Abstract
Although many developing countries use harvested rainwater (HRW) for drinking and other household purposes, its quality is seldom monitored. Continuous assessment of the microbial quality of HRW would ensure the safety of users of such water. The current study investigated the prevalence of pathogenic Escherichia coli strains and their antimicrobial resistance patterns in HRW tanks in the Eastern Cape, South Africa. Rainwater samples were collected weekly between June and September 2016 from 11 tanks in various areas of the province. Enumeration of E. coli was performed using the Colilert®18/Quanti-Tray® 2000 method. E. coli isolates were obtained and screened for their virulence potentials using polymerase chain reaction (PCR), and subsequently tested for antibiotic resistance using the disc-diffusion method against 11 antibiotics. The pathotype most detected was the neonatal meningitis E. coli (NMEC) (ibeA 28%) while pathotype enteroaggregative E. coli (EAEC) was not detected. The highest resistance of the E. coli isolates was observed against Cephalothin (76%). All tested pathotypes were susceptible to Gentamicin, and 52% demonstrated multiple-antibiotic resistance (MAR). The results of the current study are of public health concern since the use of untreated harvested rainwater for potable purposes may pose a risk of transmission of pathogenic and antimicrobial-resistant E. coli.
Keywords: antimicrobial resistance, pathogenic E. coli, harvested rainwater, public health, Sub-Saharan Africa, alternative water source
1. Introduction
Several countries around the world, including South Africa, make use of harvested rainwater (HRW) to meet their daily water needs. However, the most significant issue relating to the use of harvested rainwater is the potential health risk associated with the presence of various pathogenic organisms in such water [1]. Indicator organisms like E. coli have been used to determine the microbiological safety of water meant for drinking and other human needs. Although most E. coli strains are non-pathogenic, certain strains may be pathogenic and carry virulence genes (VGs) [2]. Pathogenic E. coli strains which can cause diseases in both humans and animals are categorised as intestinal pathogenic E. coli (InPEC) and extraintestinal pathogenic E. coli (ExPEC) [3]. Intestinal strains are mostly referred to as diarrhoeagenic Escherichia coli (DEC) due to their ability to cause diarrhoea using diverse mechanisms [4]. The ExPEC strains have been reported to cause diseases such as urinary tract infections, neonatal meningitis, sepsis and wound infections and some examples include neonatal meningitis Escherichia coli (NMEC) and uropathogenic E. coli (UPEC) [3].
Six groups of DEC strains known to cause intestinal infections include enterotoxigenic E. coli (ETEC), enteropathogenic E. coli (EPEC), enterohemorrhagic E. coli (EHEC), enteroaggregative E. coli (EAEC), diffusely adherent E. coli (DAEC) and enteroinvasive E. coli (EIEC). Among all E. coli pathotypes, ETEC strains cause a cholera-like diarrhoeal disease and are the most common cause of childhood and travellers’ diarrhoea in developing countries [5]. Diffusely adherent E. coli pathotypes were previously implicated in intestinal infections (diarrhoea in children between the ages of 18 months and 5 years) and extraintestinal infections (urinary tract infections and pregnancy complications) [6]. EIEC shows pathogenic phenotypic and genetic similarities with Shigella spp. and can be identified by their epithelial cell invasiveness mediated in part by the ipaH and virF genes and association with dysentery [7]. EHEC is associated with bloody diarrhoea and haemolytic uremic syndrome and expresses one or two Shiga-like toxin-encoding genes stx1 and stx2 [8].
Several virulence genes in these E. coli pathotypes are responsible for a wide array of infections such as diarrhoea or haemolytic colitis, neonatal meningitis, nosocomial septicaemia, haemolytic-uraemic syndrome and urinary tract infections [9]. Current molecular-based techniques such as polymerase chain reaction (PCR) allow for the identification of these VGs by amplifying specific target regions [10]. Virulence genes associated with these pathogenic strains have been isolated in diverse environments in South Africa. For example, the presence of DEC virulence genes in 60% of samples collected from the Apies River (water and sediments) was reported by Abia et al. [11]. In another study, a high prevalence of virulence genes associated with four pathogenic E. coli types (EAEC, EHEC, EPEC, and EIEC) in domestic rainwater harvesting tanks in Kleinmond, Cape Town was documented by Dobrowsky et al. [12]. Apart from being pathogenic, some of these microorganisms have developed resistance to many of the drugs designed to treat the infections they cause. For example, the antimicrobial resistance patterns of E. coli isolates in outpatient urinary tract infections in South Africa was studied and the results revealed that the isolated E. coli were resistant to trimethoprim-sulfamethoxazole (TMP-SMX; 68%), amoxicillin (65%) and ciprofloxacin (41%) [13]. Another study focused on the hospital, and community isolates of uropathogens at a tertiary hospital in South Africa and results revealed that the most isolated bacterial pathogen was E. coli (39%) [14]. Furthermore, levels of E. coli resistance to amoxicillin and co-trimoxazole ranged from 43–100% and 29–90%, respectively. The presence of such drug-resistant bacteria in human settings has placed constraints on the choice of safe, effective and inexpensive antibiotics, especially for low- and middle-income countries [15]. As such, the progression of resistant bacteria and the increasing incidence of antibiotic resistance genes (ARGs) are thus of significant public health concern [16].
Although studies have been carried out on the presence of virulence genes and antibiotic-resistant bacteria in various water sources such as wastewater effluents, taps, wells and boreholes in South Africa, very few studies have investigated their presence in harvested rainwater [12,17,18,19]. This study aimed at reporting on the prevalence of pathogenic E. coli strains and their antibiotic resistance patterns in harvested rainwater collected from tanks in the Eastern Cape Province of South Africa. Such results would highlight the need for appropriate development and implementation of effective household water treatment methods, thereby protecting the lives of populations using such water for their daily needs. Moreover, results of the current study will also add to existing research databases which report on the circulating strains of antimicrobial-resistant organisms.
2. Materials and Methods
2.1. Study Site and Sample Collection
Rooftop-harvested rainwater samples were collected from 11 rainwater-harvesting systems situated at various sites around Grahamstown west, Rhodes University campus and Kenton-on-sea in the Eastern Cape Province, South Africa. The distance between Rhodes University (33°31’36” S, 26°51’63” E) and Grahamstown west (33°18’36” S; 26°31”36” E) is approximately 4 km while the distance between Rhodes University and Kenton-on-sea (33°42’0” S, 26°41’0” E) is about 59.2 km. Mean annual rainfall in Grahamstown is 650 mm, with bimodal peaks in October–November and again in March–April. All the sites were selected based on the diversity in environmental conditions (e.g., presence of foliage and birds) as well as the various uses of the water stored in the tanks. A total of 110 water samples were collected from the 11 selected tanks from June 2016 to September 2016 and tested for E. coli. Sterile 5 L bottles were used to collect rainwater samples weekly by first rinsing the tap connected to the tanks with 70% ethanol and letting the tap run for 30 s before collection. Rainwater samples were taken from the same tanks once a week. Samples were then transported to Rhodes University laboratory on ice for microbial analysis within 6 h.
2.2. Enumeration and Isolation of E. coli
Enumeration of E. coli was carried out using the Colilert-18® Quanti-tray®/2000 (IDEXX Laboratories, Inc., Johannesburg, South Africa). The test was performed following the manufacturer’s instructions. After incubation at 37 °C for 18–24 h, presumptive E. coli isolates were obtained from fluorescent quanti-tray wells as described by Abia et al. [20]. The Colilert method has a detection limit ranging from <1 MPN/100 mL to >2419.6 MPN/100 mL. E. coli ATCC® 25922 was used as a positive control and Pseudomonas aeruginosa ATCC 49189 as a negative control. One hundred (100) E. coli isolates were then selected from the various tanks. Of the 100 isolates selected, 66 isolates were chosen from T1-T6 (11 isolates from each tank), 20 isolates were from T7 and T8 (10 isolates from each tank) and 14 isolates from T9 and T11. T10 was excluded from further analysis due to poor growth of the selected isolates from the culture media.
2.3. Identification of Pathogenic Escherichia coli Strains Using Polymerase Chain Reaction (PCR)
DNA Extraction and Detection of Virulence Genes in E. coli Isolates
One hundred (100) presumptive E. coli isolates were randomly selected and inoculated separately into 5 mL Erlenmeyer flasks containing 2 mL nutrient broth (Merck, Johannesburg, South Africa). The flasks were incubated overnight at 37 °C on a rotary shaker at 100 rpm. DNA was extracted from 1 mL of the overnight culture using the InstaGeneTM Matrix (Bio-Rad Laboratories, Johannesburg, South Africa) following the manufacturer’s instruction. The template DNA was stored at −20 °C for PCR assays. All selected samples were first confirmed as E. coli by testing for the presence of the malate dehydrogenase (mdh) gene which is found in most E. coli strains [21]. After that, the presence of a total of eight VGs (eaeA (EPEC/EHEC), eagg (EAEC), ipaH (EIEC), ST (ETEC), ibeA (NMEC), stx1 (EHEC), stx2 (EHEC) and flicH7 (EHEC)) were investigated. The primer sequences and the PCR-cycling conditions for the identification of the various VGs were as previously described by Abia et al. [19]. Both multiplex and singleplex PCR assays were performed for the target genes. Multiplex PCR assays were divided into 3 sets where set 1 contained eaeA, eagg and ipaH, set 2 contained flicH7 and Stx1 and finally set 3 contained ST and ibeA genes [19,22,23]. Singleplex real-time PCR assays were performed for the mdh and stx2 target genes [24,25].
2.4. Screening for Antibiotic-Resistant E. coli
The remaining 1 mL from the overnight culture was used for antibiotic resistance analysis using the disk-diffusion method [26]. Briefly, 100 µL of overnight E. coli culture was spread on Mueller–Hinton agar (Lasec, Cape Town, South Africa) and antibiotic mastrings (Davies diagnostics, Johannesburg, South Africa) were carefully placed onto inoculated plates, incubated at 37 °C for 18–20 h. Following incubation, the diameters (in millimetres) of clear zones of growth inhibition around the antibiotic disks were measured using a ruler and compared with the Clinical Laboratory Standard Institute (CLSI) 2013 reference values. The different phenotypic profiles (resistant, intermediate or susceptible) of the isolates were then determined following the interpretation of the zones of inhibition. A total of 11 antibiotics were selected for this study (Table 1). The antibiotics were chosen for their frequent use in the treatment of bacterial infections in South Africa Both positive (E. coli strain ATTC 25922) and negative controls (E. coli strain ATTC 35218) were included in the experiments.
Table 1.
Antibiotics used to determine antibiotic resistance of E. coli isolates.
| Class | Antibiotic | Abbreviation | Concentration (µg) |
|---|---|---|---|
| β-Lactams | Ampicillin | AP | 10 |
| Cephalothin | KF | 5 | |
| Polypeptides | Colistin sulphate | CO | 25 |
| Aminoglycosides | Gentamicin | GM | 10 |
| Aminoglycosides | Streptomicin | S | 10 |
| Tetracyclines | Tetracycline | T | 25 |
| Folate pathway inhibitors | Cotrimoxazole | TS | 25 |
| Fluoroquinolones | Ciprofloxacin | CIP | 5 |
| Penicillin combination | Augmentin(amoxillin-clavulanate) | AUG | 30 |
| Sulfonamides | Trimethoprim | TM | 5 |
| Nitrofurans | Nitrofurantoin | NI | 300 |
2.5. Data Analysis
Data were analysed using the Statistical Package for the Social Sciences (SPSS) (Version 16.0, Prentice Hall Press Company, NJ, USA) [27]. The E. coli counts were log10 transformed before computation of the means and standard deviations. A multiple antibiotic resistance (MAR) index was performed following the procedure described by Krumperman [28]. A MAR index for an isolate was calculated using the formula: MAR = a/b where ‘a’ is the number of antibiotics from each group to which a particular isolate was resistant and ‘b’ is the total number of antibiotics against which the isolate was tested. A resistance index greater than 0.2 shows that E. coli isolates are likely to be from a high-risk source.
3. Results
3.1. Concentration of E. coli in Harvested Rainwater (HRW)
The log transformed (log10) E. coli counts and the mean E. coli counts in most probable number per 100 mL (MPN/100 mL) from individual tanks are shown in Table 2. The abundance of E. coli in the rainwater-harvesting tanks differed according to the location of the HRW system. The highest concentrations of E. coli were detected in tanks situated at Rhodes University (T1–T6).
Table 2.
Log transformed E. coli (MPN/100 mL) concentrations from various rainwater tanks.
| Tank ID | n | Minimum | Maximum | Mean ± Standard Deviation |
|---|---|---|---|---|
| T1 | 11 | 2.55 | 3.29 | 3.02 ± 0.21 |
| T2 | 11 | 1.95 | 3.11 | 2.62 ± 0.35 |
| T3 | 11 | 2.58 | 3.29 | 2.84 ± 0.25 |
| T4 | 11 | 1.64 | 2.89 | 2.52 ± 0.42 |
| T5 | 11 | 0.79 | 3.00 | 2.18 ± 0.82 |
| T6 | 11 | 2.53 | 3.04 | 2.88 ± 0.19 |
| T7 | 10 | 1.73 | 2.96 | 2.36 ± 0.37 |
| T8 | 10 | 1.78 | 2.41 | 2.09 ± 0.22 |
| T9 | 7 | 0.61 | 3.04 | 1.71 ± 0.82 |
| T10 | 10 | 0.3 | 3.19 | 1.57 ± 1.04 |
| T11 | 7 | 0.61 | 1.12 | 0.85 ± 0.26 |
3.2. Identification of Virulence Genes among E. coli Isolates
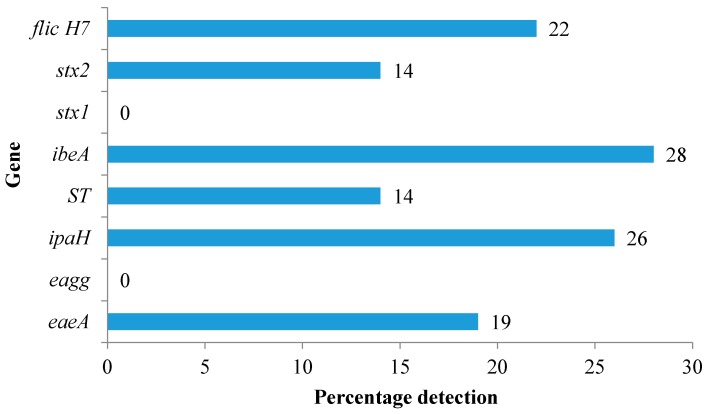
Samples which generated fluorescence from the Quanti-tray®/2000 cells were selected for the identification of the E. coli VGs. The most detected pathotypes were the NMEC and EHEC while the least detected pathotype was EAEC (Table 3). Of the 100 isolates tested for the VGs, 28% were identified as ibeA positive (Figure 1). The EAEC pathotype (eagg gene) was not detected among the tested isolates. Similarly, the Stx1 gene of EHEC was not detected in any of the isolates.
Table 3.
Number of virulence genes detected from rainwater-harvesting tanks.
| Tank Location | Tank ID | Number of E. coli Isolates Tested | EaeA (EPEC/EHEC) | Eagg (EAEC) | ipaH (EIEC) | ST (ETEC) | ibeA (NMEC) | Stx1 (EHEC) | Stx2 (EHEC) | flichH7 (EHEC) |
|---|---|---|---|---|---|---|---|---|---|---|
| Rhodes University | T1 | 11 | 6 (55%) | 0 | 4 (36%) | 0 | 4 (36%) | 0 | 2 (18%) | 4 (36%) |
| Rhodes University | T2 | 11 | 0 | 0 | 0 | 1 (9%) | 1 (9%) | 0 | 0 | 2 (18%) |
| Rhodes University | T3 | 11 | 1 (9%) | 0 | 0 | 0 | 3 (27%) | 0 | 0 | 1 (9%) |
| Rhodes University | T4 | 11 | 2 (18%) | 0 | 1 (9%) | 0 | 4 (36%) | 0 | 0 | 3 (27%) |
| Rhodes University | T5 | 11 | 1 (9%) | 0 | 0 | 0 | 4 (36%) | 0 | 0 | 2 (18%) |
| Rhodes University | T6 | 11 | 1 (9%) | 0 | 2 (18%) | 2 (18%) | 8 (72%) | 0 | 0 | 3 (27%) |
| Kenton-on-sea | T7 | 10 | 2 (20%) | 0 | 0 | 0 | 2 (20%) | 0 | 0 | 2 (20%) |
| Kenton-on-sea | T8 | 10 | 0 | 0 | 4 (40%) | 0 | 2 (20%) | 0 | 1(10%) | 2 (20%) |
| Grahamstown west | T9 | 7 | 1 (14%) | 0 | 0 | 0 | 1 (13%) | 0 | 0 | 0 |
| Grahamstown west | T11 | 7 | 0 | 0 | 0 | 0 | 1 (13%) | 0 | 0 | 0 |
Note: EPEC = Enteropathogenic E. coli, EHEC = Enterohemorrhagic E. coli, EAEC = Enteroaggregative E. coli, EIEC = Enteroinvasive E. coli, NMEC = Neonatal meningitis E. coli.
Figure 1.
Overall prevalence of virulence genes in isolated E. coli from harvested rainwater (HRW) tanks.
3.3. Antibiotic-Resistance Profiles of E. coli Isolated from the Harvested-Rainwater Samples
3.3.1. Overall Antibiotic Resistance Profiles of the E. coli
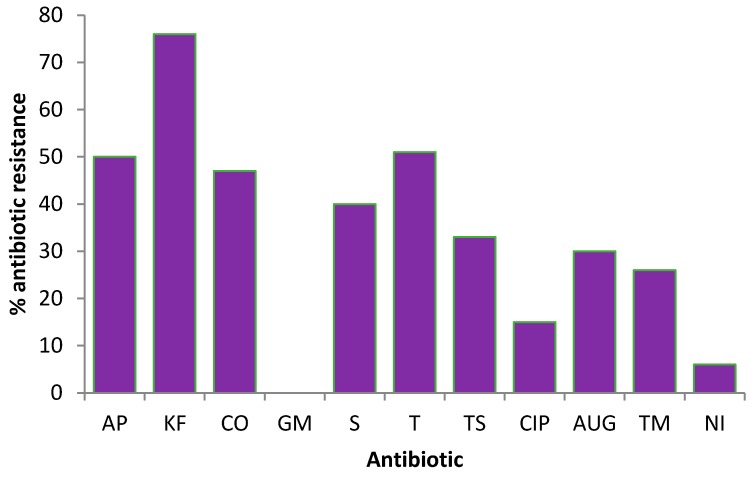
All the 100 E. coli isolates tested for the presence of VGs were further tested for antibiotic resistance. Of the 11 antibiotics tested, the highest resistance displayed by E. coli isolates was against Cephalothin (76%) while complete susceptibility (100%) was observed to Gentamycin. The overall percentage of antibiotic resistance found in the tested isolates is shown in Figure 2. E. coli isolates were resistant to 10 of the 11 antibiotics used in this study with the resistant rate ranging from 9% to 76%. Furthermore, a low percentage of the isolates showed resistance to Ciprofloxacin (15%) and Nitrofurantoin (9%).
Figure 2.
Percentage antibiotic resistance of E. coli isolates to selected antibiotics.
The bacterial resistance rate in individual tanks is shown in Table 4. Resistance to Nitrofurantoin was only observed in T1 and T2, while resistance to Augmentin was seen in all the tanks studied. Some of the selected isolates showed the presence of multiple-antibiotic resistance (MAR) where simultaneous resistance ranged from 3 to 9 antibiotics.
Table 4.
Antibiotic resistance among E. coli strains isolated from various rainwater tanks.
| Tank ID | n | % Resistance | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| AP | KF | CO | GM | S | T | TS | CIP | AUG | TM | NI | ||
| T1 | 11 | 72 | 91 | 63 | 0 | 45 | 72 | 39 | 27 | 27 | 63 | 9 |
| T2 | 11 | 72 | 81 | 36 | 0 | 27 | 45 | 45 | 9 | 54 | 45 | 18 |
| T3 | 11 | 27 | 36 | 27 | 0 | 45 | 18 | 27 | 18 | 9 | 18 | 0 |
| T4 | 11 | 36 | 90 | 54 | 0 | 36 | 36 | 27 | 0 | 9 | 27 | 0 |
| T5 | 11 | 36 | 100 | 54 | 0 | 0 | 54 | 36 | 27 | 18 | 45 | 0 |
| T6 | 11 | 45 | 100 | 45 | 0 | 18 | 36 | 36 | 0 | 9 | 27 | 0 |
| T7 | 10 | 30 | 30 | 20 | 0 | 100 | 20 | 30 | 20 | 30 | 30 | 0 |
| T8 | 10 | 30 | 100 | 10 | 0 | 20 | 10 | 0 | 0 | 40 | 0 | 0 |
| T9 | 7 | 75 | 87 | 75 | 0 | 62 | 100 | 87 | 0 | 75 | 50 | 0 |
| T11 | 7 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 40 | 0 | 0 |
3.3.2. Prevalence of Multiple-Antibiotic Resistance
The presence of MAR was also observed for most isolates. Multiple-antibiotic resistance in this study was defined as the resistance of bacterial strains to three or more antibiotics [20]. Of the 100 isolates tested, more than half (52%) were MAR (Table 5). Ten of the 52 MAR isolates demonstrated simultaneous resistance to up to nine antibiotics. A total of 24 different MAR phenotypes were identified in this study.
Table 5.
Multiple-antibiotic-resistant phenotypes of E. coli isolated from different rainwater tanks.
| T1 | T2 | ||
|---|---|---|---|
| MAR Phenotype | Number of Isolates | MAR Phenotype | Number of Isolates |
| AP-KF-CO-T | 1 | KF-T-NI | 1 |
| AP-KF-CO-T-TM | 1 | AP-KF-AUG | 1 |
| AP-KF-CO-S-T-TS-TM | 1 | AP-KF-NI | 1 |
| AP-KF-CO-S-T-TS-CIP-AUG-TM | 1 | AP-KF-S-T-TM | 1 |
| AP-KF-CO-S-T-TS-CIP-TM | 1 | AP-KF-CO-T-AUG | 1 |
| KF-CO-S-T-TS-AUG-TM | 1 | AP-KF-CO-S-T-TS-TM | 1 |
| AP-KF-CO-S-TS-CIP-TM | 1 | AP-KF-CO-S-T-TS-CIP-AUG-TM | 1 |
| AP-KF-CO-S-T-TS-AUG-TM | 1 | AP-KF-CO-T-TS-CIP-AUG-TM | 1 |
| AP-KF-CO-S-T-TS-AUG-TM | 1 | ||
| T3 | T4 | ||
| MAR Phenotype | Number of Isolates | MAR Phenotype | Number of Isolates |
| AP-KF-CO-S-TS-CIP-TM | 1 | KF-ST-AUG | 1 |
| AP-KF-CO-S-T-TS-TM | 1 | KF-T-NI | 1 |
| AP-KF-CO-S-T-TS-CIP-AUG-TM | 1 | AP-KF-CO-S-T-TS-TM | 2 |
| AP-KF-CO-S-T-TS-CIP-TM | 1 | ||
| AP-KF-CO-S-T-TS-CIP-AUG-TM | 2 | ||
| T5 | T6 | ||
| MAR Phenotype | Number of Isolates | MAR Phenotype | Number of Isolates |
| AP-KF-CO-S-T-TS-CIP-TM | 1 | AP-KF-AUG | 1 |
| AP-KF-CO-T-TS-TM | 1 | KF-CO-S-T-TS-AUG-TM | 1 |
| AP-KF-CO-S-T-TS-CIP-AUG-TM | 1 | AP-KF-CO-S-TS-TM | 1 |
| KF-CO-T-TS-AUG-TM | 1 | AP-KF-CO-T-TS-TM-NI | 1 |
| AP-KF-CO-S-T-TS-CIP-AUG-TM | 1 | AP-KF-CO-S-T-TS-CIP-AUG-TM | 2 |
| T7 | T8 | ||
| MAR Phenotype | Number of Isolates | MAR Phenotype | Number of Isolates |
| AP-KF-CO-S-T-TS-CIP-AUG-TM | 1 | AP-KF-T | 1 |
| AP-KF-CO-S-TS-AUG-TM | 1 | KF-CO-S-TS | 1 |
| AP-KF-CO-S-T-CIP-AUG-TM | 1 | AP-KF-CO-S-T-TS-CIP-AUG-TM | 2 |
| AP-KF-CO-S-T-TS-AUG-TM | 1 | ||
| T9 | T11 | ||
| MAR Phenotype | Number of Isolates | MAR Phenotype | Number of Isolates |
| AP-KF-CO-S-T-TS-CIP-AUG-TM | 1 | AP-KF-CO-S-T-TS-AUG-TM | 2 |
| AP-KF-CO-S-T-TS-AUG-TM | 2 | AP-KF-CO-S-T-TS-TM | 1 |
4. Discussion
4.1. Concentration of E. coli in Harvested Rainwater
Faecal coliform bacteria such as E. coli have been widely used as indicator organisms to assess the possibility of pathogen presence in water [29]. Therefore, the presence of E. coli in roof-harvested rainwater in the Eastern Cape, South Africa, was monitored. All the 11 tanks monitored in this study were contaminated with varying concentrations of E. coli (0.85 ± 0.26–3.02 ± 0.21 MPN/100 mL). Other scholars have previously reported on the high detection of E. coli from roof-harvested rainwater (2 to 986 CFU/100 mL; 1 to 99 MPN/100 mL and 0 to 41 CFU/100 mL) [1,30,31]. None of the tanks monitored in this study met the guidelines for drinking-water quality, as the E. coli amounts exceeded the South African drinking-water quality guidelines of 0 CFU/100 mL. The considerable amounts of E. coli in the harvested rainwater samples indicate possible faecal contamination.
The variations in the number of E. coli contamination in different HRW systems could be attributed to the fact that some of the HRW systems (Rhodes University) had a constant presence of birds which could have landed and dropped faecal matter on the roof, thereby contaminating tank water. Bird faecal droppings may negatively impact roof-harvested rainwater quality due to the presence of zoonotic pathogens [32]. A study conducted in South Africa investigated antibiotic resistance in E. coli isolates from roof-harvested rainwater tanks and urban pigeon faeces as the likely source of contamination and concluded that urban pigeons, the most likely source of HRW contamination, are also reservoirs of multiple antibiotic-resistant bacteria [33]. The findings of the South African study on bird faeces and antibiotic-resistant E. coli have a similar conclusion to our study where bird faecal matter was suspected to contribute to the contamination of HRW. In cases where the sources of faecal pollution in rainwater tanks are suspected to be from birds, the application of bird faecal markers may have the potential to confirm the sources of faecal contamination in a rainwater tank [32]. In another study to identify the likely sources of potential clinically significant E. coli in rainwater tanks, a source-tracking approach was used where a biochemical-fingerprinting method for typing of E. coli strains revealed that of the 43 strains from rainwater tank samples, 14 (from 7 tanks) and 9 (from 6 tanks) had identical biochemical phenotypes to those found in bird and possum faecal samples, respectively [34]. Furthermore, five strains from 4 rainwater tanks were identical to those isolated from both bird and possum faecal samples [34].
The rainwater tanks in the current study are used for various purposes such drinking and toilet flushing (for tanks situated at Rhodes University). Tanks situated at Grahamstown west were mainly used for gardening and sometimes drinking, depending on the availability of the municipal supply, while Kenton-on-sea tanks were used for indoor potable uses such dish-washing and laundry. In order to reduce or limit the risk of pathogenic and antimicrobial resistant E. coli, constant cleaning and maintenance of the catchment area may significantly improve the quality of the HRW, as the catchment area is suspected to contribute largely to the deterioration of the HRW in the Eastern Cape due to birds landing on the roof. Installation of first flush diverters may also help to improve the quality of the HRW. A study conducted in South Africa on the quality of HRW reported that 100% of the samples tested for E. coli exceeded the recommended standard of 0 CFU/100 mL [12]. Their results were similar to the ones observed in this study where all of the samples showed high levels of E. coli. In the Eastern Cape, where harvested rainwater is used for various household purposes including drinking, the presence of E. coli in the rainwater tanks is a major health concern as the presence of E. coli could imply the presence of other bacterial pathogens which may be detrimental to the health of rainwater users. The findings of the current study are of significant health concern as antibiotic-resistant pathogenic E. coli isolates may cause diseases if the users of the HRW consume the water without treatment. Furthermore, resistance of the isolated pathogenic E. coli to commonly used antibiotics in South Africa may lead to antibiotic treatment failure with serious public health implications for the population and the country.
4.2. Identification of Virulence Genes among E. coli Isolates
Pathogenic E. coli strains are a major cause of infections worldwide, the most common of which are diarrhoeal diseases. All the 100 E. coli isolates from the tanks tested positive for one or more VGs. The most detected pathotype was the NMEC (ibeA; 28%) which is responsible for neonatal meningitis and endothelial cell invasion [35]. The ibeA gene is also reportedly found in avian pathogenic E. coli (APEC) and causes avian colibacillosis, which is the most significant infectious bacterial disease of poultry worldwide [35]. The detection of the ibeA-positive strains in this study possibly indicates that the observed pathotype may be due to the presence of birds around the HRW systems. Although the present study did not investigate whether the ibeA gene detected was of human or avian origin, the presence of ibeA-positive isolates in the HRW systems is still of health concern given that there could be a possibility of zoonotic infections arising from the consumption of untreated rainwater containing these strains. Genes pertaining to other pathotypes of public health concern were also detected in the present study. For example, the flicH7 (22%) and Stx2 (14%) genes of EHEC were also detected in the isolates. Members of the EHEC group have been involved in many diarrhoeal disease outbreaks around the world, and they are known to cause hemorrhagic colitis and hemolytic uremic syndrome in humans [36].
The EHEC pathotype showed high prevalence across all the sampling sites except for the sites located in Grahamstown west. Both T1 and T6 which yielded a high percentage in VGs detection were situated at Rhodes University. Prevalence of the virulence gene ipaH (26%) (pathotype EIEC) was also noticeable in 4 tanks; 3 of the tanks were situated on campus and 1 in Kenton-on-sea. A previous study conducted in Cape Town, South Africa, reported that EPEC and EHEC (3% each) were detected in lower numbers, whereas EIEC was not identified in any of the rainwater tanks tested in their study [12]. The results differ from the findings of the current study where EIEC (26%) was the second most detected pathotype. This shows that the location of the tank could affect the pathotypes detected. Due to the detection of E. coli pathotypes in the current study, there is a great need to create awareness on household treatment technologies among users of HRW. Available treatment options which have proven to be successful in the treatment of HRW such as boiling, closed-couple solar pasteurizer, and solar disinfection (SODIS) can be used to decontaminate HRW [37,38,39]. In this study, all the rainwater tanks did not have any treatment option fitted, such as first-flush diverters and filters, except for T5 which had a chlorinator. However, due to limited maintenance of the rainwater-harvesting systems, the chlorinator in T5 was clogged in the middle of the sampling season and the E. coli counts increased going forward. The interruption of the treatment option observed in this study is also a clear indication of lack of proper maintenance of the HRW systems.
4.3. Detection of Antibiotic-Resistant E. coli in Harvested Rainwater
Results of the antibiotic-resistance profiling of the isolates from harvested rainwater analysed in the current study revealed that most of the E. coli isolates were resistant to the commonly prescribed antibiotics in South Africa. In areas such as the Eastern Cape where most of the population rely on harvested rainwater, exposure to antibiotic-resistant bacteria can further increase the health risk, particularly to children, the elderly and immune-compromised individuals. Antibiotic resistance is on the increase worldwide as most microorganisms now exhibit resistance to a large number of known antibiotics. The E. coli isolates from harvested rainwater in this study revealed resistance to Cephalothin (76%), Tetracyclines (51%), Colistin sulphate (47%), Ampicillin (50%) and Streptomycin (40%). The antibiotics most used in South Africa are the penicillins (Cephalothin) and fluoroquinolones, (Ciprofloxacin and glycopeptides) [40]. Tetracyclines and trimethoprim are also extensively used in the treatment of bacterial infections in both human and animals [41].
Cephalothin belongs to the β-Lactam class of antibiotics which are characterised by a β-lactam ring in their molecular structure [42]. Resistance to beta-lactam antibiotics has been highly documented as bacterial strains that produce extended-spectrum beta-lactamases have become more common [43]. Extended-spectrum beta-lactamase (ESBL)-producing E. coli are highly resistant to an array of antibiotics and infections by these strains are difficult to treat [43]. Furthermore, genes for ESBLs are most often encoded on plasmids, which can readily be transferred between bacteria [44]. Given that most of the isolates carrying virulence genes, especially the ibeA gene, were also resistant to Cephalothin, this could suggest that most of the isolated E. coli strains may carry the ESBL genes with the possibility of transfer to related organisms within the rainwater tanks. However, it is necessary to conduct further studies to ascertain such ARGs’ transfer within harvested-rainwater systems. Results of such studies would highlight the need for implementation of appropriate treatment options and better policies for the safe use of harvested rainwater, especially where such water is the main source of water for personal and household uses, thus protecting the lives of users of harvested rainwater.
In the current study, the tested E. coli isolates showed resistance to one or more antibiotics with the highest E. coli resistance recorded against Cephalothin, Ampicillin and Tetracyclines. Also, there was evidence of MAR E. coli in almost all the HRW systems with some isolates showing simultaneous resistance to a panel of up to nine antibiotics. These results indicate that in the case of infections occurring due to the consumption of contaminated harvested rainwater, treatment may fail because of the persistent resistance of the E. coli isolates detected in the HRW systems. A similar study carried out in Pretoria and Johannesburg, South Africa, showed that the resistances most encountered were against Ampicillin, Gentamicin, Amikacin and Tetracyclines [34]. These results were not in agreement with our findings, where E. coli isolates were resistant to Cephalothin and 100% susceptible to Gentamicin, although the same method and concentration was used for Gentamicin in both studies. The difference in antibiotic resistance results from the two studies could be attributed to the fact that roof-harvested rainwater samples were collected from different locations (Gauteng and Eastern Cape). Our findings were, however, similar to the those of Chidamba and Korsten [34] in that the authors also reported a substantial prevalence of MAR. All the isolates tested in this study showed a MAR index greater than 0.2, suggesting that a greater proportion of the isolates were likely to be from a high-risk source such as faecal material. These results and the differences observed with other studies could inform those implementing antibiotic-resistance surveillance schemes that would address different geographical locations. Also, the presence of MAR E. coli in harvested rainwater could pose a severe health risk to the public in general, as antibiotic resistance decreases the efficiency of antibiotics used in the treatment of infections. These findings are of major concern, as more households are now reported to be using harvested rainwater for their daily water needs.
5. Conclusions
Rainwater samples tested in this study showed contamination with varying concentrations of pathogenic E. coli strains. The outcome of the study further demonstrates that HRW tanks could serve as reservoirs for not only pathogenic but also antibiotic-resistant E. coli strains including MAR strains. These findings suggest that the tested harvested rainwater was not fit for human consumption and, therefore, should not be used for potable purposes without appropriate treatment. Furthermore, routine monitoring and treatment are essential to ensure that harvested rainwater is fit for intended use as well as to stimulate the need for strategies (e.g., maintenance of HRW systems, constant cleaning of the roof, and installation of first-flush diverters to minimise faecal contamination) that would prevent the spread of antibiotic-resistant bacteria.
Acknowledgments
This study was supported by funds from the Parliamentary Grant of the Council for Scientific and Industrial Research (CSIR Project ECHS043) and the Water Research Commission (WRC project K5/2593). Rhodes University provided access to monitoring sites and laboratory space for sample analysis. CSIR colleague Lisa Schaefer is thanked for advice on laboratory PCR analysis.
Author Contributions
A.L.K.A. and E.U.-J. conceived and designed the experiments as well as editing the manuscript; M.S.M. performed the experiments, analyzed the data and wrote the manuscript; R.T. and B.Z. contributed reagents/materials/analysis tools, sample collection as well as laboratory analysis and input on manuscript write-up; and J.-M.M.K. acquired the financial support for the project leading to this publication, supervised the project, and edited the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Ahmed W., Hodgers L., Masters N., Sidhu J.P.S., Katouli M., Toze S. Occurrence of intestinal and extraintestinal virulence genes in Escherichia coli isolates from rainwater tanks in Southeast Queensland, Australia. Appl. Environ. Microbiol. 2011;77:7394–7400. doi: 10.1128/AEM.06047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Masters N., Wiegand A., Ahmed W., Katouli M. Escherichia coli virulence genes profile of surface waters as an indicator of water quality. Water Res. 2011;45:6321–6333. doi: 10.1016/j.watres.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 3.Russo T.A., Johnson J.R. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 2000;181:1753–1754. doi: 10.1086/315418. [DOI] [PubMed] [Google Scholar]
- 4.Canizalez-Roman A., Gonzalez-Nuñez E., Vidal J.E., Flores-Villaseñor H., León-Sicairos N. Prevalence and antibiotic resistance profiles of diarrheagenic Escherichia coli strains isolated from food items in northwestern Mexico. Int. J. Food Microbiol. 2013;164:36–45. doi: 10.1016/j.ijfoodmicro.2013.03.020. [DOI] [PubMed] [Google Scholar]
- 5.Jafari A., Aslani M.M., Bouzari S. Escherichia coli: A brief review of diarrheagenic pathotypes and their role in diarrheal diseases in Iran. Iran. J. Microbiol. 2012;4:102–117. [PMC free article] [PubMed] [Google Scholar]
- 6.Servin A.L. Pathogenesis of human diffusely adhering Escherichia coli expressing Afa/Dr adhesins (Afa/Dr DAEC): Current insights and future challenges. Clin. Microbiol. Rev. 2014;27:823–869. doi: 10.1128/CMR.00036-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karmali M.A., Gannon V., Sargeant J.M. Verocytotoxin-producing Escherichia coli (VTEC) Vet. Microbiol. 2010;140:360–370. doi: 10.1016/j.vetmic.2009.04.011. [DOI] [PubMed] [Google Scholar]
- 8.Viazis S., Diez-Gonzalez F. The Twentieth Century’s Emerging Foodborne Pathogen: A Review. 1st ed. Volume 111 Elsevier Inc.; New York, NY, USA: 2011. Enterohemorrhagic Escherichia coli. [Google Scholar]
- 9.Anastasi E.M., Matthews B., Gundogdu A., Vollmerhausen T.L., Ramos N.L., Stratton H., Ahmed W., Katouli M. Prevalence and persistence of Escherichia coli strains with uropathogenic virulence characteristics in sewage treatment plants. Appl. Environ. Microbiol. 2010;76:5882–5886. doi: 10.1128/AEM.00141-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa C.F.M., Monteiro Neto V., Santos B.R.d.C., Costa B.R.R., Azevedo A., Serra J.L., Mendes H.B.R., Nascimento A.R., Mendes M.B.P., Kuppinger O. Enterobacteria identification and detection of diarrheagenic Escherichia coli in a Port Complex. Braz. J. Microbiol. 2014;45:945–952. doi: 10.1590/S1517-83822014000300026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Abia A.L.K., Ubomba-Jaswa E., Momba M.N.B. Occurrence of diarrhoeagenic Escherichia coli virulence genes in water and bed sediments of a river used by communities in Gauteng, South Africa. Environ. Sci. Pollut. Res. 2016;23:15665–15674. doi: 10.1007/s11356-016-6762-6. [DOI] [PubMed] [Google Scholar]
- 12.Dobrowsky P.H., van Deventer A., De Kwaadsteniet M., Ndlovu T., Khan S., Cloete T.E., Khan W. Prevalence of virulence genes associated with pathogenic Escherichia coli strains isolated from domestically harvested rainwater during low- and high-rainfall periods. Appl. Environ. Microbiol. 2014;80:1633–1638. doi: 10.1128/AEM.03061-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bosch F.J., van Vuuren C., Joubert G. Antimicrobial resistance patterns in outpatient urinary tractinfections—The constant need to revise prescribing habits. S. Afr. Med. J. 2011;101:328–331. doi: 10.7196/SAMJ.4346. [DOI] [PubMed] [Google Scholar]
- 14.Habte T.M., Dube S., Ismail N., Hoosen A.A. Hospital and community isolates of uropathogens at a tertiary hospital in South Africa. S. Afr. Med. J. 2009;99:584–587. [PubMed] [Google Scholar]
- 15.Detels R., Gulliford M., Karim Q.A., Tan C.C., Press O.U. In: Oxford Textbook of Global Public Health: The Practice of Public Health. 6th ed. Detels R., Beaglehole R., Lansang M., Gulliford M., editors. Volume 3 Oxford University; New York, NY, USA: 2015. [Google Scholar]
- 16.Xu Y., Guo C., Luo Y., Lv J., Zhang Y., Lin H., Wang L., Xu J. Occurrence and distribution of antibiotics, antibiotic resistance genes in the urban rivers in Beijing, China. Environ. Pollut. 2016;213:833–840. doi: 10.1016/j.envpol.2016.03.054. [DOI] [PubMed] [Google Scholar]
- 17.Kinge W.C.N., Ateba C.N., Kawadza D.T. Antibiotic resistance profiles of Escherichia coli isolated from different water sources in the Mmabatho locality, Northwest Province, South Africa. S. Afr. J. Sci. 2010;106:44–49. [Google Scholar]
- 18.Adefisoye M.A., Okoh A.I. Identification and antimicrobial resistance prevalence of pathogenic Escherichia coli strains from treated wastewater effluents in Eastern Cape, South Africa. Microbiologyopen. 2016;5:143–151. doi: 10.1002/mbo3.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Abia A.L.K., Schaefer L., Ubomba-Jaswa E., Le Roux W. Abundance of pathogenic Escherichia coli virulence-associated genes in well and borehole water used for domestic purposes in a peri-urban community of South Africa. Int. J. Environ. Res. Public Health. 2017;14:320. doi: 10.3390/ijerph14030320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abia A.L.K., Ubomba-Jaswa E., Momba M.N.B. High prevalence of multiple-antibiotic-resistant (MAR) Escherichia coli in river bed sediments of the Apies River, South Africa. Environ. Monit. Assess. 2015;187:652. doi: 10.1007/s10661-015-4879-6. [DOI] [PubMed] [Google Scholar]
- 21.Hsu S.C., Tsen H.Y. PCR primers designed from malic acid dehydrogenase gene and their use for detection of Escherichia coli in water and milk samples. Int. J. Food Microbiol. 2001;64:1–11. doi: 10.1016/S0168-1605(00)00425-6. [DOI] [PubMed] [Google Scholar]
- 22.Caine L., Nwodo U., Okoh A., Ndip R., Green E. Occurrence of virulence genes associated with diarrheagenic Escherichia coli isolated from raw cow’s milk from two commercial dairy farms in the Eastern Cape Province, South Africa. Int. J. Environ. Res. Public Health. 2014;11:11950–11963. doi: 10.3390/ijerph111111950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Titilawo Y., Obi L., Okoh A. Occurrence of virulence gene signatures associated with diarrhoeagenic and non-diarrhoeagenic pathovars of Escherichia coli isolates from some selected rivers in South-Western Nigeria. BMC Microbiol. 2015;15:204. doi: 10.1186/s12866-015-0540-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Omar K.B., Barnard T.G. Detection of diarrhoeagenic Escherichia coli in clinical and environmental water sources in South Africa using single-step 11-gene m-PCR. World J. Microbiol. Biotechnol. 2014;30:2663–2671. doi: 10.1007/s11274-014-1690-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Omar K.B., Potgieter N., Barnard T.G. Development of a rapid screening method for the detection of pathogenic Escherichia coli using a combination of Colilert® Quanti-Trays/2000 and PCR. Water Sci. Technol. Water Supply. 2010;10:7–13. doi: 10.2166/ws.2010.862. [DOI] [Google Scholar]
- 26.Stange C., Sidhu J.P.S., Tiehm A., Toze S. Antibiotic resistance and virulence genes in coliform water isolates. Int. J. Hyg. Environ. Health. 2016;219:823–831. doi: 10.1016/j.ijheh.2016.07.015. [DOI] [PubMed] [Google Scholar]
- 27.Norusis M. SPSS 16.0 Statistical Procedures Companion. Prentice Hall Press; Upper Saddle River, NJ, USA: 2008. [Google Scholar]
- 28.Krumperman P.H. Multiple antibiotic resistance indexing of Escherichia coli to indentify high-risk sources of fecal contamination of foods. Appl. Environ. Microbiol. 1983;46:165–170. doi: 10.1128/aem.46.1.165-170.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hachich E.M., Di Bari M., Christ A.P.G., Lamparelli C.C., Ramos S.S., Sato M.I.Z. Comparison of thermotolerant coliforms and Escherichia coli densities in freshwater bodies. Braz. J. Microbiol. 2012;43:675–681. doi: 10.1590/S1517-83822012000200032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spinks J., Phillips S., Robinson P., Van Buynder P. Bushfires and tank rainwater quality: A cause for concern? J. Water Health. 2006;4:21–28. [PubMed] [Google Scholar]
- 31.Sazakli E., Alexopoulos A., Leotsinidis M. Rainwater harvesting, quality assessment and utilization in Kefalonia Island, Greece. Water Res. 2007;41:2039–2047. doi: 10.1016/j.watres.2007.01.037. [DOI] [PubMed] [Google Scholar]
- 32.Ahmed W., Sidhu J.P.S., Toze S. An attempt to identify the likely sources of Escherichia coli harboring toxin genes in rainwater tanks. Environ. Sci. Technol. 2012;46:5193–5197. doi: 10.1021/es300292y. [DOI] [PubMed] [Google Scholar]
- 33.Chidamba L., Korsten L. Antibiotic resistance in Escherichia coli isolates from roof-harvested rainwater tanks and urban pigeon faeces as the likely source of contamination. Environ. Monit. Assess. 2015;187:405. doi: 10.1007/s10661-015-4636-x. [DOI] [PubMed] [Google Scholar]
- 34.Ahmed W., Hamilton K., Gyawali P., Toze S., Haas C. Evidence of avian and possum fecal contamination in rainwater tanks as determined by microbial source tracking approaches. Appl. Environ. Microbiol. 2016 doi: 10.1128/AEM.00892-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson T.J., Wannemuehler Y., Kariyawasam S., Johnson J.R., Logue C.M., Nolan L.K. Prevalence of avian-pathogenic Escherichia coli strain O1 genomic islands among extraintestinal and commensal E. coli isolates. J. Bacteriol. 2012;194:2846–2853. doi: 10.1128/JB.06375-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamilton M.J., Hadi A.Z., Griffith J.F., Ishii S., Sadowsky M.J. Large scale analysis of virulence genes in Escherichia coli strains isolated from Avalon Bay, CA. Water Res. 2010;44:5463–5473. doi: 10.1016/j.watres.2010.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spinks A.T., Dunstan R.H., Coombes P., Kuczera G. Thermal destruction analyses of water related pathogens at domestic hot water system temperatures; Proceedings of the 28th International Hydrology and Water Resources Symposium; Wollongong, Australia. 10–13 November 2003. [Google Scholar]
- 38.Dobrowsky P.H., Carstens M., De Villiers J., Cloete T.E., Khan W. Efficiency of a closed-coupled solar pasteurization system in treating roof harvested rainwater. Sci. Total Environ. 2015;536:206–214. doi: 10.1016/j.scitotenv.2015.06.126. [DOI] [PubMed] [Google Scholar]
- 39.Amin M.T., Nawaz M., Amin M.N., Han M. Solar disinfection of Pseudomonas aeruginosa in harvested rainwater: A step towards potability of rainwater. PLoS ONE. 2014;9:e90743. doi: 10.1371/journal.pone.0090743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Essack S.Y., Schellack N., Pople T., van der Merwe L., Suleman F., Meyer J.C., Gous A.G.S., Benjamin D. Part III. GARP: Antibiotic supply chain and management in human health. S. Afr. Med. J. 2011;101:562–566. [PubMed] [Google Scholar]
- 41.Ruhe J.J., Menon A. Tetracyclines as an oral treatment option for patients with community onset skin and soft tissue infections caused by methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2007;51:3298–3303. doi: 10.1128/AAC.00262-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Beceiro A., Tomás M., Bou G. Antimicrobial resistance and virulence: A successful or deleterious association in the bacterial world? Clin. Microbiol. Rev. 2013;26:185–230. doi: 10.1128/CMR.00059-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Paterson D.L., Bonomo R.A. Extended-Spectrum beta-Lactamases: A clinical update. Clin. Microbiol. Rev. 2005;18:657–686. doi: 10.1128/CMR.18.4.657-686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Haenni M., Châtre P., Madec J.Y. Emergence of Escherichia coli producing extended-spectrum AmpC β-lactamases (ESAC) in animals. Front. Microbiol. 2014;5:1–7. doi: 10.3389/fmicb.2014.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]