Lond. B Biol. Sci. 1998;265:415–420. doi: 10.1098/rspb.1998.0310. [DOI] [Google Scholar]
- 50.Lynch JA, Olesnicky EC, Desplan C. Regulation and function of tailless in the long germ wasp Nasonia vitripennis. Dev. Genes Evol. 2006;216:493–498. doi: 10.1007/s00427-006-0076-5. [DOI] [PubMed] [Google Scholar]
- 51.Funk DJ, Omland KE. Species-level paraphyly and polyphyly: Frequency, causes, and consequences, with insights from animal mitochondrial DNA. Annu. Rev. Ecol. Evol. Syst. 2003;34:397–423. doi: 10.1146/annurev.ecolsys.34.011802.132421. [DOI] [Google Scholar]
- 52.Fouquet A, et al. Underestimation of species richness in neotropical frogs revealed by mtDNA analyses. PloS One. 2007;2:1–10. doi: 10.1371/journal.pone.0001109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kathleen J. et al. Population genetics of ecological communities with DNA barcodes: An example from New Guinea Lepidoptera. Proc. Natl. Acad. Sci. USA107, 5041–5046 (2010). [DOI] [PMC free article] [PubMed]
- 54.Guarnizo CE, et al. DNA barcoding survey of anurans across the Eastern Cordillera of Colombia and the impact of the andes on cryptic diversity. PLoS One. 2015;10:1–20. doi: 10.1371/journal.pone.0127312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Borrell YJ, Piñera JA, Sánchez Prado JA, Blanco G. Mitochondrial DNA and microsatellite genetic differentiation in the European anchovy Engraulis encrasicolus L. ICES J. Mar. Sci. 2012;69:1357–1371. doi: 10.1093/icesjms/fss129. [DOI] [Google Scholar]
- 56.Lah L, et al. Spatially explicit analysis of genome-wide SNPs detects subtle population structure in a mobile marine mammal, the harbor porpoise. PLoS One. 2016;11:1–23. doi: 10.1371/journal.pone.0162792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Wyngaarden M, et al. Identifying patterns of dispersal, connectivity and selection in the sea scallop, Placopecten magellanicus, using RADseq-derived SNPs. Evol. Appl. 2017;10:102–117. doi: 10.1111/eva.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu, S. et al. Genomic evidence for local adaptation in the ovoviviparous marine fish Sebastiscus marmoratus with a background of population homogeneity. Sci. Rep. 7, 1562 (2017). [DOI] [PMC free article] [PubMed]
- 59.DeWoody JA, Avise JC. Microsatellite variation in marine, freshwater and anadromous fishes compared with other animals. J. Fish Biol. 2000;56:461–473. doi: 10.1111/j.1095-8649.2000.tb00748.x. [DOI] [Google Scholar]
- 60.Baldwin RE, Rew MB, Johansson ML, Banks MA, Jacobson KC. Population structure of three species of Anisakis nematodes recovered from Pacific sardines (Sardinops sagax) distributed throughout the California Current system. J. Parasitol. 2011;97:545–554. doi: 10.1645/GE-2690.1. [DOI] [PubMed] [Google Scholar]
- 61.Sunnucks P. Efficient genetic markers for population biology. Trends Ecol. Evol. 2000;15:199–203. doi: 10.1016/S0169-5347(00)01825-5. [DOI] [PubMed] [Google Scholar]
- 62.Reusch TBH, Häberli MA, Aeschlimann PB, Milinski M. Female sticklebacks count alleles in a strategy of sexual selection explaining MHC polymorphism. Nature. 2001;414:300. doi: 10.1038/35104547. [DOI] [PubMed] [Google Scholar]
- 63.Ronfort J, et al. Microsatellite diversity and broad scale geographic structure in a model legume: building a set of nested core collection for studying naturally occurring variation in Medicago truncatula. BMC Plant Biol. 2006;6:28. doi: 10.1186/1471-2229-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Belanger C, et al. Global environmental predictors of benthic marine biogeographic structure. Proc. Natl. Acad. Sci. USA. 2012;109:14046–14051. doi: 10.1073/pnas.1212381109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zardi GI, Nicastro KR, McQuaid CD, Hancke L, Helmuth B. The combination of selection and dispersal helps explain genetic structure in intertidal mussels. Oecologia. 2011;165:947–958. doi: 10.1007/s00442-010-1788-9. [DOI] [PubMed] [Google Scholar]
- 66.Waters JM. Driven by the West Wind Drift? A synthesis of southern temperate marine biogeography, with new directions for dispersalism. J. Biogeogr. 2008;35:417–427. doi: 10.1111/j.1365-2699.2007.01724.x. [DOI] [Google Scholar]
- 67.York KL, Blacket MJ, Appleton BR. The Bassian Isthmus and the major ocean currents of southeast Australia influence the phylogeography and population structure of a southern Australian intertidal barnacle Catomerus polymerus (Darwin) Mol. Ecol. 2008;17:1948–1961. doi: 10.1111/j.1365-294X.2008.03735.x. [DOI] [PubMed] [Google Scholar]
- 68.Brante A, Fernández M, Viard F. Phylogeography and biogeography concordance in the marine gastropod Crepipatella dilatata (Calyptraeidae) along the southeastern Pacific coast. J. Hered. 2012;103:630–637. doi: 10.1093/jhered/ess030. [DOI] [PubMed] [Google Scholar]
- 69.Haye PA, et al. Phylogeographic structure in benthic marine invertebrates of the southeast Pacific coast of Chile with differing dispersal potential. PLoS One. 2014;9:e88613. doi: 10.1371/journal.pone.0088613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Patarnello T, Volckaert FA, Castilho R. Pillars of Hercules: is the Atlantic–Mediterranean transition a phylogeographical break? Mol. Ecol. 2007;16:4426–4444. doi: 10.1111/j.1365-294X.2007.03477.x. [DOI] [PubMed] [Google Scholar]
- 71.Lourenço CR, et al. Evidence for rangewide panmixia despite multiple barriers to dispersal in a marine mussel. Sci. Rep. 2017;7:10279. doi: 10.1038/s41598-017-10753-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brown WM, George M, Wilson AC. Rapid evolution of animal mitochondrial DNA. Proc. Natl. Acad. Sci. 1979;76:1967. doi: 10.1073/pnas.76.4.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Stewart JB, Freyer C, Elson JL, Larsson N-G. Purifying selection of mtDNA and its implications for understanding evolution and mitochondrial disease. Nat. Rev. Genet. 2008;9:657. doi: 10.1038/nrg2396. [DOI] [PubMed] [Google Scholar]
- 74.Huang YM, Xia MY, Huang S. Evolutionary process unveiled by the maximum genetic diversity hypothesis. Yi Chuan. 2013;35:599–606. doi: 10.3724/SP.J.1005.2013.00599. [DOI] [PubMed] [Google Scholar]
- 75.Huang S. New thoughts on an old riddle: What determines genetic diversity within and between species? Genomics. 2016;108:3–10. doi: 10.1016/j.ygeno.2016.01.008. [DOI] [PubMed] [Google Scholar]
- 76.Stoeckle MY, Thaler DS. DNA barcoding works in practice but not in (neutral) theory. PLoS One. 2014;9:e100755. doi: 10.1371/journal.pone.0100755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gillespie JH. The neutral theory in an infinite population. Gene. 2000;261:11–18. doi: 10.1016/S0378-1119(00)00485-6. [DOI] [PubMed] [Google Scholar]
- 78.Chamary JV, Parmley JL, Hurst LD. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat. Rev. Genet. 2006;7:98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- 79.Stergachis AB, et al. Exonic transcription factor binding directs codon choice and affects protein evolution. Science. 2013;342:1367–1372. doi: 10.1126/science.1243490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kazancıoğlu E, Arnqvist G. The maintenance of mitochondrial genetic variation by negative frequency‐dependent selection. Ecol. Lett. 2014;17:22–27. doi: 10.1111/ele.12195. [DOI] [PubMed] [Google Scholar]
- 81.Smith JM, Haigh J. The hitch-hiking effect of a favourable gene. Genet. Res. 1974;23:23–35. doi: 10.1017/S0016672300014634. [DOI] [PubMed] [Google Scholar]
- 82.Charlesworth B, Morgan MT, Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134:1289–1303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kaplan NL, Hudson RR, Langley CH. The “hitchhiking effect” revisited. Genetics. 1989;123:887–899. doi: 10.1093/genetics/123.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Leffler EM, et al. Revisiting an old riddle: what determines genetic diversity levels within species? PLoS Biol. 2012;10:e1001388. doi: 10.1371/journal.pbio.1001388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Duret L. Evolution of synonymous codon usage in metazoans. Curr. Opin. Genet. Dev. 2002;12:640–649. doi: 10.1016/S0959-437X(02)00353-2. [DOI] [PubMed] [Google Scholar]
- 86.Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. [DOI] [PubMed] [Google Scholar]
- 87.Galtier N, Nabholz B, Glémin S, Hurst GDD. Mitochondrial DNA as a marker of molecular diversity: a reappraisal. Mol. Ecol. 2009;18:4541–4550. doi: 10.1111/j.1365-294X.2009.04380.x. [DOI] [PubMed] [Google Scholar]
- 88.Palumbi SR. Genetic divergence, reproductive isolation, and marine speciation. Annu. Rev. Ecol. Syst. 1994;25:547–572. doi: 10.1146/annurev.es.25.110194.002555. [DOI] [Google Scholar]
- 89.Ward JF. The complexity of DNA damage: relevance to biological consequences. Int. J. Radiat. Biol. 1994;66:427–432. doi: 10.1080/09553009414551401. [DOI] [PubMed] [Google Scholar]
- 90.Solé-Cava AM, Thorpe JP. High levels of genetic variation in natural populations of marine lower invertebrates. Biol. J. Linn. Soc. 1991;44:65–80. doi: 10.1111/j.1095-8312.1991.tb00607.x. [DOI] [Google Scholar]
- 91.Bazin E, Glémin S, Galtier N. Population size does not influence mitochondrial genetic diversity in animals. Science. 2006;312:570–572. doi: 10.1126/science.1122033. [DOI] [PubMed] [Google Scholar]
- 92.Therkildsen NO, Nielsen EE, Swain DP, Pedersen JS. Large effective population size and temporal genetic stability in Atlantic cod (Gadus morhua) in the southern Gulf of St. Lawrence. Can. J. Fish. Aquat. Sci. 2010;67:1585–1595. doi: 10.1139/F10-084. [DOI] [Google Scholar]
- 93.Avise, J. C. Phylogeography: the history and formation of species. (Harvard University Press, 2000).
- 94.Hajibabaei M, Janzen DH, Burns JM, Hallwachs W, Hebert PD. DNA barcodes distinguish species of tropical Lepidoptera. Proc. Natl. Acad. Sci. USA. 2006;103:968–971. doi: 10.1073/pnas.0510466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ward RD, Zemlak TS, Innes BH, Last PR, Hebert PD. DNA barcoding Australia’s fish species. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005;360:1847–1857. doi: 10.1098/rstb.2005.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hebert PD, Stoeckle MY, Zemlak TS, Francis CM. Identification of birds through DNA barcodes. PLoS Biol. 2004;2:e312. doi: 10.1371/journal.pbio.0020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hebert PD, Ratnasingham S, de Waard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. Lond. B Biol. Sci. 2003;270:S96–S99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Avise JC, et al. Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu. Rev. Ecol. Syst. 1987;18:489–522. doi: 10.1146/annurev.es.18.110187.002421. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analysed during the current study are available from the corresponding author on reasonable request.