Abstract
Biodiesel is an alternative energy source which has attracted increasing attention lately. Although algae-based biodiesel production has many benefits, it is still far from industrial application. Research suggests that improving lipid quality and production through genetic engineering of metabolic pathways will be the most promising way. To enhance lipid content, both lysophosphatidic acyltransferase gene (c-lpaat) and glycerol-3-phosphate dehydrogenase gene (c-gpd1), optimized according to the codon bias of Chlamydomonas reinhardtii, were inserted into the genomic DNA of model microalga C. reinhardtii by the glass bead method. Transgenic algae were screened by zeomycin resistance and RT-PCR. The transcription levels of inserted genes and the fatty acid content were significantly increased after intermittent heat shock. Most of all, the transcription levels of c-lpaat and c-gpd1 were increased 5.3 and 8.6 times after triple heat shocks, resulting in an increase of 44.5 and 67.5% lipid content, respectively. Furthermore, the content of long-chain saturated fatty acids and monounsaturated fatty acids, especially C18 and C18:1t, notably increased, while unsaturated fatty acids dramatically decreased. The results of this study offer a new strategy combining genetic manipulation and intermittent heat shock to enhance lipid production, especially the production of long-chain saturated fatty acids, using C. reinhardtii.
Electronic supplementary material
The online version of this article (10.1007/s10811-017-1349-2) contains supplementary material, which is available to authorized users.
Keywords: Chlamydomonas reinhardtii, Lipid content, Triacylglycerol synthesis, Transgenic algae, Intermittent heat shock
Introduction
Biodiesel is an alternative and relatively clean energy source which attracts increasing attention because the combustion of fossil fuels releases large amounts of CO2 and pollutants (Amaro et al. 2012; Mubarak et al. 2015). Microalgae are one of the most promising sources for the production of biodiesel as they possess short life cycle, perform photosynthesis, occupy less land, and absorb a large amount of CO2 (Mata et al. 2010; Talebi et al. 2013). The same as most plants, microalgae store lipid in the form of triacylglycerol (TAG), which is the main component of biodiesel (Mubarak et al. 2015). Found in microalgae, the lipid synthesis is mainly through two pathways, fatty acid synthesis and TAG synthesis, which happen in the chloroplast and the endoplasmic reticulum, respectively. Acetyl-coenzyme A (AcCoA) forms free fatty acids (FFAs) under the catalysis of Acetyl-CoA carboxylase (ACCase) and fatty acid synthase (FAS) (Liang and Jiang 2013; Kirchner et al. 2016). In addition, four enzymes are involved in the synthesis of TAG, including glycerol-3-phosphate dehydrogenase (GPDH), lysophosphatidic acyltransferase (LPAAT), diacylglycerol acyltransferase (DGAT), and glycerol-3-phosphate acyltransferase (GPAT) (Griffiths and Harrison 2009; Kirchner et al. 2016). Therefore, increasing the expression of those genes may achieve the aim of improving lipid content.
In the past few decades, researchers have screened algae through strain screening and mutagenesis with the aim of improving lipid content (Banerjee et al. 2016; Kirchner et al. 2016). Though research shows that lipid content of microalgae increases under nitrogen or phosphate starvation, the growth rate of algae is reduced (Hu et al. 2008; Griffiths and Harrison 2009; Himanshu et al. 2016). Nowadays, genetic engineering enables us to modify genes related to fatty acid synthesis to obtain strains with high lipid content in Chlamydomonas. However, expressing ACCase and FAS genes from plants in microalgae has not achieved the aim of significantly increasing oil content (Dunahay et al. 1995; Dehesh et al. 2001). Nevertheless, it is still hopeful to promote lipid content through enhancing key enzyme activity of TAG synthesis. In the meantime, transcriptome analysis of Chlamydomonas reinhardtii with high lipid accumulation reveals that GPDH and LPAAT are significantly upregulated, indicating a positive correlation between the transcription of those genes and cellular lipid accumulation (Lv et al. 2013; Fan et al. 2014). Introducing CrGPDH gene from C. reinhardtii into mutated yeast reveals that it causes higher glycerol production (Casais-Molina et al. 2016). When introducing glycerol-3-phosphate dehydrogenase gene (GPD1) from yeast to oil rapeseed, lipid content in the rapeseed is increased by 40% (Vigeolas et al. 2007). Moreover, overexpression of LPAAT in Brassica napus or GPAT in Arabidopsis thaliana enhances lipid content and TAG accumulation (Maisonneuve et al. 2010; Liang and Jiang 2013). The above results imply that changing fatty acid synthesis pathway or key enzyme transcription in chloroplast is not a plausible way to enhance lipid production in algae. On the other hand, enhancing key enzyme activity of TAG synthesis may promote TAG synthesis by accelerating the transport of FFAs from the chloroplast to the endoplasmic reticulum. Currently, there are a limited number of reports using genetic engineering to modify TAG synthesis pathway to enhance lipid production in microalgae. For example, it has been reported that overexpression of DGAT in C. reinhardtii has not significantly changed TAG composition or accumulation despite detecting a high level of transcription (La Russa et al. 2012). Therefore, more studies are needed to investigate the feasibility of enhancing lipid production in algae through genetic engineering of key enzymes in the TAG pathway.
Firstly, a strong and inducible promoter is needed to overexpress lipid-producing genes in C. reinhardtii. The Hsp70A-RBCS2 promoter has been previously studied and has shown that it could improve the transformation efficiency of a foreign gene and overexpress the foreign gene under 40 °C heat shock (Schroda et al. 2000, 2002). Hence, whether inserting this promoter in front of the lipid-producing gene combined with heat shock can improve gene expression and lipid production is worth investigating.
In this study, we used C. reinhardtii as a model to investigate the effect of GPDH and LPAAT on lipid production in microalgae. Chlamydomonas reinhardtii is a single-cell green alga which has been widely used as a biological model for photosynthesis and lipid metabolism (Ahmad et al. 2015). It has well-established genetic engineering systems in its nucleus, chloroplast, and mitochondria and has been used as a model to study exogenous gene expression and secondary metabolite synthesis. Here, LPAAT gene from Brassica napus and GPD1 gene from Saccharomyces cerevisiae were resynthesized, according to the codon bias of C. reinhardtii, and then inserted into its genomic DNA to obtain gene overexpression. We also inserted Hsp70A-RBCS2 promoter and used intermittent heat shocks to find out whether lipid production in C. reinhardtii can be enhanced and to assess the feasibility of adjusting key enzymes of the TAG synthesis pathway through genetic engineering to enhance lipid production.
Materials and methods
Strains and growth conditions
Chlamydomonas reinhardtii strain CC-849 was purchased from the Chlamydomonas Center in the USA and maintained in Tris-acetate-phosphate (TAP) medium (Gorman and Levine 1965), under the temperature of 22 °C and continuous light at 20 μmol photons m−2 s−1 (normal condition). Escherichia coli TOP10 was taken from the bacterial stocks in our laboratory. Plasmid pH124 carrying the ble conferring zeomycin and phleomycin resistance in host cells was constructed and kept in our laboratory (Wu et al. 2008). Tranclpaat and Trancgpd1 were transgenic algae with introduced c-lpaat and c-gpd1, respectively.
To apply heat shock (HS), 400 mL cells (wild-type (WT) and transgenic algae) in the late logarithmic phase were incubated at 40 °C and light intensity of 20 μmol photons m−2 s−1 for 15 min. Then, the conditions were restored to normal. As for multiple HSs, a 4-h recovery period was placed between each HS. The sampling time points for a single heat shock (HS1) were as follows: before HS, 0 min after HS, and 30, 60, 90, and 120 min after recovery. The sampling time points for triple heat shocks (HS3) were as follows: before HS, 0 min after each HS, and 30, 120, and 240 min after each recovery. The WT strain CC-849 without c-lpaat and c-gpd1 was used as the control. After the treatment, algal cells (WT and transgenic algae) were subjected to DNA/RNA extraction and GC-MS analysis.
Construction of vector
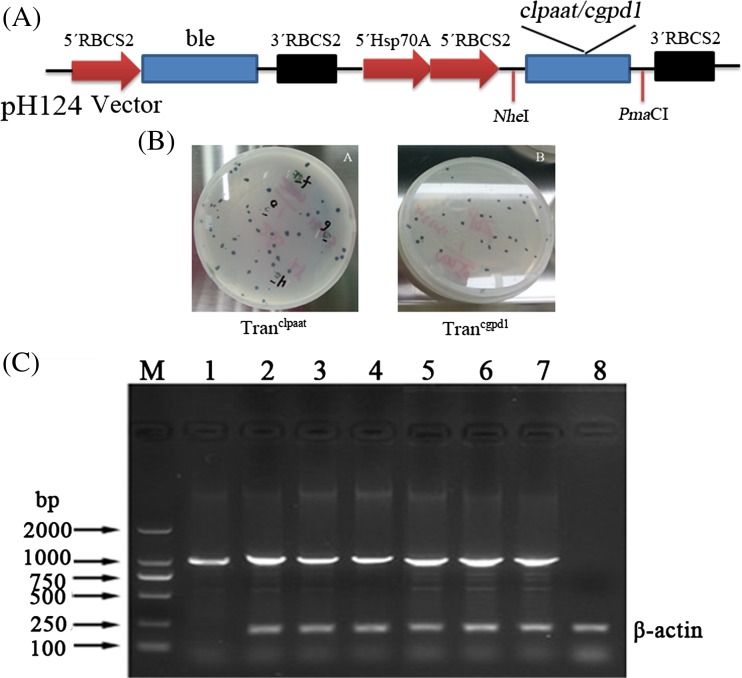
LPAAT gene from Brassica napus (GenBank: AY616009) and GPD1 gene from Saccharomyces cerevisiae (GenBank: Z24454) were resynthesized as c-lpaat and c-gpd1 according to the codon bias of C. reinhardtii and cloned in plasmid pUC57 (Sangon Biotech Co., Ltd., China). Fragments of c-lpaat and c-gpd1 were obtained after cleavage, using restriction endonucleases Nhe I and PmaC I, and then were inserted into pH124 vector to obtain pH124-c-lpaat and pH 124-c-gpd1, respectively. The pH124 vector already contained Hsp70A-RBCS2 promoter, RBCS2 terminator, and ble gene (Fig. 1a).
Fig. 1.
Obtainment of transgenic algae with c-lpaat and c-gpd1. a Construction of transforming plasmids. c-lpaat and c-gpd1 were inserted into pH124 that contained the ampicillin- and zeomycin-resistant genes and a strong heat-inducible promoter (Hsp70A promoter). b Screening transformants through zeomycin resistance (10 μg mL−1). Green clones were visible after 2–3 weeks, and transformants were kept in TAP containing 10 μg mL−1 zeomycin. c RT-PCR verification of transgenic algae with c-lpaat and c-gpd1. Specific fragments of 1176 bp were amplified from the total RNA of transformants. M DL 2000, 1 positive control, PCR fragment of 1176 bp, 2–4 transgenic algae with c-lpaat, 5–7 transgenic algae with c-gpd1, 8 negative control, PCR fragment of β-actin from wild-type CC-849. No c-lpaat and c-gpd1 products were detected in WT
Nuclear transformation of C. reinhardtii
Genetic transformation of C. reinhardtii CC-849 was carried out using the “glass bead method,” according to the protocol described by Kindle (1990) and Wang et al. (2017).
RNA extraction
Total RNAs were isolated using a RNA extraction kit (FAST200) (Fastagen Biotechnology Co., Shanghai, China) with DNase I treatment to eliminate possible DNA contamination according to the manufacturer’s instructions.
RT-PCR verification of transgenic algae
Total RNAs extracted from WT C. reinhardtii and transgenic lines were used as the templates for RT-PCR reactions. The primer sets c-lpaat-F/c-lpaat-R and c-gpd1-F/c-gpd1-R were used to amplify the c-lpaat and c-gpd1 genes. The β-actin gene (as the internal control) was amplified with the primer sets Actin-F/Actin-R (Table 1). PCR program was set as follows: initial denaturation at 95 °C for 2 min, followed by 30 cycles of incubation at 95 °C for 30 s, at 60 °C for 15 s, at 72 °C for 15 s, and a final extension at 72 °C for 5 min.
Table 1.
List of primer sets used in RT-PCR
| Primer name | Primer sequences | Product (bp) |
|---|---|---|
| c-lpaat-F | 5′ CCAAGGTGGCTCGTGACTC 3′ | 1176 |
| c-lpaat-R | 5′ ACTCGCCTCTGTGCCTGTT 3′ | |
| c-gpd1-F | 5′ CGCCGACCGCCTGAACCT 3′ | 1176 |
| c-gpd1-R | 5′ CGCCGACCGCCTGAACCT 3′ | |
| Actin-F | 5′ ACCCGTGCTGACTG 3′ | 240 |
| Actin-R | 5′ ACGTTGAAGGTCTCGAACA 3′ |
Gene expression profiling: real-time RT-PCR
Total RNAs in WT and transgenic algae were extracted from cells in the late logarithmic phase. Real-time RT-PCR analysis was performed on an ABI PRISM 7900 sequence detection system (Applied Biosystems, USA) following the protocol previously described using β-actin gene as the internal control and SYBR Premix Ex Taq kit (Takara, Japan). To obtain cDNA, 1-μg extracted RNA was reversely transcribed into cDNA and then, an equal amount of cDNA was selected as template to perform qRT-PCR. Primer sets for c-lpaat, c-gpd1, and β-actin genes are described in Table 2. PCR conditions were as follows: one step of 95 °C incubation for 30, followed by 40 cycles of 95 °C for 5 and 55 °C for 30s. Data with an R 2 above 0.998 was analyzed using the 2−ΔΔCt program (Lei et al. 2012). Three technical replicates and two biological replicates were used. The transcription value of c-lpaat and c-gpd1 before HS was defined as 1, and then, we obtained the other transcription values and expressed them as relative ratios compared against the before-HS value.
Table 2.
List of primer sets used in real-time RT-PCR
| Gene name | Primer name | Primer sequences |
|---|---|---|
| c-lpaat | Q-c-lpaat-F | 5′ CCTGTGGCTGGAGCTGGTGT 3′ |
| Q-c-lpaat-R | 5′ ATGTCCGAGCGGTGGTTGG 3′ | |
| c-gpd1 | Q-c-gpd1-F | 5′ GTGGTGGCCGAGAACTGCA 3′ |
| Q-c-gpd1-R | 5′ TGGTGGCGGGTGTTGATG 3′ | |
| β-actin | Actin-F | 5′ ACCCGTGCTGCTGATG 3′ |
| Actin-R | 5′ACGTTGAAGGTCTCGAACA 3′ |
Fatty acid methyl ester transformation and FAME analysis
Total lipid extraction was performed as previously described (Lu et al. 2012) with slight modifications. Freeze-dried algal powder of 15 mg was weighed. C19 (500 μg mL−1, ANPEL Laboratory Technologies (Shanghai) Inc.) was added into the sample as the internal standard. Qualification and quantification of fatty acid methyl esters (FAMEs) were performed on a Thermo Trace GC Ultra gas chromatograph coupled to Thermo Polaris Q mass spectrometry which was equipped with a HP-5MS column (30 m × 0.32 mm id, film thickness 0.25 μm). The temperature of the injector was maintained at 250 °C. Helium was used as the carrier gas, ions were generated by a 70-V electron beam, and the mass range scanned was 50 to 650 m/z at a rate of 2 scans s−1. The oven temperature for FAME analysis was initially maintained at 70 °C for 4 min, followed by a temperature increment rate of 5 °C min−1 to 195 °C; held for 5 min, followed by 3 °C min−1 increase to 205 °C; held for 2 min, followed by 8 °C min−1 increment to 230 °C; and then held for 1 min. GC-MS transmitting line temperature was maintained at 250 °C. Peak identification was performed by matching the mass spectra of each compound with the National Institute of Standards and Technology mass spectral library. Automatic peak deconvolution was processed with Masslynx software (V4.1, Waters Corp.,USA) (Lei et al. 2012). The datasets of FAME profiling for further analysis were obtained by normalization against the internal standard in the same chromatogram.
Protein extraction and quantification
When algal growth reached the exponential phase, algal culture was collected and centrifuged at 5000×g and 22 °C for 5 min. The supernatant was discarded, and plant total protein extraction kit (Sangon Biotech (Shanghai) Co., Ltd.) was used to extract total protein from the remaining algal cells. Bovine serum albumin (BSA) standard curve was constructed to measure the protein content according to the above protein-measuring kit’s instruction.
Statistical analysis
All experiments were repeated three times independently, and data were recorded as the mean with standard deviation (SD). For gene expression experiments, quantitative real-time PCR analysis was performed using the BioRAD iQ5 software. For each gene, the fold change was expressed as the mean ± SD (% control) and was calculated using the standard curve with approximation corrected for primer efficiency and normalized to housekeeping gene β-actin expression values. Statistical analyses were performed using the Student’s t test (SPSS19.0). For all analyses, a p value < 0.05 was considered statistically significant.
Results
Vector construction and transgenic alga obtainment
The LPAAT and GPD1 genes were resynthesized according to the codon bias of C. reinhardtii to obtain c-lpaat and c-gpd1 genes suitable for C. reinhardtii expression (Stevens et al. 1996) (see supplementary material 1). When treating with 42 °C HS, both c-lpaat and c-gpd1 genes under the control of Hsp70A promoter were overexpressed (Fig. 1a). Through zeomycin resistance and RT-PCR screening, transgenic algal cells were obtained. Transgenic algae inserted with c-lpaat and c-gpd1 were named as Tranclpaat and Trancgpd1, respectively (Fig. 1b, c). Transformation efficiency by the glass bead method was 8.4 × 10−7 and 4 × 10−7 cells mL−1 for c-lpaat and c-gpd1, respectively.
Overexpressing c-lpaat and c-gpd1 in transgenic C. reinhardtii
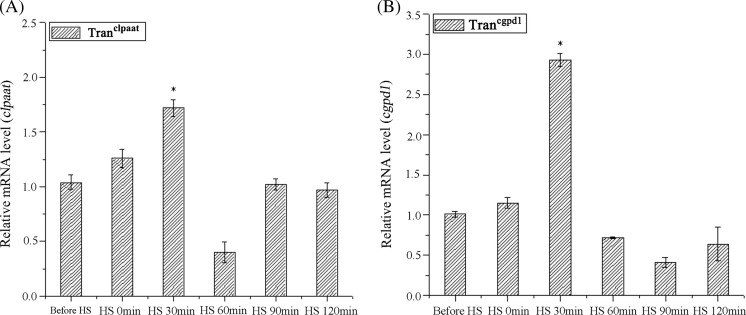
Both c-lpaat and c-gpd1 controlled by Hsp70A promoter were overexpressed under HS. The transcription levels of target genes were unchanged during HS but increased quickly after algae cells were cooled down to the normal condition and reached the highest level after 30 min, with c-lpaat increased 1.93 times and c-gpd1 increased 2.98 times (Fig. 2a, b).
Fig. 2.
Single heat shock-enhanced transcription level of transformed genes in transgenic C. reinhardtii (HS heat shock, under 40 °C and 20 μmol photons m−2 s−1 light for 15 min, HS 0 min sampling immediately after heat stimulation. *P < 0.05 compared with before heat shock)
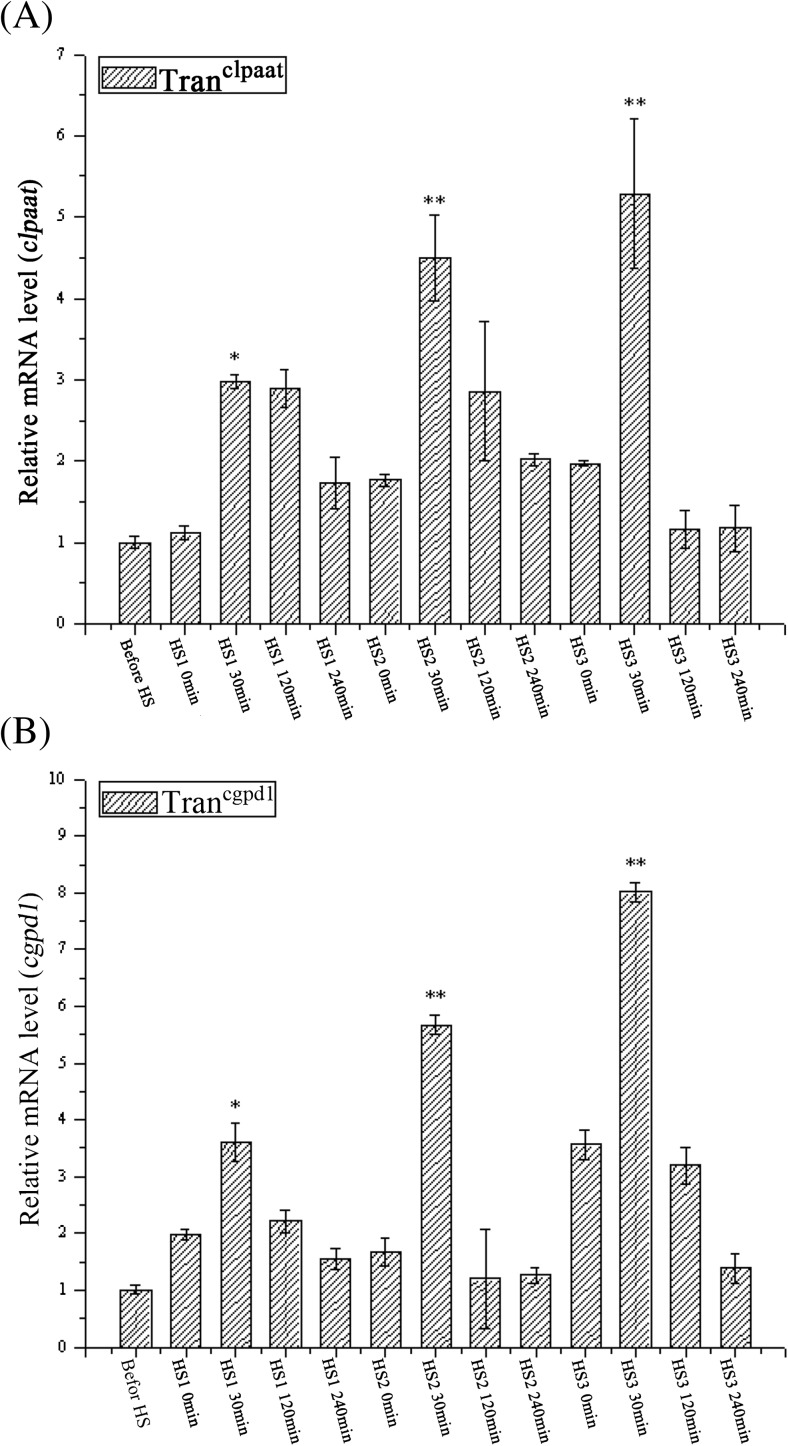
Interestingly, the transcription levels of c-lpaat and c-gpd1 were further improved when treated with intermittent HS. For instance, the transcription levels of c-lpaat in Tranclpaat were increased 3.01, 4.46, and 5.3 times after three HSs, respectively (Fig. 3a). The transcription levels of c-gpd1 in Trancgpd1 were increased 3.6, 5.42, and 8.58 times, respectively (Fig. 3b). The transcription levels of both genes reached the peak at 30 min after HS (Fig. 3a, b). Based on the data analysis, it was confirmed that the transcription levels of target genes were dramatically increased after HS.
Fig. 3.
Triple heat shocks enhanced a c-lpaat and b c-gpd1 gene transcriptions in transgenic C. reinhardtii. (*P < 0.05; **P < 0.01 compared with before heat shock)
Heat shock enhances fatty acid content in transgenic C. reinhardtii
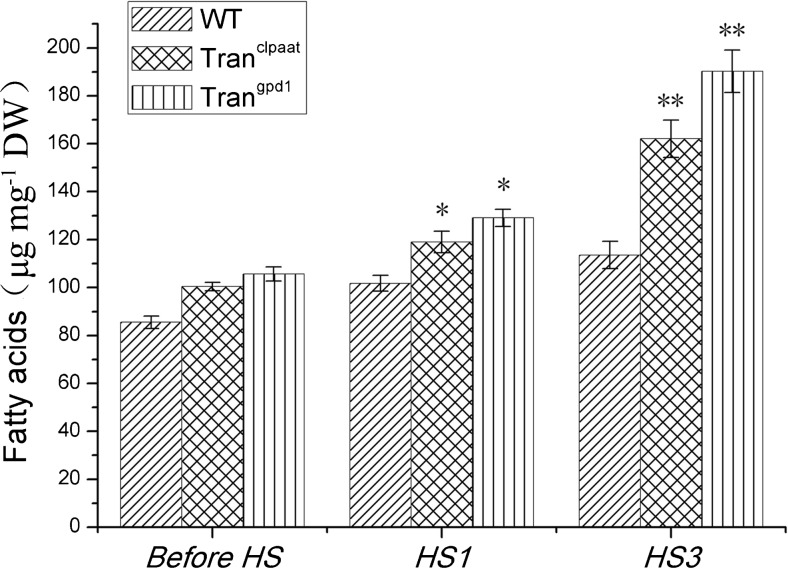
As shown above, transcription levels of c-lpaat and c-gpd1 in transgenic C. reinhardtii could be enhanced significantly after HS. Fatty acids (FAs) were extracted from WT and all transgenic algal cells under normal condition and after HS. Compared to the WT, under the normal culture condition, the total fatty acid (TFA) contents of transgenic alga strains including Tranclpaat and Trancgpd1 strains increased by 17.4 and 23.6%, respectively. The HS treatment further increased the TFA in transgenic algae. The TFA contents in WT were 101.79 and 113.57 μg mg−1 dry weight (DW) after one heat shock (HS × 1) and three heat shocks (HS × 3), respectively. TFA of Tranclpaat and Trancgpd1 increased 16.8 and 26.7%, respectively, after HS × 1, compared with WT cells treated with HS × 1 (Fig. 4). Transgenic algae had significantly higher TFA than WT cells after HS × 3, with an increase of 44.5% (Tranclpaat) or 67.5% (Trancgpd1) (Fig. 4). These results were consistent with the high level of gene transcriptions of c-lpaat and c-gpd1, indicating that overexpression of those genes could enhance lipid accumulation in transformed C. reinhardtii cells.
Fig. 4.
Lipid content change of transgenic C. reinhardtii after heat shock (HS1 and HS3 stand for single heat shock and triple heat shocks, respectively.). After heat shock, total fatty acids of Tranclpaat and Trancgpd1 significantly increased when compared with wild-type CC-849
Change in lipid composition of transgenic algae
The fatty acid profile in algae was analyzed by using GC-MS. WT and transformed algae have similar fatty acids as shown by GC-MS peaks (data not shown). The increase in TFA content of transgenic algae was contributed by the increase of nearly all types of fatty acids. For example, C18:1t in Tranclpaat and Trancgpd1 cells had increases ranging from 177.3 to 270.9% after HS × 1, compared to the WT. The most abundant component is still C18:3n3. In Tranclpaat, C16:0, C18:0, and C18:2t had increases ranging from 33 to 38%. As to Trancgpd1, C16:0, C16:1, C18:0, and C18:2t had increases ranging from 43 to 73% (see Table 3 for more details).
Table 3.
Change of algal fatty acid content and composition after one heat shock
| Fatty acid | Wild type (μg mg−1 DW) | Tranclpaat (μg mg−1 DW) | Increase (%) | Trancgpd1 (μg mg−1 DW) | Increase (%) |
|---|---|---|---|---|---|
| 16:0 | 15.05 ± 0.06 | 20.03 ± 0.09* | 33.09 | 21.87 ± 0.08* | 45.32 |
| 16:1 | 1.98 ± 0.16 | 1.99 ± 0.51 | 0.51 | 2.85 ± 0.19* | 43.94 |
| 16:4 | 27.71 ± 0.38 | 29.03 ± 0.11 | 4.76 | 31.25 ± 0.40 | 12.78 |
| 18:0 | 1.55 ± 0.06 | 2.15 ± 0.06* | 38.71 | 2.27 ± 0.07* | 46.45 |
| 18:1t | 1.10 ± 0.12 | 3.05 ± 0.09** | 177.27 | 4.08 ± 0.13** | 270.9 |
| 18:2t | 7.47 ± 0.35 | 10.06 ± 0.13* | 34.67 | 12.96 ± 0.27* | 73.49 |
| 18:3 | 9.83 ± 0.09 | 10.68 ± 0.29 | 8.65 | 10.78 ± 0.16 | 9.66 |
| 18:3n3 | 37.10 ± 0.08 | 41.99 ± 0.18 | 13.18 | 43.01 ± 0.29 | 15.93 |
Average of all the observations of four repeated tests, in the form of mean ± standard error of representation; wild-type CC-849 represents a control. All data unit is in milligram per gram (dry weight). Statistical analysis software SPSS 19.0
After HS × 3, in Tranclpaat cells, the content of C18:0 and C18:1t fatty acids increased by 355.3 and 220.1%, respectively, compared to the WT. In Trancgpd1 cells, the content of C18:0 and C18:1 t fatty acids increased by 428.2 and 394.2%, respectively, compared to the WT (see Table 4 for more details). Results showed that the increased fatty acids were mainly C18 and the most abundant component is still C18:3n3. However, in Tranclpaat and Trancgpd1 cells, C18:3n3 percentage decreased from 34 to 27%, while C18:0 increased from 2 to 7%. The content of C16:0 and C16:1 fatty acids increased after HS × 3 in Tranclpaat and Trancgpd1. Hence, it is clear that introducing c-lpaat and c-gpd1 to C. reinhardtii can significantly increase cellular lipid accumulation, in particular enhancing the production of monounsaturated fatty acids and long-chain saturated fatty acids, which could be beneficial for biodiesel production.
Table 4.
Change of algal fatty acid content and composition after triple heat shocks
| Fatty acid | Wild type (μg mg−1 DW) | Tranclpaat (μg mg−1 DW) | Increase (%) | Trancgpd1 (μg mg−1 DW) | Increase (%) |
|---|---|---|---|---|---|
| 16:0 | 18.05 ± 0.10 | 29.83 ± 0.06* | 65.26 | 35.39 ± 0.04* | 96.07 |
| 16:1 | 2.98 ± 0.06 | 4.55 ± 0.51* | 52.68 | 6.93 ± 0.21* | 132.55 |
| 16:4 | 29.71 ± 0.78 | 40.99 ± 0.11 | 37.97 | 39.65 ± 1.16 | 33.46 |
| 18:0 | 2.55 ± 0.22 | 11.61 ± 0.36** | 355.29 | 13.47 ± 0.25** | 428.24 |
| 18:1t | 1.89 ± 0.06 | 6.05 ± 0.09** | 220.11 | 9.34 ± 0.46** | 394.18 |
| 18:2t | 8.47 ± 0.13 | 13.06 ± 0.13* | 54.19 | 18.72 ± 0.37* | 121.02 |
| 18:3 | 10.83 ± 0.16 | 11.98 ± 0.50 | 10.62 | 15.36 ± 0.11* | 41.83 |
| 18:3n3 | 39.09 ± 0.19 | 44.04 ± 0.07 | 12.66 | 51.37 ± 0.21 | 31.41 |
Average of all the observations of four repeated tests, in the form of mean ± standard error of representation; wild-type CC-849 represents a control. All data unit is in milligram per gram (dry weight). Statistical analysis software SPSS 19.0
Introducing c-lpaat and c-gpd1 to C. reinhardtii reduces protein synthesis
The growth of WT and transgenic algae was similar, and the highest concentration was around 5.8 × 106 cells mL−1 for all strains, indicating that the transformed genes had no effect on the growth of algal cells (see supplementary material 2).
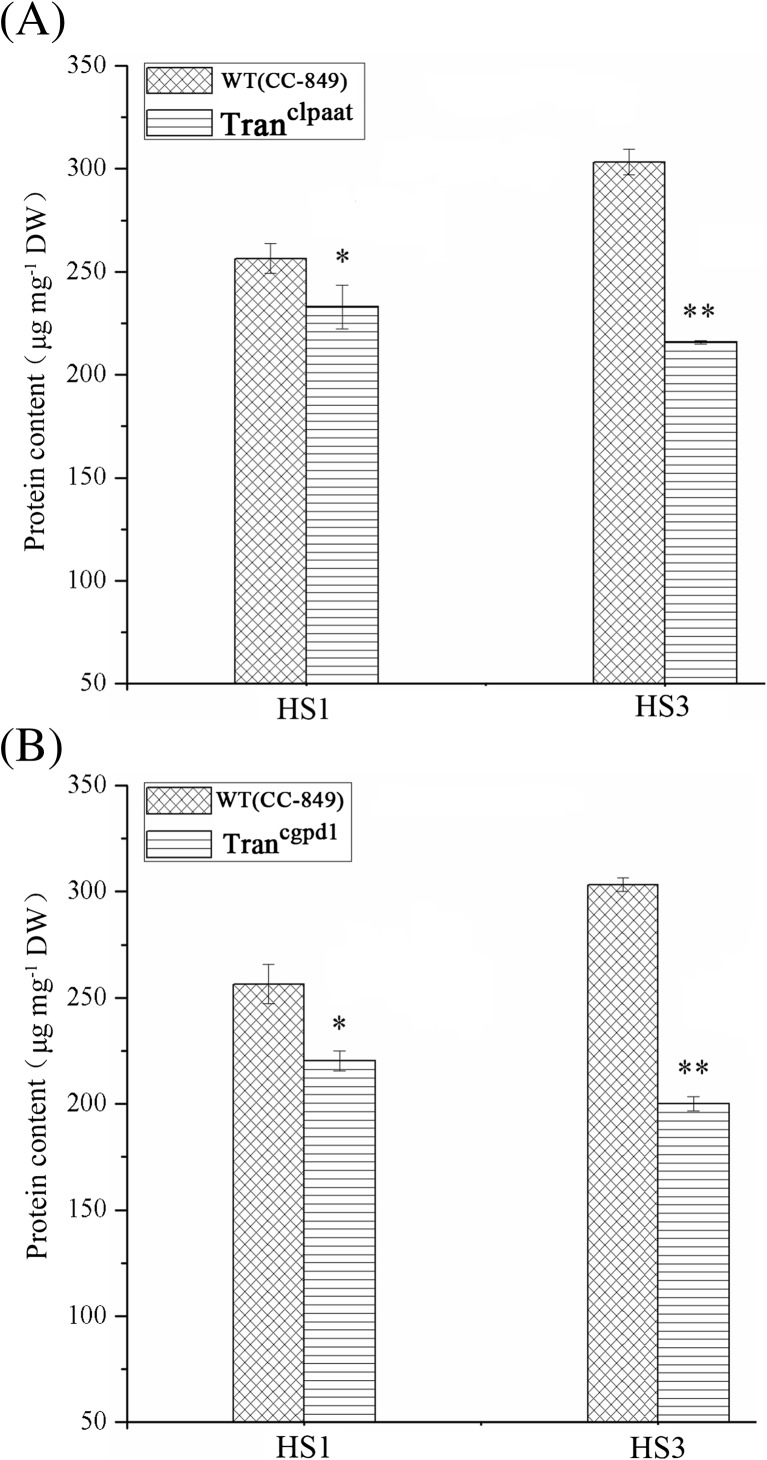
Chlamydomonas reinhardtii produces protein and lipid needed for cell function via photosynthesis. Therefore, the carbon amount is relatively fixed and kept in balance in the cellular system. There is a competitive relationship between protein and lipid syntheses in alga cells, and there are reports showing that inhibition of phosphoenolpyruvate carboxylase can enhance cellular fatty acid content in algae (Deng et al. 2014). After HS × 1, protein content of Tranclpaat and Trancgpd1cells decreased by 9.2 and 14.1%, respectively, compared to WT. Furthermore, protein contents of Tranclpaat and Trancgpd1 cells decreased by 29 and 34%, respectively, after HS × 3 (Fig. 5a, b). Interestingly, the protein content in WT showed increase after HS × 1 and HS × 3, suggesting that HS in this study had no negative effect on the growth of algae.
Fig. 5.
Protein content change after heat shock in a Tranclpaat and b Trancgpd1 cells. (HS heat shock, under 40 °C and 20 μmol photons m−2 s−1 light for 15 min, HS 0 min sampling immediately after heat stimulation. *P < 0.05 compared with the wild type)
All of the above demonstrate that introducing c-lpaat and c-gpd1 to C. reinhardtii changed carbon flow direction, which increased lipid production but reduced protein synthesis. Introducing c-gpd1 rendered higher efficiency than introducing c-lpaat, as Trancgpd1 had 23% more lipid content but only had 5.2% lower protein level.
Discussion
Recent reports show that lipid production in C. reinhardtii can be enhanced under nitrogen starvation (James et al. 2011). Transcriptome analysis shows that the expression of more than 2500 genes is upregulated, among which the transcription of enzyme genes in TAG pathway is significantly increased. One of them, GPDH, increases 5.8 times, and another gene, LPAAT, increases 3.6 times (Lv et al. 2013). GPDH catalyzes the synthesis of glycerol-3-phosphate, and LPAAT catalyzes the synthesis of phosphatidic acid, both supply substrates for TAG synthesis (Deng et al. 2011). Hence, in this study, those two genes were introduced into C. reinhardtii to investigate their ability to enhance lipid production. High-level transcription of LPAAT and GPD1 was detected, with increases of 5.3 and 8.6 times. We successfully used intermittent HS strategy to enhance lipid production for up to 67.5% more than the WT.
Currently known key TAG synthesis enzymes are GPDH, GPAT, LPAAT, and DGAT (Lv et al. 2013). Inhibition of LPAAT and DGAT at transcription levels results in lipid reduction (Zhang et al. 2005; Lv et al. 2013). On the other hand, introducing and overexpressing genes related to TAG synthesis cause lipid content to increase (Jain et al. 2000; Zhang et al. 2005). Those results suggest that key enzymes of TAG synthesis play important roles in lipid production. Our approach of introducing LPAAT and GPD1 into C. reinhardtii is capable of enhancing lipid production significantly under HS. This approach has the potential to be used in high-oil-production algae, such as diatoms. Successful application of our strategy may reduce the cost of biodiesel production.
In our study, Hps70A promoter has been used to drive c-gpd1 and c-lpaat expressions. This promoter can greatly enhance gene expression under heat stimulation (Schroda et al. 2002). Previous reports show that the transcription level of target genes increases 3–26 times when employing Hsp70A promoter to express them (Schroda et al. 2000, 2002; Wu et al. 2008). We attempted intermittent multiple HS strategy to increase gene expression without affecting the growth of algal cells. Our intermittent triple-HS method has been proven to be successful, where gene expression has been increased significantly compared with a single HS. Since mRNA is unlikely to accumulate after expression and HS and TATA-binding factors can bind to the cis-elements of Hsp70A promoter such as HSE sequence and TATA-box (Schroda et al. 2000, 2002), we suggest that after the first HS, those regulatory proteins should be retained in the cells, which bind to the promoter faster at the second and third HSs, which in turn results in fast enhancement of gene expression. The enhanced expression of c-gpd1 and c-lpaat increases the TAG synthesis. The 67.5% increase in lipid production resulting from the above is significantly higher than previous reports in other microalgae (La Russa et al. 2012; Ibáñez-Salazar et al. 2014; Kang et al. 2015).
In C. reinhardtii, the fatty acids are fairly evenly split between 16-carbon chains (two thirds of which are C16:0) and 18-carbon chains (predominantly C18:3). Non-saturated fatty acid content is higher than saturated fatty acid content, and the main fatty acids include C16:0, C16:4, and C18:3n3 (James et al. 2011). Change of fatty acid synthesis enzymes may change the composition of fatty acids as well. In particular, GPDH catalyzes dihydroxyacetone phosphatein to glycerol-3-phosphate, while LPAAT catalyzes the synthesis of phosphatidic acid (La Russa et al. 2012). When introducing diacylglycerol acyltransferase gene from B. napus to C. reinhardtii, there is no major change in the main fatty acid components. However, the levels of saturated fatty acids in the transformed algae decrease to about 7% while unsaturated fatty acids increase proportionately. Polyunsaturated fatty acids, especially α-linolenic acid, increase up to 12% in the transformed line. Interestingly, C18:1–3 slightly decreases in the DAGAT mutant strain in C. reinhardtii, while C16:0 slightly increases (La Russa et al. 2012). In yeast with introduced LPAAT, the proportion of C12:0 and C14:0 fatty acids increases, while in N. tabacum with introduced LPAAT, that proportion actually decreases (Yuan et al. 2015). Hence, the same gene expressed in different species may result in a different outcome. There is little information about the role of TAG synthesis enzymes in microalga lipid production. In this study, we have shown that with introduced LPAAT or GPD1, saturated and monounsaturated fatty acids (C16:0, C18:0, C18:1t, and C18:2 t) have increased significantly. This is consistent with a previous report (Mentewab and Stewart 2005). We also have observed that polyunsaturated fatty acids decreased significantly. Therefore, our genetic manipulation has increased long-chain saturated fatty acids, predominantly C16–C18, which make genetically modified C. reinhardtii an attractive source for biodiesel production with relatively low costs.
In conclusion, we have established a C. reinhardtii genetically modified model that uses Hsp70A promoter to overexpress LPAAT gene from B. napus and GPD1 gene from S. cerevisiae. Applying intermittent multiple-HS strategy, this model can increase lipid production by up to 67.5% and the introduced c-lpaat and c-gpd1 gene expressions can be enhanced by 5.3 and 8.6 times, respectively. We also have proven that overexpression of c-lpaat and c-gpd1 genes is the cause of high lipid production, which in the meantime reduces protein content. Our results suggest that it is possible to create new strains of microalgae with high lipid production ability, and this method can reduce the cost of biodiesel production, which has high technological and economic potentials.
Electronic supplementary material
(DOCX 15.5 kb)
(DOCX 94.3 kb)
(DOCX 15.9 kb)
Acknowledgements
We would like to thank the anonymous reviewers for their constructive suggestions.
Funding information
This work was supported by the National Natural Science Foundation of China (Grant Nos. 31470389, 31470431), Guangdong Natural Science Foundation (2014A030308017, 2016A030313052), Project of DEGP (2015KTSCX125), and Shenzhen Grant Plan for Science & Technology (JCYJ20160422171614147).
Footnotes
Electronic supplementary material
The online version of this article (10.1007/s10811-017-1349-2) contains supplementary material, which is available to authorized users.
References
- Ahmad AI, Sharma AK, Daniell H, Kumar S. Altered lipid composition and enhanced lipid production in green microalga by introduction of Brassica diacylglycerol acyltransferase 2. Plant Biotechnol J. 2015;13:540–550. doi: 10.1111/pbi.12278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaro HM, Ângela CM, Malcata FX. Microalgae: an alternative as sustainable source of biofuels? Energy. 2012;44:158–166. doi: 10.1016/j.energy.2012.05.006. [DOI] [Google Scholar]
- Banerjee C, Singh PK, Shukla P. Microalgal bioengineering for sustainable energy development: recent transgenesis and metabolic engineering strategies. Biotechnol J. 2016;41:355–363. doi: 10.1002/biot.201500284. [DOI] [PubMed] [Google Scholar]
- Dehesh K, Tai H, Edwards P, Byrne J, Jaworski JG. Overexpression of 3-ketoacyl-acyl-carrier protein synthase IIIs in plants reduces the rate of lipid synthesis. Plant Physiol. 2001;125:1103–1114. doi: 10.1104/pp.125.2.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Cai J, Li Y, Fei X. Expression and knockdown of the pepc1, gene affect carbon flux in the biosynthesis of triacylglycerols by the green alga Chlamydomonas reinhardtii. Biotechnol Lett. 2014;36:449–568. doi: 10.1007/s10529-014-1593-3. [DOI] [PubMed] [Google Scholar]
- Deng X, Li Y, Fei X. The mRNA abundance of pepc2, gene is negatively correlated with oil content in Chlamydomonas reinhardtii. Biomass Bioenergy. 2011;35:1811–1817. doi: 10.1016/j.biombioe.2011.01.005. [DOI] [Google Scholar]
- Dunahay TG, Jarvis EE, Roessler PG. Genetic transformation of the diatoms Cyclotella cryptica and Navicula saprophila. J Phycol. 1995;31:1004–1012. doi: 10.1111/j.0022-3646.1995.01004.x. [DOI] [Google Scholar]
- Fan J, Cui Y, Wan M, Wang W, Li Y. Lipid accumulation and biosynthesis genes response of the oleaginous Chlorella pyrenoidosa under three nutrition stressors. Biotechnol Biofuels. 2014;7:1–14. doi: 10.1186/1754-6834-7-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman DS, Levine RP. Cytochrome f and plastocyanin: their sequence in the photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc Natl Acad Sci U S A. 1965;54:1665–1669. doi: 10.1073/pnas.54.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths MJ, Harrison STL. Lipid productivity as a key characteristic for choosing algal species for biodiesel production. J Appl Phycol. 2009;21:493–507. doi: 10.1007/s10811-008-9392-7. [DOI] [Google Scholar]
- Himanshu S, Manish RS, Basuthkar JR, Kandala VRC. Regulation of starch, lipids and amino acids upon nitrogen sensing in Chlamydomonas reinhardtii. Algal Res. 2016;18:33–44. doi: 10.1016/j.algal.2016.05.028. [DOI] [Google Scholar]
- Ibáñez-Salazar A, Rosales-Mendoza S, Rocha-Uribe A, Ramírez-Alonso JI, Lara-Hernández I, Hernández-Torres A, Paz-Maldonado LMT, Silva-Ramirez AS, Banuelos-Hernandez B, Martinez-Salgado JL, Soria-Guerra RE. Over-expression of Dof-type transcription factor increases lipid production in Chlamydomonas reinhardtii. J Biotech. 2014;184:27–38. doi: 10.1016/j.jbiotec.2014.05.003. [DOI] [PubMed] [Google Scholar]
- Jain RK, Coffey M, Lai K, Kumar A, Mackenzie SL. Enhancement of seed oil content by expression of glycerol-3-phosphate acyltransferase genes. Biochem Soc Trans. 2000;28:958–961. doi: 10.1042/bst0280958. [DOI] [PubMed] [Google Scholar]
- James GO, Hocart CH, Hillier W, Chen H, Kordbacheh F, Price GD, Djordjevic MA. Fatty acid profiling of Chlamydomonas reinhardtii, under nitrogen deprivation. Bioresour Technol. 2011;102:3343–3351. doi: 10.1016/j.biortech.2010.11.051. [DOI] [PubMed] [Google Scholar]
- Kang NK, Jeon S, Kwon S, Koh HG, Shin SE, Lee B, Choi GG, Yang JW, Jeong BR, Chang YK. Effects of overexpression of a bHLH transcription factor on biomass and lipid production in Nannochloropsis salina. Biotechnol Biofuels. 2015;8:1–13. doi: 10.1186/s13068-015-0386-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kindle KL. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 1990;87:1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner L, Wirshing A, Kurt L, Reinard T, Glick J, Cram EJ, Jacobsen H-J, Lee-Parsons CWT. Identification, characterization, and expression of diacylgylcerol acyltransferase type-1 from Chlorella vulgaris. Algal Res. 2016;13:167–181. doi: 10.1016/j.algal.2015.10.017. [DOI] [Google Scholar]
- La Russa M, Bogen C, Uhmeyer A, Doebbe A, Filippone E, Kruse O, Mussgnug JH. Functional analysis of three type-2 DGAT homologue genes for triacylglycerol production in the green microalga Chlamydomonas reinhardtii. J Biotech. 2012;162:13–20. doi: 10.1016/j.jbiotec.2012.04.006. [DOI] [PubMed] [Google Scholar]
- Lei AP, Chen H, Shen GM, ZL H, Chen L, Wang JX. Expression of fatty acid synthesis genes and fatty acid accumulation in Haematococcus pluvialis under different stressors. Biotechnol Biofuels. 2012;5:1–11. doi: 10.1186/1754-6834-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang MH, Jiang JG. Advancing oleaginous microorganisms to produce lipid via metabolic engineering technology. Prog Lipid Res. 2013;52:395–408. doi: 10.1016/j.plipres.2013.05.002. [DOI] [PubMed] [Google Scholar]
- Lu S, Wang J, Niu Y, Jie Y, Jian Z, Yuan Y. Metabolic profiling reveals growth related fame productivity and quality of Chlorella sorokiniana, with different inoculum sizes. Biotechnol Bioeng. 2012;109:1651–1662. doi: 10.1002/bit.24447. [DOI] [PubMed] [Google Scholar]
- Lv H, Qu G, Qi X, Lu L, Tian C, Ma Y. Transcriptome analysis of Chlamydomonas reinhardtii during the process of lipid accumulation. Genomics. 2013;101:229–237. doi: 10.1016/j.ygeno.2013.01.004. [DOI] [PubMed] [Google Scholar]
- Maisonneuve S, Bessoule JJ, Lessire R, Delseny M, Roscoe TJ. Expression of rapeseed microsomal lysophosphatidic acid acyltransferase isozymes enhances seed oil content in Arabidopsis. Plant Physiol. 2010;152:670–684. doi: 10.1104/pp.109.148247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata TM, Martins AA, Caetano NS. Microalgae for biodiesel production and other applications: a review. Renew Sust Energy Rev. 2010;14:217–232. doi: 10.1016/j.rser.2009.07.020. [DOI] [Google Scholar]
- Casais-Molina ML, Peraza-Echeverria S, Echevarría-Machado I, Herrera-Valencia VA. Expression of Chlamydomonas reinhardtii CrGPDH2, and CrGPDH3, cDNAs in yeast reveals that they encode functional glycerol-3-phosphate dehydrogenases involved in glycerol production and osmotic stress tolerance. J Appl Phycol. 2016;28:219–226. doi: 10.1007/s10811-015-0588-3. [DOI] [Google Scholar]
- Mentewab A, Stewart CN. Overexpression of an Arabidopsis thaliana ABC transporter confers kanamycin resistance to transgenic plants. Nat Biotechnol. 2005;23:1177–1180. doi: 10.1038/nbt1134. [DOI] [PubMed] [Google Scholar]
- Mubarak M, Shaija A, Suchithra TV. A review on the extraction of lipid from microalgae for biodiesel production. Algal Res. 2015;7:117–123. doi: 10.1016/j.algal.2014.10.008. [DOI] [Google Scholar]
- Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M, Darzins A. Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 2008;54:621–639. doi: 10.1111/j.1365-313X.2008.03492.x. [DOI] [PubMed] [Google Scholar]
- Schroda M, Beck CF, Vallon O. Sequence elements within an HSP70 promoter counteract transcriptional transgene silencing in Chlamydomonas. Plant J. 2002;31:445–455. doi: 10.1046/j.1365-313X.2002.01371.x. [DOI] [PubMed] [Google Scholar]
- Schroda M, Blöcker D, Beck CF. The HSP70A promoter as a tool for the improved expression of transgenes in Chlamydomonas. Plant J. 2000;21:121–131. doi: 10.1046/j.1365-313x.2000.00652.x. [DOI] [PubMed] [Google Scholar]
- Stevens DR, Rochaix J-D, Purton S. The bacterial phleomycin resistance gene ble as a dominant selectable marker in Chlamydomonas. Mol Gen Genet. 1996;251(1):23–30. doi: 10.1007/BF02174340. [DOI] [PubMed] [Google Scholar]
- Talebi AF, Mohtashami SK, Tabatabaei M, Tohidfar M, Bagheri A, Zeinalabedini M, Mirzaei HH, Mirzajanzadeh M, Shafaroudi SM, Bakhtiari S. Fatty acids profiling: a selective criterion for screening microalgae strains for biodiesel production. Algal Res. 2013;2:258–267. doi: 10.1016/j.algal.2013.04.003. [DOI] [Google Scholar]
- Vigeolas H, Waldeck P, Zank T, Geigenberger P. Increasing seed oil content in oil-seed rape (Brassica napus L.) by over-expression of a yeast glycerol-3-phosphate dehydrogenase under the control of a seed-specific promoter. Plant Biotechnol J. 2007;5:431–441. doi: 10.1111/j.1467-7652.2007.00252.x. [DOI] [PubMed] [Google Scholar]
- Wang CG, Chen X, Li H, Wang JX, ZL H. Artificial miRNA inhibition of phosphoenolpyruvate carboxylase increases fatty acid production in a green microalga Chlamydomonas reinhardtii. Biotechnol Biofuels. 2017;10:91. doi: 10.1186/s13068-017-0779-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JX, ZL H, Wang CG, Li SF, Lei AP. Efficient expression of green fluorescent protein (gfp) mediated by a chimeric promoter in Chlamydomonas reinhardtii. Chin J Oceanol Limnol. 2008;26:242–247. doi: 10.1007/s00343-008-0242-x. [DOI] [Google Scholar]
- Yuan Y, Liang Y, Gao L, Sun R, Zheng Y, Li D. Functional heterologous expression of a lysophosphatidic acid acyltransferase from coconut (Cocos nucifera L.) endosperm in Saccharomyces cerevisiae and Nicotiana tabacum. Scientia Hort. 2015;192:224–230. doi: 10.1016/j.scienta.2015.06.003. [DOI] [Google Scholar]
- Zhang FY, Yang MF, YN X. Silencing of DGAT1 in tobacco causes a reduction in seed oil content. Plant Sci. 2005;169:689–694. doi: 10.1016/j.plantsci.2005.05.019. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 15.5 kb)
(DOCX 94.3 kb)
(DOCX 15.9 kb)