Abstract
Lotus predominantly accumulates benzylisoquinoline alkaloids (BIAs), but their biosynthesis and regulation remain unclear. Here, we investigated structural and regulatory genes involved in BIA accumulation in lotus. Two clustered CYP80 genes were identified to be responsible for the biosynthesis of bis-BIAs and aporphine-type BIAs, respectively, and their tissue-specific expression causes divergence in alkaloid component between leaf and embryo. In contrast with the common (S)-reticuline precursor for most BIAs, aporphine alkaloids in lotus leaf may result from the (S)-N-methylcoclaurine precursor. Structural diversity of BIA alkaloids in the leaf is attributed to enzymatic modifications, including intramolecular C–C phenol coupling on ring A and methylation and demethylation at certain positions. Additionally, most BIA biosynthetic pathway genes show higher levels of expression in the leaf of high-BIA cultivar compared with low-BIA cultivar, suggesting transcriptional regulation of BIA accumulation in lotus. Five transcription factors, including three MYBs, one ethylene-responsive factor, and one basic helix–loop–helix (bHLH), were identified to be candidate regulators of BIA biosynthesis in lotus. Our study reveals a BIA biosynthetic pathway and its transcriptional regulation in lotus, which will enable a deeper understanding of BIA biosynthesis in plants.
Synthesis of medicinal compounds in lotus
Researchers at the Chinese Academy of Sciences have discovered the genes in lotus responsible for the synthesis of benzylisoquinole alkaloids (BIAs), a diverse set of pharmacologically significant plant metabolites. By comparing gene expression in two lotus varieties, a high- and low-BIA cultivar, they identified the genes that were active during BIA biosynthesis as well as five transcription factors which regulate the process. One of the biosynthesis genes is expressed more strongly in the embryo and another in the leaves, leading to the accumulation of different types of BIAs in these tissues. Although many of these genes are related to known BIA synthesis genes in other species, identifying the lotus equivalents not only reveals the particulars of the process in this species but also expands our understanding of BIA synthesis in general.
Introduction
Lotus (Nelumbo nucifera) belongs to Nelumbonaceae, a family of basal eudicots that is composed of only one genus, Nelumbo. As an aquatic perennial, lotus is widely grown in Asian countries both as an ornamental and for its edible rhizome and seed. In addition, all parts of the lotus plant have been used in traditional medicine to treat diseases such as cough, fever, hematemesis, hepatopathy, small pox, dysentery, and cholera1. Recently, various bioactive compounds in lotus have been identified and their significant pharmacological effects have been testified, including anti-oxidant2, anti-obesity3, anticancer4, antivirus5, and hepato-protection6. The main bioactive components in lotus are benzylisoquinoline alkaloids (BIAs), which are richly concentrated in tissues such as leaf and embryo7,8.
BIA alkaloids are a diverse class of nitrogen-containing plant secondary metabolites. Unlike terpenoids and phenylpropanoids that are commonly found in most higher plants, BIAs are restricted to certain plant families, including Magnoliaceae, Ranunculaceae, Papaveraceae, and Berberidaceae9. BIAs are structurally diverse, with approximated 2500 known BIAs, including pharmacological significant morphine and codeine (analgesic painkiller), sanguinarine and berberine (anti-microbial), noscapine (antitussive and potentially antineoplastic), and tubocuraine (muscle relaxant)10,11. Several plant species, such as opium poppy (Papaver somniferum), California poppy (Eschscholzia californica), Japanese goldthread (Coptis japonica), and yellow meadow rue (Thalictrum flavum), have served as models for studies of BIA biosynthesis12–19.
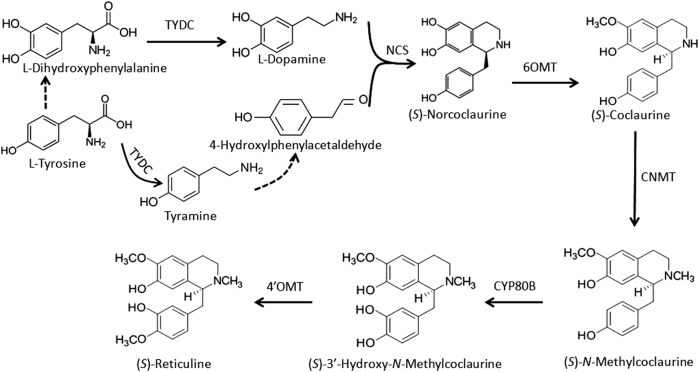
(S)-reticuline is well known to be the common precursor to the majority of BIAs, and its biosynthesis begins with the decarboxylation of tyrosine or l-dihydroxyphenylalanine (DOPA) to yield 4-hydroxyphenylacetaldehyde (4-HPAA) and l-dopamine, respectively, under the catalyzation of tyrosine/DOPA decarboxylase (TYDC) (Fig. 1). Subsequently, norcoclaurine synthase (NCS) catalyzes condensation of l-dopamine and 4-HPAA to form (S)-norcoclaurine, the first BIA scaffold in plants20. Four additional enzymatic steps, 6-O-methylation, N-methylation, 3′-hydoxylation and 4′-O-methylation, catalyzed by norcoclaurine 6-O-methyltransferase (6OMT), (S)-coclaurine N-methyltransferase (CNMT), (S)-N-methylcoclaurine 3′-hydroxylase (CYP80B), and 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase (4′OMT), respectively, yield the central intermediate (S)-reticuline. Later, a variety of additional reactions, such as internal carbon–carbon, carbon–oxygen phenol coupling, and functional group substitutions, result in the formation of different types of BIAs. To date, significant efforts have been made to investigate biosynthetic pathways of BIAs, and enzymes involved in the biosynthesis of structurally diverse BIAs, such as morphine, berberine, and sanguinarine, have been nearly completely identified11,21. More recently, high-throughput sequencing approaches have accelerated isolation of novel BIA biosynthetic and regulatory genes in plants22,23.
Fig. 1. Biosynthetic pathway of the common precursor to BIAs in plants.
The dash line indicates unknown reaction. TYDC tyrosine/DOPA decarboxylase, NCS norcoclaurine synthase, 6OMT norcoclaurine 6-O-methyltransferase, CNMT (S)-coclaurine N-methyltransferase, CYP80B, (S)-N-methylcoclaurine 3′-hydroxylase, 4′OMT 3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase
In lotus, BIA alkaloids represent the major active components and a total of 45 BIAs have been so far isolated7. Our recent study revealed that two organs, laminae and embryos, contain the highest amount of BIAs, followed by petals and petioles8. Moreover, significant variation in BIA component has been also observed between different organs24,25. Overall, lotus lamina predominantly accumulates aporphine-type BIAs such as nuciferine and its derivates, whereas embryo mainly contains dimeric bis-BIAs (bis-BIAs), such as liensinine, isoliensinine, and neferine. The total concentrations of BIAs in the leaf and embryo can reach 1.41% and 2.43% on the dry weight basis, respectively8,26.
Lotus BIA alkaloids are well known for a variety of pharmacological effects and have been used to treat various diseases. However, little is known about the molecular mechanisms underlying the biosynthesis and regulation of BIAs in lotus. Although enzymes catalyzing the conversion of tyrosine to (S)-reticuline are common to all BIAs, the BIA biosynthetic pathway is flexible in plants. Both structural and regulatory genes involved in BIA biosynthesis remain largely unknown in lotus. Recently, our evaluation of lotus germplasm for alkaloid content revealed that BIA content in the leaf varies considerably among cultivars and nuciferine is the major component of alkaloids in the leaf8. In this study, comparative transcriptome analysis was conducted between a high-BIA cultivar Luming (LM) and a low-BIA cultivar WD40 (WD). The contents of BIAs and nuciferine in mature leaf of cv. LM are approximately 4-fold and 6-fold higher than in mature leaf of cv. WD, respectively. Our aim was to explore structural genes and transcription factors (TFs) involved in BIA biosynthesis in lotus. The results will facilitate our deeper understanding of the biosynthetic pathway of BIAs and regulatory mechanism of BIA accumulation in plants.
Materials and methods
Plant materials
Two lotus cultivars used in this study, LM and WD, were grown separately in a square concrete pond at Wuhan Botanical Garden of the Chinese Academy of Sciences (Wuhan, Hubei province, China). Leaf samples at different developmental stages8 were collected during the summer of 2016, frozen immediately in liquid nitrogen, and stored in −80 °C freezer until use.
RNA extraction, RNA-Seq library construction, and sequencing
Leaves at two developmental stages of two cultivars were subjected to RNA-Seq library construction. For each stage, leaf samples represented a pool of five leaves, each of which was collected from different plants. Frozen leaf samples were ground into fine powder in liquid nitrogen with a mortar and pestle, and total RNA was isolated using Tiangen RNA Extraction Kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions. RNA integrity was evaluated using 1% agarose gel electrophoresis. Purified RNA was quantified using a Nanodrop Lite spectrophotometer (Thermo Scientific, CA, USA) and analyzed using a Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). All samples showed a 260/280 ratio >1.8, with the RNA integrity number >8.0. Finally, RNA concentrations were measured using Quibit 2.0® Flurometer (Life Technologies, Carlsbad, CA, USA) and the QubitTM RNA HS Assay Kit.
The RNA-Seq library was constructed using the NEB-Next Ultra Directional RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, MA, USA) according to the manufacturer’s instructions. Briefly, poly(A)-containing mRNAs were purified from 3 µg of total RNA using oligo (dT)-attached magnetic beads (Life Technologies, Carlsbad, CA, USA), and then fragmented using divalent cations under elevated temperature in NEB-Next First-Strand Synthesis Reaction Buffer (5×). M-MuLV reverse transcriptase (RNase H) and random hexamers were used for first-strand cDNA synthesis and the second-strand cDNA synthesis was subsequently synthesized using DNA polymerize I and RNAse H. The double-stranded cDNA was purified with AMPure XP beads (Beckman Coulter, Beverly, MA, USA), end repaired, and poly(A) end added at the 3′ end. cDNA fragments of approximately 200 bp in size were finally selected for PCR enrichment that was performed using the NEB Universal PCR Primer and Index primer. The constructed library was quantified using the Agilent Bioanalyzer 2100 system.
The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumia) according to the manufacturer’s instructions. RNA-Seq libraries were sequenced on an Illumina Hiseq platform (Novogene, Beijing, China) and 150-bp paired-end reads were generated.
Analysis of RNA-Seq data
The raw reads were cleaned by removing adapter sequences and low-quality regions. Clean reads were mapped to the genome sequence of the sacred lotus27 using TopHat v.2.0.1228. The number of read counts per gene locus was summarized by the HTSeq v0.6.129. Gene expression levels were calculated based on fragments per kilobase of transcript per million fragments mapped. The read counts were standardized between samples by scaling the number of reads in a given library to a common value across all sequenced libraries in this study30.
Differentially expressed genes were identified using the DEGSeq R package (ver. 2.1.0). A false discovery rate threshold of <0.05 and a log 2 fold change of 1 were used to determine differentially expressed genes. Annotation of differentially expressed genes was performed according to the results of a Blast query against the NCBI RefSeq nucleotide database and the Swiss-Prot and UniPro protein databases, followed by the pathway annotation pipelines, including Gene ontology (GO) (http://www.geneontology.org) and Kyoto Encyclopedia of Genes and Genomes (www.genome.jp/kegg/).
Quantitative real-time PCR
Total RNA was extracted using RNAprep Pure Plant Kit (Tiangen Biotec, Beijing, China) according to the manufacturer’s instructions. RNA samples were treated with DNase I (TaKaRa, Dalian, China) to remove any genomic DNA contamination. cDNA synthesis were conducted using cDNA Synthesis SuperMix (TransGen Biotech, Beijing, China) according to the manufacturer’s instructions. A SYBR Green-based real-time PCR assay was performed in Applied Biosystems StepOnePlusTM Real-Time PCR System (Applied Biosystems), with a total volume of 25 μl reaction mixture containing 12.5 μl of 2× SYBR Green I Master Mix (Takara, Dalian, China), 0.2 μM of each primer, and 100 ng of template cDNA. The amplification program consisted of one cycle of 95 °C for 10 min, followed by 40 cycles of 95 °C for 30 s and 60 °C for 30 s. Fluorescence readings were consecutively collected during the melting process from 60 to 90 °C at a heating rate of 0.5 °C/s. An actin gene was used as a constitutive control31. The relative expression of genes was calculated using the 2inu−∆∆ct method32. All analyses were performed in three biological replicates. The primer sequences are listed in Table S1.
Lotus alkaloid extraction and quantification
Alkaloid extraction was performed according to our previously reported protocol8. Briefly, fresh samples were ground to fine powder in liquid nitrogen and approximately 600 mg of powder were soaked in 7 ml of extraction buffer (0.3 M HCl-methanol, 1:1, v/v). The mixture was sonicated for 30 min at room temperature and then centrifuged at 11,000 × g for 10 min. The supernatant was transferred to an eppendorf tube, and the pellet was extracted again with 5 ml of extraction buffer. The supernatants were combined and diluted with extraction buffer to a final volume of 15 ml. The alkaloid extracts were filtered through 0.22 µm membranes (Shanghai New Asia Purification Device Factory, Shanghai, China) and then subjected to the concentration measurement using high-performance liquid chromatography according to the protocol described by Deng et al 8.
Dual luciferase reporter assay
Transient dual luciferase reporter assay was conducted in tobacco (Nicotiana benthamiana) according to our previously reported protocol33. Briefly, promoter regions upstream from the start codon of four lotus gene, NnTYDC1 (1.8 kb), NnNCS1 (2.2 kb), NnCYP80G (1.4 kb), and Nn7OMT2 (2.1 kb), were isolated and inserted individually into the MCS of vector pGreenII 0800-LUC. Full CDS of five lotus TFs, NnMYB6, NnMYB12, NnMYB113, NnbHLH1, and NnRAV1, were isolated and inserted into pSAK277 vectors under the 35S promoter. Genomic DNA and RNA templates were prepared from young leaves of cv. LM. The sequences of primers used for vector construction are listed in Table S1. All constructs were transformed individually into Agrobacterium tumefaciens strain GV3101, and incubated at 28 °C for 2 days. The confluent bacteria were resuspended in 10 ml infiltration buffer containing 10 mM MgCl2, 50 μM acetosyringone, and 10 mM 2-(N-morpholine)-ethanesulfonic acid (pH = 5.7). Before infiltration, the bacteria were incubated at room temperature without shaking for at least 2 h. Transient transformation was conducted by mixing Agrobacterium GV3101 culture transformed with the reporter cassette with an equal volume Agrobacterium culture transformed with a cassette containing NnMYB6, NnMYB12, NnMYB113, NnbHLH1, or NnRAV1 fused to the 35S promoter. All tests were repeated at least three times using biological replicates. Firefly luciferase (Luc) and Renilla luciferase (Ren) activity were measured 3 days after infiltration using Dual-Glo® Luciferase Assay System (Promega, Madison, WI, USA) on an Infinite M200 luminometer (Tecan, Mannerdorf, Switzerland). Promoter activation ability was shown as the ratio of Luc to Ren activity.
Results
Dynamic alkaloid concentration in lotus leaf throughout the whole developmental stages
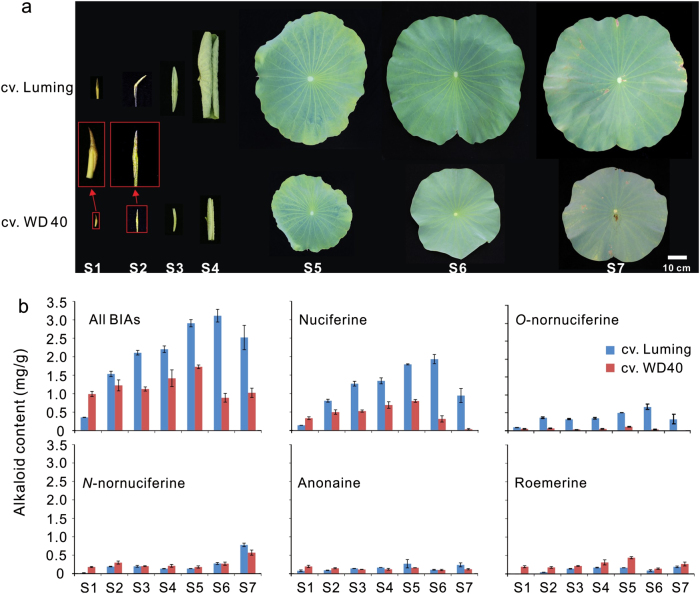
In this study, we mainly focused on BIA biosynthesis in lotus leaf as it produces a considerable amount of alkaloids. Our recent study revealed that two N. nucifera cultivars, LM, and WD, contain relatively high to low levels of BIAs in the leaf, respectively8. Overall, cv. LM with larger lamina discs accumulated higher levels of alkaloids in the leaf through all the seven developmental stages, ranging from the bud stage to the senescence stage, compared with cv. WD (Fig. 2a). Total alkaloid amount in the two cultivars increased steadily during the early stages, reached a peak at stages 5 or 6, and decreased slightly after the mature stage (Fig. 2b). In addition, two major BIA components, nuciferine and O-nornuciferine, showed a great difference in concentration between the two cultivars, with the highest fold change of 26.3 at stage 7 or 24.1 at stage 6, respectively. By contrast, the rest three components, N-nornuciferine, anonaine, and roemerine, had very low concentration in the leaf throughout the whole developmental stages, and showed a small difference between the two cultivars. Given the fact that nuciferine is the major component of BIAs in lotus leaf, these two cultivars are deemed to be suitable for comparative transcriptome study.
Fig. 2. Profile of lotus alkaloids in leaves at different developmental stages.
Leaves at seven developmental stages (a) and their corresponding alkaloid contents (b) of two cultivars Luming and WD40. Stage 1, leaf is under the soil and covered by sheath, with fusiform shape and pale yellow color interspersed with deep purple pots; stage 2, leaf has fusiform shape and tender green color and is out of the soil and still under the water surface; stage 3, leaf sticks out of the water surface, with fusiform shape and tender green color; stage 4, leaf is half-opened, with curly edge and tender green color; stage 5, leaf is fully opened with disk shape, with the upper epidermis being tender green color; stage 6, leaf is fully opened with disk shape, with the upper epidermis being dark green; stage 7, leaf starts to show senescence symptoms, with part of leaf edges changing color from green to yellow.
Our previous study shows that the NnNCS7 gene involved in the catalyzation of the first committed step in the alkaloid biosynthetic pathway is predominantly expressed in lotus leaf and its expression level is significantly correlated with alkaloid content34. To determine the leaf developmental stages that are suitable for comparative transcriptome study, we investigated the expression profile of NnNCS7 in the leaf. The NnNCS7 gene showed similar expression profiles between cvs. LM and WD, and its expression level reached a peak in the leaf at stage 4 (Fig. S1). This suggests that the expression of the alkaloid pathway genes reached the peak earlier than did alkaloid accumulation. In addition, alkaloid content in the leaf at stage 1 was significantly lower compared with that in the leaf at stage 4 (Fig. 2b). Thus, leaf samples of cvs. LM and WD at stage 1 and stage 4 were selected and subjected to later comparative transcriptome analysis.
Global gene expression patterns in leaves at different developmental stages
Four RNA-Seq libraries, designated LM_S1, LM_S4, WD_S1, and WD_S4, were constructed and sequenced for leaf samples of cvs. LM and WD at stage 1 (S1) and stage 4 (S4), respectively. Overall, the raw reads for each library ranged from 44.10 to 53.77 million reads, with an average of 48.20 million reads (Table 1). The raw reads were trimmed by removing adapter, empty reads, and low-quality sequences. As a result, an average of 47.14 million clean reads was generated for each library, with 7.07 Gb in size and approximately 45.6% of GC content. Approximately 75.2% of clean reads were mapped to the lotus reference genome27. Of the clean reads mapped, 99.2% and 0.8% were single-mapped and multiple-mapped reads, respectively.
Table 1.
Summary of lotus leaf transcriptome data
| Sample | No. of reads (million) | No. of mapped clean reads (million) | GC content of clean reads | No. of transcripts | ||||
|---|---|---|---|---|---|---|---|---|
| Raw | Clean | Single-mapped | Multiple-mapped | Total | Total | Novel | ||
| LM_S1 | 53.77 | 52.71 | 40.29 | 0.34 | 40.63 | 44.95% | 24728 | 1079 |
| LM_S4 | 48.55 | 47.54 | 35.88 | 0.26 | 36.14 | 45.55% | 24667 | 1105 |
| WD_S1 | 46.39 | 45.45 | 34.06 | 0.31 | 34.37 | 45.95% | 24782 | 1056 |
| WD_S4 | 44.10 | 42.85 | 30.44 | 0.26 | 30.70 | 45.77% | 24640 | 1063 |
The mapped reads were subjected to transcript discovery, and 29,489 genes were identified from all four samples tested. Of the 29,489 genes, 28,074 were previously predicted in the lotus reference gene set27, while the remaining 1415 genes were not previously included in the lotus reference gene set, thus representing novel genes. Moreover, 24,728, 24,667, 24,782, and 24,641 genes were expressed in LM_S1, LM_S4, WD_S1, and WD_S4 samples, respectively, while 22,800 genes were commonly expressed in all four samples tested. By contrast, 297, 215, 239, and 279 genes were exclusively expressed in LM_S1, LM_S4, WD_S1, and WD_S4 samples, respectively.
Mining of candidate genes involved in BIA biosynthesis in lotus
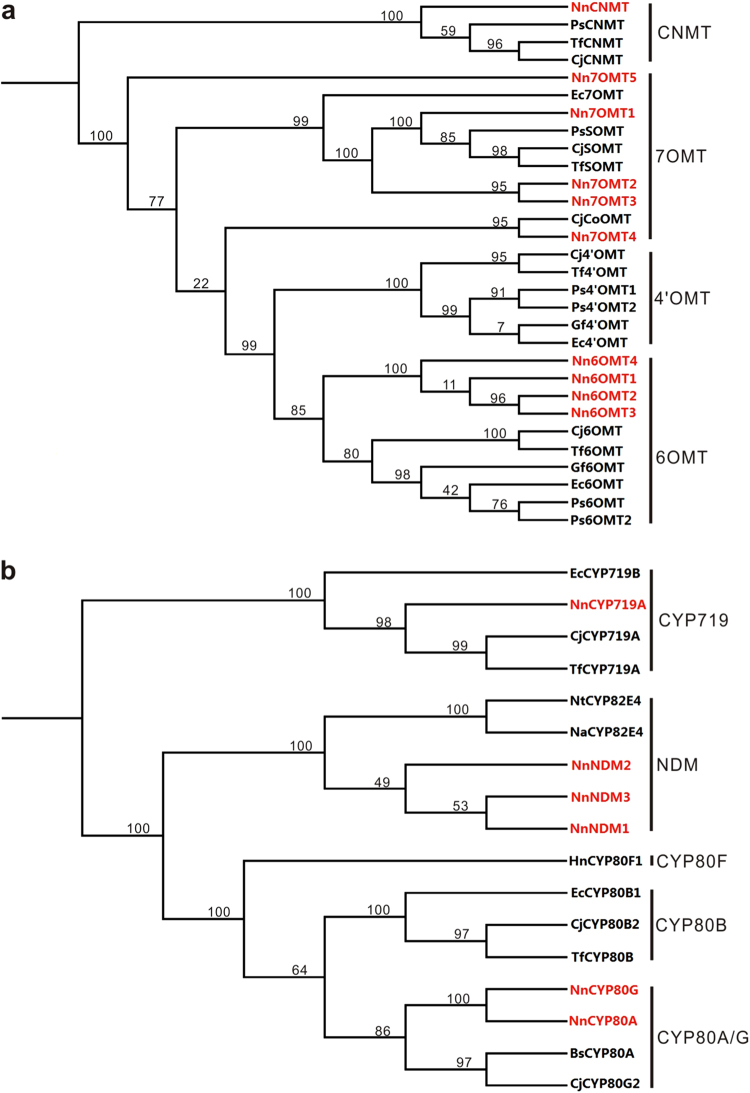
To identify BIA biosynthetic pathway genes in lotus, coding DNA sequences of previously reported BIA biosynthesis genes were compared against those of the 29,489 genes expressed in lotus leaf. Initially, we investigated structural genes involved in the biosynthesis of the BIA precursor (S)-reticuline. Five, four, and one copies of TYDC, NCS, and CNMT genes, respectively, were identified (Table S2) and the amino acid sequence alignment of each gene family is shown in Supplementary data 1. Seven copies of genes encoding OMT showed high similarity to both 6OMT and 7OMT in Papaver somniferum35–37 and Coptis japonica38. Phylogenetic analysis showed that all these OMT genes were closely related to either the 6OMT or 7OMT gene clusters, but distinct from the 4′OMT gene cluster (Fig. 3a). Likewise, three copies of genes encoding BIA-related cytochrome P450 (CYP) were identified, but none of them was phylogenetically close to CYP80B genes (Fig. 3b).
Fig. 3. Phylogenetic trees derived from amino acid sequences of genes encoding methyltransferase and CYP protein, respectively, which are expressed in the leaf of lotus.
The genes identified in this study are highlighted in red color
Lotus leaf accumulates five types of BIAs, nuciferine, N-nornuciferine, O-nornuciferine, roemerine, and anonaine, with nuciferine as the predominant type8. The conversion of the (S)-reticuline precursor to nuciferine consists of at least two reactions, an intramolecular C–C phenol coupling and a 7-O-methylation, under the catalyzation of CYP80G39 and 7-O-methyltransferase35, respectively. Likewise, an intramolecular C–C phenol-coupling reaction together with a methylenedioxy bridge-forming reaction catalyzed by CYP719A40 are required for the biosynthesis of roemerine. Additional N-demethylation and O-demethylation reactions that are catalyzed by N-demethylase (NDM) or O-demethylase (ODM), respectively41,42, result in structurally diversified BIAs. Analysis of the expressed genes in the leaf revealed one, three, one, three, and four copies of CYP80G, 7OMT, CYP719A, NDM, and ODM genes, respectively.
The NnCYP80A and NnCYP80G genes shared approximately 88.8% identity in coding DNA sequence, and they were phylogenetically related to previously reported CYP80A and CYP80G genes, but separated from the CYP80B gene cluster (Fig. 3b). CYP80A is well known to catalyze (S)-N-methylcoclaurine to form bis-BIAs43,44, while CYP80G is involved in the aporphine-type BIA biosynthesis39. Lotus embryo and leaf predominantly accumulates bis-BIAs and aporphine-type BIAs, respectively8. Hence, NnCYP80A and NnCYP80G are likely associated with BIA accumulation in the embryo and leaf, respectively. To confirm this hypothesis, we conducted quantitative real-time PCR (qRT-PCR) analysis. The results showed that NnCYP80A was highly expressed in the embryo, but with an extremely low expression in the leaf (Fig. S2). In contrast, NnCYP80G was highly expressed in the leaf, but its transcripts were nearly undetectable in the embryo. This spatial expression pattern is consistent with the accumulation of different types of BIAs in the embryo and leaf.
Difference in expression of BIA biosynthetic pathway genes in the leaf between different developmental stages or between cultivars
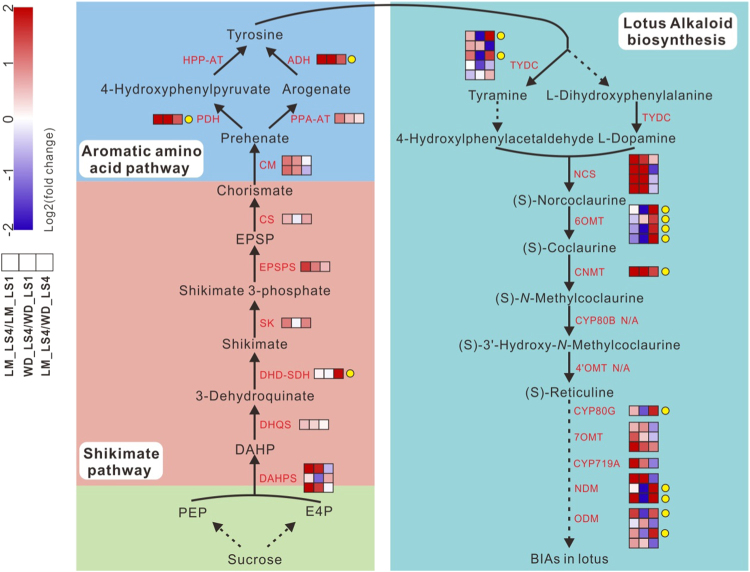
To uncover the mechanism underlying leaf alkaloid accumulation, we further investigated the digital expression profiles of BIA biosynthetic pathway genes in leaves of “LM” and “WD” at two developmental stages. For cv. LM, almost all the BIA pathway genes except NnTYDC5, Nn6OMT2, Nn6OMT3, Nn6OMT4, NnCYP80A, and NnNDM3 were up-regulated in the leaf at stage 4 compared with stage 1 (Fig. 4). For example, the expression levels of NnNCS genes in stage 4 showed 5-fold to 30-fold higher than those in stage 1 (Table S2). The NnCNMT gene showed an approximately 9-fold increase in expression level at stage 4 compared with stage 1. In addition, we also analyzed the dynamic expression patterns of genes involved in shikimate and aromatic amino acid pathways, which supply the tyrosine source for BIA biosynthesis. All genes in these two pathways were up-regulated in the leaf of “LM” at stage 4 compared with stage 1. For example, two copies of genes encoding 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase, a rate-controlling enzyme in the shikimate pathway45, showed a 4-fold increase at stage 4 compared with stage 1. The gene encoding agrogenate/prephenate that converts arogenate to tyrosine displayed an over 26-fold increase.
Fig. 4. Schematic diagram of fold change in expression level of structural genes in BIA, shikimate, and aromatic amino acid pathways in lotus leaf between two stages of the same cultivar or between cultivars with the same stage 4.
Three boxes that are arranged in the same row indicate comparisons of LM_LS4/LM_LS1, WD_LS4/WD_LS1, and LM_LS4/WD_LS4, respectively. The layers of boxes represent different gene copies, which are listed in a numerical order from top to bottom. LM_LS4/LM_LS1 and WD_LS4/WD_LS1 represent comparison of gene expression levels in leaves of cvs. LM and WD, respectively, between stages 1 and 4, while LM_LS4/ WD_LS4 indicates comparison of gene expression levels at stage 4 between cvs. LM and WD. The arrows with broken lines indicate multi-enzymatic reactions. PEP phosphoenolpyruvate, E4P d-erythrose 4-phosphate, DAHPS 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase, DHQS 3-dehydroquinate synthase, DHD 3-dehydroquinate dehydratase, SDH shikimate dehydrogenase, SK shikimate kinase, PESPS 5-enolpyruvylshikimate-3-phosphate synthase, CS chorismate synthase, CM chorismate mutase, PPA-AT prephenate aminotransferase, ADH arogenate dehygrogenase, PDH prephenate dehydrogenase, HPP-AT 4-hydroxyphenylpyruvate aminotransferase
For cv. WD, approximately half of the BIA pathway genes were up-regulated or down-regulated in the leaf at stage 4 compared with stage 1. Moreover, eight shikimate related and four aromatic amino acid-related pathway genes were up-regulated and down-regulated, respectively, in the leaf at stage 4 compared with stage 1. Almost all the up-regulated genes in cv. WD had smaller fold-change values than in cv. LM.
In the leaf at stage 4, 16 BIA pathway genes showed higher levels of expression in cv. LM compared with cv. WD, with a 4.9-fold increase on average. However, the remaining 11 genes had lower levels of expression in cv. LM than in cv. WD. Moreover, most genes involved in shikimate and aromatic amino acid pathways were expressed at higher levels in cv. LM than in cv. WD.
In summary, a set of BIA biosynthetic pathway genes were up-regulated in the leaf at stage 4 compared with stage 1 in both cultivars tested. In the leaf at stage 4, multiple BIA biosynthetic pathway genes also showed higher level expression in high-BIA cultivar than in low-BIA cultivar. These findings suggest the possibility of transcriptional regulation mechanism controlling BIA biosynthesis in lotus.
Mining of candidate regulatory genes controlling BIA accumulation in lotus
As mentioned above, the high-BIA “LM” and the low-BIA “WD” cultivars showed a significant difference in alkaloid content of leaves at stage 4, but with a relatively similar low content at stage 1. Moreover, “LM” showed a great variation of alkaloid content in leaves between stages 1 and 4. Thus, comparative analysis of two paired transcriptome profiles, LM_S4 vs. LM_S1 and LM_S4 vs. WD_S4, was conducted to identify differentially expressed regulatory genes related to BIA accumulation. However, the comparison of WD_S4 vs. WD_S1 was not included in this study because “WD” had relatively low levels of BIA accumulation in the leaf, which is probably due to mutation of key regulatory gene(s) involved in BIA biosynthesis.
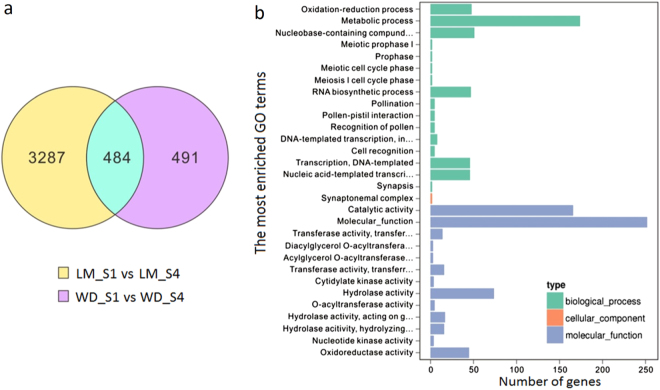
A total of 4072 genes were identified to be differentially expressed in leaves of cv. LM, with 2145 and 1927 up-regulated and down-regulated, respectively, when stage 4 was compared with stage 1. Similarly, 976 genes were differentially expressed in leaves at stage 4 between cvs. LM and WD, with 523 and 453 genes up-regulated and down-regulated, respectively. Of these differentially expressed genes (DEGs), 484 (Supplementary data 2) were commonly up-regulated or down-regulated among the two comparisons (Fig. 5a), and they were deemed to contain candidate regulatory genes for BIA biosynthesis in the leaf. GO enrichment analysis indicated that all the 484 DEGs had one or more GO annotations, including 450 biological process, 2 cellular component, and 619 molecular function terms (Fig. 5b).
Fig. 5. Identification of candidate transcription factors regulating BIA accumulation in lotus.
a Venn diagrams showing the numbers of genes commonly and exclusively up-regulated or down-regulated in leaves of each cultivar tested between stages 1 and 4. b Gene ontology (GO) enrichment analysis of the 484 genes commonly differentially expressed in leaves of cultivars LM and WD between stages 1 and 4
Of the 484 differentially expressed genes, 24 were TFs encoding AP2/ethylene-responsive factor (ERF), WRKY, MYB, helix–loop–helix (bHLH), NAC, and zinc-finger protein (Table 2). Previous studies showed that several TFs, such as bHLH, WRKY, and ORCA3, are involved in the regulation of BIA biosynthesis in plants46,47. Therefore, sixteen TFs, including six NnWRKYs, five NnERFs, four NnMYBs, and one NnbHLH, were deemed to be candidate regulators responsible for BIA biosynthesis in lotus.
Table 2.
Transcription factors potentially involved in the regulation of BIA biosynthesis in lotus
| Number | Name | ID | Type | Fold change* | ||
|---|---|---|---|---|---|---|
| LM_S4/LM/S1 | WD_S4/WD_S1 | LM_S4/WD_S4 | ||||
| 1 | NnERF81 | 104599450 | AP2/ERF | 1.87 | 0.41 | 3.21 |
| 2 | NnRAV1 | 104603036 | AP2/ERF | 12.23 | 1.27 | 3.65 |
| 3 | NnRAP23 | 104606214 | AP2/ERF | 3.82 | 0.89 | 2.97 |
| 4 | NnERF92 | 104610030 | AP2/ERF | 3.15 | 0.69 | 2.78 |
| 5 | NnERF2 | 104610490 | AP2/ERF | 3.07 | 0.69 | 2.41 |
| 6 | NnWRKY6 | 104588301 | WRKY | 4.15 | 0.66 | 2.21 |
| 7 | NnWRKY701 | 104593755 | WRKY | 3.67 | 0.49 | 3.96 |
| 8 | NnWRKY702 | 104594468 | WRKY | 1.91 | 0.73 | 2.47 |
| 9 | NnWRKY31 | 104594045 | WRKY | 75.32 | 3.77 | 2.85 |
| 9 | NnWRKY401 | 104603312 | WRKY | 4.73 | 0.35 | 3.00 |
| 10 | NnWRKY402 | 104610723 | WRKY | 13.44 | 1.12 | 2.24 |
| 11 | NnMYB6 | 104589500 | MYB | 5.92 | 1.25 | 2.39 |
| 12 | NnMYB113 | 104599741 | MYB | 3.94 | 3.23 | 2.18 |
| 13 | NnMYB12 | 104605119 | MYB | 4.73 | 1.22 | 4.03 |
| 14 | NnMYB4 | 104610016 | MYB | 14.62 | 0.55 | 2.94 |
| 15 | NnZHD11 | 104587730 | Zinc-finger | 2.54 | 2.98 | 2.04 |
| 16 | NnZAT8 | 104594613 | Zinc-finger | 2.56 | 0.19 | 3.35 |
| 17 | NnZAT10 | 104608146 | Zinc-finger | 2.34 | 0.47 | 2.02 |
| 18 | NnbHLH1 | 104606713 | bHLH | 3.27 | 3.38 | 13.41 |
| 19 | NnNAC72 | 104612551 | Nac | 26.85 | 2.11 | 3.97 |
| 20 | NnPHRF1 | 104588985 | PHD | 1.92 | 0.76 | 2.36 |
| 22 | NnDIV | 104589256 | DIVARICATA | 2.30 | 0.34 | 2.42 |
| 23 | NnAPRR5 | 104590049 | PRR | 3.16 | 0.90 | 3.28 |
| 24 | NnJM706 | 104588821 | JMJ | 1.75 | 0.761 | 2.82 |
* LM_LS4/LM_LS1 and WD_LS4/WD_LS1 represent fold-change values derived from comparison of gene expression levels in leaves of cvs. LM and WD, respectively, between stages 1 and 4, while LM_LS4/WD_LS4 indicates fold-change value for the comparison of gene expression levels at stage 4 between cvs. LM and WD.
Expression consistency between BIA biosynthetic pathway genes and their candidate regulators in the leaf of two tested cultivars
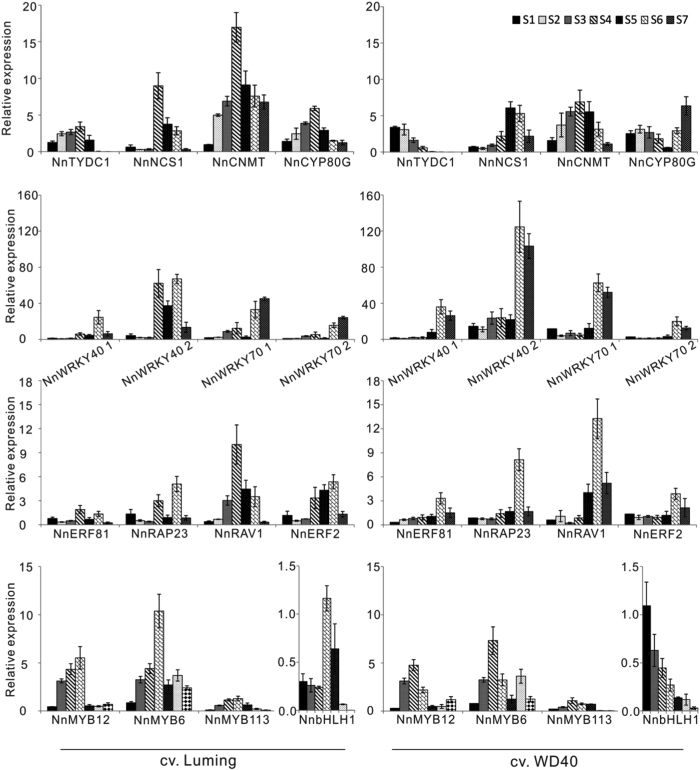
To verify regulatory role of the above-mentioned candidate TFs, their expression profiles in the leaf throughout whole developmental stages were compared with those of BIA biosynthetic pathway genes. Overall, all the four structural genes tested, NnTYDC1, NnNCS1, NnCNMT, and NnCYP80G, showed higher levels of expression in the leaf of cv. LM compared with cv. WD (Fig. 6a). The expression levels of all the tested structural genes in the leaf of cv. LM increased stably during the early stages, reached a peak at stage 4, and decreased gradually during the later stages. Similar expression profile was also observed for structural genes NnNCS1 and NnCNMT in cv. WD. However, the expression level of NnTYDC1 showed a decrease in the leaf of cv. WD throughout the whole developmental stages, with a highest expression at stage 1. By contrast, the expression level of NnCYP80G showed a peak at stage 2, and decreased until stage 5, followed by a substantial increase during the last two stages. For easy description, the expression profile of the structural genes in cv. LM was designated the classic upside-down V pattern.
Fig. 6.
qRT-PCR analysis of BIA biosynthetic pathway genes and candidate regulators in leaves of two lotus cultivars throughout the whole developmental stages
Of the 16 candidate regulators, four, NnERF92, NnWRKY6, NnWRKY31, and NnMYB4, showed a very low expression in leaves of two cultivars throughout the whole development stages, suggesting that they play little role in the regulation of BIA accumulation. Three MYB genes, NnMYB12, NnMYB6, and NnMYB113, had the classic upside-down V pattern of expression in both “WD” and “LM,” with a peak at stage 4 (Fig. 6b). This was consistent with the expression profile of all the four structural genes tested in cv. LM and two structural genes NnNCS1 and NnCNMT in cv. WD. Two genes, NnbHLH1 and NnRAV1, showed the classic upside-down V pattern of expression in cv. LM, but not in cv. WD. However, the rest candidate regulators showed no classic upside-down V pattern of expression in either “LM” or “WD.”
The above results suggest that five TFs, NnMYB12, NnMYB6, NnMYB113, NnbHLH1, and NnRAV1, are deemed to be candidate regulators of BIA accumulation in lotus.
Functional analysis of candidate TFs involved in BIA regulation using dual luciferase assay
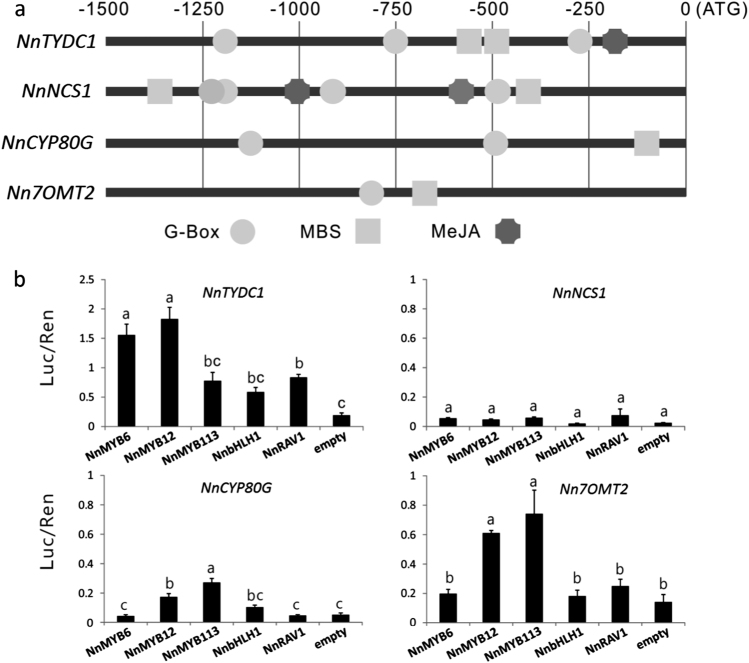
To gain an insight into regulatory mechanism controlling BIA biosynthesis in lotus, cis-regulatory elements were scanned in the promoter region of four BIA structural genes, including NnTYDC1, NnNCS1, NnCYP80G, and Nn7OMT2. A 1.5-kb DNA fragment upstream of translational start codon was scanned using the PlantCARE software48. All the promoters tested contained putative cis-elements, MBS and G-box, which are the binding sites of MYB and AP2/ERF TFs, respectively (Fig. 7a). Two genes, NnTYDC1 and NnNCS1, contained one or two, respectively, putative MeJA-response cis-elements (MeJA), the possible binding site of bHLH and AP2/ERF TFs. These results suggest that the five regulators mentioned above, NnMYB12, NnMYB6, NnMYB113, NnbHLH1, and NnRAV1, could bind to the promoters of BIA pathway genes to induce their transcription, resulting in the accumulation of BIAs in lotus leaf.
Fig. 7. Relationship between regulatory genes and BIA biosynthetic pathway genes in lotus.
a Cis-elements in the promoter regions 1.5-kb upstream of the translation initiation codon of BIA biosynthetic pathway genes in lotus. ATG, start codon. MeJA methyl jasmonate, MBS MYB binding site. b Estimation of effect of TFs on activation of the promoter of BIA pathway genes using transient dual luciferase assay. Error bars show SE of three biological replicates. Different lowercase letters indicate significant difference at P < 0.05
To validate whether these regulators had direct activation on transcription of structural genes, dual luciferase assay was further carried out (Fig. 7b). Of the three MYB regulators, NnMYB12 was well able to activate the promoters of three structural genes, NnTYDC1, NnCYP80G, and Nn7OMT2. NnMYB6 was well able to activate the promoter of NnTYDC1, while NnMYB113 showed a relative strong activation on the promoters of NnCYP80G and Nn7OMT2. NnRAV1 was able to activate the NnTYDC1 promoter, but NnbHLH1 showed no activation on the promoters of all structural genes tested. These results indicate that the three MYB TFs are likely to play an important role in BIA regulation.
In addition, NCS is crucial for BIA accumulation as it catalyzes condensation of dopamine and 4-HPAA to form (s)-norcoclaurine, a central precursor of BIAs. However, dual luciferase assay showed that the NnNCS1 promoter could not be activated by all the five candidate regulators. Interestingly, NnbHLH1 co-infiltrated with NnRAV1 showed a strong activation on the NnNCS1 promoter (Fig. S3), suggesting a potential interaction between the NnbHLH1 and NnRAV1 regulators. In contrast, NnbHLH1 co-infiltrated with NnMYB113 showed no activation on the NnNCS1 promoter although the interaction between MYB and bHLH TFs has been well characterized. In addition, NnbHLH1 co-infiltrated with NnRAV1 or NnMYB113 did not increase significantly the activation on the NnTYDC promoter compared with those infiltrated with NnbHLH1, NnRAV1, or NnMYB113 alone.
Taken together, the results presented above indicate that all the five candidate regulators may act in an independent or cooperative manner to induce the transcription of BIA biosynthetic pathway genes, resulting in a complex network of transcriptional regulation controlling BIA accumulation in lotus.
Discussion
The BIA biosynthetic pathway and its regulation in lotus
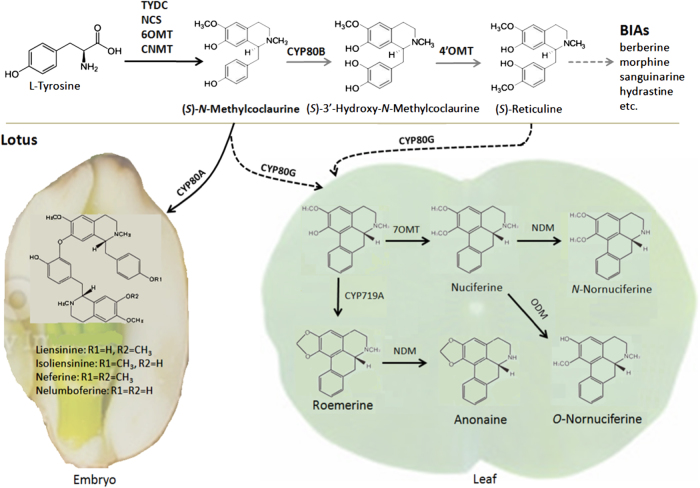
(S)-reticuline is well known to be the common precursor to BIA alkaloids, such as morphine, sanguinarine, papaverine, and berberine. However, an exception has been noted in dimeric bis-BIA alkaloids such as berbamunine in Berberis stolonifera, which result from the regioselective and stereoselective oxidative carbon–oxygen phenol coupling of (S)-N-methylcoclaurine under the catalyzation of CYP80A43. In this study, a CYP80A homolog NnCYP80A was also identified in lotus leaf transcriptome, although its expression was extremely low. However, the NnCYP80A gene is highly expressed in lotus embryo. In lotus, bis-BIA alkaloids are predominantly accumulated in embryos, but almost undetectable in the leaf. These results suggest that (S)-N-methylcoclaurine is the precursor to bis-BIAs in lotus embryos. However, more studies are still needed to address whether this reaction is catalyzed by NnCYP80A.
Conversion of (S)-N-methylcoclaurine to (S)-reticuline requires two enzymatic steps catalyzed by CYP80B and 4′OMT49. Although OMTs involved in BIA biosynthesis have extensive amino acid homology, they can be divided into distinct clades, such as 6OMT, 4′OMT, 7OMT, and 9OMT, based on phylogenetic analysis50,51. Here, nine OMT genes were identified in the lotus leaf transcriptome, but none of them was phylogenetically related to previously reported 4′OMT genes. Similarly, two CYP80 genes were found in the lotus leaf transcriptome, but neither of them appeared in the CYP80B cluster. In addition, CYP80B and 4′OMT are responsible for hydroxylation and methylation at the C-3′ and C-4′ position, respectively52,53. However, the aporphine-type BIAs in lotus leaf do not contain any hydroxyl or methyl groups at the C-4′ and C-3′ position, which suggests that the two enzymatic steps catalyzed by CYP80B and 4′OMT are not crucial for the biosynthesis of aporphine-type BIAs in lotus. Given the finding that neither CYP80B nor 4′OMT genes are found in the leaf transcriptome, we speculate that (S)-N-methylcoclaurine may serve as the precursor to both aporphine and bio-BIA alkaloids in lotus. However, one thing to keep in mind is that the lotus leaf transcriptome may contain other genes with functionality similar to CYP80B and 4′OMT, resulting in a possible pathway that uses (S)-reticuline as precursor for BIA synthesis, similar like most of other BIA-producing plants such as opium poppy and California poppy. Therefore, we propose a model to illuminate the BIA biosynthetic pathway in lotus (Fig. 8). In addition, it is worth noting that conversion of (S)-N-methylcoclaurine or (S)-reticuline to lotus aporphine alkaloids requires enzymatic steps, dehydroxylation at the C-4′ position, and/or demethylation at C-3′ position. Thus, more studies are still needed to clarify enzymatic reactions that convert (S)-N-methylcoclaurine or (S)-reticuline into aporphine alkaloids in lotus.
Fig. 8. Schematic diagram of a proposed biosynthetic pathway of BIAs derived from the precursor (S)-N-methylcoclaurine in lotus.
Lotus leaf accumulates aporphine-type BIAs, while embryo accumulating bis-BIAs. The gray line indicates the common biosynthetic pathway of BIAs derived from the precursor (S)-reticuline. The black dash line indicates the potential biosynthetic pathway of BIAs in the leaf. Dotted lines represent multiple enzymatic steps
Increasing evidences show that alkaloid accumulation is controlled by TFs such as WRKY, bHLH, and ERF at the transcriptional level54–58. Such transcriptional regulation was also observed in our study, with a set of BIA biosynthetic pathway genes showing higher expression in the leaf of high-BIA cv. LM compared with low-BIA cv. WD. Two TFs, NnbHLH1 and NnRAV1, both show an expression profile that is consistent with those of BIA structural genes in the leaf of cv. LM, but not in the leaf of cv. WD. The G-box or MeJA binding motifs for NnbHLH1 and NnRAV1 were found in the promoter of BIA structural genes. Dual luciferase assay shows that NnbHLH1 and NnRAV1 may be interacted with each other to form a transcriptional regulation for alkaloid biosynthesis in lotus.
The WRKY1 gene has been reported to control BIA accumulation by activating the 4′OMT gene in C. japonica59 and California poppy60. In this study, two homologs of CrWRKY1, NnWRKY701, and NnWRKY702, together with four additional WRKY TFs, were identified in lotus leaf transcriptome. However, none of these NnWRKY TFs shows a consistent expression profile with BIA structural genes in the leaf of both “LM” and “WD.” It is unclear whether this inconsistency is attributed to the lack of the 4′OMT gene in the leaf transcriptome. Interestingly, three MYB regulators, NnMYB6, NnMYB12, and NnMYB113, show a consistent expression profile in the leaf of both “LM” and “WD” with BIA structural genes. The activation of these MYB genes on BIA structural genes was confirmed by dual luciferase assay. In addition, MYB binding motifs have been found in the promoters of all the four BIA structural genes tested. Thus, these results suggest that NnMYB6/NnMYB12/NnMYB113, like NnbHLH1 and NnRAV1, have a potential to induce BIA structural gene transcription by binding the MBS motif in their promoters. To our knowledge, this study reveals for the first time that MYB TFs are involved in the regulation of BIA accumulation. In addition, recent studies have shown that MYB TFs can interact with AP2/ERF TFs to regulate anthocyanin and lignin biosynthesis in fruits61,62. Thus, it is worthy of further study to ascertain whether NnMYB6/NnMYB12/NnMYB113 regulate lotus BIA biosynthesis through interacting with AP2/ERF TFs in lotus.
NnbHLH1 alone is unable to induce BIA structural gene transcription, but it may interact with NnRAV1 to induce the transcription of NnNCS1, which cannot be activated by NnMYB genes. Moreover, NnRAV1 alone shows a relative weak activation on the NnTYDC transcription. Unlike NnbHLH1 and NnRAV1, the NnMYB genes are able to individually activate three out of the four BIA structural genes tested. These results not only show that NnMYBs are likely to key regulators controlling BIA accumulation in lotus but also suggest a complex network of transcriptional regulation involved in lotus BIA biosynthesis. Moreover, a bHLH TF CrMYBC2 is well known to function as the major activator of the expression of the AP2/ERF-domain TF ORCA3 in Catharanthus roseus63. In this study, the G-box of bHLH binding motif was found in the promoter of NnMYB6/NnMYB12 and NnRAV1. It is worthy of further study to clarify whether NnbHLH1 has ability to active transcription of NnMYB6/NnMYB12 and NnRAV1 by binding the G-box motif in their promoters, resulting in hierarchical interaction between regulators involved in BIA accumulation.
Tandem duplication of BIA biosynthesis genes and its impact on structural diversity of BIAs in lotus
Gene duplication is a major driving force for recruitment of genes involved in secondary metabolism, which contributes to structural diversify of secondary metabolites in plants. Duplicated genes involved in secondary metabolism often appear to cluster within the plant genome64, and tandem duplication is frequently observed for genes involved in biosynthesis of secondary metabolites, such as flavonoids65, volatiles66, caffeine67, and BIA68.
Our recent study reveals a cluster of five NnNCS genes within an 83-kb region on linkage group 8 of sacred lotus30. Only one member, NnNCS1 out of the five NnNCS genes, was expressed in the leaf, which suggests that they have undergone divergence in expression after duplication69. Here, three more clusters of BIA structural genes were also observed. First, two CYP80 genes, NnCYP80A and NnCYP80G, are located closely to each other within a 20-kb region on scaffold 1227, which suggests that they are evolved from a common ancestor. As mentioned above, their spatiotemporal expression results in difference in alkaloid component between organs in lotus. This indicates that NnCYP80A and NnCYP80G have functionally diverged after duplication. Second, two 6OMT genes, Nn6OMT2 and N6OMT3, are located in a 42-kb region on scaffold 43227. These two OMT genes are both expressed in the leaf with similar expression level. Last, three ODM genes, NnODM2, NnODM3, and NnODM4, are tightly clustered within an 85-kb region on scaffold 8027. These three ODM genes are all expressed in the leaf, but show a divergence in expression level, with NnODM2 having the highest expression level, followed by NnODM3 and NnODM4. Thus, it seems that the clustered Nn6OMT and NnODM genes may be functionally redundant.
In summary, tandem duplication occurs often among BIA structural genes, and the duplicated genes may have undergone divergence in enzymatic activity and/or expression in lotus. The functional divergence of two duplicated CYP80 genes, NnCYP80A and NnCYP80G, play an important role in structural divergence of two groups of alkaloids in lotus, aporphine-type BIAs and bis-BIAs. In addition, lotus leaf is becoming more and more attractive because it provides a resource of the bioactive BIA ingredients with diversified health benefits. Our study reveals the biosynthetic pathway and transcriptional regulation of BIAs in lotus, which not only provides a deeper understanding of BIA biosynthesis in plants but will also be helpful for development of new cultivars with high level of BIAs in lotus breeding programs.
Electronic supplementary material
Acknowledgements
This project was supported by funds received from the National Natural Science Foundation of China (Grants 31450110420 and 31272195) and the Overseas Construction Plan for Science and Education Base, China-Africa Center for Research and Education, Chinese Academy of Sciences (Grant No. SAJC201327).
Author contributions
Y.H. and X.D. conceived and designed the experiments. X.D., L.Z., Y.X. T.F., and D.Y. performed the experiments. Y.H. and X.D. wrote the paper. C.O., S.V., and Y.L. revised the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41438-018-0035-0).
References
- 1.Mukherjee PK, Mukherjee D, Maji AK, Rai S, Heinrich M. The sacred lotus (Nelumbo nucifera)—phytochemical and therapeutic profile. J. Pharm. Pharmacol. 2009;61:407–422. doi: 10.1211/jpp.61.04.0001. [DOI] [PubMed] [Google Scholar]
- 2.Zhao X, Shen J, Chang KJ, Kim SH. Comparative analysis of antioxidant activity and functional components of the ethanol extract of lotus (Nelumbo nucifera) from various growing regions. J. Agric. Food Chem. 2014;62:6227–6235. doi: 10.1021/jf501644t. [DOI] [PubMed] [Google Scholar]
- 3.Ono Y, Hattori E, Fukaya Y, Imai S, Ohizumi Y. Antiobesity effect of Nemumbo nucifera leaves extract in mice and rats. J. Ethnopharmacol. 2006;106:238–244. doi: 10.1016/j.jep.2005.12.036. [DOI] [PubMed] [Google Scholar]
- 4.Poornima P, Weng CF, Padma VV. Neferine, an alkaloid from lotus seed embryo inhibits human lung cancer cell growth by MAPK activation and cycle arrest. Biofactors. 2014;40:121–131. doi: 10.1002/biof.1115. [DOI] [PubMed] [Google Scholar]
- 5.Kashiwada Y, et al. Anti-HIV benzylisoquinoline alkaloids and flavonoids from the leaves of Nelumbo nucifera, and structure activity correlations with related alkaloids. Bioorg. Med. Chem. 2015;13:443–448. doi: 10.1016/j.bmc.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 6.Huang B, et al. Hepatoprotective and antioxidant activity of ethanolic extracts of edible lotus (Nelumbo nucifera Gaertn.) leaves. Food Chem. 2010;1:873–878. doi: 10.1016/j.foodchem.2009.11.020. [DOI] [Google Scholar]
- 7.Sharma BR, Gautam LMS, Adhikari D, Karki R. A comprehensive review on chemical profiling of Nelumbo nucifera: potential for drug development. Phytother. Res. 2017;31:3–26. doi: 10.1002/ptr.5732. [DOI] [PubMed] [Google Scholar]
- 8.Deng X, et al. Analysis of isoquinoline alkaloid composition and wound-induced variation in Nelumbo using HPLC-MS/MS. J. Agric. Food Chem. 2016;64:1130–1136. doi: 10.1021/acs.jafc.5b06099. [DOI] [PubMed] [Google Scholar]
- 9.Liscombe DK, Macleod BP, Loukanina N, Nandi OL, Facchini PJ. Evidence for the monophyletic evolution of benzylisoquinoline alkaloid biosynthesis in angiosperms. Phytochemistry. 2005;66:1374–1393. doi: 10.1016/j.phytochem.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 10.Facchini PJ. Alkaloid biosynthesis in plants: biochemistry, cell biology, molecular regulation, and metabolic engineering applications. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:29–66. doi: 10.1146/annurev.arplant.52.1.29. [DOI] [PubMed] [Google Scholar]
- 11.Liscombe DK, Facchini PJ. Evolutionary and cellular webs in benzylisoquinoline alkaloid biosynthesis. Curr. Opin. Biotechnol. 2008;19:173–180. doi: 10.1016/j.copbio.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 12.Facchini PJ, De Luca V. Opium poppy and Madagascar periwinkle: model and non-model systems to investigate alkaloid biosynthesis in plants. Plant J. 2008;54:763–784. doi: 10.1111/j.1365-313X.2008.03438.x. [DOI] [PubMed] [Google Scholar]
- 13.Onoyovwe A, et al. Morphine biosynthesis in opium poppy involves two cell types: sieve elements and laticifers. Plant Cell. 2013;25:4110–4122. doi: 10.1105/tpc.113.115113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boke H, et al. Regulation of the alkaloid biosynthesis by miRNA in opium poppy. Plant Biotechnol. J. 2015;13:409–420. doi: 10.1111/pbi.12346. [DOI] [PubMed] [Google Scholar]
- 15.Winzer T, et al. Morphinan biosynthesis in opium poppy requires a P450-oxidoreductase fusion protein. Science. 2015;340:309–312. doi: 10.1126/science.aab1852. [DOI] [PubMed] [Google Scholar]
- 16.Otani M, et al. Characterization of vacuolar transport of the endogenous alkaloid berberine in Coptis japonica. Plant Physiol. 2005;138:1939–1946. doi: 10.1104/pp.105.064352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamada Y, Motomura Y, Sato F. CjbHLH1 homologs regulate sanguinarine biosynthesis in Eschscholzia californica cells. Plant Cell Physiol. 2015;56:1019–1030. doi: 10.1093/pcp/pcv027. [DOI] [PubMed] [Google Scholar]
- 18.Torres MA, et al. Structural and functional studies of pavine N-methyltransferase from Thalictrum flavum reveal novel insights into substrate recognition and catalytic mechanism. J. Biol. Chem. 2016;291:23403–23415. doi: 10.1074/jbc.M116.747261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Robin AY, Giustini C, Graindorge M, Matringe M, Dumas R. Crystal structure of norcoclaurine-6-O-methyltransferase, a key rate-limiting step in the synthesis of benzylisoquinoline alkaloids. Plant J. 2016;87:641–653. doi: 10.1111/tpj.13225. [DOI] [PubMed] [Google Scholar]
- 20.Hagel JM, Facchini PJ. Benzylisoquinoline alkaloid metabolism: a century of discovery and a brave new world. Plant Cell Physiol. 2013;54:647–672. doi: 10.1093/pcp/pct020. [DOI] [PubMed] [Google Scholar]
- 21.Ziegler J, Facchini PJ. Alkaloid biosynthesis: metabolism and trafficking. Annu. Rev. Plant Biol. 2008;59:735–769. doi: 10.1146/annurev.arplant.59.032607.092730. [DOI] [PubMed] [Google Scholar]
- 22.Farrow SC, Hagel JM, Facchini PJ. Transcript and metabolite profiling in cell cultures of 18 plant species that produce benzylisoquinoline alkaloids. Phytochemistry. 2012;77:79–88. doi: 10.1016/j.phytochem.2012.02.014. [DOI] [PubMed] [Google Scholar]
- 23.Hagel JM, et al. Transcriptome analysis of 20 taxonomically related benzylisoquinoline alkaloid-producing plants. BMC Plant Biol. 2015;15:227. doi: 10.1186/s12870-015-0596-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xiao J, et al. Supercritical fluid extraction and identification of isoquinoline alkaloids from leaves of Nelumbo nucifera Gaertn. Eur. Food Res. Technol. 2010;231:407–414. doi: 10.1007/s00217-010-1290-y. [DOI] [Google Scholar]
- 25.Zhou M, et al. Identification and comparison of anti-inflammatory ingredients from different organs of lotus Nelumbo by UPLC/Q-TOF and PCA coupled with a NF-κB reporter gene assay. PLoS ONE. 2013;8:e81971. doi: 10.1371/journal.pone.0081971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Do TCMV, Nguyen TD, Tran H, Stuppner H, Ganzera M. Analysis of alkaloids in lotus (Nelumbo nucifera Gaertn.) leaves by nonaqueous capillary electrophoresis using ultraviolet and mass spectrometric detection. J. Chromatogr. A. 2013;1302:174–180. doi: 10.1016/j.chroma.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 27.Ming R, et al. Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.) Genome Biol. 2013;14:R41. doi: 10.1186/gb-2013-14-5-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C, et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C, et al. Transcript assembly and quantification by RNA-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gu C, et al. Characterization of genes encoding granule-bound starch synthase in sacred lotus reveals phylogenetic affinity of Nelumbo to Proteales. Plant Mol. Biol. Rep. 2013;31:1157–1165. doi: 10.1007/s11105-013-0605-0. [DOI] [Google Scholar]
- 32.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 33.Zhou H, et al. Multiple R2R3-MYB transcription factors involved in the regulation of anthocyanin accumulation in peach flower. Front. Plant Sci. 2016;7:1557. doi: 10.3389/fpls.2016.01557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vimolmangkang S, et al. Evolutionary origin of the NCSI gene subfamily encoding norcoclaurine synthase is associated with the biosynthesis of benzylisoquinoline alkaloids in plants. Sci. Rep. 2016;6:26323. doi: 10.1038/srep26323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ounaroon A, Decker G, Schmidt J, Lottspeich F, Kutchan TM. (R,S)-Reticuline 7-O-methyltransferase and (R,S)-norcoclaurine 6-O-methyltransferase of Papaver somniferum: cDNA cloning and characterization of methyl transfer enzymes of alkaloid biosynthesis in opium poppy. Plant J. 2003;36:808–819. doi: 10.1046/j.1365-313X.2003.01928.x. [DOI] [PubMed] [Google Scholar]
- 36.Ziegler J, Diaz-Chávez ML, Kramell R, Ammer C, Kutchan TM. Comparative macroarray analysis of morphine containing Papaver somniferum and eight morphine free Papaver species identifies an O-methyltransferase involved in benzylisoquinoline biosynthesis. Planta. 2005;222:458–471. doi: 10.1007/s00425-005-1550-4. [DOI] [PubMed] [Google Scholar]
- 37.Pienkny S, Brandt W, Schmidt J, Kramell R, Ziegler J. Functional characterization of a novel benzylisoquinoline O-methyltransferase suggests its involvement in papaverine biosynthesis in opium poppy (Papaver somniferum L) Plant J. 2009;60:56–67. doi: 10.1111/j.1365-313X.2009.03937.x. [DOI] [PubMed] [Google Scholar]
- 38.Morishige T, Tsujita T, Yamada Y, Sato F. Molecular characterization of the S-adenosyl-l-methionine:3′-hydroxy-N-methylcoclaurine 4′-O-methyltransferase involved in isoquinoline alkaloid biosynthesis in Coptis japonica. J. Biol. Chem. 2000;275:23398–23405. doi: 10.1074/jbc.M002439200. [DOI] [PubMed] [Google Scholar]
- 39.Ikezawa N, Iwasa K, Sato F. Molecular cloning and characterization of CYP80G2, a cytochrome P450 that catalyzes an intramolecular C–C phenol coupling of (S)-reticuline in magnoflorine biosynthesis, from cultured Coptis japonica cells. J. Biol. Chem. 2008;283:8810–8821. doi: 10.1074/jbc.M705082200. [DOI] [PubMed] [Google Scholar]
- 40.Ikezawa N, et al. Molecular cloning and characterization of CYP719, a methylenedioxy bridge-forming enzyme that belongs to a novel P450 family, from cultured Coptis japonica cells. J. Biol. Chem. 2003;278:38557–38565. doi: 10.1074/jbc.M302470200. [DOI] [PubMed] [Google Scholar]
- 41.Runguphan W, Glenn WS, O’Connor SE. Redesign of a dioxygenase in morphine biosynthesis. Chem. Biol. 2012;19:674–678. doi: 10.1016/j.chembiol.2012.04.017. [DOI] [PubMed] [Google Scholar]
- 42.Liedschulte V, et al. Identification of CYP82E21 as a functional nicotine N-demethylase in tobacco flowers. Phytochemistry. 2016;131:9–16. doi: 10.1016/j.phytochem.2016.08.004. [DOI] [PubMed] [Google Scholar]
- 43.Kraus PF, Kutchan TM. Molecular cloning and heterologous expression of a cDNA encoding berbamunine synthase, a C–O phenol-coupling cytochrome P450 from the higher plant Berberis stolonifera. Proc. Natl. Acad. Sci. USA. 1995;92:2071–2075. doi: 10.1073/pnas.92.6.2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mizutani M, Sato F. Unusual P450 reactions in plant secondary metabolism. Arch. Biochem. Biophys. 2011;507:194–203. doi: 10.1016/j.abb.2010.09.026. [DOI] [PubMed] [Google Scholar]
- 45.Tzin V, et al. Expression of a bacterial feedback-insensitive 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase of the shikimate pathway in Arabidopsis elucidates potential metabolic bottlenecks between primary and secondary metabolism. New Phytol. 2012;194:430–439. doi: 10.1111/j.1469-8137.2012.04052.x. [DOI] [PubMed] [Google Scholar]
- 46.Patra B, Schluttenhofer C, Wu Y, Pattanaik S, Yuan L. Transcriptional regulation of secondary metabolite biosynthesis in plants. Biochim. Biophys. Acta. 2013;1829:1236–1247. doi: 10.1016/j.bbagrm.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 47.Zhou M, Memelink J. Jasmonate-responsive transcription factors regulating plant secondary metabolism. Biotechnol. Adv. 2016;34:441–449. doi: 10.1016/j.biotechadv.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 48.Lescot M, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dang TT, Onoyovwe A, Farrow SC, Facchini PJ. Biochemical genomics for gene discovery in benzylisoquinoline alkaloid biosynthesis in opium poppy and related species. Methods Enzymol. 2012;515:231–226. doi: 10.1016/B978-0-12-394290-6.00011-2. [DOI] [PubMed] [Google Scholar]
- 50.Dang TT, Facchini PJ. Characterization of three O-methyltransferases involved in noscapine biosynthesis in opium poppy. Plant Physiol. 2012;159:618–631. doi: 10.1104/pp.112.194886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Purwanto R, Hori K, Yamada Y, Sato F. Unraveling additional O-methylation steps in benzylisoquinoline alkaloid biosynthesis in California poppy (Eschscholzia californica) Plant Cell Physiol. 2017;58:1528–1540. doi: 10.1093/pcp/pcx093. [DOI] [PubMed] [Google Scholar]
- 52.Pauli HH, Kutchan TM. Molecular cloning and functional heterologous expression of two alleles encoding (S)-N-methylcoclaurine 3′-hydroxylase (CYP80B1), a new methyl jasmonate-inducible cytochrome P-450-dependent mono-oxygenase of benzylisoquinoline alkaloid biosynthesis. Plant J. 1998;13:193–801. doi: 10.1046/j.1365-313X.1998.00085.x. [DOI] [PubMed] [Google Scholar]
- 53.Gurkok T, et al. Functional characterization of 4′OMT and 7OMT genes in BIA biosynthesis. Front. Plant Sci. 2016;7:98. doi: 10.3389/fpls.2016.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Memelink J, Gantet P. Transcription factors involved in terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Phytochem. Rev. 2007;6:353. doi: 10.1007/s11101-006-9051-z. [DOI] [Google Scholar]
- 55.Shoji T, Kajikawa M, Hashimoto T. Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. Plant Cell. 2010;22:3390–3409. doi: 10.1105/tpc.110.078543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Suttipanta N, et al. The transcription factor CrWRKY1 positively regulates the terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Physiol. 2011;157:2081–2093. doi: 10.1104/pp.111.181834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Moerkercke A, et al. The bHLH transcription factor BIS1 controls the iridoid branch of the monoterpenoid indole alkaloid pathway in Catharanthus roseus. Proc. Natl. Acad. Sci. USA. 2015;112:8130–9135. doi: 10.1073/pnas.1504951112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Moerkercke A, et al. The basic helix–loop–helix transcription factor BIS2 is essential for monoterpenoid indole alkaloid production in the medicinal plant Catharanthus roseus. Plant J. 2016;88:3–12. doi: 10.1111/tpj.13230. [DOI] [PubMed] [Google Scholar]
- 59.Kato N, et al. Identification of a WRKY protein as a transcriptional regulator of benzylisoquinoline alkaloid biosynthesis in Coptis japonica. Plant Cell Physiol. 2007;48:8–18. doi: 10.1093/pcp/pcl041. [DOI] [PubMed] [Google Scholar]
- 60.Apuya NR, et al. Enhancement of alkaloid production in opium and California poppy by transactivation using heterologous regulatory factors. Plant Biotechnol. J. 2008;6:160–175. doi: 10.1111/j.1467-7652.2007.00302.x. [DOI] [PubMed] [Google Scholar]
- 61.Yao G, et al. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 2017;92:437–451. doi: 10.1111/tpj.13666. [DOI] [PubMed] [Google Scholar]
- 62.Zeng JK, et al. EjAP2-1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnol. J. 2015;13:1325–1334. doi: 10.1111/pbi.12351. [DOI] [PubMed] [Google Scholar]
- 63.Zhang H, et al. The basic helix–loop–helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J. 2011;67:61–71. doi: 10.1111/j.1365-313X.2011.04575.x. [DOI] [PubMed] [Google Scholar]
- 64.Ober D. Seeing double—gene duplication and diversification in plant secondary metabolism. Trends Plant Sci. 2005;10:444–449. doi: 10.1016/j.tplants.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 65.Cheng J, et al. Unraveling the mechanism underlying the glycosylation and methylation of anthocyanins in peach. Plant Physiol. 2014;166:1044–1058. doi: 10.1104/pp.114.246876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matsuba Y, et al. Evolution of a complex locus for terpene biosynthesis in solanum. Plant Cell. 2013;25:2022–2036. doi: 10.1105/tpc.113.111013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Denoeud F, et al. The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science. 2014;345:1181–1184. doi: 10.1126/science.1255274. [DOI] [PubMed] [Google Scholar]
- 68.Hori K, et al. Mining of the uncharacterized cytochrome p450 genes involved in alkaloid biosynthesis in California poppy using a draft genome sequence. Plant Cell Physiol. 2018;59:222–233. doi: 10.1093/pcp/pcx210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Marais DD, Rausher MD. Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature. 2008;454:762–765. doi: 10.1038/nature07092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.