Summary
Osteoarthritis (OA) is the most common degenerative joint disorder. OA was conceived as a “wear and tear” problem of articular cartilage, yet there is a lack of treatment options to delay or rescue articular cartilage degeneration once it is established. Actually, the degradation of articular cartilage is related to a complex network of biochemical pathways involving the diffusion of catabolic factors within and between different joint tissues and particularly bone and cartilage. Advanced proteomics technology provides a powerful tool to allow us to build up a library of such factors. Factors that govern the bone-cartilage interplay could be the candidate diagnostic biomarkers and therapeutic targets for OA. Currently, a growing body of proteomic studies has been done to unveil a number of inflammatory cytokines, proteases, and cartilaginous matrix cleavages in the blood serum, synovial fluid, and articular cartilage from OA patients. Little information is available regarding the protein profiles of disturbances at subchondral bone in the pathophysiology of OA. The technical difficulties in protein extraction from tissues particularly bone and quantitative analyses of protein profile are discussed; cellular proteomics of the defective osteoblasts and secretomics for the osteoblasts–chondrocytes crosstalk are proposed to supplement the information obtained from the bone tissue proteomics.
Keywords: articular cartilage, osteoarthritis, proteomics, subchondral bone
Introduction
Osteoarthritis (OA) is a prevalent debilitating whole-joint disorder, which commonly afflicts the load-bearing joints such as knees and hips [1]. OA is a major cause for joint pain and disability in elderly people. The hallmark of OA is loss of articular cartilage, which cushions the joint during movement. Yet the integrity of articular cartilage relies on the interplay with other joint tissues, particularly subchondral bone [2]. Given the lack of treatment options to delay or rescue degradation of articular cartilage, bone antiresorptives and anabolics are recently the candidate treatments for OA [3]. Yet the factors or mediators that govern the bone-cartilage interactions in the pathogenesis of OA remain largely unknown.
Proteomics, a large-scale analysis of proteins that involves isolation, purification, and mass spectrometry of proteins of interest, makes it possible to search such factors or mediators in a systemic fashion. Proteomic technologies have been widely adopted in studying various rheumatic diseases [4], [5], [6], [7], [8], [9], [10]. Development of high-throughput and high sensitivity mass spectrometry has opened a door to look into the proteins and peptides contained in body fluids and joint tissues [11], [12]. Synovial fluid and serum are the frequently studied specimens whereas synovial and cartilaginous tissues were studied in only a handful of studies. To the best of our knowledge, OA subchondral bones have yet to be studied due to some technique challenges in the protein extraction, purification, and identification process. With the advancement of proteomic technologies, it allows us to study the proteins or peptides inside bone that are likely to participate in the interplay between bone and cartilage in the pathogenesis of OA.
To the best of our knowledge, little information is available regarding the challenges and perspectives of subchondral bone and cell proteomics in the context of OA pathophysiology, which is the motivation of this review article.
Tissue proteomics in OA
The past decade has witnessed the values of proteomic technology in identification of biomarkers and therapeutic targets for various arthritis and rheumatic disorders. Proteomics, i.e., establishing a library of proteins and peptides of interest, enables us to delineate the diseased status from their healthy counterparts. Synovial fluid is the most commonly studied specimen in the field of arthritis and rheumatology research. As a dialysate of plasma, synovial fluid contains a much lower concentration of high molecular weight proteins than plasma, and its total amount of proteins is also 30% lower than plasma in physiological conditions [13]. In an inflamed joint, proteins in the blood stream can enter synovial fluid freely due to an increase in blood vessel numbers and permeability. Over 100 inflammation-related proteins such as apolipoproteins, complements, and fibrinogens have already been identified in synovial fluid samples from OA patients [14], [15], [16]. Their biochemical functions could be grouped into three dominant pathways: acute phase response signalling, complement pathway, and coagulation pathway. Apolipoproteins might originate from systemic metabolism and complements could be produced locally by synoviocytes. A degraded derivative of complement 3f (C3f) was identified in synovial fluid (SF) of OA patients [15]. C3f, a plasma zinc metalloproteinase, was known as an inflammatory regulator and is related to vascular involvement in another rheumatic disorder — systemic sclerosis [17]. Ligands for toll-like receptors such as hyaluronic acid, fibronectin, and alarmins (S100 proteins) were also detected in OA synovial fluid [18]. They induce macrophages and synoviocytes to produce inflammatory cytokines, mediating the catabolic responses in degenerative process of articular cartilage. Inflammation-related proteins are not only produced in the late-stage, but also in the early stages of OA. This implies that innate immunity might contribute to the onset and progression of OA. Meanwhile, it was not surprising to note a decrease in cysteine proteases inhibitors level in OA synovial fluid [14], which failed to protect aggrecan from degradation. As a consequence, the level of extracellular matrix proteins, e.g., aggrecan and cartilage oligomatrix protein, were significantly increased in OA synovial fluid [9].
Although some meaningful results have been produced, the technical challenges in proteomic studies on OA synovial fluid have to be addressed. Firstly, the abundant proteins in synovial fluid such as various extracellular matrix components and albumins may mask the minutely present proteins, e.g., inflammatory cytokines. Structural proteins could be depleted via immunodepletion or two-dimensional cleanup kits [10], [15]. Acetone precipitation, despite being a fairly commonly used technique for proteome study, should not be employed because it lowers the overall protein concentration, including the target proteins [19]. Multiple fractionations involving SDS–polyacrylamide gel electrophoresis (PAGE) for protein level and SCX-Offgel at peptide level could significantly reduce the complexity of the sample [20] and hugely increased the number of newly identified novel proteins. The greatest hurdle to be overcome, however, is that it is very difficult to find a healthy control for OA proteomics study. Usually, synovial fluid samples from rheumatoid arthritis (RA) patients are being compared, which may not expand our understanding in OA.
Synovial fluid is partly produced by synovium, a thin lining in the joint cavity responsible for homeostasis and joint functions. Fibroblast-like synoviocytes is the major cell type in synovium, which can synthesize hyaluronic acid to stabilize water content in synovial fluid and lubricate the articular surfaces during joint movements. Synovial tissues were often investigated in RA, and OA synovium usually serves as a “noninflammatory” control. A transcriptome-proteome combined study on RA and OA synovial tissue demonstrated that many gene expression changes did not coincide at transcription and protein levels, again verifying the need of a more powerful proteome study [8]. To further investigate the physical distribution of proteins, a study involving Matrix-assisted laser desorption/ionization mass spectrometry (MALDI–MS) imaging was performed and this technique involves the use of digital photography of stained histological sections of synovium and MALDI mass spectrometry. Interestingly, the expression of thymosin beta-4, responsible for T-lymphocyte maturation, increased in synovial lining of both RA and OA samples. This finding did suggest the involvement of lymphoid cells in the synovial pathologies of both RA and OA.
Proteomic analysis of articular cartilage will provide a direct insight into the pathogenesis of OA; yet articular cartilage is mainly composed of extracellular matrix including collagens and proteoglycans and dominance of such components limits identification of the less abundant signalling proteins produced by articular chondrocytes. Various robust and reliable extraction–separation protocols were proposed including the use of urea-free solvent and high molecular weight filter, etc. [21], [22], [23]. However, only proteins of ∼ 100 kDa were collected, suggesting a need for optimizing the existing extraction–separation protocols. Despite exhaustive removal processes, abundant proteins are still present as contaminants. For example, highly anionic macromolecules such as aggrecan and hyaluronic acid are exceptionally hard to completely remove for proteomic analysis of articular cartilage. The technical limitations aside, proteomic study of OA cartilage uncovered a similar protein profile as in synovial fluid including the complements, immunoglobulin chains, thrombopoietin, fibrinogen, etc. [21]. Besides, a comparative study was performed on lateral and medial articular chondrocytes from OA patients [24]. Chondrocytes from the less damaged lateral side produced more proteins responsible for actin cytoskeleton organization, glucose metabolic process, and antiapoptosis. On the severely damaged medial side, chondrocytes tended to express more inflammatory cytokines and acute response phase mediators. This indicates that degradation of articular cartilage occurs in the inflammatory environment of synovial fluid.
Proteomic analyses of serum are the most challenging among all the previously performed OA proteome studies. The serum protein profile is affected by an assortment of variables such as the donors' health conditions, age, and physique; thus making it very hard to study without interference from the said constraints. To make matters more complicated, serum is rich in “abundant proteins”, just the “top eight” highly abundant proteins make up 85% of the protein content, and 14 other moderately abundant proteins make up 14%, meaning the remaining make up only 1% by mass. Therefore, immunodepletion of abundant protein via affinity columns is commonly adopted. A separate depletion step for albumin with a high capacity immunodepletion column prior to the depletion of the other seven highly abundant proteins was found to yield better results than just performing in one single column; but because the depletion step results in the dilution of proteins, a concentration step is required after the immunodepletion. A chemical sequential depletion method using Dithiothreitol (DTT) to remove proteins rich in disulphide bonds such as albumin followed by acetonitrile depletion of high molecular weight proteins of over 75 kDa is also viable [25]. De Seny et al [26] had utilized gene chip arrays in his investigation, hence eliminating the need for exhaustive sample treatments altogether. The downside of Surface-enhanced laser desorption/ionization (SELDI) though is that the protein chip can only detect a limited number of proteins of certain biochemical properties and functions instead of the entire protein profile, hence requiring multiple chips to cover a wider spectrum.
Numerous potential serum biomarkers for OA have been named in several proteome studies [27], which can be categorized into metabolic and immune regulators such as apolipoproteins, adiponectins, haptoglobin truncated proteins, interleukin 1β and 6; or metabolites of extracellular matrix, e.g., C-terminal telopeptide of collagen type II (CTX-II) and cartilage oligomatrix protein (COMP). Poor reliability of CTX-II and COMP as OA biomarkers, in spite of their close associations with the disease severity of OA, stops them from being extensively used for OA screening [28]. Glycosylation and other posttranslational modifications of proteins are also important in the discovery of biomarkers. Fukuda et al [28] had performed the first glycoproteomic study on OA using the N-Glycoproteomic 2D-LC-MALDI approach, in which glycoproteins in plasma were specifically concentrated using Affi-Gel Hz hydrazide gel and the trypsin-digested glycoproteins were subjected to comprehensive analysis by 2D-LC-MALDI. It is worth mentioning that glycoproteomic studies do not only evaluate the amount of proteins, but also the level of glycosylation of that protein. For example, ELISA did not detect any significant difference on acute-phase inflammatory response proteins such as clusterin and hemopexin between progressive and nonprogressive OA samples; but the significantly increased glycosylation level of these proteins in progressive OA samples implicated the glycosylation of proteins as the biomarkers for disease progression.
Bone and osteoblasts proteomics in OA
It has been well received that the homeostasis of articular cartilage relies on the biochemical and biomechanical interplay with subchondral bone [2]. It was proposed several decades ago that hardening of subchondral bone would increase the “wear and tear” risk of articular cartilage [29]. The expansion of subchondral bone size correlated with cartilage loss in OA patients [30]. The relationship between bone and cartilage in pathogenesis of OA may be described as “shoe” and “foot”. The disproportional changes of subchondral bone (“shoe”) and overlying cartilage (“foot”) will lead to a “wear and tear” problem. Yet the factors or mediators that govern the hypertrophic changes of subchondral bone in OA remain largely unknown.
Bone protein profiles provided a unique tool to differentiate osteopenia from OA as compared with serum proteins [31]. As identified in proteomic analyses of bone from femoral neck, metabolic enzymes, such as carbonic anhydrase I and phosphoglycerate kinase 1, were the most significant variations of proteins in patients with various bone disorders. In addition, the amount of transforming growth factor-β1 (TGFβ1), insulin-like growth factor (IGF-1), and osteocalcin that embedded in bone matrices from the iliac crest were much higher in patients with generalized OA than healthy patients [32]. Among these factors, a high level of TGF-β1 had been proven to account for OA osteoblasts dysfunctions and subchondral bone disturbances [33], [34]. Serum level of active TGFβ1 was identified as a prognostic biomarker for the progression of OA in animal models [35], although it remained controversial [36]. Collectively, all these data indicated the need for bone proteomics in OA research.
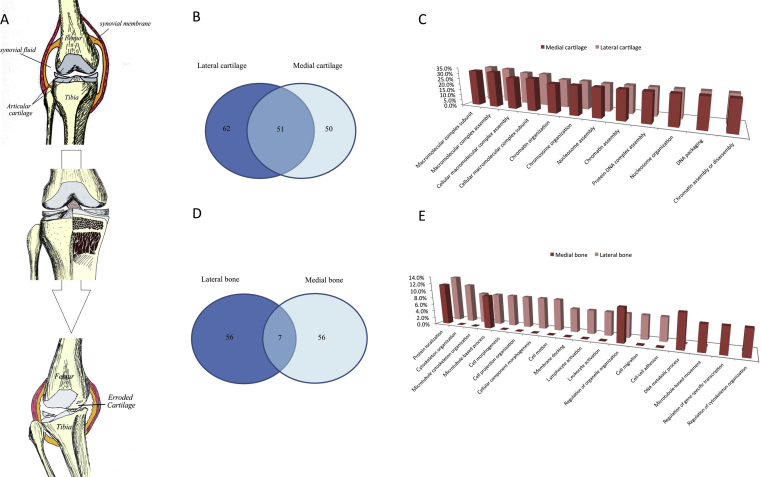
It is technically challenging to extract the proteins of interest from bone that contains the greatest abundance of collagens and minerals [37]. Hydrochloric acid (HCl) was employed to demineralize bone tissue in an attempt to improve the efficiency of protein extraction [37]. We have adopted a similar strategy, using the HCl demineralization procedure as the first step followed by urea-based protein extraction from OA subchondral bone in a trial. Samples were sent for MS analysis by LC-MS/MS LTQ-Orbitrap (LTQ-Orbitrap Velos, Thermo Scientific, MA, USA) and data were analyzed with Maxquant against human database using default settings (version 1.3.0.5). A major lesson learned from the trial is that deep frozen samples kept at −80°C for > 1 week failed to yield any significant signal, suggesting that fresh samples are a must for effective protein extraction. As shown in Fig. 1, a total of 163 and 119 proteins have been successfully identified in fresh articular cartilage and subchondral bone respectively. This protein extraction strategy seems to work for both cartilage and bone tissues. It is very interesting to distinguish patterns of protein profiles in lateral and medial compartments (less or severely damaged parts) of osteoarthritic joints. Less and severely damaged cartilage shared nearly one-third of the proteins identified, whereas 62 and 50 distinct proteins were found only in either the lateral or medial compartments respectively. The striking different protein expression pattern was in the subchondral bone, where the lateral and medial sides only shared 7 proteins.
Figure 1.
A trial for subchondral bone proteomics in osteoarthritis. Proteomics will provide a snapshot for the status of disease during its progression process (A). In our recent trial for subchondral bone proteomics in osteoarthritis samples, we identified differential protein profiles of articular cartilage (B, C) and subchondral bone (D, E) between the lateral and medial compartments of diseased joints, which is subject to varied mechanical loading and also exhibited different severities of joint damage.
The differential protein expression pattern suggests a distinguished signalling mechanism in the corresponding bone region echo to the different stage or severity of OA. However, the data warrants a further validation study. While the pilot data seems promising, we still need to point out that most of the proteins identified are still extracellular matrix (ECM) proteins and we cannot exclude the possibility that the variation in different regions is simply due to experimental error. As a result, further experiments need to confirm the unique protein expression patterns and we should also address the issues regarding the high-abundance of ECM proteins in future experiments. The urea-based extraction buffer made the depletion of high-abundance proteins impossible. SDS–PAGE molecular weight-based fractionation step is, however, possible. This added dimension of separation is expected to increase the number of proteins identified and reduce the effect of highly abundant proteins. Meanwhile we also noticed a recent report showing that phenol extraction could effectively reduce the amount of hyaluronan and proteoglycans [38]. We are going to adopt this method to further improve our current protocol in order to generate much more meaningful information regarding protein expression pattern in different regions of cartilage/bones of OA samples.
Osteoblast dysfunction has been documented in the pathogenesis of disturbances at subchondral bone and OA. It was demonstrated that osteoblasts derived from sclerotic bone region overproduced collagen type I with the wrong composition and poor mineralization [39], and also expressed a high level of inflammatory cytokines such as TGFβ1, prostaglandin E2, interleukin 1β and 6, etc. [33]. They could induce a procatabolic phenotype in healthy chondrocytes, by increasing the expression of MMP-3 and 13 and reducing the production of aggrecan [40], [41]. However, there is little information regarding a complete protein expression profile of defective osteoblasts in OA whereas two proteomic studies into the OA bone marrow derived mesenchymal progenitors, which give rise to osteoblasts [43], [42]. As reported, a high percentage of metabolic enzymes were increased and most of the proteins related to cytoskeleton/motility were decreased in mesenchymal progenitors of OA. However, it could not explain the enhanced migration response of mesenchymal progenitors to platelet-derived growth factor-BB with decreased cytoskeleton/motility proteins in the same study [42]. In the other proteomic study, the increased cytoskeletal proteins, such as beta actin and alpha tubulin were identified in OA compared with RA [43].
Besides bone cell proteomics, a bone–cartilage communication model is also much needed in the search for soluble mediators released by loaded osteoblasts/osteocytes to induce a procatabolic phenotype of articular chondrocytes [44]. It was postulated that OA bone cells when subjected to mechanical stimuli secrete novel soluble mediators, which activate chondrocytes to produce degradation enzymes. In order to decipher which soluble proteins were involved in this crosstalk, a sophisticated technology, Isobaric tags for relative and absolute quantitation (iTRAQ) secretomic approach, was adopted to identify which proteins were differentially present when a mechanical stress was applied on bone cells. Although the preliminary results were encouraging and a soluble protein (14-3-3ε) has been identified, the role of this protein in the pathophysiology of OA remains to be elucidated. Proteomic studies of key mediators for the osteoblast–chondrocyte communications will provide a new insight into the pathogenesis of OA. Limited to the slow growth rate of primarily cultured osteoblasts, pooling multiple samples for a single run may be necessary to obtain sufficient proteins for a meaningful proteomic analysis of osteoblasts or their secretory proteins in a conditioned medium.
Concluding remarks and perspectives
Proteomics has been proven to be a powerful tool for OA research, enabling researchers to identify the novel diagnostic biomarkers and therapeutic targets at an unprecedented pace. Yet the unsolved technical difficulties in protein extraction and separation from OA tissues and cells are the major issues that impede proteomic data from being used clinically. The most common technical challenge the researchers confront is the presence of abundant proteins that mask the signals of their minutely present counterparts. Various techniques have been employed to remove these massive signals of little interest, yet some other proteins of interest might also be removed during the process. For example, the interacting proteins were depleted when prefractionation was done under nondissociating conditions. This phenomenon is particularly pronounced when abundant proteins such as immunoglobulins and albumin are removed from serum and synovial fluid by extensive affinity chromatographic prefractionations, leading to poor data reproducibility which diminishes the value of proteomic studies.
The protein profile of OA is far more complex than previously thought. The posttranslational modifications of proteins such as glycosylation should be worthy of further investigation for OA. Some inflammation related proteins were not differentially present and instead differentially glycosylated in progressive or nonprogressive OA, which were overlooked in conventional proteomic studies [28]. In addition, glycoproteomics can identify a much higher number of proteins [20]. It opens a door to hunt for more biomarkers by using this technique, despite the fact that glyco- or phosphoproteomic studies will cost more than plain proteome studies. We do believe that the glyco- or phosphoproteomic should be the next generation tools for identifying differentially present proteins.
As there are difficulties in protein extraction and separation at tissue level, cellular proteomics may be an alternative choice for studying OA. There is mounting evidence showing the defects in OA osteoblast function by producing proinflammatory cytokines and the collagen and mineral with altered composition and ratio [39]. It may account for the disturbances at OA subchondral bone. Cellular and molecular proteomics into the respective cellular pathways may give us hints to reveal the pathomechanism underlying defective osteoblasts in OA. Secretory proteins mediating osteoblast–chondrocyte crosstalk could also be investigated by secretome analysis of a conditioned medium. Pooling of samples can effectively reduce the variations and improve the repeatability. More accurate protein quantification approaches by means of stable isotope labeling by/with amino acids in cell culture (SILAC) and QconCAT for secretomics, or iTRAQ for typical proteomics [12], [46], [47], [45] may potentially yield more meaningful and insightful data for biological pathway analyses. The choice of the control is another important issue in data interpretation of cellular proteomics. Osteoblasts derived from RA or osteoporotic bone may serve as a disease positive control, and those from fractured and biopsy from young adults should be regarded as a negative control. Review articles regarding the proteomics in OA mainly focused on the technical issues, in particular. Here, we aim to interpret the existing proteomic data from OA patients for identification of information gaps, in order to shed light on the future direction of proteomics research for OA.
Conflicts of interest
The authors declare that they have no competing interests.
Acknowledgements
The authors acknowledge the support from Li Shu Fan Medical Foundation Endowed Professorship in Orthopaedic Surgery and Hong Kong Research Grant Council General Research Fund (HKU-M17105314).
Contributor Information
Chun Yi Wen, Email: paulwen@hku.hk.
Kwong Yuen Chiu, Email: pkychiu@hku.hk.
References
- 1.Zhang Y., Jordan J.M. Epidemiology of osteoarthritis. Clin Geriatr Med. 2010;6:355–369. doi: 10.1016/j.cger.2010.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lories R.J., Luyten F.P. The bone-cartilage unit in osteoarthritis. Nat Rev Rheumatol. 2011;7:43–49. doi: 10.1038/nrrheum.2010.197. [DOI] [PubMed] [Google Scholar]
- 3.Karsdal M.A., Bay-Jensen A.C., Lories R.J., Abramson S., Spector T., Pastoureau P. The coupling of bone and cartilage turnover in osteoarthritis: opportunities for bone antiresorptives and anabolics as potential treatments? Ann Rheum Dis. 2014;73:336–348. doi: 10.1136/annrheumdis-2013-204111. [DOI] [PubMed] [Google Scholar]
- 4.Lambrecht S., Tilleman K., Elewaut D., Deforce D. Proteomics in rheumatology: the beginning of a fairy tale? Proteomics Clin Appl. 2008;2:411–419. doi: 10.1002/prca.200780084. [DOI] [PubMed] [Google Scholar]
- 5.Vanarsa K., Mohan C. Proteomics in rheumatology: the dawn of a new era. F1000 Med Rep. 2010;2:87. doi: 10.3410/M2-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baillet A., Trocme C., Berthier S., Arlotto M., Grange L., Chenau J. Synovial fluid proteomic fingerprint: S100A8, S100A9 and S100A12 proteins discriminate rheumatoid arthritis from other inflammatory joint diseases. Rheumatol. 2010;49:671–682. doi: 10.1093/rheumatology/kep452. [DOI] [PubMed] [Google Scholar]
- 7.Bo G.P., Zhou L.N., He W.F., Luo G.X., Jia X.F., Gan C.J. Analyses of differential proteome of human synovial fibroblasts obtained from arthritis. Clin Rheumatol. 2009;28:191–199. doi: 10.1007/s10067-008-1013-y. [DOI] [PubMed] [Google Scholar]
- 8.Lorenz P., Ruschpler P., Koczan D., Stiehl P., Thiesen H.J. From transcriptome to proteome: differentially expressed proteins identified in synovial tissue of patients suffering from rheumatoid arthritis and osteoarthritis by an initial screen with a panel of 791 antibodies. Proteomics. 2003;3:991–1002. doi: 10.1002/pmic.200300412. [DOI] [PubMed] [Google Scholar]
- 9.Mateos J., Lourido L., Fernandez-Puente P., Calamia V., Fernandez-Lopez C., Oreiro N. Differential protein profiling of synovial fluid from rheumatoid arthritis and osteoarthritis patients using LC-MALDI TOF/TOF. J Proteomics. 2012;75:2869–2878. doi: 10.1016/j.jprot.2011.12.042. [DOI] [PubMed] [Google Scholar]
- 10.Sinz A., Bantscheff M., Mikkat S., Ringel B., Drynda S., Kekow J. Mass spectrometric proteome analyses of synovial fluids and plasmas from patients suffering from rheumatoid arthritis and comparison to reactive arthritis or osteoarthritis. Electrophoresis. 2002;23:3445–3456. doi: 10.1002/1522-2683(200210)23:19<3445::AID-ELPS3445>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 11.Cretu D., Diamandis E.P., Chandran V. Delineating the synovial fluid proteome: recent advancements and ongoing challenges in biomarker research. Crit Rev Clin Lab Sci. 2013;50:51–63. doi: 10.3109/10408363.2013.802408. [DOI] [PubMed] [Google Scholar]
- 12.Wilson R., Bateman J.F. Cartilage proteomics: challenges, solutions and recent advances. Proteomics Clin Appl. 2008;2:251–263. doi: 10.1002/prca.200780007. [DOI] [PubMed] [Google Scholar]
- 13.Gibson D.S., Rooney M.E. The human synovial fluid proteome: a key factor in the pathology of joint disease. Proteomics Clin Appl. 2007 Aug;1(8):889–899. doi: 10.1002/prca.200700044. [DOI] [PubMed] [Google Scholar]
- 14.Gobezie R., Kho A., Krastins B., Sarracino D.A., Thornhill T.S., Chase M. High abundance synovial fluid proteome: distinct profiles in health and osteoarthritis. Arthritis Res Ther. 2007;9:R36. doi: 10.1186/ar2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan X., Huang L., Chen J., Dai Y., Chen X. Analysis of synovial fluid in knee joint of osteoarthritis:5 proteome patterns of joint inflammation based on matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Int Orthop. 2012;36:57–64. doi: 10.1007/s00264-011-1258-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ritter S.Y., Subbaiah R., Bebek G., Crish J., Scanzello C.R., Krastins B. Proteomic analysis of synovial fluid from the osteoarthritic knee: comparison with transcriptome analyses of joint tissues. Arthritis Rheum. 2013;65:981–992. doi: 10.1002/art.37823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xiang Y., Matsui T., Matsuo K., Shimada K., Tohma S., Nakamura H. Comprehensive investigation of disease-specific short peptides in sera from patients with systemic sclerosis — complement C3f-des-arginine, detected predominantly in systemic sclerosis sera, enhances proliferation of vascular endothelial cells. Arthritis Rheum. 2007;56:2018–2030. doi: 10.1002/art.22645. [DOI] [PubMed] [Google Scholar]
- 18.Han M., Dai J., Zhang Y., Lin Q., Jiang M., Xu X. Identification of osteoarthritis biomarkers by proteomic analysis of synovial fluid. J Int Med Res. 2012;40:2243–2250. doi: 10.1177/030006051204000622. [DOI] [PubMed] [Google Scholar]
- 19.Chen C.P., Hsu C.C., Yeh W.L., Lin H.C., Hsieh S.Y., Lin S.C. Optimizing human synovial fluid preparation for two-dimensional gel electrophoresis. Proteome Sci. 2011;9:65. doi: 10.1186/1477-5956-9-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Balakrishnan L., Nirujogi R.S., Ahmad S., Bhattacharjee M., Manda S.S., Renuse S. Proteomic analysis of human osteoarthritis synovial fluid. Clin Proteomics. 2014;11:6. doi: 10.1186/1559-0275-11-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Ceuninck F., Marcheteau E., Berger S., Caliez A., Dumont V., Raes M. Assessment of some tools for the characterization of the human osteoarthritic cartilage proteome. J Biomol Tech. 2005;16:256–265. [PMC free article] [PubMed] [Google Scholar]
- 22.Vincourt J.B., Lionneton F., Kratassiouk G., Guillemin F., Netter P., Mainard D. Establishment of a reliable method for direct proteome characterization of human articular cartilage. Mol Cell Proteomics. 2006;5:1984–1995. doi: 10.1074/mcp.T600007-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Garcia B.A., Platt M.D., Born T.L., Shabanowitz J., Marcus N.A., Hunt D.F. Protein profile of osteoarthritic human articular cartilage using tandem mass spectrometry. Rapid Commun Mass Spectrom. 2006;20:2999–3006. doi: 10.1002/rcm.2692. [DOI] [PubMed] [Google Scholar]
- 24.Stenberg J., Ruetschi U., Skioldebrand E., Karrholm J., Lindahl A. Quantitative proteomics reveals regulatory differences in the chondrocyte secretome from human medial and lateral femoral condyles in osteoarthritic patients. Proteome Sci. 2013;11:43. doi: 10.1186/1477-5956-11-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez-Costa C., Calamia V., Fernandez-Puente P., Capelo-Martinez J.L., Ruiz-Romero C., Blanco F.J. Sequential depletion of human serum for the search of osteoarthritis biomarkers. Proteome Sci. 2012;10:55. doi: 10.1186/1477-5956-10-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Seny D., Sharif M., Fillet M., Cobraiville G., Meuwis M.A., Maree R. Discovery and biochemical characterisation of four novel biomarkers for osteoarthritis. Ann Rheum Dis. 2011;70:1144–1152. doi: 10.1136/ard.2010.135541. [DOI] [PubMed] [Google Scholar]
- 27.Lotz M., Martel-Pelletier J., Christiansen C., Brandi M.L., Bruyere O., Chapurlat R. Value of biomarkers in osteoarthritis: current status and perspectives. Ann Rheum Dis. 2013;72:1756–1763. doi: 10.1136/annrheumdis-2013-203726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fukuda I., Ishihara T., Ohmachi S., Sakikawa I., Morita A., Ikeda M. Potential plasma biomarkers for progression of knee osteoarthritis using glycoproteomic analysis coupled with a 2D-LC-MALDI system. Proteome Sci. 2012;10:9. doi: 10.1186/1477-5956-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radin E.L., Paul I.L., Rose R.M. Role of mechanical factors in pathogenesis of primary osteoarthritis. Lancet. 1972;1:519–522. doi: 10.1016/s0140-6736(72)90179-1. [DOI] [PubMed] [Google Scholar]
- 30.Ding C., Cicuttini F., Jones G. Tibial subchondral bone size and knee cartilage defects: relevance to knee osteoarthritis. Osteoarthritis Cartilage. 2007;15:479–486. doi: 10.1016/j.joca.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 31.Chaput C.D., Dangott L.J., Rahm M.D., Hitt K.D., Stewart D.S., Wayne Sampson H. A proteomic study of protein variation between osteopenic and age-matched control bone tissue. Exp Biol Med. 2012;237:491–498. doi: 10.1258/ebm.2012.011374. [DOI] [PubMed] [Google Scholar]
- 32.Dequeker J., Mohan S., Finkelman R.D., Aerssens J., Baylink D.J. Generalized osteoarthritis associated with increased insulin-like growth factor types I and II and transforming growth factor beta in cortical bone from the iliac crest. Possible mechanism of increased bone density and protection against osteoporosis. Arthritis Rheum. 1993;36:1702–1708. doi: 10.1002/art.1780361209. [DOI] [PubMed] [Google Scholar]
- 33.Massicotte F., Lajeunesse D., Benderdour M., Pelletier J.P., Hilal G., Duval N. Can altered production of interleukin-1beta, interleukin-6, transforming growth factor-beta and prostaglandin E(2) by isolated human subchondral osteoblasts identify two subgroups of osteoarthritic patients. Osteoarthritis Cartilage. 2002;10:491–500. doi: 10.1053/joca.2002.0528. [DOI] [PubMed] [Google Scholar]
- 34.Yu J., Taylor L., Wilson J., Comhair S., Erzurum S., Polgar P. Altered expression and signal transduction of endothelin-1 receptors in heritable and idiopathic pulmonary arterial hypertension. J Cell Physio. 2013;228:322–329. doi: 10.1002/jcp.24132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fahlgren A., Andersson B., Messner K. TGF-beta1 as a prognostic factor in the process of early osteoarthrosis in the rabbit knee. Osteoarthritis Cartilage. 2001;9:195–202. doi: 10.1053/joca.2000.0376. [DOI] [PubMed] [Google Scholar]
- 36.Nelson A.E., Golightly Y.M., Kraus V.B., Stabler T., Renner J.B., Helmick C.G. Serum transforming growth factor-beta 1 is not a robust biomarker of incident and progressive radiographic osteoarthritis at the hip and knee: the Johnston County Osteoarthritis Project. Osteoarthritis Cartilage. 2010;18:825–829. doi: 10.1016/j.joca.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang X., Ye M., Jiang X., Liu G., Feng S., Cui L. Method development of efficient protein extraction in bone tissue for proteome analysis. J Proteome Res. 2007;6:2287–2294. doi: 10.1021/pr070056t. [DOI] [PubMed] [Google Scholar]
- 38.Desjardin C., Balliau T., Valot B., Zivy M., Wimel L., Guerin G. A method for proteomic analysis of equine subchondral bone and epiphyseal cartilage. Proteomics. 2012;12:1870–1874. doi: 10.1002/pmic.201100366. [DOI] [PubMed] [Google Scholar]
- 39.Couchourel D., Aubry I., Delalandre A., Lavigne M., Martel-Pelletier J., Pelletier J.P. Altered mineralization of human osteoarthritic osteoblasts is attributable to abnormal type I collagen production. Arthritis Rheum. 2009;60:1438–1450. doi: 10.1002/art.24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prasadam I., Crawford R., Xiao Y. Aggravation of ADAMTS and matrix metalloproteinase production and role of ERK1/2 pathway in the interaction of osteoarthritic subchondral bone osteoblasts and articular cartilage chondrocytes — possible pathogenic role in osteoarthritis. J Rheumatol. 2012;39:621–634. doi: 10.3899/jrheum.110777. [DOI] [PubMed] [Google Scholar]
- 41.Sanchez C., Deberg M.A., Piccardi N., Msika P., Reginster J.Y., Henrotin Y.E. Subchondral bone osteoblasts induce phenotypic changes in human osteoarthritic chondrocytes. Osteoarthritis Cartilage. 2005;13:988–997. doi: 10.1016/j.joca.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 42.Rollin R., Marco F., Camafeita E., Calvo E., Lopez-Duran L., Jover J.A. Differential proteome of bone marrow mesenchymal stem cells from osteoarthritis patients. Osteoarthritis Cartilage. 2008;16:929–935. doi: 10.1016/j.joca.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 43.Kamada T., Kurokawa M.S., Kato T., Takenouchi K., Takahashi K., Yoshioka T. Proteomic analysis of bone marrow-adherent cells in rheumatoid arthritis and osteoarthritis. Int J Rheum Dis. 2012;15:169–178. doi: 10.1111/j.1756-185X.2012.01702.x. [DOI] [PubMed] [Google Scholar]
- 44.Priam S., Bougault C., Houard X., Gosset M., Salvat C., Berenbaum F. Identification of soluble 14-3-3 as a novel subchondral bone mediator involved in cartilage degradation in osteoarthritis. Arthritis Rheum. 2013;65:1831–1842. doi: 10.1002/art.37951. [DOI] [PubMed] [Google Scholar]
- 45.Onnerfjord P., Khabut A., Reinholt F.P., Svensson O., Heinegard D. Quantitative proteomic analysis of eight cartilaginous tissues reveals characteristic differences as well as similarities between subgroups. J Biol Chem. 2012;287:18913–18924. doi: 10.1074/jbc.M111.298968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Peffers M.J., Beynon R.J., Clegg P.D. Absolute quantification of selected proteins in the human osteoarthritic secretome. Int J Mol Sci. 2013;14:20658–20681. doi: 10.3390/ijms141020658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fernandez-Puente P., Mateos J., Fernandez-Costa C., Oreiro N., Fernandez-Lopez C., Ruiz-Romero C. Identification of a panel of novel serum osteoarthritis biomarkers. J Proteome Res. 2011;10:5095–5101. doi: 10.1021/pr200695p. [DOI] [PubMed] [Google Scholar]