Summary
The aim of the study was to establish an induced pluripotent stem cell line from urine-derived cells (UiPSCs) from a patient with phenylketonuria (PKU) in order to provide a useful research tool with which to examine the pathology of this rare genetic metabolic disease. Urine-derived epithelial cells (UCs) from a 15-year-old male patient with PKU were isolated and reprogrammed with integration-free episomal vectors carrying an OCT4, SOX2, KLF4, and miR-302-367 cluster. PKU-UiPSCs were verified as correct using alkaline phosphatase staining. Pluripotency markers were detected with real-time PCR and flow cytometry. Promoter methylation in two pluripotent genes, NANOG and OCT4, was analyzed using bisulphite sequencing. An embryoid body (EB) formation assay was also performed. An induced pluripotent stem cell line (iPSC) was generated from epithelial cells in urine from a patient with PKU. This cell line had increased expression of stem cell biomarkers, it efficiently formed EBs, it stained positive for alkaline phosphatase (ALP), and it had a marked decrease in promoter methylation in the NANOG and OCT4 genes. The PKU-UiPSCs created here had typical characteristics and are suitable for further differentiation.
Keywords: Phenylketonuria, induced pluripotent stem cells, urinary cells, integration-free, disease model
1. Introduction
The high molecular weight melanoma-associated Phenylketonuria (PKU, OMIM 261600) is a common inherited metabolic disease due to mutations in the phenylalanine hydroxylase (PAH) gene that cause accumulation of phenylalanine in the blood and brain, leading to multiple clinical manifestations. These manifestations are mainly development and mental retardation and occasionally include epilepsy, eczema, skeletal fragility, corneal softening, and congenital heart disease (1,2). Restrictions on the intake of phenylalanine are the main treatment for PKU, but the effectiveness of that treatment differs among individuals (3,4). More importantly, such strategies do not target the molecular pathways caused by mutations in the PAH gene, so they cannot reverse every symptom. Therefore, the pathology of PKU needs to be ascertained more accurately in order to identify more potential therapeutic targets (5).
Currently, study on the pathogenesis of PKU is limited, which is partly due to the lack of appropriate disease models, and in particular those that can mimic the pathology of the disease in vitro and in vivo (1,6). In clinical settings, obtaining cells associated with PKU, like neural cells, is also difficult.
The recent advent of induced somatic cell reprogramming with a combination of factors has allowed different disease-related cells to be obtained from induced pluripotent stem cells (iPSCs) (7), thus providing a new "disease model in a dish" strategy with which to study the biological characteristics of many diseases, and rare genetic disorders in particular (8,9). iPSCs can be differentiated into different types of cells and iPSCs obtained from patients share the same genetic background, so iPSC-based models can mimic almost the entire process of pathogenesis. These aspects explain why iPSCs have garnered considerable attention (10-12).
The current study developed a noninvasive and integration-free approach to generating PKU-UiPSCs from urine-derived cells, allowing a continuous supply of those cells for in vitro research on mechanisms and treatments of PKU.
2. Materials and Methods
2.1. The patient with PKU and ethical approval
Urine samples were collected from a 15-year-old male Han Chinese patient with PKU after informed consent was obtained. PKU was diagnosed during neonatal screening based on a blood Phe concentration of 22 mg/dL. Once the diagnosis was confirmed, the patient was subject to dietary restrictions to prevent any irreversible neurological damage. The patient has no significant symptoms at this time. This study was approved by the ethics committee of Shandong Medical Biotechnological Center.
2.2. Culture of epithelial cells from urine
Cells were isolated from urine using the method described by Zhou et al. (13). Briefly, 500 mL of urine was collected from the patient and then centrifuged at 400 g for 10 min. The supernatant was discarded, and the cells were washed with 10-15 mL of PBS containing 1% penicillin and streptomycin (Gibco, USA). Cells were centrifuged again and then resuspended in 3 mL of urine cell medium (1:1 mixture of Dulbecco's Modified Eagle Medium (DMEM) high glucose (Life Technologies, USA) with 10% (vol/vol) fetal bovine serum (FBS) (Gibco, USA), 1% 100× nonessential amino acid (Gibco, USA), 1% 100× L-GlutaMax (Gibco, USA) and Renal Cell Growth Medium (REGM) SingleQuot kit supplements (Lonza)). Cells were seeded in one well of a 6-well plate (Sorfa, China) coated with 0.1% gelatin (Millipore, USA). The medium was then removed and replaced with urine cell medium. Subsequently, half of the culture medium was replaced daily. Upon reaching 90% confluence, cells were passaged using Trypsin (Gibco, USA) with a split ratio of 1:2-1:3 and subcultured for a maximum of 3 passages.
2.3. Reprogramming and iPSC culture
Integration-free episomal vectors carrying the OCT4, SOX2, KLF4, and miR-302-367 cluster were used to perform reprogramming. Cultured cells at a density of 105-106 cells were electrotransfected (Lonza, Switzerland). Cells were seeded in 6-well plates coated Matrigel (Coring, USA) in urine cell medium. After 48 h, the medium was replaced with IM1 medium (Osinglay, China), and cells were cultured for 10-16 d while replacing the medium every day until iPSC colonies appeared. Individual colonies were removed using a glass needle and expanded in Biociso medium (Osinglay, China) on plates coated with Matrigel. The iPSCs were passaged every 3-5 d using 0.5 mM EDTA (Life Technologies, USA) in DPBS (Gibco, USA) at a ratio of 1:3-1:5.
2.4. Confirmation of the absence of the reprogramming vectors
After 10 passages, iPSCs were tested for the absence of programming vectors. A real-time quantitative polymerase chain reaction (RT-PCR) was performed to detect vector genomes and transgenes. To that end, total DNA was isolated using a genomic DNA kit (Tiangen, China) in accordance with the manufacturer's instructions. RT-PCR was performed as suggested by the manufacturer of the Taq-HS PCR Forest Mix (Nova, China): 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 1 min, and finally 72°C for 5 min. Primer sequences are shown in Table 1. PCR products were analyzed using 1% agarose gel electrophoresis.
Table 1. List of primers for vectors.
| Target genes | Primer sequence |
|---|---|
| OCT4 | |
| Forward | AGTGAGAGGCAACCTGGAGA |
| Reverse | AGGAACTGCTTCCTTCACGA |
| SOX2 | |
| Forward | ACCAGCTCGCAGACCTACAT |
| Reverse | CCCCCTGAACCTGAAACATA |
| KLF4 | |
| Forward | CCCACACAGGTGAGAAACCT |
| Reverse | CCCCCTGAACCTGAAACATA |
| SV40LT | |
| Forward | TGGGGAGAAGAACATGGAAG |
| Reverse | AGGAACTGCTTCCTTCACGA |
| oriP | |
| Forward | TTCCACGAGGGTAGTGAACC |
| Reverse | TCGGGGGTGTTAGAGACAAC |
| EBNA-1 | |
| Forward | ATCGTCAAAGCTGCACACAG |
| Reverse | CCCAGGAGTCCCAGTAGTCA |
| miRNA-302 | |
| Forward | TTTCCAAAATGTCGTAATAACCCCG |
| Reverse | CTCCCAAAGAGTCCTGTTCTGTCCT |
2.5. ALP staining
The BCIP/NBT Kit (CoWin Biosciences, China) was used to perform ALP staining to preliminarily identify pluripotency. The cells in one well of a 6-well plate were washed twice with PBS and then 4% polyoxymethylene was used to fix the cells. Afterwards, the well was washed twice using TBST buffer (20 mM Tris-HCl, 150 mM NaCl, 0.05% Tween 20). A color reagent was then prepared according to the manufacturer's instructions and added to the well for 15 min.
2.6. Detection of the pluripotency of markers
Real-time quantitative PCR or RT-qPCR was used to analyze the endogenous pluripotency genes OCT4 and NANOG in iPSCs. Quantitative PCR reactions were performed using 5 ng of reverse-transcribed cDNA with 5 μL of Sybr Green Realtime PCR Master Mix (TOYOBO, Japan) and the primers listed in Table 2. The cycle program was as follows: 95°C for 1 min, 45 cycles of 95°C for 10 s, 60°C for 15 s, and 72°C for 20 s. Each reaction was run in technical triplicates on the Light Cycler®480 (Roche, Switzerland) and normalized to β-actin as an endogenous control. All data were calculated using the ΔΔCp method.
Table 2. List of primers for pluripotency and differentiation markers.
| Target genes | Primer sequence |
|---|---|
| OCT4 | |
| Forward | CCTCACTTCACTGCACTGTA |
| Reverse | CAGGTTTTCTTTCCCTAGCT |
| SOX2 | |
| Forward | CCCAGCAGACTTCACATGT |
| Reverse | CCTCCCATTTCCCTCGTTTT |
| NANOG | |
| Forward | AAGGTCCCGGTCAAGAAACAG |
| Reverse | CTTCTGCGTCACACCATTGC |
| Actin | |
| Forward | CCCAGAGCAAGAGAGG |
| Reverse | GTCCAGACGCAGGATG |
| FOXA2 | |
| Forward | CCAACCCCACAAAATGGA |
| Reverse | ATAATGGGCCGGGAGTACA |
| SOX17 | |
| Forward | ACCGCACGGAATTTGAAC |
| Reverse | GCAGTAATATACCGCGGAGC |
| BRACHYURY | |
| Forward | CCCTATGCTCATCGGAACA |
| Reverse | TTCCAAGGCTGGACCAAT |
| MSX1 | |
| Forward | TCCGCAAACACAAGACGA |
| Reverse | ACTGCTTCTGGCGGAACTT |
| MAP2 | |
| Forward | TGAAGCAAAGGCACCTCAC |
| Reverse | TATGGGAATCCATTGGCG |
| PAX6 | |
| Forward | TTGCTTGGGAAATCCGAG |
| Reverse | TGCCCGTTCAACATCCTT |
| GAPDH | |
| Forward | GGTGGTCTCCTCTGACTTC |
| Reverse | CTCTTCCTCTTGTGCTCTTG |
2.7. Flow cytometry of cell surface markers
Harvested iPSCs were resuspended in 100 uL of FACs buffer (PBS with 2% FBS). Antibodies (BD, USA) against nuclear transcription factor OCT3/4 and the surface markers TRA-1-60, TRA-1-81, and SSEA-4 as described in Table 3 were added in accordance with the manufacturer's instructions. Cells were incubated at 4ºC for 40 min. Permeabilization buffer was added after centrifugation. Cells were centrifuged for 1,500 rpm at 10 min and then resuspended in 100 uL of FACs buffer prior to sorting on the BD FACS Aria II (BD, USA).
Table 3. List of antibodies for flow cytometry.
| Target | Antibody | Volume |
|---|---|---|
| TRA-1-60 | PerCP-Cy™5.5 Mouse Anti-Human TRA-1-60Antigen |
5 uL |
| TRA-1-81 | FITC Mouse Anti-Human TRA-1-81 Antigen |
20 uL |
| SSEA-4 | Alexa Fluor® 647 Mouse anti-SSEA4 | 20 uL |
| OCT3/4 | PE Mouse anti-Oct3/4 | 20 uL |
2.8. Bisulfite promoter sequencing
A DNA Methylation-Direct Kit (EZ, USA) was used to perform bisulfite treatment in accordance with the manufacturer's protocol, and the partial sequence of the promoter region was amplified with nested PCR. The primer sequences are shown in Table 4. The products of amplification with PCR were ligated into a T-vector and cloned into DH5α bacteria. Ten clonal colonies were selected and sequenced.
Table 4. List of primers for bisulfite promoter sequencing.
| Target genes | Primer sequence |
|---|---|
| BSP-OCT4-1 | |
| Forward | AGGTGTGGGAGTGATTTTAGATAGT |
| Reverse | AAACCTTAAAAACTTAACCAAATC |
| BSP-OCT4-2 | |
| Forward | GAGGTTGGAGTAGAAGGATTGTTTTGG |
| Reverse | CCCCCCTAACCCATCACCTCCACCACC |
| BSP-NANOG-1 | |
| Forward | TTGTTGTTTAGGTTGGAGTATAGTGG |
| Reverse | CCTAACGAACACACCCCCTACT |
| BSP-NANOG-2 | |
| Forward | TGGTTAGGTTGGTTTTAAATTTTTG |
| Reverse | AACCCACCCTTATAAATTCTCAATTA |
2.9. Formation of embryoid bodies
Embryoid bodies (EBs) formed once UiPSCs reached 95% confluence. Cells were washed with DMEM/ F12 (BI, Israel) and then PDE1 (Osinglay, China) was applied for 9 min. A fine-tip Pasteur pipette was used to draw grid lines and then collect cells. The cells were resuspended in EB differentiation medium. Cells were plated onto ultra-low attachment plates for 7 d. Medium was replaced every other day. On day 8, EBs were collected and then replated on Matrigel-coated 6-well culture plates for 7 d. Afterwards, total RNA was extracted to detect 3 germ layers using RT-qPCR. The primer sequences are shown in Table 2.
3. Results
3.1. Derivation of PKU-UiPSCs
Integration-free episomal vectors with the OCT4, SOX2, KLF4, and miRNA 302-307 cluster were transfected into PKU urine cells (Figure 1A). The plasmid was added, and the cells began several stages of growth (Figure 1). After first day of electrotransfection, a large number of cells failed to adhere and died. On days 3-5 of electrotransfection, the cells grew stably, and the number of cells increased markedly (Figure 1B). After days 5-7, the medium contained numerous cells. On day 10, embryonic stem cell (ESC) like clones were evident (Figure 1C). On day 21, the clones reached confluence and matured. The clones were separated from surrounding cells with a glass needle and then transferred to a new culture plate (Figure 1D). After 3-5 passages, the differentiated cells gradually decreased until they disappeared (Figures 1E and 1F). Embryonic stem cell-like colonies were stably cultured and passaged.
Figure 1.
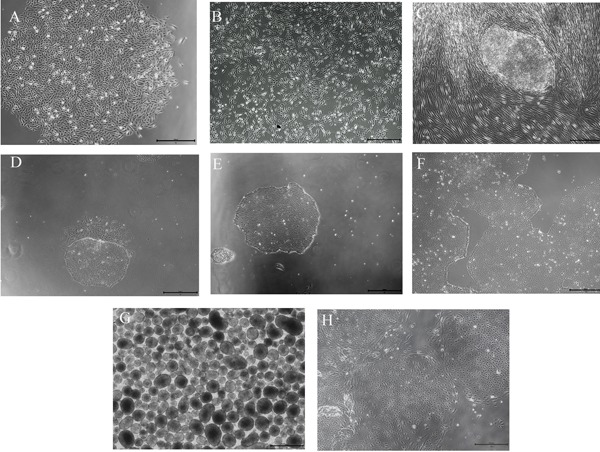
Main types of cell culture (Scale bar = 500 μm). (A) Morphology of urine cells. (B) Urine cells on day 3 after electrotransfection. (C) iPSC colonies. (D) iPSC colonies after transition to feeder-free conditions with differentiated cells. (E) Purified colonies of iPSCs. (F) Stable growth and rapid proliferation of iPSC colonies are evident. (G) EBs are suspended. (H) Change in the morphology of EBs with adherence.
3.2. Verification of the absence of reprogramming vectors
After purification, PKU-UiPSCs displayed typical phenotypes like ESCs. After 10 passages, reprogramming vectors were detected. As shown in Figure 2A, no exogenous genes were amplified in the established PKU-UiPSCs, indicating the integrity of iPSCs after reprogramming.
Figure 2.
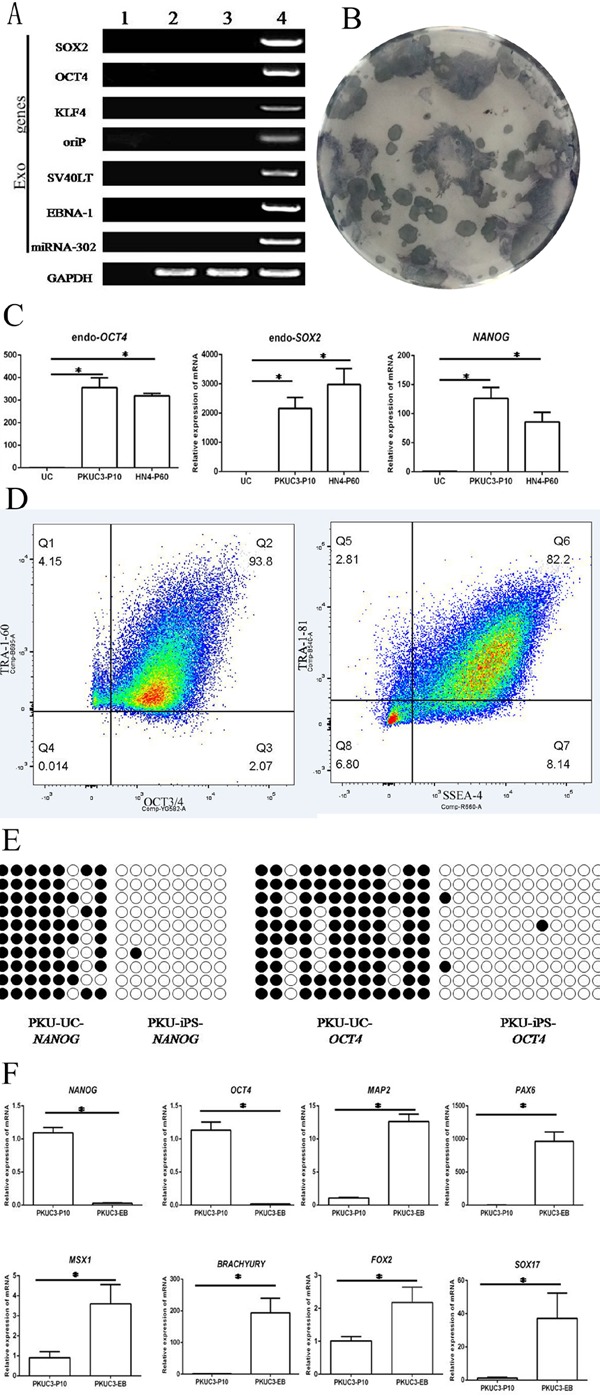
iPSCs from a patient with phenylketonuria are pluripotent. (A) Test for absence of reprogramming vectors. RT-PCR of PKU-UiPSCs after 2 passages in (4) the positive control, urine cells (UCs) in (2) the negative control, (3) PKU-UiPSCs after 12 passages, H2O in (1). (B) Alkaline phosphatase-positive ESC-like colonies (blue) from the PKU UCs (white). (C) Pluripotent UiPSCs (PKUC3-P10) and ESCs (HN4P60) expressed higher levels of the pluripotency markers OCT4, NANOG, and SOX2 in comparison to UCs (p < 0.05). (D) Flow cytometry of the pluripotency markers anti-stage specific embryonic antigen 4 (SSEA-4) (cell surface), anti-Tra-1-60 (cell surface), anti-Oct3/4 (nuclear), and anti- Tra-1-81 (cell surface). (E) Bisulfite genomic sequencing of the key pluripotent gene NANOG and OCT4 promoter area indicated a marked decrease in methylation in UiPSCs. The open and closed circles represent unmethylated and methylated CpGs, respectively. (F) Differentiated cultures of 7-day-old EBs (PKUC3-EB) expressed higher levels of 3 germ layer markers and lower levels of pluripotency markers in comparison to UiPSCs (PKUC3-P10) (p < 0.05).
3.3. ALP staining
ALP was highly expressed in ESCs and iPSCs but slightly expressed or not expressed in differentiated cells. After staining, alkaline phosphatase-positive ESC-like colonies were evident because they stained distinctly blue while PKU urine cells were barely stained (Figure 2B).
3.4. Detection of pluripotency markers
Pluripotency markers (endoOCT4, endoSOX2, and NANOG) were assayed in PKU-UiPSCs (Figure 2C). Expression of endoOCT4 was 400-fold that in PKU urine cells. Expression of endoSOX2 in PKU-UiPSCs was 3,000-fold higher, and expression of NANOG was about 150-fold higher. Expression differed little between PKU-UiPSCs and human ESCs.
3.5. Flow cytometry
Flow cytometry detected all of the markers in PKU-UiPSCs (Figure 2D). OCT3/4 and TRA-1-60 were detected in 93.8% of iPSCs (the Q2 region). In the Q6 region, about 82.2% of cells were positive for SSEA-4 and TRA-1-81.
3.6. Bisulfite promoter sequencing
Bisulfite genomic sequencing indicated that cytosine guanine dinucleotides (CpG) in the promoter regions of OCT4 and NANOG were highly unmethylated in iPSCs in comparison to the urine cells from the same donor (Figure 2E). This indicates that the promoters were reactivated in iPSCs and that the urine cells were reprogrammed by reprogramming factors.
3.7. EB formation
iPSCs formed 3 germ layers in EBs (Figures 1G and 1H). Genetic markers of pluripotency and the 3 germ layers were detected with RT-qPCR (Figure 2F). Using PKU-UiPSCs from the same donor as negative controls, PKU-EBs had about a 5-1,000-fold increase in levels of markers of the endoderm (FOXA2, SOX17), mesoderm (BRACHYURY, MSX1), and ectoderm (MAP2, PAX6), RT-qPCR also revealed a considerable decrease in expression of pluripotency markers (OCT4 and NANOG), indicating that iPSCs were devoted to
developing the 3 germ layers.
4. Discussion
The induction of iPSCs provides a new tool with which to study rare diseases. The current study collected urine from a patient with PKU and it then obtained UiPSCs. Several experiments demonstrated the pluripotency of those PKU-UiPSCs. ALP staining revealed a high level of alkaline phosphatase expression. RT-qPCR and flow cytometry indicated that markers of pluripotency were highly expressed. Methylation sequencing revealed a marked change in methylation of the promoter region of pluripotent markers in UCs and UiPSCs. iPSCs were able to differentiate into all three germ layers in vitro through EB formation. In conclusion, PKU-UiPSCs were generated from urine-derived cells, and in principle the generated cells have the ability to differentiate into specialized cells and tissues.
Significant advances have been made in determining the pathogenesis of many rare diseases, drug screening, and regenerative medicine as a result of studies using patient-specific iPSCs (14-17). In comparison to a previous study that generated PKU-iPSCs from peripheral blood mononuclear cells (PBMCs) (16), UiPSCs in the current study were generated from kidney epithelial cells in urine, and the latter approach has clear advantages (18). First, urine-derived cells can be generated less expensively than cells from other sources such as PBMCs and fibroblasts, and this generation is simple, consistent, and safe for researchers. Second, urine cells can be readily obtained from patients because ample urine is naturally excreted by the body and urine collection poses no burden to the patient. Third, UiPSCs have a high level of reprogramming efficiency (19). In short, UiPSCs are currently the best way to create a bank of PKU-associated cells with different types of gene mutations.
In addition, several studies have found that UiPSCs preferentially differentiate into neurons and that even epithelial-like cells from human urine can directly differentiate into neural progenitor cells (20,21). Although studies have suggested that irreversible nerve damage due to a high concentration of phenylalanine in the blood may be related to brain-derived neurotrophic factor (22,23), no studies have thoroughly examined the mechanism whereby hyperphenylalaninemia results in mental retardation. PKU-UiPSCs provide a useful approach with which to understand the neurological effects of PKU since UiPSCs differentiate into neurons and the ability to create key cell types not directly available from patients with PKU.
Moreover, the ability of iPSCs to differentiate into cells such as osteoblasts (8) and osteoclasts (24) means that PKU-UiPSCs could be highly useful in the study of the biological mechanisms underlying bone impairment in PKU (25,26). Disease models using UiPSCs from patients with PKU offer an unprecedented opportunity to develop new treatments such as enzyme substitution therapy using Phe ammonia lyase (PAL) before conducting clinical experiments (27) and to identify targets in order to correct PAH misfolding or to develop new therapeutic candidates (28).
The current study generated iPSCs from urine-derived cells from a patient with PKU and it verified the pluripotency of those cells. The UiPSC line could be used in advanced research and to provide insight into the clinical variability of the disease, the effects of genetic background, and epigenetic influences in humans.
Acknowledgements
This study was supported by the Innovation Project of the Shandong Academy of Medical Sciences & Key Projects in the National Science & Technology Pillar Program under the Twelfth Five-year Plan (2013BAI07B00).
References
- 1. Al Hafid N, Christodoulou J. Phenylketonuria: A review of current and future treatments. Transl Pediatr. 2015; 4:304-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Blau N, van Spronsen FJ, Levy HL. Phenylketonuria. Lancet. 2010; 376:1417-1427. [DOI] [PubMed] [Google Scholar]
- 3. Andrade F, López-Suárez O, Llarena M, Couce ML, Aldámiz-Echevarría L. Influence of phenylketonuria's diet on dimethylated arginines and methylation cycle. Medicine (Baltimore). 2017; 96:e7392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Evans S, Daly A, Chahal S, Ashmore C, MacDonald J, MacDonald A. The influence of parental food preference and neophobia on children with phenylketonuria (PKU). Mol Genet Metab Rep. 2017; 14:10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Enns GM, Koch R, Brumm V, Blakely E, Suter R, Jurecki E. Suboptimal outcomes in patients with PKU treated early with diet alone: Revisiting the evidence. Mol Genet Metab. 2010; 101:99-109. [DOI] [PubMed] [Google Scholar]
- 6. van Calcar SC, MacLeod EL, Gleason ST, Etzel MR, Clayton MK, Wolff JA, Ney DM. Improved nutritional management of phenylketonuria by using a diet containing glycomacropeptide compared with amino acids. Am J Clin Nutr. 2009; 89:1068-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007; 131:861-872. [DOI] [PubMed] [Google Scholar]
- 8. Matsumoto Y, Hayashi Y, Schlieve C R, et al. Induced pluripotent stem cells from patients with human fibrodysplasia ossificans progressiva show increased mineralization and cartilage formation. Orphanet J Rare Dis. 2013; 8:190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dimos JT1, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, Wichterle H, Henderson CE, Eggan K. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008; 321:1218-1221. [DOI] [PubMed] [Google Scholar]
- 10. Barruet E, Hsiao EC. Using human induced pluripotent stem cells to model skeletal diseases. Methods Mol Biol. 2016; 1353:101-118. [DOI] [PubMed] [Google Scholar]
- 11. Cyranoski D. Stem-cell pioneer banks on future therapies. Nature. 2012; 488:139. [DOI] [PubMed] [Google Scholar]
- 12. Cai J, Orlova VV, Cai X, Eekhoff EM, Zhang K, Pei D, Pan G, Mummery CL, ten Dijke P. Induced pluripotent stem cells to model human fibrodysplasia ossificans progressiva. Stem Cell Reports. 2015; 5:963-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou T, Benda C, Dunzinger S, Huang Y, Ho JC, Yang J, Wang Y, Zhang Y, Zhuang Q, Li Y, Bao X, Tse HF, Grillari J, Grillari-Voglauer R, Pei D, Esteban MA. Generation of human induced pluripotent stem cells from urine samples. Nat Protoc. 2012; 7:2080-2089. [DOI] [PubMed] [Google Scholar]
- 14. Hamasaki M, Hashizume Y, Yamada Y, Katayama T, Hohjoh H, Fusaki N, Nakashima Y, Furuya H, Haga N, Takami Y, Era T. Pathogenic mutation of ALK2 inhibits induced pluripotent stem cell reprogramming and maintenance: Mechanisms of reprogramming and strategy for drug identification. Stem Cells. 2012; 30:2437-2749. [DOI] [PubMed] [Google Scholar]
- 15. Kim BY, Jeong S, Lee SY, Lee SM, Gweon EJ, Ahn H, Kim J, Chung SK. Concurrent progress of reprogramming and gene correction to overcome therapeutic limitation of mutant ALK2-iPSCs. Exp Mol Med. 2016; 48:e237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu T, Liang D, Zhang J, Ji X, Hu H, Sun Y, Jiang T, Wang X, Hu P, Xu Z. Generation of integration-free induced pluripotent stem cell line (NJMUi001-A) from a phenylketonuria patient. Stem Cell Res. 2017; 25:179-182. [DOI] [PubMed] [Google Scholar]
- 17. Fleischer A, Lorenzo IM, Palomino E, Aasen T, Gómez F, Servera M, Asensio VJ, Gálvez V, Izpisúa-Belmonte JC, Bachiller D. Generation of two induced pluripotent stem cell (iPSC) lines from p.F508del cystic fibrosis patients. Stem Cell Res. 2018; 29:1-5. [DOI] [PubMed] [Google Scholar]
- 18. Felix JS, Sun TT, Littlefield JW. Human epithelial cells cultured from urine: Growth properties and keratin staining. In Vitro. 1980; 16:866-874. [DOI] [PubMed] [Google Scholar]
- 19. Liang Shi, Yazhou Cui, Jing Luan, Xiaoyan Zhou, Jinxiang Han. Urine-derived induced pluripotent stem cells as a modeling tool to study rare human diseases. Intractable Rare Dis Res. 2016; 5:192-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kim JB, Greber B, Araúzo-Bravo MJ, Meyer J, Park KI, Zaehres H, Schöler HR. Direct reprogramming of human neural stem cells by OCT4. Nature. 2009; 461:649-653. [DOI] [PubMed] [Google Scholar]
- 21. Shi L, Cui Y, Zhou X, Luan J, Wang L, Han J. Comparative transcriptomic analysis identifies reprogramming and differentiation genes differentially expressed in UiPSCs and ESCs. Biosci Trends. 2017; 11:355-359. [DOI] [PubMed] [Google Scholar]
- 22. Li D, Gu X, Lu L, Liang L. Effects of phenylalanine on the survival and neurite outgrowth of rat cortical neurons in primary cultures: Possible involvement of brain-derived neurotrophic factor. Mol Cell Biochem. 2010; 339:1-7. [DOI] [PubMed] [Google Scholar]
- 23. Lu L, Ben X, Xiao L, Peng M, Zhang YJ. AMP-activated protein kinase activation in mediating phenylalanine-induced neurotoxicity in experimental models of phenylketonuria. J Inherit Metab Dis. 2017. doi: 10.1007/s10545-017-0115-6. [DOI] [PubMed] [Google Scholar]
- 24. Grigoriadis AE, Kennedy M, Bozec A, Brunton F, Stenbeck G, Park IH, Wagner EF, Keller GM. Directed differentiation of hematopoietic precursors and functional osteoclasts from human ES and iPS cells. Blood. 2010; 115:2769-2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roato I, Porta F, Mussa A, D'Amico L, Fiore L, Garelli D, Spada M, Ferracini R. Bone impairment in phenylketonuria is characterized by circulating osteoclast precursors and activated T cell increase. PLoS One. 2010; 5:e14167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Porta F, Roato I, Mussa A, Repici M, Gorassini E, Spada M, Ferracini R. Increased spontaneous osteoclastogenesis from p e r i p h e r a l blood mononuclear c e l l s i n phenylketonuria. J Inherit Metab Dis. 2008; 31 Suppl 2:S339-S342. [DOI] [PubMed] [Google Scholar]
- 27. BabaoğluAydaş S, Şirin S, Aslim B. Biochemical analysis of Centaurea depressa phenylalanine ammonia lyase (PAL) for biotechnological applications in phenylketonuria (PKU). Pharm Biol. 2016; 54:2838-2844. [DOI] [PubMed] [Google Scholar]
- 28. Underhaug J, Aubi O, Martinez A. Phenylalanine hydroxylase misfolding and pharmacological chaperones. Curr Top Med Chem. 2012; 12:2534-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]


