Abstract
S1 is the largest ribosomal protein, present in the small subunit of the bacterial ribosome. It has a pivotal role in stabilizing the mRNA on the ribosome. Thus far, S1 has eluded structural determination. We have identified the S1 protein mass in the cryo-electron microscopic map of the Escherichia coli ribosome by comparing the map with a recent x-ray crystallographic structure of the 30S subunit, which lacks S1. According to our finding, S1 is located at the junction of head, platform, and main body of the 30S subunit, thus explaining all existing biochemical and crosslinking data. Protein S1 as identified in our map has a complex, elongated shape with two holes in its central portion. The N-terminal domain, forming one of the extensions, penetrates into the head of the 30S subunit. Evidence for direct interaction of S1 with 11 nucleotides of the mRNA, immediately upstream of the Shine–Dalgarno sequence, explains the protein's role in the recognition of the 5′ region of mRNA.
Protein S1 is the largest ribosomal protein, 68 kDa (1), present in the small subunit of the Escherichia coli 70S ribosome. It has been implicated in the selection of the translation start site on the 30S subunit (2). It has been suggested that a pyrimidine-rich region upstream of the Shine–Dalgarno (SD) sequence of mRNA interacts with protein S1 and serves as one of the ribosome recognition sites (3). Protein S1 has been reported to be necessary in some cases for translation initiation (4) and for translation elongation (5). It is the only ribosomal protein that has a high affinity for mRNA (6). As a ribosomal protein, S1 is strikingly atypical. Association of S1 to the ribosome is weak and reversible (ribosome preparations often contain less than stoichiometric amounts of protein S1; ref. 7), whereas most other ribosomal proteins are strongly bound. Furthermore, S1 is an acidic protein, whereas all other ribosomal proteins, with the exception of protein L7/L12, are basic.
The S1 protein consists of 557 amino acid residues. Mild trypsin digestion cleaves S1 into two fragments (8), whereas cleavage with cyanogen bromide yields three different fragments (9). The N-terminal fragment from the latter cleavage contains amino acids 1–193, the middle fragment contains amino acids 224–309, and the C-terminal fragment contains amino acids 332–547 (9). The N-terminal domain binds to the platform region of the 30S subunit (10), whereas the C-terminal domain has been suggested to interact with RNAs (11). S1 has been reported to have an elongated shape in solution (12, 13). Neutron scattering (11) and fluorescence anisotropy experiments (14) indicate that the protein maintains its elongated shape even in its ribosome-bound state.
So far, no x-ray crystallographic study has been reported for S1. However, an NMR structure of a 75-aa fragment of S1-type polynucleotide phosphorylase of E. coli, which corresponds to the C-terminal domain of S1, has been determined (15). The structure consists of a five-stranded antiparallel β-barrel, a structural motif known to bind RNAs (15). This motif is found in a number of other translational protein factors, such as chloroplast and prokaryotic initiation factor 1 and eukaryotic initiation factor 2α. Sequence analysis of the E. coli S1 protein indicates that a major portion of S1 consists of six similar structural motifs of ≈70 aa each (16). Four of these repeats (amino acid residues 193–272, 278–360, 365–448, and 452–534), which lie on the C-terminal side, are expected to have higher structural similarity among themselves than to the other two repeats (amino acid residues 22–101 and 106–185), which lie on the N-terminal side.
The tentative location of S1 on the 30S subunit has been previously reported by immunoelectron microscopy (IEM; refs. 17 and 18) and neutron scattering experiments (11, 20, 21) to be on the platform, near the junction of head and main body. S1–RNA crosslinking experiments placed the protein in the same region, close to the cleft of the 30S subunit (22). However, because of the large size of S1, the question was left open whether S1 would span an even larger region of the 30S subunit. To date, all x-ray crystallographic studies of the Thermus thermophilus 30S subunit (23–26), carried out independently by two groups, have failed to visualize the S1 protein. One group (25) intentionally removed S1 from the 30S subunit before crystallization to obtain better-diffracting crystals. We have located the protein on the 30S subunit of the 11.5-Å resolution cryoelectron microscopy (cryo-EM) map of the E. coli 70S ribosome. We found the location by comparing the 30S subunit portion of that map, which contains S1, with the x-ray map of the S1-depleted T. thermophilus 30S subunit (25). The position derived from this study is consistent with results from IEM (17, 18, 19), neutron scattering (11, 20, 21), and S1–rRNA crosslinking (22) experiments, and it helps to explain all existing biochemical data (27, 28). Our study shows that S1 is an elongated structure that makes several contacts with components of head, platform, and main body of the 30S subunit. Furthermore, the study reveals that the central portion of the protein makes direct contact with the 5′ region of the ribosome-bound mRNA.
Materials and Methods
A three-dimensional map of the 30S subunit was derived from the 11.5-Å resolution cryo-EM map of the E. coli 70S ribosome of an initiation-like fMet-tRNA –ribosome complex programmed with a 46-nucleotide mRNA fragment (29), by using SPIDER (30) and IRIS EXPLORER (Numerical Algorithms Group, Downers Grove, IL). The atomic coordinates of the x-ray crystal structure (25) of the T. thermophilus 30S subunit were fitted into the 30S cryo-EM map and converted into an electron density map by computing averaged densities within volume elements scale-matched to those of the cryo-EM map, i.e., by using a pixel size of 2.93 Å. The x-ray map was then filtered to the resolution of the cryo-EM map. The two maps were aligned with respect to one another and a difference map was computed. Another difference map was calculated between the protein-only portion of the cryo-EM map of the 30S subunit (isolated in this lab by differentiating proteins from nucleic acids based on density histogram information and a technique of region growing; see ref. 31), and the fitted, filtered protein portion of the x-ray map (25). To determine the position of the mRNA, a third difference map was computed by subtracting cryo-EM maps of 70S ribosomes with and without mRNA (see ref. 32). Computation of difference maps, separation of clusters, and calculation of molecular mass of the isolated clusters were done by using SPIDER. Docking of x-ray crystal structures into the cryo-EM map was done with O (33) and visualization of the density maps was done with IRIS EXPLORER.
–ribosome complex programmed with a 46-nucleotide mRNA fragment (29), by using SPIDER (30) and IRIS EXPLORER (Numerical Algorithms Group, Downers Grove, IL). The atomic coordinates of the x-ray crystal structure (25) of the T. thermophilus 30S subunit were fitted into the 30S cryo-EM map and converted into an electron density map by computing averaged densities within volume elements scale-matched to those of the cryo-EM map, i.e., by using a pixel size of 2.93 Å. The x-ray map was then filtered to the resolution of the cryo-EM map. The two maps were aligned with respect to one another and a difference map was computed. Another difference map was calculated between the protein-only portion of the cryo-EM map of the 30S subunit (isolated in this lab by differentiating proteins from nucleic acids based on density histogram information and a technique of region growing; see ref. 31), and the fitted, filtered protein portion of the x-ray map (25). To determine the position of the mRNA, a third difference map was computed by subtracting cryo-EM maps of 70S ribosomes with and without mRNA (see ref. 32). Computation of difference maps, separation of clusters, and calculation of molecular mass of the isolated clusters were done by using SPIDER. Docking of x-ray crystal structures into the cryo-EM map was done with O (33) and visualization of the density maps was done with IRIS EXPLORER.
Results and Discussion
Isolation and Identification of S1 Volume from 30S Subunit.
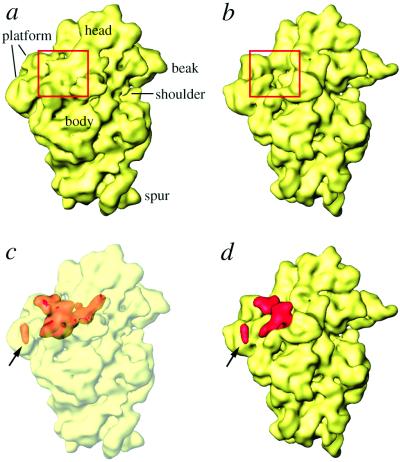
The 11.5-Å cryo-EM map (29) of the E. coli 70S ribosome was used to find the S1 mass. Because the ribosomes for this map were isolated under very low salt conditions, and were never washed with any salt, the preparation retains all of the proteins in close-to-stoichiometric amounts (see ref. 34). We made use of the separation of the 11.5-Å cryo-EM map into a “protein only” and an “RNA only” map, achieved on the basis of histogram information and region growing (see Materials and Methods and ref. 31). In the protein-only map that we obtained by using this method, protein masses occur in clusters. We calculated a difference map between this cryo-EM protein map and a map computed by assembling all proteins from the x-ray structure of the 30S subunit from T. thermophilus (25). This latter structure was obtained from ribosomes crystallized after intentional removal of S1. The difference map shows a large, complex mass in the region of the cleft between head and platform of the 30S subunit (Fig. 1) and scattered relatively small pieces in the head and lower body regions of the 30S subunit (not shown). The latter we attribute mostly to conformational changes between 50S-bound (cryo-EM map) and unbound (x-ray map) states of the 30S subunit (35, 36), and to minor species-related differences between the ribosomes studied by cryo-EM (E. coli) and x-ray crystallography (T. thermophilus). The location of the major mass in the cleft region matches the previous placement of S1 by IEM (17, 18), neutron scattering (11, 20, 21), and S1–RNA crosslinking (22) experiments.
Figure 1.
Location of S1 within the 30S subunit. (a) Surface representation of the 11.5-Å resolution cryo-EM map (29). (b) X-ray structure of the 30S subunit (25), filtered to the resolution of the cryo-EM map and shown in the same solvent-side orientation. The area highlighted with a rectangle shows a large, extra mass of density in the cryo-EM map (a). (c and d) The difference map (red), obtained by subtracting the masses corresponding to only 30S ribosomal proteins in the x-ray (25) and cryo-EM (31) maps, is superimposed on the cryo-EM map (c) and on the filtered x-ray map of the 30S subunit (d). In c, the 30S map is shown as a semitransparent surface, in which the difference density map is embedded. The S21 mass is marked with an arrow in c and d.
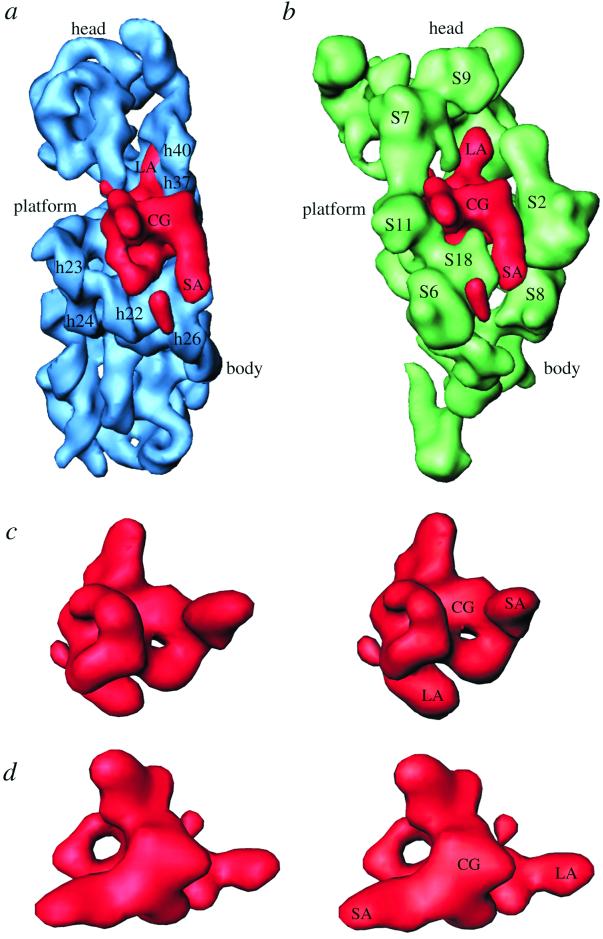
The extra mass has a complex shape, with a central globular (CG) portion having two holes of unequal size, with their openings lying perpendicular to each other (Fig. 2), and two stretched arm-like extensions of unequal lengths. We will refer to these extensions as short arm (SA, ≈28 Å long) and long arm (LA, ≈45 Å long). Distance between the ends of the longest axis, involving tips of LA and SA, is ≈87 Å. LA penetrates into the head of the 30S subunit, whereas SA is located on the solvent side, facing protein S2 (see ref. 25 for structural reference, and Figs. 2 a and b and 3 a and b). The larger of the two holes (Fig. 2d) found in the CG portion is located on the platform side, allowing a direct connection between the cleft of the 30S subunit and the 50S subunit. The small hole (Fig. 2c), in contrast, is partially blocked by protein S18 from the platform. Thus, the mass makes numerous contacts with the rest of the ribosome, and also has a large solvent-exposed surface.
Figure 2.
Neighborhood and structure of protein S1. (a) Position of protein S1 with respect to the16S rRNA, and (b) with respect to other small-subunit ribosomal proteins of the 30S x-ray structure (25). Components of the x-ray structure were filtered to the resolution of the S1 map. Some of the relevant 16S-rRNA helices and small-subunit proteins are labeled. c and d are stereo representations of S1 mass in two orientations: c showing the smaller hole; and d showing the bigger hole within the central globular (CG) region. SA and LA identify the short and long arms, respectively, of S1.
Figure 3.
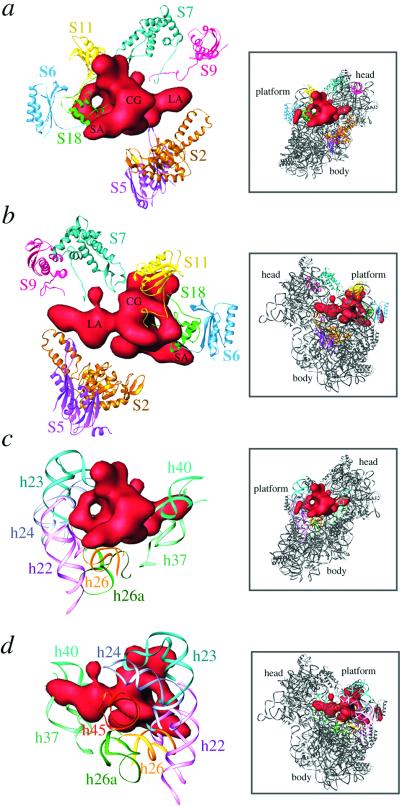
Molecular environment of S1. S1 is shown in red. (a and b) Neighboring proteins. (c and d) Neighboring16S-rRNA helices. Atomic coordinates were taken from Wimberly and coworkers (25) and are represented by using RIBBONS (54). Boxed areas to the right of each panel represent the 30S x-ray structure in the corresponding views, as an aid to orientation. The 30S subunit is viewed from the solvent side (a and c) and from the 50S-interface side (b and d). The bigger hole within the CG portion of the S1 mass is clearly visible in both views. Landmarks are the same as in Fig. 2.
Going by the size of the enclosed volume, the extra mass is equivalent to ≈83 kDa, which is somewhat higher than the known molecular mass of S1 (≈68 kDa). We can attribute this discrepancy to the presence of two additional components in the cryo-EM map that are absent from the T. thermophilus x-ray map: a 46-nucleotide mRNA structure, part of which would be in the immediate vicinity of S1 (see ref. 32), and S21, a protein adjacent to S1 in E. coli that is absent from T. thermophilus (25). Apparently, masses corresponding to both a portion of the mRNA and protein S21 (≈8.5 kDa), the smallest 30S subunit protein, were included in the same group of clusters of the protein-only cryo-EM difference map because of their close proximity to S1. Thus, we attribute the separate, small globular mass (Fig. 1, arrow) to protein S21, which is known to be located in the immediate neighborhood of the S6–S11–S18 protein cluster (see Fig. 2b) on the platform (11, 37, 38). Indeed, a fluorescence resonance energy transfer (FRET) measurement study has suggested that S21 should be ≈60 Å from the 3′ end of the 16S RNA (39). Our placement of S21 fully satisfies the FRET data. Based on the previous visualization of mRNA in the cryo-EM work (32), we attribute a small portion of the CG region of the extra mass to the density contributed by an unstructured upstream region of mRNA (≈11 nucleotides), which makes direct contact with S1 (see Fig. 4). An estimate of the mass of the volumes contributed by S21 and mRNA, when subtracted from the total extra mass, gives a value that closely matches 68 kDa for the remaining mass attributable to S1.
Figure 4.
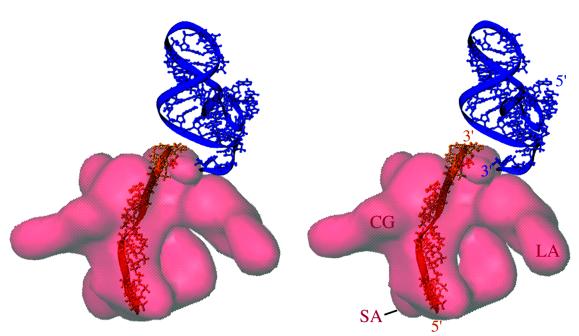
Stereo presentation of the relative positions of S1, helix 45 of 16S rRNA, and mRNA segment. The mRNA segment (orange) was derived from a previous study (32) by computing a difference map between cryo-EM maps of ribosomes with and without bound mRNA and by fitting the atomic coordinates of a single-stranded 12-nucleotide stretch by using polypyrimidine. The mRNA segment corresponding to the 11-nucleotide stretch is seen embedded on the surface of the CG region of the S1 (see text). The 3′ end of the mRNA lies near the 3′ end of helix 45 (blue), from which the anti-SD sequence extends. The actual anti-SD region of the 16S rRNA is not shown because of conflicting positions in the x-ray maps of the isolated 30S subunit (25) and the 70S ribosome (53) (see text). The S1 mass is shown as semitransparent red. Landmarks are the same as in Fig. 2.
Other Ribosomal Proteins in the Immediate Neighborhood of S1 Protein.
We have determined the specific regions of other small-subunit ribosomal proteins that interact with the S1 protein (Figs. 2b and 3 a and b). S1 interacts with proteins from head, platform, and main-body regions on the solvent side of the 30S subunit. In addition to S1 and S21, proteins S6, S11, and S18 cluster on the platform. Of these, S11 and S18 participate in direct interactions with S1 (Fig. 3 a and b). Amino acid residues 89–119 of S11, containing the unstructured C terminus, a β-sheet, and a long α-helix (residues 89–102), contact the CG region near the bigger hole of S1 from the platform side (Fig. 3b), whereas S18, as fitted into our cryo-EM protein map (31), makes contact with both the CG domain and the base of SA through its long α-helix, involving residues 59–77. These residues partially block the opening of the small hole in the CG region from the cleft side of the platform. Protein S6 faces S1 with a surface made of four β-sheets and an unstructured end that reaches near S1 (Fig. 3 a and b). These observations are consistent with previous IEM (18) and neutron scattering (11, 21) data that suggest that these proteins are situated in the immediate neighborhood of S1. Also, our finding corroborates the results of crosslinking experiments between S1 and other ribosomal proteins (40). LA, which penetrates into the 30S-subunit head, is surrounded by proteins S7 and S9 from the head side and by protein S5 from the shoulder side (Fig. 3 a and b). The C-terminal regions of proteins S7 and S9 and the N-terminal loop region (residues 20–25) of S5 are oriented toward S1. The solvent side of S1 faces the large concave surface of protein S2 (Figs. 2b and 3 a and b).
Sillers and Moore (11) proposed that S1 has two active sites, namely an N-terminal ribosome-binding site, physically located on the ribosome in the region of protein S9, which is known to play an important role in S1 binding to the ribosome (41), and an mRNA interaction site, comprising a portion of the C-terminal half of the molecule, located on the S6 side (Fig. 3 a and b). Accordingly, LA would represent the N-terminal portion, and SA, facing toward the protein S6 and the solvent side of 30S, would represent the C-terminal portion (Fig. 3 a and b). It is likely that the CG part contains a portion of the N-terminal half and a major portion of the C-terminal half.
16S rRNA Helices in the Immediate Vicinity of S1 Protein.
In addition to making several contacts with other ribosomal proteins, S1 also interacts with several different 16S-rRNA regions (Figs. 2a and 3 c and d). The 16S rRNA helices 22, 23, 24, and 45 approach S1 from the platform side, whereas helices 25, 26, and 26a approach from the solvent side. Most of these contacts involve the CG region of the S1 protein. The tip of LA (Fig. 3 c and d) is surrounded by helices 34, 35, 37, and 40 of the 16S rRNA. We have found that the tail region of helix 45 (Figs. 3d and 4) and the middle portion of helix 26 make direct contacts with the S1 (Fig. 3 c and d). S1 is known to interact with helix 45 (terminal 49 nucleotides) (42) and helix 26 (nucleotides 846–851) (43). S1 is known to form crosslinks (together with protein S21) in situ near the 3′ end of 16S rRNA (44). It can also be crosslinked to nucleotides 861–889 (helices 25 and 26a) (45) in a region in the immediate neighborhood where S18 crosslinks to the 16S RNA (nucleotides 845–851, helix 26) (43). All of these interactions are in full agreement with the position of S1 derived in the present study.
The positions of some of the 16S rRNA helices (e.g., helices 23a, 26, 37, and 45) surrounding the S1 mass are slightly different in the x-ray structure (25) than their positions in the cryo-EM map (29, 31). In fact, a portion of helix 37 in the x-ray structure partially overlaps with LA (Fig. 3 c and d). Helices 23 (its 23a portion) and 45 appear to have moved into a groove-like structure formed within the CG mass of S1. We believe that these helices move into the empty space (in the x-ray structure, Fig. 1b) that is occupied by S1 in the cryo-EM map (Fig. 1a); the different positions of these helices could be due either to the absence of protein S1 in the x-ray crystal (25) or to an effect of crystal packing.
S1 and mRNA.
Our previous study (32) showed that the 5′ end of the mRNA is located in the cleft region, where we now find a portion of CG of the S1 mass. We superimposed the cryo-EM mass corresponding to the mRNA onto the S1 mass and found that a linear stretch of the upper region (solvent side) of the CG actually corresponds to an ≈11-nucleotide-long fragment of the 5′ region of the mRNA (Fig. 4). It is well known that the 5′ end of the mRNA makes contact with the anti-SD sequence (nucleotides 1535–1540), a pyrimidine-rich extension from helix 45 of the 16S rRNA. We find helix 45 immediately beside S1, on the 3′ end of the mRNA stretch identified on the CG. Thus, a contiguous 5′ stretch of mRNA can make contacts with both S1 (on the upstream side) and the anti-SD (on the downstream side). This finding is strongly supported by evidence for interactions of mRNA with anti-SD and S1 (3, 46).
S1 is known to be directly involved in the process of mRNA recognition and binding (47, 48). Two RNA-binding sites on S1 have been identified (49), and it was suggested that both sites could bind 16S rRNA and mRNA, either simultaneously or sequentially, and that S1 facilitates proper binding of mRNA to the ribosome during translation. S1-depleted ribosomes were found to be governed by base pairing of mRNA to the 3′ end of 16S rRNA (anti-SD sequence), whereas binding to the intact 30S ribosome was dominated by S1 protein (28). However, the possibility of a simultaneous interaction of mRNA with S1 and anti-SD was not ruled out (3). Loechel and coworkers (50) described an mRNA element that enhances translational initiation independent of the SD sequence, and Melancon and coworkers (51) showed that ribosomes lacking the 3′ end of 16S rRNA correctly select the translational start site in some natural mRNAs, suggesting that the mRNA–S1 interaction is an important determinant of the rate of formation of the translational initiation complex. Also, protein S1 utilizes its RNA-binding domain for binding of mRNAs that are lacking the strong SD sequence (52).
The recent 70S T. thermophilus x-ray structure (53) places the single-stranded 3′ end of the 16S rRNA (containing the anti-SD sequence) into the groove region of the platform, partially overlapping the position of S1. The anti-SD region in the x-ray map is far away from the experimentally derived position of mRNA (32), and hence does not allow interaction between the 5′ upstream region of mRNA and anti-SD. The anti-SD position derived in the x-ray study is in conflict with the requirement that three simultaneous interactions must take place: between codon and anticodon at the ribosomal P site, SD and anti-SD, and 5′ upstream region of mRNA and S1. Furthermore, the same single-stranded 3′ end of the 16S rRNA was found in completely different positions in the x-ray structure of the isolated 30S subunit (25), where a portion of it was interpreted as an mRNA fragment in the P site, and in the 70S ribosome (53). These x-ray studies thus suggest high flexibility of the 3′ region of the 16S rRNA. It is possible that the single-stranded anti-SD region has flipped into the groove of the platform in the 70S ribosome (53) because of the absence of S1 in the x-ray study, or alternatively, that its different position is an effect of crystal packing.
The reactivity of S1-specific antibodies was shown to be 3-fold higher for the 70S ribosome than for the 30S subunit (18). The interpretation given for the increased reactivity was that protein S1 has different conformational states in the 30S subunit and the 70S ribosome. However, in a cryo-EM study (35, 36), it was found that the platform is curled inwards toward the head of 30S subunit in the 50S-bound (70S) state. This suggests, as an alternative explanation, that the narrowing of the cleft region promotes tighter binding of S1, and hence increased reactivity to antibodies, within the 70S ribosome.
Direct visualization of S1, and the revelation of its proximity to the 5′ upstream region of mRNA and to helix 45 in the present study, explain all previous biochemical data and demonstrate the direct involvement of S1 in mRNA binding. It is most likely that during the elongation cycle, when SD interaction is not warranted, a nonspecific interaction of the 5′ upstream region of mRNA with S1 helps in stabilizing and maintaining the reading frame on the ribosome.
Acknowledgments
We are grateful to Michael Watters and Yu Chen for the preparation of the illustrations. The work was supported by National Institutes of Health Grants R37 GM29169, R01 GM55440, P41 RR01219, NSF BIR 9219043 (to J.F.), and R01 GM61576 (to R.K.A.).
Abbreviations
- IEM
immunoelectron microscopy
- CG
central globular
- LA
long arm
- SA
short arm
- SD
Shine–Dalgarno sequence
- FRET
fluorescence resonance energy transfer
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Wittmann H G. In: Ribosomes. Namura M, Tissieres A, Lengyel P, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1974. pp. 93–114. [Google Scholar]
- 2.Subramanian A R. Trends Biochem Sci. 1984;9:491–494. [Google Scholar]
- 3.Boni I V, Isaeva D M, Musychenko M L, Tzareva N V. Nucleic Acids Res. 1991;19:155–162. doi: 10.1093/nar/19.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tzareva N V, Makhno V I, Boni I V. FEBS Lett. 1994;337:189–194. doi: 10.1016/0014-5793(94)80271-8. [DOI] [PubMed] [Google Scholar]
- 5.Potapov A P, Subramanian A R. Biochem Int. 1992;27:745–753. [PubMed] [Google Scholar]
- 6.Draper D E, van Hippel P H. J Mol Biol. 1978;122:321–338. doi: 10.1016/0022-2836(78)90193-6. [DOI] [PubMed] [Google Scholar]
- 7.Subramanian A R, van Duin J. Mol Gen Genet. 1977;258:1–9. doi: 10.1007/BF00455113. [DOI] [PubMed] [Google Scholar]
- 8.Suryanarayana T, Subramanian A R. J Mol Biol. 1979;127:41–54. doi: 10.1016/0022-2836(79)90458-3. [DOI] [PubMed] [Google Scholar]
- 9.Subramanian A R. Prog Nucleic Acids Res Mol Biol. 1983;28:101–142. doi: 10.1016/s0079-6603(08)60085-9. [DOI] [PubMed] [Google Scholar]
- 10.Giorginis S, Subramanian A R. J Mol Biol. 1980;141:393–408. doi: 10.1016/0022-2836(80)90253-3. [DOI] [PubMed] [Google Scholar]
- 11.Sillers I Y, Moore P B. J Mol Biol. 1981;153:761–780. doi: 10.1016/0022-2836(81)90417-4. [DOI] [PubMed] [Google Scholar]
- 12.Laughrea M, Moore P B. J Mol Biol. 1977;112:399–421. doi: 10.1016/s0022-2836(77)80189-7. [DOI] [PubMed] [Google Scholar]
- 13.Labischinski M, Subramanian A R. Eur J Biochem. 1979;95:359–366. doi: 10.1111/j.1432-1033.1979.tb12973.x. [DOI] [PubMed] [Google Scholar]
- 14.Odom O W, Deng H-Y, Subramanian A R, Hardesty R. Arch Biochem Biophys. 1984;230:178–193. doi: 10.1016/0003-9861(84)90099-7. [DOI] [PubMed] [Google Scholar]
- 15.Bycroft M, Hubbard T J P, Proctor M, Freund S M V, Murzin A G. Cell. 1997;88:235–242. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- 16.Gribskov M. Gene. 1992;119:107–111. doi: 10.1016/0378-1119(92)90073-x. [DOI] [PubMed] [Google Scholar]
- 17.Politz S M, Glitz D G. Proc Natl Acad Sci USA. 1977;74:1468–1472. doi: 10.1073/pnas.74.4.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walleczek J, Albrecht-Ehrlich R, Stöffler G, Stöffler-Meilicke M. J Biol Chem. 1990;265:11338–11344. [PubMed] [Google Scholar]
- 19.Scheinman A, Atha T, Aguinaldo A M, Kahan L, Shankweiler G, Lake J A. Biochimie. 1992;74:307–317. doi: 10.1016/0300-9084(92)90108-q. [DOI] [PubMed] [Google Scholar]
- 20.Capel M S, Kjeldgaard M, Engelman D M, Moore P B. J Mol Biol. 1988;200:65–87. doi: 10.1016/0022-2836(88)90334-8. [DOI] [PubMed] [Google Scholar]
- 21.Ramakrishnan V, Yabuki S, Sillers I Y, Schindler D G, Engelman D M, Moore P B. J Mol Biol. 1981;153:739–760. doi: 10.1016/0022-2836(81)90416-2. [DOI] [PubMed] [Google Scholar]
- 22.Steitz J A, Wahba A J, Laughrea M, Moore P B. Nucl Acids Res. 1977;4:1–15. doi: 10.1093/nar/4.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clemons W M, Jr, May J L, Wimberly B T, McCutcheon J P, Capel M S, Ramakrishnan V. Nature (London) 1999;400:833–840. doi: 10.1038/23631. [DOI] [PubMed] [Google Scholar]
- 24.Tocilj A, Schlünzen F, Janell D, Glühmann M, Hansen H A, Harms J, Bashan A, Bartels H, Agmon I, Franceschi F, Yonath A. Proc Natl Acad Sci USA. 1999;96:14252–14257. doi: 10.1073/pnas.96.25.14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wimberly B T, Brodersen D E, Clemons Jr W, Morgan-Warren R, Carter A P, Vonrhein C, Hartsch T, Ramakrishnan V. Nature (London) 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 26.Schlüenzen F, Tocilj A, Zarivach R, Harms J, Glühmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, Yonath A. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 27.Boni I V, Zlatkin I V, Budowsky E I. Eur J Biochem. 1982;121:371–376. doi: 10.1111/j.1432-1033.1982.tb05796.x. [DOI] [PubMed] [Google Scholar]
- 28.Ringquist S, Jones T, Snyder E E, Gibson T, Boni I, Gold L. Biochemistry. 1995;34:3640–3648. doi: 10.1021/bi00011a019. [DOI] [PubMed] [Google Scholar]
- 29.Gabashvili I S, Agrawal R K, Spahn C M T, Grassucci R, Svergun D I, Frank J, Penczek P. Cell. 2000;100:537–549. doi: 10.1016/s0092-8674(00)80690-x. [DOI] [PubMed] [Google Scholar]
- 30.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 31.Spahn C M T, Penczek P A, Leith A, Frank J. Structure (London) 2000;8:937–948. doi: 10.1016/s0969-2126(00)00185-4. [DOI] [PubMed] [Google Scholar]
- 32.Frank J, Penczek P, Grassucci R, Heagle A, Spahn C M T, Agrawal R K. In: The Ribosome: Structure, Function, Antibiotics and Cellular Interactions. Garrett R A, Douthwaite S R, Liljas A, Moore P B, Noller H F, editors. Washington, DC: Am. Soc. Microbiol.; 2000. pp. 45–57. [Google Scholar]
- 33.Jones T A, Zhou J Y, Cowan S W, Kjeldgaard M. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 34.Bommer U, Burkhardt N, Jünemann R, Spahn C M T, Triana-Alonso F J, Nierhaus K H. In: Subcellular Fractionation: A Practical Approach. Graham J, Rickwood D, editors. Washington, DC: IRL; 1997. pp. 271–301. [Google Scholar]
- 35.Lata K R, Agrawal R K, Penczek P, Grassucci R, Zhu J, Frank J. J Mol Biol. 1996;262:43–52. doi: 10.1006/jmbi.1996.0497. [DOI] [PubMed] [Google Scholar]
- 36.Agrawal R K, Lata K R, Frank J. Int J Biochem Cell Biol. 1999;31:243–254. doi: 10.1016/s1357-2725(98)00149-6. [DOI] [PubMed] [Google Scholar]
- 37.Odom O W, Deng H-Y, Dabbs E R, Hardesty R. Biochemistry. 1984;23:5069–5076. doi: 10.1021/bi00316a037. [DOI] [PubMed] [Google Scholar]
- 38.Breitenrenter G, Lotti M, Stöffler-Meilicke M, Stöffler G. Mol Gen Genet. 1984;197:189–195. doi: 10.1007/BF00330962. [DOI] [PubMed] [Google Scholar]
- 39.Odom O W, Stöffler G, Hardesty B. FEBS Lett. 1984;173:155–158. doi: 10.1016/0014-5793(84)81037-6. [DOI] [PubMed] [Google Scholar]
- 40.Traut R R, Lambert J M, Boileau G, Kenny J W. In: Ribosomes: Structure, Function and Genetics. Chambliss G, Craven G R, Davies J, Davis K, Kahan L, Nomura M, editors. Baltimore: University Park Press; 1980. pp. 89–110. [Google Scholar]
- 41.Laughrea M, Moore P B. J Mol Biol. 1978;122:109–112. doi: 10.1016/0022-2836(78)90111-0. [DOI] [PubMed] [Google Scholar]
- 42.Dahlberg A E, Dahlberg J E. Proc Natl Acad Sci USA. 1975;72:2940–2944. doi: 10.1073/pnas.72.8.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greuer B, Osswald M, Brimacombe R, Stöffler G. Nucleic Acids Res. 1987;15:3241–3255. doi: 10.1093/nar/15.8.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Czernilofsky A P, Kurland C G, Stöffler G. FEBS Lett. 1975;58:281–289. doi: 10.1016/0014-5793(75)80279-1. [DOI] [PubMed] [Google Scholar]
- 45.Golinska B, Millon R, Backendorf C, Olomucki M, Ebel J P, Ehresmann B. Eur J Biochem. 1981;115:479–484. doi: 10.1111/j.1432-1033.1981.tb06227.x. [DOI] [PubMed] [Google Scholar]
- 46.Moll I, Resch A, Bläsi U. FEBS Lett. 1998;436:213–217. doi: 10.1016/s0014-5793(98)01131-4. [DOI] [PubMed] [Google Scholar]
- 47.Suryanarayana T, Subramanian A R. Biochemistry. 1983;22:2715–2719. doi: 10.1021/bi00280a020. [DOI] [PubMed] [Google Scholar]
- 48.Suryanarayana T, Subramanian A R. Biochemistry. 1984;23:1047–1051. doi: 10.1021/bi00301a002. [DOI] [PubMed] [Google Scholar]
- 49.Draper D E, Pratt C W, van Hippel P H. Proc Natl Acad Sci USA. 1977;74:4786–4790. doi: 10.1073/pnas.74.11.4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loechel S, Inamine J M, Hu P-C. Nucleic Acids Res. 1992;19:6905–6911. doi: 10.1093/nar/19.24.6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Melancon P, Leclerc D, Destroismaisons N, Brakies-Gingras L. Biochemistry. 1990;29:3402–3407. doi: 10.1021/bi00465a037. [DOI] [PubMed] [Google Scholar]
- 52.Sorensen M A, Fricke J, Pedersen S. J Mol Biol. 1998;280:561–569. doi: 10.1006/jmbi.1998.1909. [DOI] [PubMed] [Google Scholar]
- 53.Yusupov M, Yusupova G Z, Boucom A, Lieberman K, Earnest T N, Cate J H D, Noller H. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 54.Carson M. Acta Crystallogr B. 1997;277:493–505. [Google Scholar]