Abstract
Papillomaviridae is a diverse family of circular, double-stranded DNA (dsDNA) viruses that infect a broad range of mammalian, avian and fish hosts. While papillomaviruses have been characterized most extensively in humans, the study of non-human papillomaviruses has contributed greatly to our understanding of their pathogenicity and evolution. Using high-throughput sequencing approaches, we identified 7 novel papillomaviruses from vaginal swabs collected from 81 adult female Weddell seals (Leptonychotes weddellii) in the Ross Sea of Antarctica between 2014–2017. These seven papillomavirus genomes were amplified from seven individual seals, and six of the seven genomes represented novel species with distinct evolutionary lineages. This highlights the diversity of papillomaviruses among the relatively small number of Weddell seal samples tested. Viruses associated with large vertebrates are poorly studied in Antarctica, and this study adds information about papillomaviruses associated with Weddell seals and contributes to our understanding of the evolutionary history of papillomaviruses.
Keywords: papillomavirus, Leptonychotes weddellii, Antarctic, Carnivora
Introduction
Papillomaviridae is a family of circular, double-stranded DNA viruses that have ~7–8 kb genomes. There are >350 distinct papillomavirus types that infect skin, squamous and mucosal epithelial cells [1] in a wide range of hosts, including mammals, birds, reptiles and fish. Papillomaviruses have a genome organization that can be divided into three major regions: early, late and a long control region (LCR), which is involved in viral replication [2]. Upon infection, the early genes (E1, E2 and E4) are expressed and involved in DNA replication and transcription regulation. Most papillomaviruses encode at least one additional early protein (E5, E6 or E7) that is involved in manipulating the cellular environment. The expression of these proteins compromises the regulation processes of the cell cycle and leads to the proliferation of infected cells. In certain papillomaviruses, these proteins are oncogenic in their hosts [3–6], likely in part due to their ability to inactivate p53 and pRb proteins that have an essential role in modulating the cell cycle. The viral late region includes two structural proteins that form the viral capsid (L1 and L2) and are expressed later, once infected cells have proliferated to the squamous and mucosal epithelial layer. The LCR is involved in viral replication [2].
Most of the papillomaviruses identified to date have been associated with humans. The human papillomaviruses (HPVs) are classified in five genera, Alpha-, Beta-, Mu-, Nu- and Gammapapillomavirus. Non-human animal papillomaviruses have been characterized in hosts across 18 taxonomic orders [1, 7]. Consistent with the co-evolution hypothesis, phylogenetic analyses have shown the divergence of mammalian papillomaviruses from avian and reptile papillomaviruses [8]. Better understanding of the diversity of papillomaviruses and their evolutionary history will help to elucidate their impact on the health of wild animal populations. This has important implications for wildlife conservation management.
Just as viral diversity is better understood in humans than in other species, it is also better understood in regions where there is a long-established human presence. In more remote regions, such as polar ecosystems, little is known about viruses and associated diseases. Yet these areas are just those that are predicted to change most and to understand the polar disease ecology it essential to collect baseline data on viruses and other microbes circulating within these ecosystems. For example, very little is known about the viruses circulating amongst Antarctic animals [9], but there is evidence that endemic Antarctic species are infected with a similar viral diversity to that in other regions. The handful of viral genomes from Antarctic animals that have been identified include those that belong to the viral families Adenoviridae [10–12], Anelloviridae [13], Orthomyxoviridae [14, 15], Papillomaviridae [8, 16], Paramyxoviridae [17–20], Polyomaviridae [21–23], Poxviridae [24] and Togavirdae [25, 26].
The four Antarctic seals, Weddell, leopard (Hydrurga leptonyx), crabeater (Lobodon carcinophaga) and Ross (Ommatophoca rossii), are classified in the taxonomic tribe Lobodontini within the family Phocidae. Pinnipeds form a clade within the order Carnivora and diverged most recently from the family Ursidae. Within the carnivore order, pinnipeds diverged after the split of Feliformia and Caniformia, and therefore seals are also closely related to canines and mustelids [27, 28]. Weddell seals are the most southerly distributed pinniped, breeding on the fast ice of Antarctica. They remain at high latitudes year round by maintaining breathing holes along tidal cracks in the fast ice throughout the winter months, and consume a diverse diet [29–31]. Building on the first identification of a papillomavirus in pinnipeds (Zalophus californianus papillomavirus 1; ZcPV1) from California sea lions (Zalophus californianus) [32], this study aimed to assess whether the southernmost mammal, living in the isolated and unique polar habitat of the Ross Sea, Antarctica, is associated with unique and diverse papillomaviruses.
Results and discussion
Papillomaviruses associated with Weddell seals
We identified and recovered seven papillomavirus genomes that ranged in size from 7392 to 7836 nts from individual Weddell seals (aged 10–19 years at the time of sampling) from the three seasons (Fig. 1; Table 1). The seals were deemed clinically healthy at the time of sampling, and no lesions or papillomas were noted.
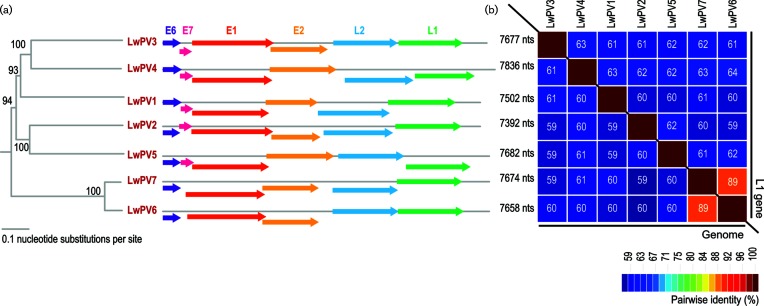
Fig. 1.
(a) Neighbour-joining phylogenetic tree of the tree inferred from the aligned genome sequences of LwPV1-7 with the genome organization for each genotype showing E6, E7, E2, L2 and L1 ORFs coupled with genome size. (b) Pairwise identity plot with percentage pairwise identities provided in coloured boxes for the genome and the L1 nucleotide sequences.
Table 1. Sample data for LwPV1-7, including the GenBank accession number (MG571089-MG571095), the season the LwPV was identified, the specimen number (SPENO) for the individual seal in which the respective virus was identified, the age of the individual at the time of sampling, the date the sample was taken and the abutting primer pair used to recover the genome.
| Name | Accession no. | Season | SPENO | Age at sampling | Sampling date | Primers |
|---|---|---|---|---|---|---|
| Leptonychotes weddellii papillomavirus 1 | MG571090 | 2014/2015 | 13 241 | 16 | 19 November 2014 | F: 5′-GCCTTATACTCATCAGGCTTATATGGATTTTGGG-3′ R: 5′-CTACCTAAGGCATACAGAGGAATATGACATTGCAT-3′ |
| Leptonychotes weddellii papillomavirus 2 | MG571089 | 2014/2015 | 11 445 | 19 | 18 January 2015 | F: 5′-TAATTTCAGAAACCTGTGATGCTGGAAGTGTTTG-3′ R: 5′-ATACTAAGCATGCTATTATGATGAAGTTGGTTTT-3′ |
| Leptonychotes weddellii papillomavirus 3 | MG571093 | 2015/2016 | 17 181 | 10 | 30 November 2015 | F: 5′-TATCCCTTTAATGATGAAGGACAGCCCACATATCT-3′ R: 5′-CTCATTTTTAAAGGTAAAGCACTGCAGTCTGCTG-3′ |
| Leptonychotes weddellii papillomavirus 4 | MG571095 | 2014/2015 | 12 091 | 18 | 28 January 2015 | F: 5′-AAAGTGTTGCCTTCCACTTTATTTACAACATCATC-3′ R: 5′-TGCAGATAGATTTTTAAAATGGGCAAGCTCTTTTC-3′ |
| Leptonychotes weddellii papillomavirus 5 | MG571094 | 2016/2017 | 12 975 | 18 | 24 November 2016 | F: 5′-TAAACAGTGACACACAGCTGTTTAACAGGCCTTTT-3′ R: 5′-CTAGAGAACCGCTGGGAGTTCCATAGTAGATAGAA-3′ |
| Leptonychotes weddellii papillomavirus 6 | MG571091 | 2016/2017 | 17 495 | 10 | 8 December 2016 | F: 5′-CATATCCAATAAAGTCAGATGATTCAGGAGGTAGC-3′ R: 5′-CTGTGTATAGTGACTTCTATTATGACCCTAGCCTT-3′ |
| Leptonychotes weddellii papillomavirus 7 | MG571092 | 2016/2017 | 17 629 | 10 | 19 January 2017 | F: 5′-GGCTATACTAACCTGATTTAGTATCCATTTTGGCC-3′ R: 5′-CACATATCACAGGACAGTACACCATTTGAACTATC-3′ |
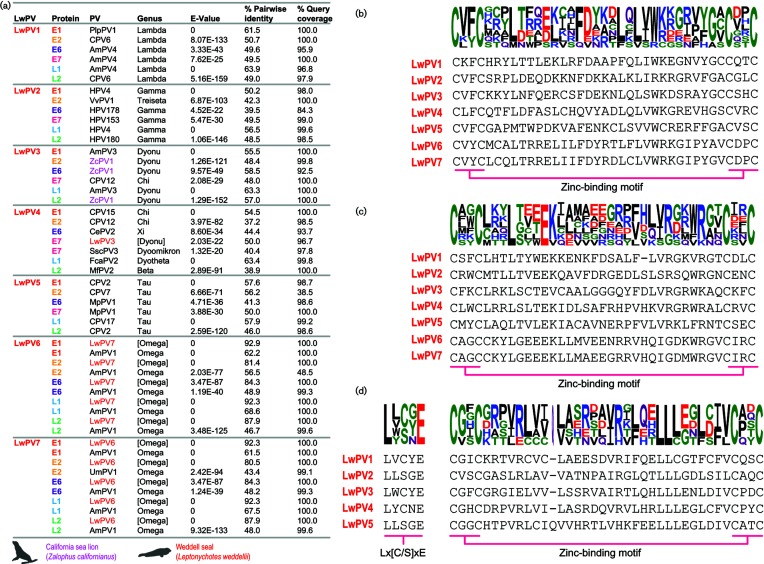
The L1 gene is relatively conserved among papillomaviruses and is the current basis for classification within this family, with the International Committee on Taxonomy of Viruses (ICTV) recommending that L1 gene sequences that share <90 % pairwise identity with those previously classified should be considered unique papillomavirus types. The L1 sequences of the seven recovered papillomaviruses from Weddell seals share <89 % pairwise identity (Fig. 1). When compared to all other papillomavirus L1 sequences, the ones from this study share <67 % pairwise identity. Thus, the seven papillomaviruses are all new types and have been named Leptonychotes weddellii papillomavirus 1–7 (LwPV1-7). A summary of the protein sequence similarities as determined by blastp [33] is provided in Fig. 2. LwPV6 and −7 which are most closely related, sharing 89 % genome-wide identity, both lack an E7 open reading frame (ORF) (Fig. 1). Both the E6 and E7 protein sequences of LwPV1-5 contain canonical zinc-binding motifs (CX2CX21–23CX2C metal-binding motif) in both the E6 and E7 protein sequences. In the E7 sequences of three LwPVs (LwPV1, −3 and −4) we identified the conserved pRB LxCxE binding motif (Fig. 2). LwPV2 and LwPV5 have a slightly modified LxSxE motif. In other papillomaviruses, similarly belonging to the Gamma- and Taupapillomaviridae, this modified motif is not involved in binding to and degradation of pRb [34].
Fig. 2.
(a) blastp results for each of the proteins encoded by LwPV1-7 with Weddell seal papillomaviruses highlighted in red and California sea lion papillomavirus 1 (ZcPV1) given in pink. Protein sequence of first (b) and second (c) zinc-binding motifs (CX2CX21–23CX2C) present in the L6 ORF of LwPV1-7. (d) Protein sequence of pRB-binding motif (Lx[C/S]xE) and zinc-binding motif in E7 ORF of LwPV1-5.
Sequences that share <60 % L1 nucleotide sequence identity are classified into different genera, while novel species isolates have 60–70 % sequence identity and types differ by at least 10 % sequence identity [35]. Based on pairwise L1 nucleotide sequence comparisons and L1 phylogenetic tree support (Fig. S1, available in the online version of this article), LwPV1-5 are likely members of new species within existing genera (LwPV1, Lambdapapillomavirus; LwPV2, Trisetapapimmomavirus; LwPV3, Dyonupapillomavirus; LwPV4, Dyothetapapillomavirus; LwPV5, Taupapillomavirus). Furthermore, it is apparent that LwPV6 and −7 share ~68–71 % pairwise identity and cluster with Ailuropoda melanoleuca papillomavirus (AmPV) 1–2 and Ursus maritimus papillomavirus 1 (UmPV1) in the genus Omegapapillomavirus (Fig. S1). Thus, LwPV6 and −7 are likely to be assigned to a new species in the genus Omegapapillomavirus.
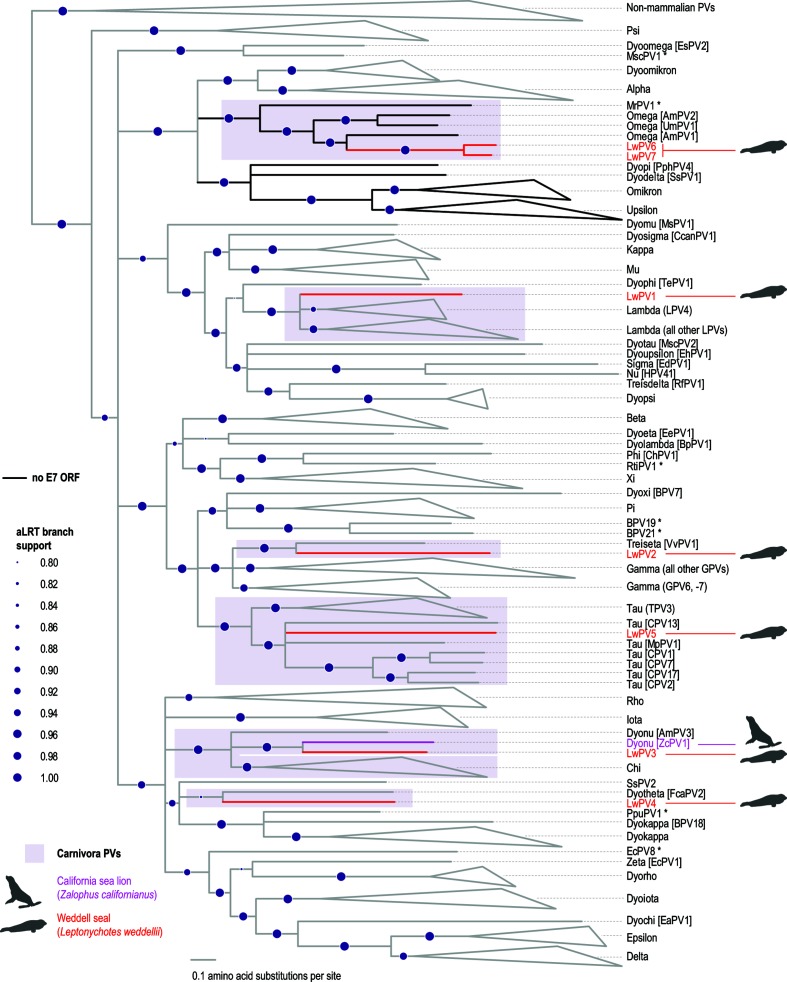
To investigate the evolutionary history of the seven novel Weddell seal papillomaviruses we constructed a phylogenetic tree based on the conserved E1, E2 and L1 ORFs [36] (Fig. 3). With regard to the seven novel Weddell seal papillomaviruses, this tree is congruent with the L1 nucleotide tree (Fig. S1). LwPV1 clusters with lambdapapillomaviruses (Figs 3 and S1), with the highest L1 amino acid pairwise identity (63.9 %) being with the L1 sequence of AmPV4 (Fig. 2). Phylogenetically, LwPV3 is most closely related to ZcPV1, despite sharing the highest L1 amino acid pairwise identity with AmPV3 (63.3 %). LwPV2 shares the highest amino acid pairwise identity (50 %) with human papillomavirus (HPV) 4 in the genus Gammapapillomavirus, but phylogenetically it is most closely related to VvPV1, the sole member of the genus Treisetapapillomavirus (Figs 3 and S1). The E7 protein of LwPV3 shares 50 % pairwise identity with that of LwPV4 but in general is most closely related to ZcPV1 (Figs 2, 3 and S1). The L1 protein of LwPV4 shared 63.4 % pairwise identity with that of Felis catus papillomavirus (FcaPV2). LwPV5 clusters with taupapillomaviruses, sharing 57.9, 57.6 and 56.2% amino acid pairwise identity with the L1, E1 and E2 sequences of canine papillomavirus (CPV) 17, CPV2 and CPV7, respectively. LwPV6-7 are most closely related to AmPV1 in the genus Omegapapillomavirus, with an L1 amino acid pairwise identity of 68–69 %.
Fig. 3.
Maximum-likelihood phylogenetic tree inferred using concatenated protein sequences of E1, E2 and L1. LwPV1-7 are highlighted in red and California sea lion papillomavirus 1 (ZcPV1) is highlighted in pink. Branches in black indicate lineages that have no recognizable E7 ORF. Branches with <0.75 aLRT branch support have been collapsed. Branch support values are given in purple circle size gradients. Papillomaviruses marked with asterisks are unclassified.
Papillomaviruses lacking the E7 gene
The oncoprotein E7 is not encoded by all papillomaviruses and studies based on the high-risk HPVs have shown that this oncoprotein associates with cellular pRB through the conserved LxCxE motif [37]. All classified members of the genera Omega- (including LwPV6-7), Omikron- and Upsilonpapillomavirus, as well as Myotis ricketti papillomavirus (MrPV1) and Sus scrofa papillomavirus (SsPV1), lack the E7 ORF [38–42]. The E7 ORF is also absent from all characterized cetacean papillomaviruses [43–46].
The bold branches in the phylogenetic trees presented in Figs 3 and S1 show lineages lacking an E7 gene. As depicted in the partition tree, the bold clades have diverged from a recent common ancestor shared with alpha- and dyoomikronpapillomaviruses. Multiple gene loss events have been reported in the evolution of certain papillomavirus clades. Loss of the E6 gene has occurred on at least two occasions in the evolution of papillomaviruses. Gamma-6 papillomavirus species all lack E6, while it is present in all other gammapapillomaviruses, indicating a loss of E6 in the shared ancestor of Gammapapillomaviruses 6 [47]. Furthermore, loss of E6 has been hypothesized to have occurred twice in the evolution of Xipapillomavirus genera: once in the divergence of bovine papillomavirus 12 (BPV12) and again in the Xipapillomvirus 1 species [47].
Papillomaviruses associated with the order Carnivora
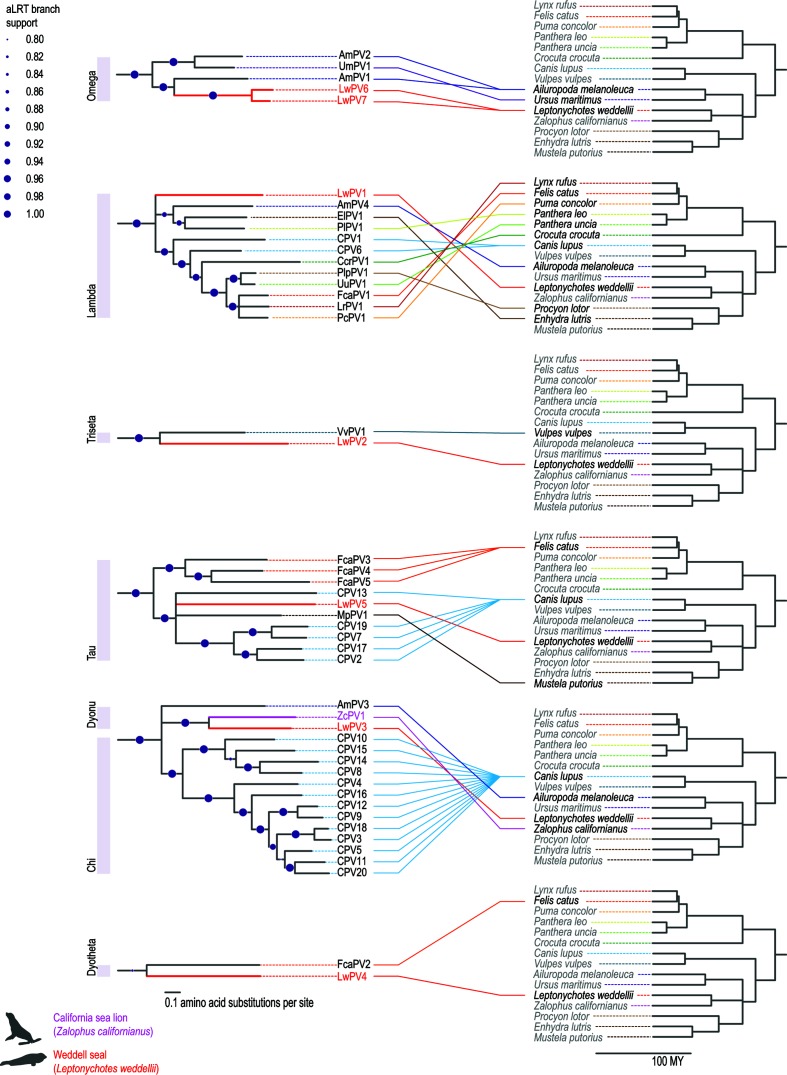
Papillomaviruses in general appear to have co-evolved with their hosts, but recombination, adaptive radiation, host switching and positive selection have been shown to contribute to the evolution of papillomaviruses [36, 48–50]. Pinnipeds belong to the order Carnivora, and their most recent common ancestor with caniforms is approximately 45 mya [28, 51]. Among the Pinnipedia, the Antarctic lobodontines originated and speciated in the late Miocene to early Pliocene, when the relative isolation around Antarctica allowed for rapid diversification of those individuals that could tolerate the relatively cold climate conditions that existed south of the Antarctic Circumpolar Current [28]. To date, 40 papillomaviruses have been identified across 14 distinct carnivore hosts (Figs 4 and S2). [1, 41, 52–54]. With the exception of UmPV1 (polar bear; Omega), VvPV1 (red fox; Treiseta), FcaPV2 (domestic cat; Dyotheta), and ZcPV1 (California sea lion; Dyonu), all Carnivora-associated papillomaviruses belong to the genera Chi-, Lambda- or Taupapillomavirus. ZcPV1, the only pinniped-associated papillomavirus that was known until this study, is most closely related to chipapillomaviruses and belongs to the genus Dyonupapillomavirus [32]. All LwPVs cluster phylogenetically in clades containing papillomaviruses of closely related carnivore hosts (Figs 4 and S2). This is reminiscent of primate and artiodactyl papillomaviruses. Primate papillomaviruses form five defined clades, and within each genus the viruses recapitulate the host evolutionary history [42]. It has previously been proposed that, similar to polyomaviruses [21], papillomavirus diversity can be explained by intra-host duplication followed by episodes of co-evolution [42, 55].
Fig. 4.
Co-evolution of carnivore papillomaviruses from Omega-, Lambda-, Triesta-, Tau-, Dyonu-, Chi- and Dyothetapapillomaviruses. Clades of concatenated E1–E2–L1 maximum-likelihood phylogenetic tree (see Fig. 3). All Carnivora papillomaviruses known to date are present in the six clades and these are linked to their host phylogenies.
Conclusions
The diversity of papillomaviruses in Weddell seals supports the presence of four or five distinct clades, corresponding to four or five ancestral viruses. We hypothesize that the first terrestrial animals were infected with at least four distinct papillomaviruses. As papillomaviruses co-evolved with these hosts, viral niche adaptation allowed for intra-host duplication [56, 57], in turn resulting in radiation and further host–parasite coevolution.
It has become evident that papillomaviruses identified in a single host may originate from multiple evolutionary lineages. This is shown for human papillomaviruses, which are classified into five highly divergent genera, with the majority classified as Alpha-, Beta- and Gammapapillomavirus. Similarly, canine papillomaviruses reveal at least three evolutionary lineages that are classified into distinct genera. This work has revealed that, even in a fairly small sample set, Weddell seals are similarly infected with a diverse set of papillomaviruses that are distinct from those found in other mammals. It is highly likely that these seven papillomaviruses are benign, as no anogenital or oral cancers associated with Weddell seals were identified in this study and nor have they been previously reported.
Metagenomics and high-throughput sequencing have increased our knowledge of Antarctic animal virology exponentially over the last 5 years [9]. Determining the viral diversity in this extreme and isolated habitat is important for monitoring animal health. Furthermore, expanding our understanding of carnivore papillomaviruses in a novel host has offered strong support for a gene loss event in the evolutionary history of papillomaviruses, thus extending our knowledge of the family Papillomaviridae.
Methods
Sampling and sample processing
Across three Antarctic field seasons, vaginal swabs were taken from a total of 81 (2014/2015, n=25 ; 2015/2016, n=29; 2016/2017, n=27) individual adult female Weddell seals (Leptonychotes weddellii). All applicable international, national and institutional guidelines for the care and use of animals were followed during sampling. The vaginal swabs were stored in UTM Viral Transport Media (Copan, USA) at 4 °C for up to 6 months prior to analysis. We filtered 1 ml of the transport media through a 0.2 µm syringe filter for each sample and 200 µl of the filtrate was used for viral DNA extraction using the High Pure Viral Nucleic Acid kit (Roche Diagnostics, USA). Viral circular DNA molecules were subsequently amplified using rolling-circle amplification (RCA) with the TempliPhi kit (GE Healthcare, USA).
High-throughput sequencing and sequence analysis
The RCA products (5 µl from each sample) were pooled for each season. The three pooled RCA products were used to prepare 2×100 bp libraries and these were sequenced on an Illumina HiSeq4000 at Macrogen, Inc. (Republic of Korea). The resulting paired-end reads were de novo assembled using ABySS v2.02 (kmer=64) [58]. blastx [33] analysis of the assembled contigs >2000 nts revealed seven contigs that had similarities to papillomavirus sequences. Abutting primers (Table 1) were designed based on each PV-like de novo assembled viral contig to recover the full genomes from individual samples. Amplification of the papillomavirus-like molecules was carried out using KAPA Hifi Hotstart DNA polymerase (Kapa Biosystems, USA), the abutting primers and the RCA product as template (0.5 µl) with the following polymerase chain reaction (PCR) protocol on an Eppendorf Mastercycler: initial denaturation at 95 °C for 3 min, then 30 cycles at 95 °C for 30 s, 60 °C for 30 s, 72 °C for 8 min and a final extension at 72 °C for 8 min. The amplicons were resolved on a 0.7 % agarose gel, gel-purified and cloned into pJET1.2 plasmid (ThermoFisher, USA). The resulting recombinant plasmids were Sanger sequenced at Macrogen, Inc. (Republic of Korea) by primer walking. The Sanger-sequenced contigs were assembled using DNA Baser v4 (Heracle BioSoft SRL, Romania).
Representative annotated papillomavirus genomes (n=352; Table S1) sequences were downloaded from the PaVE database [59, 60]. From the 352 annotated sequences downloaded from PaVE and the 7 papillomaviruses genomes recovered in this study, the L1 gene, and E1, E2 and L1 protein sequences were extracted. Two datasets were created, one of aligned L1 gene sequences and the other of concatenated aligned E1, E2 and L1 protein sequences. All alignments were carried out using MAFFT [61].
The aligned L1 gene sequences were used to infer a maximum-likelihood phylogenetic tree using PhyML 3.0 [62] with GTR+I+G4 as the best fit model as determined using jModelTest [63]. A maximum-likelihood phylogenetic tree for the concatenated aligned E1, E2 and L1 proteins was inferred using PhyML 3.0 [62] and the LG+F+I+G4 model was determined as best fit model for the E1, E2 and L1 partitions using PartitionFinder 2 [64]. Branches with <0.75 aLRT branch support [65] were collapsed using TreeGraph 2 [66]. The phylogenetic trees were visualized and edited using iTOL v3 [67]. The pairwise identities of the full genomes and L1 gene were determined using SDT v1.2 [68]. The Carnivora host phylogeny was inferred with TimeTree (www.timetree.org/) [69].
Funding information
J. M. B. received funding from National Science Foundation grant ANT-1246463. Z.E.S. is supported through startup funding awarded to A.V. from The Biodesign Institute and School of Life Sciences, Arizona State University, USA. A.K. and R.B. were supported by Institutional Development Awards (IDeA) Networks of Biomedical Research Excellence Assistantships (grant number P20GM103395) from the National Institute of General Medical Sciences of the National Institutes of Health. The content is solely the responsibility of the authors and does not necessarily reflect the official views of the NIH. K.V.D. is supported by a State of Arizona Improving Health TRIF and by the National Institute of Food and Agriculture, US Department of Agriculture, Hatch NC229. The molecular work described in this study is supported by a Center of Evolution and Medicine Venture Fund (Center of Evolution and Medicine, Arizona State University, USA) grant awarded to A.V.
Acknowledgements
All of the samples were collected with logistics supplied from the US Antarctic Program. Weddell seal samples were collected under National Marine Fisheries Service Marine Mammal permit #17411, Antarctic Conservation Act permit #2014–003, and University of Alaska Anchorage’s Institutional Animal Care and Use Committee approval #419971 and #854089. Logistical support for this project in Antarctica was provided by the US National Science Foundation through the US Antarctic Program. J.M.B. contributed to this work while serving at the National Science Foundation. Any opinion, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Data availability: all sequence data reported in this study has been deposited in GenBank (https://www.ncbi.nlm.nih.gov/genbank/) under accession #MG571089 – MG571095.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
Weddell seal samples were collected under National Marine Fisheries Service Marine Mammal permit #17411, Antarctic Conservation Act permit #2014–003 and the University of Alaska Anchorage’s Institutional Animal Care and Use Committee approval #419971 and #854089. The data used for the analyses in this manuscript are publicly available in GenBank.
Supplementary Data
Footnotes
Abbreviations: aLRT, approximate likelihood ratio test; nts, nucleotides; pRB, retinoblastoma-binding protein; PV, papillomavirus PV.
Two supplementary figures and one supplementary table are available in the online version of this article.
References
- 1.Rector A, van Ranst M. Animal papillomaviruses. Virology. 2013;445:213–223. doi: 10.1016/j.virol.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 2.Doorbar J. The papillomavirus life cycle. J Clin Virol. 2005;32:7–15. doi: 10.1016/j.jcv.2004.12.006. [DOI] [PubMed] [Google Scholar]
- 3.Münger K, Scheffner M, Huibregtse JM, Howley PM. Interactions of HPV E6 and E7 oncoproteins with tumour suppressor gene products. Cancer Surv. 1992;12:197–217. [PubMed] [Google Scholar]
- 4.Münger K, Werness BA, Dyson N, Phelps WC, Harlow E, et al. Complex formation of human papillomavirus E7 proteins with the retinoblastoma tumor suppressor gene product. EMBO J. 1989;8:4099–4105. doi: 10.1002/j.1460-2075.1989.tb08594.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yew PR, Berk AJ. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature. 1992;357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- 6.Dyson N, Howley PM, Münger K, Harlow E. The human papilloma virus-16 E7 oncoprotein is able to bind to the retinoblastoma gene product. Science. 1989;243:934–937. doi: 10.1126/science.2537532. [DOI] [PubMed] [Google Scholar]
- 7.López-Bueno A, Mavian C, Labella AM, Castro D, Borrego JJ, et al. Concurrence of iridovirus, polyomavirus, and a unique member of a new group of fish papillomaviruses in lymphocystis disease-affected gilthead sea bream. J Virol. 2016;90:8768–8779. doi: 10.1128/JVI.01369-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van Doorslaer K, Ruoppolo V, Schmidt A, Lescroël A, Jongsomjit D, et al. Unique genome organization of non-mammalian papillomaviruses provides insights into the evolution of viral early proteins. Virus Evol. 2017;3:vex027. doi: 10.1093/ve/vex027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smeele ZE, Ainley DG, Varsani A. Viruses associated with Antarctic wildlife: From serology based detection to identification of genomes using high throughput sequencing. Virus Res. 2018;243:91–105. doi: 10.1016/j.virusres.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee SY, Kim JH, Park YM, Shin OS, Kim H, et al. A novel adenovirus in Chinstrap penguins (Pygoscelis antarctica) in Antarctica. Viruses. 2014;6:2052–2061. doi: 10.3390/v6052052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee SY, Kim JH, Seo TK, No JS, Kim H, et al. Genetic and molecular epidemiological characterization of a novel adenovirus in Antarctic penguins collected between 2008 and 2013. PLoS One. 2016;11:e0157032. doi: 10.1371/journal.pone.0157032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Park YM, Kim JH, Gu SH, Lee SY, Lee MG, et al. Full genome analysis of a novel adenovirus from the South Polar skua (Catharacta maccormicki) in Antarctica. Virology. 2012;422:144–150. doi: 10.1016/j.virol.2011.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fahsbender E, Burns JM, Kim S, Kraberger S, Frankfurter G, et al. Diverse and highly recombinant anelloviruses associated with Weddell seals in Antarctica. Virus Evol. 2017;3:vex017. doi: 10.1093/ve/vex017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurt AC, Su YC, Aban M, Peck H, Lau H, et al. Evidence for the introduction, reassortment, and persistence of diverse influenza a viruses in Antarctica. J Virol. 2016;90:9674–9682. doi: 10.1128/JVI.01404-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hurt AC, Vijaykrishna D, Butler J, Baas C, Maurer-Stroh S, et al. Detection of evolutionarily distinct avian influenza a viruses in Antarctica. MBio. 2014;5:e01098-14. doi: 10.1128/mBio.01098-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Varsani A, Kraberger S, Jennings S, Porzig EL, Julian L, et al. A novel papillomavirus in Adélie penguin (Pygoscelis adeliae) faeces sampled at the Cape Crozier colony, Antarctica. J Gen Virol. 2014;95:1352–1365. doi: 10.1099/vir.0.064436-0. [DOI] [PubMed] [Google Scholar]
- 17.Goraichuk IV, Dimitrov KM, Sharma P, Miller PJ, Swayne DE, et al. Complete genome sequences of four avian paramyxoviruses of serotype 10 isolated from Rockhopper Penguins on the Falkland Islands. Genome Announc. 2017;5:e00472-17. doi: 10.1128/genomeA.00472-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller PJ, Afonso CL, Spackman E, Scott MA, Pedersen JC, et al. Evidence for a new avian paramyxovirus serotype 10 detected in rockhopper penguins from the Falkland Islands. J Virol. 2010;84:11496–11504. doi: 10.1128/JVI.00822-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neira V, Tapia R, Verdugo C, Barriga G, Mor S, et al. Novel avulaviruses in penguins, Antarctica. Emerg Infect Dis. 2017;23:1212–1214. doi: 10.3201/eid2307.170054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomazelli LM, Araujo J, Oliveira DB, Sanfilippo L, Ferreira CS, et al. Newcastle disease virus in penguins from King George Island on the Antarctic region. Vet Microbiol. 2010;146:155–160. doi: 10.1016/j.vetmic.2010.05.006. [DOI] [PubMed] [Google Scholar]
- 21.Buck CB, van Doorslaer K, Peretti A, Geoghegan EM, Tisza MJ, et al. The ancient evolutionary history of polyomaviruses. PLoS Pathog. 2016;12:e1005574. doi: 10.1371/journal.ppat.1005574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Varsani A, Frankfurter G, Stainton D, Male MF, Kraberger S, et al. Identification of a polyomavirus in Weddell seal (Leptonychotes weddellii) from the Ross Sea (Antarctica) Arch Virol. 2017;162:1403–1407. doi: 10.1007/s00705-017-3239-y. [DOI] [PubMed] [Google Scholar]
- 23.Varsani A, Porzig EL, Jennings S, Kraberger S, Farkas K, et al. Identification of an avian polyomavirus associated with Adélie penguins (Pygoscelis adeliae) J Gen Virol. 2015;96:851–857. doi: 10.1099/vir.0.000038. [DOI] [PubMed] [Google Scholar]
- 24.Tryland M, Klein J, Nordøy ES, Blix AS. Isolation and partial characterization of a parapoxvirus isolated from a skin lesion of a Weddell seal. Virus Res. 2005;108:83–87. doi: 10.1016/j.virusres.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 25.Forrester NL, Palacios G, Tesh RB, Savji N, Guzman H, et al. Genome-scale phylogeny of the alphavirus genus suggests a marine origin. J Virol. 2012;86:2729–2738. doi: 10.1128/JVI.05591-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.La Linn M, Gardner J, Warrilow D, Darnell GA, McMahon CR, et al. Arbovirus of marine mammals: a new alphavirus isolated from the elephant seal louse, Lepidophthirus macrorhini. J Virol. 2001;75:4103–4109. doi: 10.1128/JVI.75.9.4103-4109.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bininda-Emonds OR, Gittleman JL, Purvis A. Building large trees by combining phylogenetic information: a complete phylogeny of the extant Carnivora (Mammalia) Biol Rev Camb Philos Soc. 1999;74:143–175. doi: 10.1017/S0006323199005307. [DOI] [PubMed] [Google Scholar]
- 28.Higdon JW, Bininda-Emonds OR, Beck RM, Ferguson SH. Phylogeny and divergence of the pinnipeds (Carnivora: Mammalia) assessed using a multigene dataset. BMC Evol Biol. 2007;7:216. doi: 10.1186/1471-2148-7-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burns JM, Trumble SJ, Castellini MA, Testa JW. The diet of Weddell seals in McMurdo Sound, Antarctica as determined from scat collections and stable isotope analysis. Polar Biol. 1998;19:272–282. doi: 10.1007/s003000050245. [DOI] [Google Scholar]
- 30.Stirling I. Ecology of the Weddell Seal in McMurdo Sound, Antarctica. Ecology. 1969;50:573–586. doi: 10.2307/1936247. [DOI] [Google Scholar]
- 31.Castellini MA, Davis RW, Kooyman GL. Annual Cycles of Diving Behavior and Ecology of the Weddell Seal. Univ of California Press; 1991. [Google Scholar]
- 32.Rivera R, Robles-Sikisaka R, Hoffman EM, Stacy BA, Jensen ED, et al. Characterization of a novel papillomavirus species (ZcPV1) from two California sea lions (Zalophus californianus) Vet Microbiol. 2012;155:257–266. doi: 10.1016/j.vetmic.2011.09.027. [DOI] [PubMed] [Google Scholar]
- 33.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 34.Wang J, Zhou D, Prabhu A, Schlegel R, Yuan H. The canine papillomavirus and gamma HPV E7 proteins use an alternative domain to bind and destabilize the retinoblastoma protein. PLoS Pathog. 2010;6:e1001089. doi: 10.1371/journal.ppat.1001089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Villiers EM, Fauquet C, Broker TR, Bernard HU, Zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 36.Gottschling M, Stamatakis A, Nindl I, Stockfleth E, Alonso A, et al. Multiple evolutionary mechanisms drive papillomavirus diversification. Mol Biol Evol. 2007;24:1242–1258. doi: 10.1093/molbev/msm039. [DOI] [PubMed] [Google Scholar]
- 37.Roman A, Munger K. The papillomavirus E7 proteins. Virology. 2013;445:138–168. doi: 10.1016/j.virol.2013.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevens H, Rector A, Bertelsen MF, Leifsson PS, van Ranst M. Novel papillomavirus isolated from the oral mucosa of a polar bear does not cluster with other papillomaviruses of carnivores. Vet Microbiol. 2008;129:108–116. doi: 10.1016/j.vetmic.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 39.Stevens H, Rector A, van der Kroght K, van Ranst M. Isolation and cloning of two variant papillomaviruses from domestic pigs: Sus scrofa papillomaviruses type 1 variants a and b. J Gen Virol. 2008;89:2475–2481. doi: 10.1099/vir.0.2008/003186-0. [DOI] [PubMed] [Google Scholar]
- 40.Wu Z, Ren X, Yang L, Hu Y, Yang J, et al. Virome analysis for identification of novel mammalian viruses in bat species from Chinese provinces. J Virol. 2012;86:10999–11012. doi: 10.1128/JVI.01394-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang W, Yang S, Shan T, Hou R, Liu Z, et al. Virome comparisons in wild-diseased and healthy captive giant pandas. Microbiome. 2017;5:90. doi: 10.1186/s40168-017-0308-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Doorslaer K. Evolution of the papillomaviridae. Virology. 2013;445:11–20. doi: 10.1016/j.virol.2013.05.012. [DOI] [PubMed] [Google Scholar]
- 43.Rector A, Stevens H, Lacave G, Lemey P, Mostmans S, et al. Genomic characterization of novel dolphin papillomaviruses provides indications for recombination within the Papillomaviridae. Virology. 2008;378:151–161. doi: 10.1016/j.virol.2008.05.020. [DOI] [PubMed] [Google Scholar]
- 44.Rehtanz M, Ghim SJ, Rector A, van Ranst M, Fair PA, et al. Isolation and characterization of the first American bottlenose dolphin papillomavirus: Tursiops truncatus papillomavirus type 2. J Gen Virol. 2006;87:3559–3565. doi: 10.1099/vir.0.82388-0. [DOI] [PubMed] [Google Scholar]
- 45.Robles-Sikisaka R, Rivera R, Nollens HH, St Leger J, Durden WN, et al. Evidence of recombination and positive selection in cetacean papillomaviruses. Virology. 2012;427:189–197. doi: 10.1016/j.virol.2012.01.039. [DOI] [PubMed] [Google Scholar]
- 46.Gottschling M, Bravo IG, Schulz E, Bracho MA, Deaville R, et al. Modular organizations of novel cetacean papillomaviruses. Mol Phylogenet Evol. 2011;59:34–42. doi: 10.1016/j.ympev.2010.12.013. [DOI] [PubMed] [Google Scholar]
- 47.van Doorslaer K, McBride AA. Molecular archeological evidence in support of the repeated loss of a papillomavirus gene. Sci Rep. 2016;6:33028. doi: 10.1038/srep33028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.García-Vallvé S, Alonso A, Bravo IG. Papillomaviruses: different genes have different histories. Trends Microbiol. 2005;13:514–521. doi: 10.1016/j.tim.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 49.Varsani A, van der Walt E, Heath L, Rybicki EP, Williamson AL, et al. Evidence of ancient papillomavirus recombination. J Gen Virol. 2006;87:2527–2531. doi: 10.1099/vir.0.81917-0. [DOI] [PubMed] [Google Scholar]
- 50.Burk RD, Harari A, Chen Z. Human papillomavirus genome variants. Virology. 2013;445:232–243. doi: 10.1016/j.virol.2013.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McKenna MC, Bell SK. Classification of Mammals: Above the Species Level. Columbia University Press; 1997. [Google Scholar]
- 52.Mengual-Chuliá B, Wittstatt U, Bravo IG. The first papillomavirus isolated from Vulpes vulpes (VvulPV1) is basal to the Gammapapillomavirus genus. Genome Announc. 2015;3:e00111-15. doi: 10.1128/genomeA.00111-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ng TF, Wheeler E, Greig D, Waltzek TB, Gulland F, et al. Metagenomic identification of a novel anellovirus in Pacific harbor seal (Phoca vitulina richardsii) lung samples and its detection in samples from multiple years. J Gen Virol. 2011;92:1318–1323. doi: 10.1099/vir.0.029678-0. [DOI] [PubMed] [Google Scholar]
- 54.Smits SL, Raj VS, Oduber MD, Schapendonk CM, Bodewes R, et al. Metagenomic analysis of the ferret fecal viral flora. PLoS One. 2013;8:e71595. doi: 10.1371/journal.pone.0071595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bravo IG, Alonso A. Phylogeny and evolution of papillomaviruses based on the E1 and E2 proteins. Virus Genes. 2007;34:249–262. doi: 10.1007/s11262-006-0017-4. [DOI] [PubMed] [Google Scholar]
- 56.Warren CJ, van Doorslaer K, Pandey A, Espinosa JM, Pyeon D. Role of the host restriction factor APOBEC3 on papillomavirus evolution. Virus Evol. 2015;1:vev015. doi: 10.1093/ve/vev015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Doorslaer K, Desalle R, Einstein MH, Burk RD. Degradation of human PDZ-proteins by human alphapapillomaviruses represents an evolutionary adaptation to a novel cellular niche. PLoS Pathog. 2015;11:e1004980. doi: 10.1371/journal.ppat.1004980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, et al. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Doorslaer K, Li Z, Xirasagar S, Maes P, Kaminsky D, et al. The Papillomavirus Episteme: a major update to the papillomavirus sequence database. Nucleic Acids Res. 2017;45:D499–D506. doi: 10.1093/nar/gkw879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.van Doorslaer K, Tan Q, Xirasagar S, Bandaru S, Gopalan V, et al. The Papillomavirus Episteme: a central resource for papillomavirus sequence data and analysis. Nucleic Acids Res. 2013;41:D571–D578. doi: 10.1093/nar/gks984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 63.Darriba D, Taboada GL, Doallo R, Posada D. jModelTest 2: more models, new heuristics and parallel computing. Nat Methods. 2012;9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B. PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol. 2017;34:772–773. doi: 10.1093/molbev/msw260. [DOI] [PubMed] [Google Scholar]
- 65.Anisimova M, Gascuel O. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol. 2006;55:539–552. doi: 10.1080/10635150600755453. [DOI] [PubMed] [Google Scholar]
- 66.Stöver BC, Müller KF. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics. 2010;11:7. doi: 10.1186/1471-2105-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Muhire BM, Varsani A, Martin DP. SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One. 2014;9:e108277. doi: 10.1371/journal.pone.0108277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumar S, Stecher G, Suleski M, Hedges SB. TimeTree: a resource for timelines, timetrees, and divergence times. Mol Biol Evol. 2017;34:1812–1819. doi: 10.1093/molbev/msx116. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.