Abstract
Purpose
Membrane fluidity to a large extent is governed by the presence of branched-chain fatty acids (BCFAs). Branched-chain α-keto acid dehydrogenase (BKD) is the key enzyme in BCFA synthesis. A Staphylococcus aureus BKD-deficient strain still produced substantial levels of BCFAs. Pyruvate dehydrogenase (PDH) with structural similarity to BKD has been speculated to contribute to BCFAs in S. aureus.
Methodology
This study was carried out using BKD-, PDH- and BKD : PDH-deficient derivatives of methicillin-resistant S. aureus strain JE2. Differences in growth kinetics were evaluated spectrophotometrically, membrane BCFAs using gas chromatography and membrane fluidity by fluorescence polarization. Carotenoid levels were estimated by measuring A465 of methanol extracts from 48 h cultures. MIC values were determined by broth microdilution.
Results/Key findings
BCFAs made up 50 % of membrane fatty acids in wild-type but only 31 % in the BKD-deficient mutant. BCFA level was ~80 % in the PDH-deficient strain and 38 % in the BKD : PDH-deficient strain. BKD-deficient mutant showed decreased membrane fluidity, the PDH-deficient mutant showed increased membrane fluidity. The BKD- and PDH-deficient strains grew slower and the BKD : PDH-deficient strain grew slowest at 37 °C. However at 20 °C, the BKD- and BKD : PDH-deficient strains grew only a little followed by autolysis of these cells. The BKD-deficient strain produced higher levels of staphyloxanthin. The PDH-deficient and BKD : PDH-deficient strains produced very little staphyloxanthin. The BKD-deficient strain showed increased susceptibility to daptomycin.
Conclusion
The BCFA composition of the cell membrane in S. aureus seems to significantly impact cell growth, membrane fluidity and resistance to daptomycin.
Keywords: Staphylococcus aureus, BCFA, BKD, PDH
Introduction
Staphylococcus aureus is a significant human pathogen with the ability to cause minor to severe life-threatening illnesses [1–5]. Clinical management of these infections is complicated due to widespread emergence of methicillin-resistant S. aureus (MRSA) strains. Some of these MRSA strains are now resistant to more than 20 different antimicrobial agents [6–9].
S. aureus encounters a variety of environmental conditions to which it must adapt in order to survive, including changing temperature [10, 11], fluctuations in pH [12], osmotic pressure [13, 14], nutrient availability and antibiotic stress [13, 15]. Specifically, membrane fluidity is a critical determinant in its adaptation [16–18]. S. aureus, like other species, maintains membrane fluidity by altering its fatty acid composition [17]. Branched-chain fatty acids (BCFAs) occupy significantly larger cross-sectional areas than saturated straight-chain fatty acids (SCFAs), disrupt the close packing of membrane fatty acids and allow the membranes to remain fluid at lower temperatures [19–21].
Recent studies have revealed that the membrane lipid composition in S. aureus is remarkably plastic, and this is largely determined by the nutritional environment [22]. When grown in artificial media, S. aureus fatty acids are a mixture of BCFAs and SCFAs [22, 23]. The percentage of BCFAs typically exceeds that of SCFAs in standard media such as brain–heart infusion and tryptic soy broth, and the proportion of BCFAs is very high in cells grown in Mueller–Hinton broth and defined medium [22, 24]. However, when grown in serum [22] or fed individual fatty acids, S. aureus can incorporate significant quantities of SCFAs and straight-chain unsaturated fatty acids (SCUFAs) [25, 26]. Additionally, the triterpenoid carotenoid ‘staphyloxanthin’ is a characteristic of S. aureus cells and its level fluctuates widely in different strains of this bacterium and under different growth conditions [22, 24].
Given the importance of BCFAs in membrane structure and function, a BCFA-deficient mutant was created by insertional inactivation of the lpdA gene of S. aureus strain SH1000, the first gene of the cluster that codes for the four polypeptides of the branched-chain α-keto acid dehydrogenase complex (BKD) [23]. BCFAs were reduced to 35.4 % from 63.5 % in the mutant compared to the wild-type. The mutant had a less fluid membrane, was more susceptible to cold, alkaline and oxidative stress and diminished survival in mice [23]. BCFAs are biosynthesized from the branched-chain amino acids isoleucine, leucine and valine via branched-chain amino acid transaminase and BKD. The biosynthetic route to the substantial percentage of BCFAs present in the BCFA-deficient S. aureus mutant is not clear.
BKD is a multi-subunit complex composed of four polypeptides: a dehydrogenase (E1α), a decarboxylase (E1β), a dihydrolipoamide acyltransferase (E2) and a dihydrolipoamide dehydrogenase (E3) [27, 28]. It is anticipated that the residual levels of BCFAs [23] in the BKD-deficient S. aureus are produced by another enzyme complex sharing structural and/or functional homology to BKD. In this context, pyruvate dehydrogenase (PDH) is also a multi-enzyme complex similar to BKD. In S. aureus, the chromosomal locus possesses four genes that encode the PDH polypeptides: pyruvate dehydrogenase E1 component (alpha subunit), pyruvate dehydrogenase E1 component (beta subunit), branched-chain alpha-keto acid dehydrogenase (E2 subunit) and dihydrolipoamide dehydrogenase (LpdA). The PDH enzyme complex indeed shares significant sequence homology with the BKD complex [29, 30], and was predicted to be a prime candidate in contributing to BCFA metabolism [30–32], as activity of PDH with some branched-chain α-keto acids has been demonstrated [33, 34].
To investigate the roles of these complexes in BCFA production, BKD- and PDH-deficient S. aureus strains were studied. Surprisingly, lack of PDH activity led to a very significant increase in BCFAs, opposite to the effect that was seen in BKD-deficient S. aureus. These changes in BCFAs also impacted physiological functions in S. aureus.
Methods
Bacterial strains, plasmids and growth conditions
The bacterial strains and plasmids used in this study are shown in Table 1. S. aureus cultures were grown in tryptic soy broth or agar (TSB or TSA; Becton Dickinson). When needed, erythromycin (10 µg ml−1), kanamycin (100 µg ml−1) and chloramphenicol (10 µg ml−1) were added to the growth medium. Mueller–Hinton broth (MHB, Becton Dickinson) was used for measuring minimum inhibitory concentrations (MICs) of antibiotics. TSB was used to culture S. aureus for the analysis of fatty acid composition. Cloning and plasmid DNA preparations were made by culturing appropriate Escherichia coli cells in Luria–Bertani broth or agar in the presence of ampicillin at 100 µg ml−1.
Table 1. Bacterial strains and plasmids used in this study.
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| Bacteria | ||
| S. aureus JE2 | Most common CA-MRSA isolate, resistant to methicillin/oxacillin | [57] |
| lpdA | JE2 with mutation in the lpdA (Kanr) | This study |
| lpdA+pCU-bkd | JE2 : lpdA complemented with pCU-bkd plasmid (Kanr, Camr) | This study |
| NE1724 | JE2 with a transposon insertion in the pdhA gene (Ermr) | [35] |
| pdhA+pCU-pdh | JE2 : pdhA complemented with pCU-pdh plasmid (Ermr, Camr) | This study |
| lpdA : pdhA | JE2 with mutation in the lpdA and pdhA genes (Kanr, Ermr) | This study |
| lpdA-pdhA+pCU-bkd | JE2Δ(pdhA+lpdA)complemented with pCU-bkd (Kanr, Ermr, Camr) | This study |
| lpdA-pdhA+pCU-pdh | JE2Δ(pdhA+lpdA)complemented with pCU-pdh (Kanr, Ermr, Camr) | This study |
| Plasmids | ||
| pGEM-T | An E. coli cloning plasmid | Promega |
| pCU1 | A shuttle vector (Ampr in E. coli, Camr in S. aureus) | [36] |
| pCU-bkd | Plasmid pCU1 containing a 5.1-kb DNA fragment containing all four genes of the S. aureus bkd locus | [23] |
| pCU-pdh | Plasmid pCU1 containing a 5.9-kb DNA fragment containing all four genes of the S. aureus pdh locus | This study |
Kanr, kanamycin resistant; Ermr, erythromycin resistant; Camr, chloramphenicol resistant.
Construction of lpdA (BKD-deficient) and pdhA (PDH-deficient) mutants
The previously reported mutation in the lpdA gene of strain SH1000 [23] was transduced in S. aureus strain JE2 using a phage transduction procedure as described previously [23]. A PDH-deficient derivative of S. aureus strain JE2 (NE1724 of the Nebraska Transposon mutant library) [35] was obtained from the Network on Antimicrobial Resistance in S. aureus (NARSA). NE1724 contains a transposon insertion in the gene coding for the pyruvate dehydrogenase E1 alpha subunit (pdhA). An lpdA : pdhA (BKD : PDH-deficient) double mutant was generated by transducing the lpdA mutation in to NE1724.
Creation of complemented stains
For complementation with the pdh locus, the entire locus starting 586 nt upstream of the first gene (pdhA) and terminating 410 nt after the fourth gene (pdhD) was PCR amplified (a 5886 bp amplicon) using appropriate primers (forward primer; 5′-GGTACCAGTTCGCGGTAAAGCAGTAAAGG-3′ and backward primer; 5′-TCTAGAGTGAGTTTTTACATCTTGAACTGC-3′) and S. aureus genomic DNA as the template. PCR was carried out using Stratagene EXL DNA polymerase (Stratagene, CA) as per the manufacturer’s instructions. This amplicon was cloned in vector pGEM-T (Promega), from where it was subcloned in vector pCU1 [36] at KpnI and XbaI sites. The pdhA mutant of S. aureus strain JE2 was subsequently transformed with this construct. For complementation of the lpdA mutant, the entire bkd locus comprising all four BKD encoding peptides on pCU1 described previously [23] was transduced into the lpdA mutant.
Analysis of fatty acid composition in the wild-type JE2 and various S. aureus mutants
To determine the membrane fatty acid composition, cultures of various mutants, complemented strains and wild-type S. aureus strains were grown in 50 ml TSB in a 250 ml flask at 37 °C to an OD600=0.6. The bacterial cells were then harvested and processed for the identification of fatty acids by the MIDI microbial identification system (Sherlock 4.5 microbial identification system) at Microbial ID, Inc. (Newark, DE, USA) as described previously [23].
Growth kinetics of wild-type S. aureus strain JE2 and its derivative mutants
Mid-exponential phase cultures (OD600=0.6) were diluted 10-fold in an Erlenmeyer flask containing 50 ml fresh TSB, with a flask-to-medium volume ratio of 6 : 1. Bacterial growth was subsequently monitored by incubating the flask in a shaking incubator (220 r.p.m.) and measuring the turbidity (OD600) of the liquid culture periodically using a Thermo Scientific Biomate 3 spectrophotometer.
Estimation of carotenoid pigments in wild-type S. aureus strain JE2 and its derivative mutants and complemented strains
The carotenoid production of wild-type and S. aureus mutant strains was determined as previously described [37]. The overnight cultures were diluted 1 : 100 in 50 ml TSB and incubated at 37 °C with shaking (220 r.p.m.) for 48 h. Cells from 5 ml of each culture were collected by centrifugation. The cells were washed three times with sterile, ice-cold distilled water and the cell pellet wet weight was determined. Cells were resuspended in 450 µl methanol and incubated at 55 °C for 3 min with intermittent vortexing. The cells were subsequently pelleted by centrifugation, and A465 of the carotenoid-containing methanol extract was measured.
Determination of membrane fluidity of wild-type S. aureus strain JE2 and its derivative mutants and complemented strains
Membrane fluidity was determined using fluorescence polarization as previously described [38], with some modifications. For these studies, overnight cultures were diluted 1 : 100 in a flask containing 50 ml fresh TSB and were grown to OD600=0.6 at 37 °C in a shaking incubator (220 r.p.m.). Bacterial cells were collected by centrifugation and washed twice with PBS. The bacterial cells were then resuspended in PBS to an OD600 to approximately 1.0. Subsequently, 2.0 ml of PBS containing 3 µM 1,6-diphenyl-1,3,5-hexatriene (DPH, Sigma-Aldrich, St. Louis, MO) was added to 1.0 ml of the bacterial suspension. DPH was initially dissolved in tetrahydofuran (THF) and added to PBS at the time of use. To the control blank, 2.0 ml PBS with an equivalent amount of THF was added to 1.0 ml of bacterial cell suspension. Both test and blank samples were incubated at 37 °C for 1 h. Fluorescence polarization was then measured in quartz cuvettes using a PerkinElmer LS55 Spectrofluorometer with excitation and emission wavelengths of 360 and 426 nm, respectively. Vertically polarized 360 nm light excites the DPH dye, which emits fluorescent light at 426 nm and is detected through polarized light filters. Polarization was calculated as described [12, 38, 39].
Determination of antimicrobial susceptibility
The antibiotic MICs for the wild-type and different mutant strains of S. aureus were determined as previously described [40, 41].
qPCR analysis of lpdA transcript levels in PDH-deficient and crtM in BKD-deficient S. aureus strain JE2
qRT-PCR assays were used to investigate whether the gene encoding the LpdA polypeptide in the pdhA mutant and the gene encoding the CrtM polypeptide in the lpdA mutant were overexpressed. Cultures of S. aureus wild-type strain JE2 and derivative mutants were grown to OD600=0.6. Total RNA was extracted from these cells as described previously [42]. cDNA from DNase-treated 0.5 µg of total RNA was synthesized in a 20 µl reverse transcription reaction containing random hexamers and SuperScript III reverse transcriptase (Invitrogen). All real-time PCR reactions were carried out with a Bio-Rad iCycler (iQ5 system). The transcript levels were quantified using specific lpdA (forward primer; 5′-CGTTCTCGGTG GAGGTAC-3′ and backward primer; 5′-GATTCACCA AGCCTTCTTGC-3′) and crtM primer pairs (forward primer; 5′-TTGAAACGGACGCTGAATTA-3′ and backward primer; 5′-AGCAGCGATTTAGTAGGAATAC-3′). Expression levels of lpdA and crtM were normalized to the expression of the gene encoding DNA gyrase using gyrA-specific primer pairs (forward primer; 5′-TCCACAAGTCGCACGTACAG-3′ and backward primer; 5′-GGAAGGCTTGCTACATCTAACG-3′) based on a previous report [43, 44]. Changes in gene expression were calculated using the formula 2-∆∆Cq as previously described [45].
Statistical analysis
Results are reported as the mean±sem of at least three independent experiments unless stated otherwise. Two sample t-tests were carried out to compare the membrane fluidity and staphyloxanthin levels in the mutant to the wild-type S. aureus. Data were analysed utilizing SAS version 9.4 (SAS Institute Inc., Cary, NC). Statistical significance was set at P≤0.05.
Results
Fatty acid compositions of S. aureus JE2 and its derivative mutants
The composition of major fatty acids present in the cytoplasmic membrane of wild-type JE2 and its various derivative strains is shown (Table 2). As expected, the total BCFA content in the lpdA mutant was significantly reduced (30.79 %) compared to the BCFA content of the wild-type (50.09 %) (Table 2). The lpdA mutant showed low levels of ante-iso fatty acids (11.56 %) compared to the wild-type JE2 (30.86 %), thus leading to a low ante-iso:iso ratio in the mutant (0.60) relative to the wild-type (1.60). Furthermore, the amounts of odd-iso and even-iso fatty acids were also found to be altered in the lpdA mutant. The wild-type JE2 had a higher odd-iso fatty acid content whereas the lpdA mutant showed higher even-iso fatty acids (Table 2). On the other hand, a mutation in pdhA leading to the inactivation of pyruvate dehydrogenase caused a drastic decrease in the SCFA content (only 19.10 %) and a drastic increase in BCFAs (79.65 %) (Table 2). However, in the pdhA knockout mutant, the ante-iso:iso ratio did not change like the lpdA mutant and was similar to the wild-type JE2, because the ante-iso and iso fatty acid contents both increased by proportional amounts (Table 2). The lpdA : pdhA double mutant had slightly higher amounts of BCFAs in comparison to the lpdA mutant. This double mutant showed an increase in both total ante-iso and total iso fatty acids, leading to an ante-iso:iso ratio similar to the lpdA mutant (Table 2).
Table 2. Fatty acid profiles of wild-type JE2 and isogenic lpdA, pdhA and lpdA-pdhA mutants and complemented strains.
|
Fatty acid |
% (wt/wt) of total fatty acids | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Wild type | lpdA | lpdA with | pdhA | pdhA with | lpdA : pdhA | ΔlpdA : pdhA with | ||||
| pCU-bkd | pCU-pdh | pCU-bkd | pCU-pdh | pCU-bkd | pCU-pdh | |||||
| Ante-iso C15 : 0 | 28.16 | 11.56 | 26.72 | 24.19 | 37.09 | 37.41 | 29.23 | 10.84 | 34.50 | 23.29 |
| Ante-iso C17 : 0 | 2.70 | nd | 2.88 | 2.99 | 10.00 | 11.16 | 2.86 | nd | 7.88 | 1.35 |
| Ante-iso C19 : 0 | nd | nd | nd | nd | 2.30 | 2.69 | nd | 3.10 | 1.88 | nd |
| Ante-iso | 30.86 | 11.56 | 29.60 | 27.18 | 49.39 | 51.26 | 32.09 | 13.94 | 44.26 | 24.64 |
| Straight even | 45.34 | 63.98 | 49.80 | 47.50 | 19.10 | 20.55 | 47.83 | 58.87 | 22.51 | 56.52 |
| Iso odd | 14.76 | 3.08 | 6.91 | 5.72 | 26.26 | 24.11 | 12.11 | nd | 17.50 | 11.25 |
| Iso even | 4.47 | 16.15 | 13.69 | 19.60 | 4.00 | 2.64 | 5.28 | 23.61 | 13.99 | 5.27 |
| Iso | 19.23 | 19.23 | 20.60 | 25.32 | 30.26 | 26.75 | 17.39 | 23.61 | 31.49 | 16.52 |
| Ante-iso : iso ratio | 1.60 | 0.60 | 1.44 | 1.07 | 1.63 | 1.92 | 1.85 | 0.59 | 1.41 | 1.49 |
| BCFA | 50.09 | 30.79 | 50.20 | 52.50 | 79.65 | 78.01 | 49.48 | 37.55 | 75.75 | 41.16 |
| SCFA | 46.89 | 63.98 | 49.80 | 47.50 | 19.10 | 20.55 | 49.08 | 58.87 | 22.51 | 57.55 |
nd, none detected.
Values indicate the average of two independent experiments.
In these studies, when the lpdA mutant of JE2 was complemented with the bkd locus in trans, it resulted in restoration of the BCFAs and other membrane fatty acids in the lpdA mutant (Table 2). The ante-iso and iso fatty acids in the BKD-complemented lpdA mutant increased to equal the wild-type JE2 levels. The iso fatty acids also increased to the wild-type amounts, but the odd-iso and even-iso pattern did not revert to the wild-type JE2 levels in the lpdA-complemented strain (Table 2). Complementation of the lpdA mutant with the pdh locus had effects similar to the complementation of this mutant with the bkd locus (Table 2). Also, when the pdhA mutant was complemented with the pdh locus, it resulted in the restoration of the membrane fatty acid composition similar to the profile of the wild-type JE2 (Table 2). In contrast, complementation of the pdhA mutant with the bkd locus had little to no impact and caused no change in the membrane fatty acid composition in the complemented strain (Table 2). Complementation with the bkd locus of the lpdA : pdhA double mutant resulted in a profile similar to the pdhA mutant in terms of a very high BCFA content (Table 2). Complementation of the lpdA : pdhA double mutant with the pdh locus increased the BCFA content relative to the lpdA and lpdA : pdhA double mutant, but it remained lower than the BCFA levels in wild-type JE2 (Table 2).
Growth characteristics of JE2 and isogenic mutants
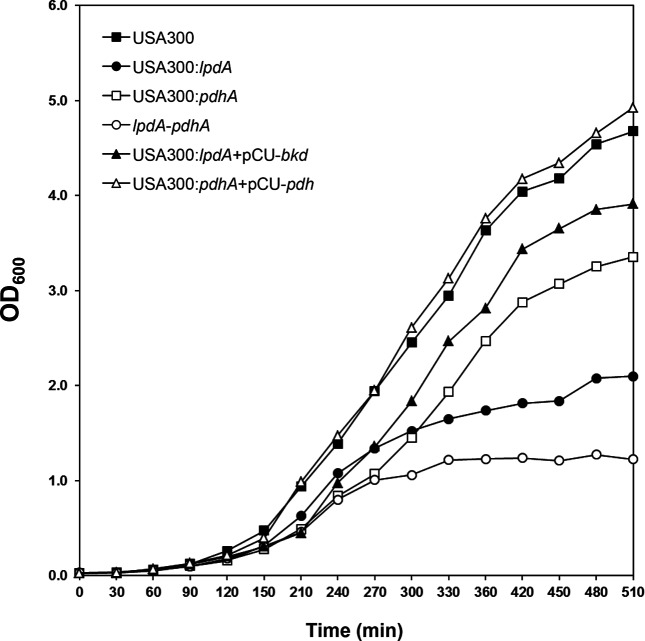
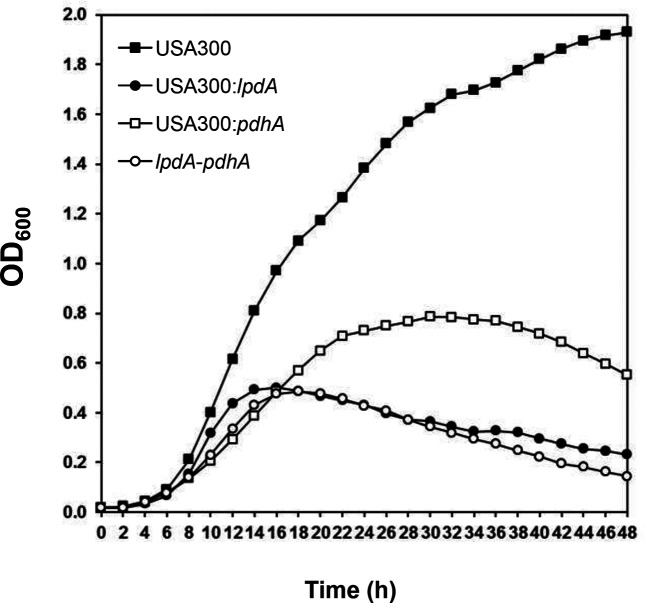
In growth kinetic studies, growth of the lpdA and pdhA mutants was slower compared to the wild-type JE2 (Fig. 1). Growth of the lpdA mutant was slower compared even to the pdhA mutant (Fig. 1). Growth of the lpdA : pdhA double mutant was slowest at 37 °C (Fig. 1). Complementation of the lpdA mutant with bkd locus and pdhA mutant with pdh locus corrected the growth defect observed in these individual mutants (Fig. 1). When these mutants were cultured at 20 °C, the pdhA mutant showed some growth but the lpdA and lpdA : pdhA double mutants failed to grow (Fig. 2).
Fig. 1.
Growth curve of the wild-type S. aureus strain JE2 and its derivative mutants in TSB at 37 °C. Values indicate the average of two independent experiments.
Fig. 2.
Growth curve of the wild-type S. aureus strain JE2 and its derivative mutants in TSB at 20 °C. Values indicate the average of two independent experiments.
Cytoplasmic membrane fluidity of JE2 and isogenic mutants as determined by fluorescence polarization
Polarization values that are inversely correlated with the fluidity of the cytoplasmic membrane showed a nice correlation with the level of BCFAs in different membranes. The pdhA mutant that showed highest levels of BCFAs and the lpdA mutant with the lowest levels of BCFAs showed the lowest and highest polarization values, respectively, and these polarization values were significantly different to the polarization values for the wild-type JE2 (P≤0.005) (Table 3). The lpdA : pdhA double mutant with BCFAs similar to the wild-type JE2 showed polarization values comparable to that of the wild-type. The complemented strains, lpdA mutant with the bkd locus and the pdhA mutant with the pdh locus, exhibited polarization values that were similar to the wild-type levels (Table 3).
Table 3. Fluidity of S. aureus strains as measured by their relative polarization.
| Strain | Polarization value |
|---|---|
| Wild-type JE2 | 0.320±0.007 |
| lpdA | 0.374±0.016* |
| pdhA | 0.274±0.001* |
| lpdA : pdhA | 0.337±0.016 |
| lpdA+pCU-BKD | 0.317±0.011 |
| pdhA+pCU-PDH | 0.322±0.009 |
*P value ≤0.005.
Staphyloxanthin (carotenoid) content of JE2 and isogenic mutants
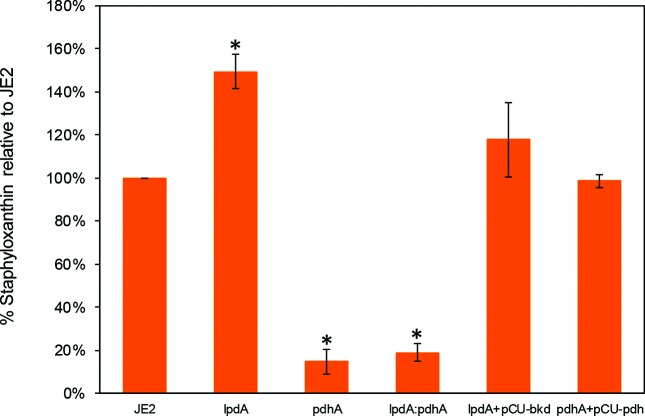
It was frequently observed that the lpdA mutant colonies were slightly darker (yellow) and the pdhA and the lpdA : pdhA double mutants were white on TSA plates. In view of these observations, the carotenoid levels in these strains were quantified. Upon quantification, the level of carotenoids showed a significant increase in the case of the lpdA mutant (Fig. 3). The levels of carotenoids were very low in the case of the pdhA mutant and the lpdA : pdhA double mutant (only 14.6 and 18.8 % compared to the wild-type JE2) (Fig. 3). When carotenoids were measured in the complemented strains, the lpdA mutant with the bkd locus and the pdhA mutant with the pdh locus showed carotenoid levels similar to the wild-type levels (Fig. 3).
Fig. 3.
Production of staphyloxanthin in the wild-type S. aureus strain JE2 and its derivative mutants. Values indicate the average of three independent experiments±standard error. 1 – Wild-type JE2; 2 – lpdA; 3 – pdhA; 4 – lpdA : pdhA; 5 – lpdA+pCU bkd; 6 – pdhA+pCU pdh.
Antibiotic susceptibility of JE2 and isogenic mutants
In these studies, no difference was observed in the susceptibility of the lpdA, pdhA or lpdA : pdhA double mutants to oxacillin and bacitracin compared to wild-type JE2 (Table 4). A twofold reduction in MIC for vancomycin and d-cycloserine was observed only in the lpdA : pdhA double mutant. The lpdA mutant showed a twofold and the lpdA : pdhA double mutant showed a fourfold reduction in daptomycin MIC (Table 4).
Table 4. Susceptibilities of S. aureus parental strain JE2 and its isogenic mutants to cell wall- and membrane-active antibiotics.
MIC values (µg ml−1) indicate average of two independent experiments.
| Strain | Antibiotic | ||||
|---|---|---|---|---|---|
| Vancomycin | Oxacillin | Bacitracin | d-cycloserine | Daptomycin | |
| Wild-type JE2 | 6.25 | 200 | 125 | 125 | 5 |
| lpdA | 6.25 | 200 | 125 | 125 | 2.5 |
| pdhA | 6.25 | 200 | 125 | 125 | 2.5 |
| lpdA : pdhA | 3.125 | 200 | 125 | 62.5 | 1.25 |
Expression of bkd and crt locus genes in pdhA and lpdA mutants of S. aureus strain JE2
Even though there was a significant increase in the amount of staphyloxanthin produced in the lpdA mutant, in qPCR assays there was only a slight increase (1.38-fold) in expression of the the crtM gene (a gene of the staphyloxanthin pathway) in the lpdA mutant compared to its expression in the wild-type JE2 (Table 5). Similarly, a dramatic increase in the level of BCFAs was noted in the pdhA mutant, but in qPCR assays there was only a slight increase (1.25-fold) in expression of the lpdA gene (a gene of the BKD locus) in the pdhA mutant compared to its expression in the wild-type JE2 (Table 5).
Table 5. Expression of lpdA and crtM genes in mutants of S. aureus strain JE2.
|
Strain |
Fold increase in expression of: | |
|---|---|---|
| crtM gene | lpdA gene | |
| lpdA | 1.375±0.161 | nd |
| pdhA | nd | 1.252±0.113 |
nd, Not done.
Values indicate averages of three independent experiments±sem.
Discussion
Considering the well-developed abilities of S. aureus to adapt to its environment, where its membrane fatty acid composition plays a critical role, pathways that are likely to contribute to the production of BCFAs in this pathogen were investigated. The enzyme, BKD, acts on deaminated branched-chain amino acids and converts these to branched-chain acyl coenzyme A molecules. These acyl coenzyme A molecules then serve as precursors for BCFA synthesis through the activity of β-ketoacyl-acyl carrier protein synthase III (FabH) [28, 46–48]. β-keto-acyl carrier protein synthase II (FabF) is responsible for subsequent rounds of elongation until the acyl chain reaches 14 to 17 carbon atoms [28, 46–48].
Previously, we studied the role of BKD in a methicillin-susceptible S. aureus strain SH1000 and determined that the lack of BKD reduces the amount of BCFAs but that they are not completely eliminated [23]. Membrane fatty acid analysis in this study indicates that the amount of SCFAs is relatively higher in methicillin-resistant S. aureus strain JE2 (46.9 %) (Table 2) compared to strain SH1000 (32.4 %), as reported earlier [22, 23]. The ante-iso:iso fatty acid ratio (1.6) was also a little higher in strain JE2 than in strain SH1000 (1.4) [23]. However, TSB-grown bacterial cells were used for determining fatty acid composition in this study compared to S. aureus SH1000 cells grown in BHI medium in the previous study [23]. This factor is not expected to alter our conclusions, since S. aureus cells grown in TSB or BHI demonstrate a similar amount of BCFAs [22]. Overall, the fatty acid composition of the cytoplasmic membrane of the lpdA mutant of JE2 is similar to that of the lpdA mutant of SH1000 [23].
The role of PDH in the production of BCFAs in this study was a bit unexpected. In S. aureus strain JE2, Tn disruption of pdhA, a gene responsible for one of the polypeptides of the PDH complex, led to a significant increase in the amount of BCFAs (78 %) and decrease in SCFAs in the cytoplasmic membrane compared to the wild-type bacterium, where the BCFAs account for about 50 %. Complementation of the mutant by the pdh locus helped restore the membrane fatty acid composition to that of the wild-type JE2. PDH acts on pyruvate and converts it to acetyl CoA, which then enters the tricarboxylic acid cycle [34]. Additionally, acetyl CoA can also lead to the production of butyryl CoA via the enzymes in the crotonyl CoA pathway [49]. FabH from S. aureus exhibits significant activity with butyryl CoA and also acetyl CoA [50], indicating that either compound could be the precursor for the synthesis of SCFAs. A comparison of the activity of PDH vs BKD [34] showed that BKD can also utilize pyruvate, albeit with lower efficiency than PDH, which helps to explain the low SCFA content in the membrane in the absence of functional PDH. The presence of both SCFAs and BCFAs in the membrane of the double mutant implies that another system is capable of supplementing the necessary precursors in the absence of both BKD and PDH. It is noteworthy that the proposed redundant system has a substrate specificity that resembles that of PDH, due to the higher amounts of even iso fatty acids and a similar ante-iso:iso ratio. The α-keto glutarate dehydrogenase (GDH) enzyme complex, with its homology to BKD and PDH complexes, was also considered to play a probable role in providing the precursors of SCFAs in the absence of BKD and PDH. However, the membrane fatty acid analysis of S. aureus JE2 lacking a functional GDH showed no alteration in composition compared to the wild-type JE2 bacterium (data not shown).
We postulated that PDH could account for the decreased BCFA that was shown in the absence of BKD, and thus we complemented the lpdA mutant with the pdh locus to investigate this hypothesis. Complementation with the pdh locus resulted in almost complete restoration of membrane BCFA content in the lpdA mutant, and a significant restoration of the BCFA content in the lpdA : pdhA double mutant. In addition, the iso even fatty acid content was determined to be higher in the strains complemented with the pdh locus in trans compared to even the wild-type levels. Kinetic characterization of PDH has shown its higher activity with α-keto iso valerate, a source of isobutyryl CoA (precursor of even-iso fatty acids) in comparison to α-keto iso caproate (source of odd-iso fatty acids). BKD on the other hand prefers α-keto iso caproate to α-keto iso valerate [34]. Thus, the membrane lipids of the lpdA mutant could reflect the intracellular acyl CoA pool resulting from the substrate specificity of PDH, while the membrane lipids of the pdhA mutant could reflect the substrate specificity of BKD.
The BCFA composition of the cell membrane in S. aureus seems to significantly impact cell growth. The BKD-deficient JE2 showed slower growth not only compared to the wild-type strain but even with respect to the PDH-deficient strain of JE2. The PDH-deficient strain is expected to have reduced ability to produce the energy needed for bacterial growth [51, 52]. An lpdA : pdhA double mutant showed the slowest growth in growth kinetic studies. When these strains were cultured at 20 °C, the lpdA and the lpdA : pdhA double mutant grew very poorly. These findings might also be indicative of reduced fitness of the mutants. A fitness issue with the lpdA mutant has also been documented in S. aureus SH1000 in regard to environmental stresses [23] and Listeria monocytogenes that showed poor survival in macrophages, in addition to its tolerance to unfavourable physiological conditions [53].
The membrane fluidity data in this study reflect the BCFA content in the cytoplasmic membrane. The pdhA mutant with highest BCFA level showed the greatest fluidity, and the lpdA mutant with smallest amount of BCFA in the cytoplasmic membrane showed the lowest fluidity. The lpdA : pdhA double mutant that showed BCFA content similar to the wild-type JE2 showed no significant change in membrane fluidity. Additionally, in a recent study it was documented that methyl-branched fatty acids reduce lipid condensation and the bi-layer thickness, resulting in the formation of kinks at the branching point that and a concomitant increase in membrane fluidity [54].
Staphyloxanthin, a unique pigment in S. aureus, is not only important in its protection from host defence but also impacts membrane fluidity [55, 56]. The lpdA mutant appeared more pigmented during routine culturing on the TSA plates. Upon quantification, this mutant with low membrane BCFAs produced significantly elevated amounts of staphyloxanthin. In contrast, the pdhA mutant with a very high level of membrane BCFAs showed very little or no measurable production of staphyloxanthin. Elevated levels of staphyloxanthin contribute to reduced membrane fluidity [56], and the lpdA mutant had the least fluid membrane. In addition, acetyl CoA is not only a precursor of SCFAs but is also the first substrate in the mevalonate pathway that eventually yields carotenoids. In the absence of a functional PDH, the limited pool of acetyl CoA is funnelled towards fatty acid biosynthesis and thus is not available for staphyloxanthin production. In contrast, in the lpdA mutant, more acetyl CoA is likely funnelled into the staphyloxanthin pathway. This speculation is also supported by the fact that there was no increase in the expression of the genes of the staphyloxanthin pathway in the lpdA mutant.
The membrane fatty acid perturbations also seem to affect antibiotic tolerance of S. aureus to some extent. The lpdA : pdhA double mutant showed some sensitivity to the cell wall-inhibiting enzymes such as vancomycin and d-cycloserine, and also to a membrane-acting antibiotic daptomycin. This reduction in antibiotic tolerance may solely be due to the growth defects of the lpdA and lpdA : pdhA double mutants.
In summary, the PDH and the BKD enzyme complexes seem to have divergent effects on membrane fatty acid composition due to their substrate preferences and their specific roles in BCFA and energy metabolism. The lack of these enzymes causes significant perturbations in S. aureus membrane associated functions.
Funding information
This work was supported in part by Warner Fermaturo and Board of Trustees Research Funds and Grant 1R15AI090680 from the National Institutes of Health to VKS, grants 1R15AI099977 BJW and CG, R21AI135351 to BJW, and a grant from KCOM Biomedical Sciences Graduate Program to RPR.
Acknowledgements
We acknowledge the Network on Antimicrobial Resistance in S. aureus (NARSA) for providing NE1724 for use in this study. We also acknowledge Shalini Bhatia of ATSU Research support for assistance with statistical analysis.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
This manuscript has been prepared in accordance with the Ethics and Research Integrity policy of the Journal of Medical Microbiology.
Footnotes
Abbreviations: BCFA, branched-chain fatty acid; BKD, branched-chain α-keto acid dehydrogenase; PDH, Pyruvate dehydrogenase; SCFA, straight-chain fatty acid; SCUFA, straight-chain unsaturated fatty acid.
References
- 1.Durai R, Ng PC, Hoque H. Methicillin-resistant Staphylococcus aureus: an update. AORN J. 2010;91:599–609. doi: 10.1016/j.aorn.2009.11.065. [DOI] [PubMed] [Google Scholar]
- 2.Karampela I, Poulakou G, Dimopoulos G. Community acquired methicillin resistant Staphylococcus aureus pneumonia: an update for the emergency and intensive care physician. Minerva Anestesiol. 2012;78:930–940. [PubMed] [Google Scholar]
- 3.Leamer NK, Clemmons NS, Jordan NN, Pacha LA. Update: community-acquired methicillin-resistant Staphylococcus aureus skin and soft tissue infection surveillance among active duty military personnel at Fort Benning GA, 2008– 2010. Mil Med. 2013;178:914–920. doi: 10.7205/MILMED-D-13-00082. [DOI] [PubMed] [Google Scholar]
- 4.Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev. 2002;15:194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Projan SJ, Novick RP. The molecular basis of pathogenicity. In: Crossley KB, Archer GL, editors. The Staphylococci in Human Disease. NY: Churchill Livinstone; 1997. pp. 55–81. (editors) [Google Scholar]
- 6.Bal AM, Gould IM. Antibiotic resistance in Staphylococcus aureus and its relevance in therapy. Expert Opin Pharmacother. 2005;6:2257–2269. doi: 10.1517/14656566.6.13.2257. [DOI] [PubMed] [Google Scholar]
- 7.Chopra I. Antibiotic resistance in Staphylococcus aureus: concerns, causes and cures. Expert Rev Anti Infect Ther. 2003;1:45–55. doi: 10.1586/14787210.1.1.45. [DOI] [PubMed] [Google Scholar]
- 8.Cui L, Iwamoto A, Lian JQ, Neoh HM, Maruyama T, et al. Novel mechanism of antibiotic resistance originating in vancomycin-intermediate Staphylococcus aureus. Antimicrob Agents Chemother. 2006;50:428–438. doi: 10.1128/AAC.50.2.428-438.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schito GC. The importance of the development of antibiotic resistance in Staphylococcus aureus. Clin Microbiol Infect. 2006;12:3–8. doi: 10.1111/j.1469-0691.2006.01343.x. [DOI] [PubMed] [Google Scholar]
- 10.Abdallah M, Chataigne G, Ferreira-Theret P, Benoliel C, Drider D, et al. Effect of growth temperature, surface type and incubation time on the resistance of Staphylococcus aureus biofilms to disinfectants. Appl Microbiol Biotechnol. 2014;98:2597–2607. doi: 10.1007/s00253-013-5479-4. [DOI] [PubMed] [Google Scholar]
- 11.Onyango LA, Dunstan RH, Gottfries J, von Eiff C, Roberts TK, et al. Effect of low temperature on growth and ultra-structure of Staphylococcus spp. PLoS One. 2012;7:e29031. doi: 10.1371/journal.pone.0029031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mykytczuk NC, Trevors JT, Leduc LG, Ferroni GD. Fluorescence polarization in studies of bacterial cytoplasmic membrane fluidity under environmental stress. Prog Biophys Mol Biol. 2007;95:60–82. doi: 10.1016/j.pbiomolbio.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 13.Crompton MJ, Dunstan RH, MacDonald MM, Gottfries J, von Eiff C, et al. Small changes in environmental parameters lead to alterations in antibiotic resistance, cell morphology and membrane fatty acid composition in Staphylococcus lugdunensis. PLoS One. 2014;9:e92296. doi: 10.1371/journal.pone.0092296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Onyango LA, Hugh Dunstan R, Roberts TK, MacDonald MM, Gottfries J. Phenotypic variants of staphylococci and their underlying population distributions following exposure to stress. PLoS One. 2013;8:e77614. doi: 10.1371/journal.pone.0077614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liebeke M, Lalk M. Staphylococcus aureus metabolic response to changing environmental conditions – a metabolomics perspective. Int J Med Microbiol. 2014;304:222–229. doi: 10.1016/j.ijmm.2013.11.017. [DOI] [PubMed] [Google Scholar]
- 16.Shireen T, Singh M, Dhawan B, Mukhopadhyay K. Characterization of cell membrane parameters of clinical isolates of Staphylococcus aureus with varied susceptibility to alpha-melanocyte stimulating hormone. Peptides. 2012;37:334–339. doi: 10.1016/j.peptides.2012.05.025. [DOI] [PubMed] [Google Scholar]
- 17.Wang LH, Wang MS, Zeng XA, Liu ZW. Temperature-mediated variations in cellular membrane fatty acid composition of Staphylococcus aureus in resistance to pulsed electric fields. Biochim Biophys Acta. 2016;1858:1791–1800. doi: 10.1016/j.bbamem.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 18.Zhu B, Xia X, Xia N, Zhang S, Guo X, et al. Modification of Fatty acids in membranes of bacteria: implication for an adaptive mechanism to the toxicity of carbon nanotubes. Environ Sci Technol. 2014;48:4086–4095. doi: 10.1021/es404359v. [DOI] [PubMed] [Google Scholar]
- 19.Cronan JE, Gelmann EP. An estimate of the minimum amount of unsaturated fatty acid required for growth of Escherichia coli. J Biol Chem. 1973;248:1188–1195. [PubMed] [Google Scholar]
- 20.Cronan JE, Gennis RB, Maloy SR. Cytoplasmic membrane. In: Neidhardt FC, Ingraham JL, Low KB, Magasanik B, Schaechter M, et al., editors. Escherichia coli and Salmonella: Cellular and Molecular biology. Washington, DC: American Society for Microbiology; 1987. pp. 31–55. (editors) [Google Scholar]
- 21.Willecke K, Pardee AB. Fatty acid-requiring mutant of bacillus subtilis defective in branched chain alpha-keto acid dehydrogenase. J Biol Chem. 1971;246:5264–5272. [PubMed] [Google Scholar]
- 22.Sen S, Sirobhushanam S, Johnson SR, Song Y, Tefft R, et al. Growth-environment dependent modulation of Staphylococcus aureus branched-chain to straight-chain fatty acid ratio and incorporation of unsaturated fatty acids. PLoS One. 2016;11:e0165300. doi: 10.1371/journal.pone.0165300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh VK, Hattangady DS, Giotis ES, Singh AK, Chamberlain NR, et al. Insertional inactivation of branched-chain alpha-keto acid dehydrogenase in Staphylococcus aureus leads to decreased branched-chain membrane fatty acid content and increased susceptibility to certain stresses. Appl Environ Microbiol. 2008;74:5882–5890. doi: 10.1128/AEM.00882-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaiser JC, Sen S, Sinha A, Wilkinson BJ, Heinrichs DE, et al. The role of two branched-chain amino acid transporters in Staphylococcus aureus growth, membrane fatty acid composition and virulence. Mol Microbiol. 2016;102:850–864. doi: 10.1111/mmi.13495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Altenbern RA. Cerulenin-inhibited cells of Staphylococcus aureus resume growth when supplemented with either a saturated or an unsaturated fatty acid. Antimicrob Agents Chemother. 1977;11:574–576. doi: 10.1128/AAC.11.3.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parsons JB, Rock CO. Bacterial lipids: metabolism and membrane homeostasis. Prog Lipid Res. 2013;52:249–276. doi: 10.1016/j.plipres.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Debarbouille M, Gardan R, Arnaud M, Rapoport G. Role of bkdR, a transcriptional activator of the sigL-dependent isoleucine and valine degradation pathway in Bacillus subtilis. J Bacteriol. 1999;181:2059–2066. doi: 10.1128/jb.181.7.2059-2066.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhu K, Bayles DO, Xiong A, Jayaswal RK, Wilkinson BJ, et al. Precursor and temperature modulation of fatty acid composition and growth of Listeria monocytogenes cold-sensitive mutants with transposon-interrupted branched-chain alpha-keto acid dehydrogenase. Microbiology. 2005;151:615–623. doi: 10.1099/mic.0.27634-0. [DOI] [PubMed] [Google Scholar]
- 29.Patel MS, Nemeria NS, Furey W, Jordan F. The pyruvate dehydrogenase complexes: structure-based function and regulation. J Biol Chem. 2014;289:16615–16623. doi: 10.1074/jbc.R114.563148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wexler ID, Hemalatha SG, Patel MS. Sequence conservation in the alpha and beta subunits of pyruvate dehydrogenase and its similarity to branched-chain alpha-keto acid dehydrogenase. FEBS Lett. 1991;282:209–213. doi: 10.1016/0014-5793(91)80479-M. [DOI] [PubMed] [Google Scholar]
- 31.Kumaran S, Patel MS, Jordan F. Nuclear magnetic resonance approaches in the study of 2-oxo acid dehydrogenase multienzyme complexes–a literature review. Molecules. 2013;18:11873–11903. doi: 10.3390/molecules181011873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vilhelmsson O, Miller KJ. Synthesis of pyruvate dehydrogenase in Staphylococcus aureus is stimulated by osmotic stress. Appl Environ Microbiol. 2002;68:2353–2358. doi: 10.1128/AEM.68.5.2353-2358.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaneda T. Biosynthesis of branched chain fatty acids. II. Microbial synthesis of branched long chain fatty acids from certain short chain fatty acid substrates. J Biol Chem. 1963;238:1229–1235. [PubMed] [Google Scholar]
- 34.Oku H, Kaneda T. Biosynthesis of branched-chain fatty acids in Bacillus subtilis. A decarboxylase is essential for branched-chain fatty acid synthetase. J Biol Chem. 1988;263:18386–18396. [PubMed] [Google Scholar]
- 35.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, et al. A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio. 2013;4:e00537-12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Augustin J, Rosenstein R, Wieland B, Schneider U, Schnell N, et al. Genetic analysis of epidermin biosynthetic genes and epidermin-negative mutants of Staphylococcus epidermidis. Eur J Biochem. 1992;204:1149–1154. doi: 10.1111/j.1432-1033.1992.tb16740.x. [DOI] [PubMed] [Google Scholar]
- 37.Davis AO, O'Leary JO, Muthaiyan A, Langevin MJ, Delgado A, et al. Characterization of Staphylococcus aureus mutants expressing reduced susceptibility to common house-cleaners. J Appl Microbiol. 2005;98:364–372. doi: 10.1111/j.1365-2672.2004.02460.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bayer AS, Prasad R, Chandra J, Koul A, Smriti M, et al. In vitro resistance of Staphylococcus aureus to thrombin-induced platelet microbicidal protein is associated with alterations in cytoplasmic membrane fluidity. Infect Immun. 2000;68:3548–3553. doi: 10.1128/IAI.68.6.3548-3553.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shinitzky M, Barenholz Y. Fluidity parameters of lipid regions determined by fluorescence polarization. Biochim Biophys Acta. 1978;515:367–394. doi: 10.1016/0304-4157(78)90010-2. [DOI] [PubMed] [Google Scholar]
- 40.Singh VK, Schmidt JL, Jayaswal RK, Wilkinson BJ. Impact of sigB mutation on Staphylococcus aureus oxacillin and vancomycin resistance varies with parental background and method of assessment. Int J Antimicrob Agents. 2003;21:256–261. doi: 10.1016/S0924-8579(02)00359-X. [DOI] [PubMed] [Google Scholar]
- 41.Singh VK, Utaida S, Jackson LS, Jayaswal RK, Wilkinson BJ, et al. Role for dnaK locus in tolerance of multiple stresses in Staphylococcus aureus. Microbiology. 2007;153:3162–3173. doi: 10.1099/mic.0.2007/009506-0. [DOI] [PubMed] [Google Scholar]
- 42.Singh VK, Syring M, Singh A, Singhal K, Dalecki A, et al. An insight into the significance of the DnaK heat shock system in Staphylococcus aureus. Int J Med Microbiol. 2012;302:242–252. doi: 10.1016/j.ijmm.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 43.Eleaume H, Jabbouri S. Comparison of two standardisation methods in real-time quantitative RT-PCR to follow Staphylococcus aureus genes expression during in vitro growth. J Microbiol Methods. 2004;59:363–370. doi: 10.1016/j.mimet.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 44.Goerke C, Campana S, Bayer MG, Döring G, Botzenhart K, et al. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect Immun. 2000;68:1304–1311. doi: 10.1128/IAI.68.3.1304-1311.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 46.Choi KH, Heath RJ, Rock CO. β-ketoacyl-acyl carrier protein synthase III (FabH) is a determining factor in branched-chain fatty acid biosynthesis. J Bacteriol. 2000;182:365–370. doi: 10.1128/JB.182.2.365-370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaneda T. Iso- and anteiso-fatty acids in bacteria: biosynthesis, function, and taxonomic significance. Microbiol Rev. 1991;55:288–302. doi: 10.1128/mr.55.2.288-302.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu YJ, Zhang YM, Rock CO. Product diversity and regulation of type II fatty acid synthases. Biochem Cell Biol. 2004;82:145–155. doi: 10.1139/o03-076. [DOI] [PubMed] [Google Scholar]
- 49.Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. MBio. 2014;5:e00889-14. doi: 10.1128/mBio.00889-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qiu X, Choudhry AE, Janson CA, Grooms M, Daines RA, et al. Crystal structure and substrate specificity of the beta-ketoacyl-acyl carrier protein synthase III (FabH) from Staphylococcus aureus. Protein Sci. 2005;14:2087–2094. doi: 10.1110/ps.051501605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pronk JT, Wenzel TJ, Luttik MA, Klaassen CC, Scheffers WA, et al. Energetic aspects of glucose metabolism in a pyruvate-dehydrogenase-negative mutant of Saccharomyces cerevisiae. Microbiology. 1994;140:601–610. doi: 10.1099/00221287-140-3-601. [DOI] [PubMed] [Google Scholar]
- 52.Wang L, Ko KW, Lucchinetti E, Zhang L, Troxler H, et al. Metabolic profiling of hearts exposed to sevoflurane and propofol reveals distinct regulation of fatty acid and glucose oxidation: CD36 and pyruvate dehydrogenase as key regulators in anesthetic-induced fuel shift. Anesthesiology. 2010;113:541–551. doi: 10.1097/ALN.0b013e3181e2c1a1. [DOI] [PubMed] [Google Scholar]
- 53.Sun Y, Wilkinson BJ, Standiford TJ, Akinbi HT, O'Riordan MX, et al. Fatty acids regulate stress resistance and virulence factor production for Listeria monocytogenes. J Bacteriol. 2012;194:5274–5284. doi: 10.1128/JB.00045-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Poger D, Caron B, Mark AE. Effect of methyl-branched fatty acids on the structure of lipid bilayers. J Phys Chem B. 2014;118:13838–13848. doi: 10.1021/jp503910r. [DOI] [PubMed] [Google Scholar]
- 55.Liu GY, Essex A, Buchanan JT, Datta V, Hoffman HM, et al. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J Exp Med. 2005;202:209–215. doi: 10.1084/jem.20050846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mishra NN, Liu GY, Yeaman MR, Nast CC, Proctor RA, et al. Carotenoid-related alteration of cell membrane fluidity impacts Staphylococcus aureus susceptibility to host defense peptides. Antimicrob Agents Chemother. 2011;55:526–531. doi: 10.1128/AAC.00680-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McDougal LK, Steward CD, Killgore GE, Chaitram JM, McAllister SK, et al. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. J Clin Microbiol. 2003;41:5113–5120. doi: 10.1128/JCM.41.11.5113-5120.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]