Abstract
Forest trees use various strategies to cope with drought stress and these strategies involve complex molecular mechanisms. Pinus halepensis Miller (Aleppo pine) is found throughout the Mediterranean basin and is one of the most drought-tolerant pine species. In order to decipher the molecular mechanisms that P. halepensis uses to withstand drought, we performed large-scale physiological and transcriptome analyses. We selected a mature tree from a semi-arid area with suboptimal growth conditions for clonal propagation through cuttings. We then used a high-throughput experimental system to continuously monitor whole-plant transpiration rates, stomatal conductance and the vapor pressure deficit. The transcriptomes of plants were examined at six physiological stages: pre-stomatal response, partial stomatal closure, minimum transpiration, post-irrigation, partial recovery and full recovery. At each stage, data from plants exposed to the drought treatment were compared with data collected from well-irrigated control plants. A drought-stressed P. halepensis transcriptome was created using paired-end RNA-seq. In total, ~6000 differentially expressed, non-redundant transcripts were identified between drought-treated and control trees. Cluster analysis has revealed stress-induced down-regulation of transcripts related to photosynthesis, reactive oxygen species (ROS)-scavenging through the ascorbic acid (AsA)-glutathione cycle, fatty acid and cell wall biosynthesis, stomatal activity, and the biosynthesis of flavonoids and terpenoids. Up-regulated processes included chlorophyll degradation, ROS-scavenging through AsA-independent thiol-mediated pathways, abscisic acid response and accumulation of heat shock proteins, thaumatin and exordium. Recovery from drought induced strong transcription of retrotransposons, especially the retrovirus-related transposon Tnt1-94. The drought-related transcriptome illustrates this species’ dynamic response to drought and recovery and unravels novel mechanisms.
Keywords: conifers, drought resistance, next generation sequencing, retrotransposable elements
Introduction
Tree response to water stress is complex at the whole-plant level, as well as at the tissue and cellular levels. To cope with drought conditions, forest trees rely on their genetic toolkit, which has developed over the course of their evolution. Drought-resistance mechanisms vary among tree species and have been shown to differ between angiosperms (broadleaf plants) and gymnosperms (needle-leaf plants) (Niinemets 2001, Choat et al. 2012, Anderegg et al. 2016, Cailleret et al. 2016). The evolutionary distance of 285 million years between conifers and angiosperms (Savard et al. 1994) suggests that these groups of plants may have both common and unique mechanisms for drought resistance. Whereas, molecular responses to drought stress have been extensively studied in broadleaf species, studies of needle-leaf species have been limited.
Response to water deficit begins with reduction in cell growth due to turgor loss, which was found to be accompanied by a high expression level of Histidine Kinase 1 (HK1) in both Arabidopsis and Eucalyptus (Liu et al. 2001, Tran et al. 2007). In order to maintain high turgor, plants make active osmotic adjustments by producing osmolytes such as free amino acids, various ions, soluble sugars and free polyamines, while the free amino acid proline is the most common osmolyte (Krasensky and Jonak 2012). In order to stabilize membranes that are negatively affected by drought stress, plants induce expression of late embryogenesis abundant (LEA) proteins, heat shock proteins (HSP) and dehydrins, all of which may have an important role in the stabilization of membranes (Harfouche et al. 2014). As drought continues, increasing level of the phytohormone abscisic acid (ABA) causes a decrease in stomatal conductance, which is inevitably accompanied by lower rates of CO2 uptake and reduced photosynthesis. Reactive oxygen species (ROS), which are produced following reductions in photosynthesis, damage many cell components but can also act as an alarm signal that triggers the plant’s defense pathways and responses. Reactive oxygen species production evokes one or more of the antioxidant defense systems such as CATALASE (CAT), the ascorbic acid-glutathione (AsA-GSH) pathway and two AsA-independent thiol-mediated pathways, the peroxiredoxin/thioredoxin (Prx/Trx) and the glutathione peroxidase/glutathione S-transferase (GPX/GST) (Noctor et al. 2014). In addition, hormonal changes seem to be involved in all of these reactions. Decrease in cytokinin levels and increased levels of ABA and auxin have been documented in angiosperms and in gymnosperms in response to drought (Peleg and Blumwald 2011, De Diego et al. 2012).
Secondary metabolites, which are known to be induced upon biotic and abiotic stresses, have been shown to be affected by drought stress. Increase in terpenoids levels was documented in Quercus ilex and in Pinus Miller halepensis under drought stress (Blanch et al. 2009). In Eucalyptus, two types of terpenes were decreased in response to drought (McKiernan et al. 2014), however, no alteration was detected in the levels of isoprenoids, mono- and sesquiterpenes (McKiernan et al. 2016). Concentrations of phenolic compounds were found to decrease in response to drought in Eucalyptus (McKiernan et al. 2014). These and other studies suggest that involvement of secondary metabolites in drought response is complex and depends on various parameters such as stress intensity and duration and tree developmental stage (Niinemets 2016).
Another aspect of response to drought stress is epigenetic changes, which refers to modifications affecting gene expression without changing the DNA sequence itself, and mainly include DNA methylation and histone modifications (Rey et al. 2016). Methylation of lysine 9 on histone 3 (H3K9me) have been shown to induce expression of drought-related genes such as RD20, which mediates stomatal closure via ABA response (Aubert et al. 2010, Kim et al. 2015). DNA methylation in Populus trichocarpa under drought stress has also been shown to occur in transposable elements (TE) (Liang et al. 2014). Transposable elements are a principal component of nuclear DNA. Their mobility induces changes in genome structure, thereby inducing changes in gene expression in the majority of eukaryotic organisms (Lippman et al. 2004, Butelli et al. 2012, Mita and Boeke 2016, Negi et al. 2016). The relevant biological significance of TEs in relation to drought response is not fully understood.
Molecular responses are controlled by multiple layers of regulation starting at the DNA level and extending through the RNA and protein-function levels. RNA concentration is the most frequently measured genetic element in most plant species and had been examined in pine species in response to drought. Chang et al. (1996) identified four differentially expressed (DE) genes out of 15,000 in a cDNA library of loblolly pine (Pinus taeda). These included: S-adenosylmethionine synthetase (sams), an intermediate in the synthesis of ethylene; an ABA-inducible gene; a type I copper-containing glycoprotein; and a glycine-rich protein similar to cell wall proteins (Chang et al., 1996). Watkinson et al. (2003) used a cDNA library of 2173 clones and demonstrated differential expression of genes encoding HSP, LEA proteins, aromatic amino acids and flavonoids upon mild drought stress in loblolly pine. Lorenz et al. (2006) succeeded in building a cDNA library of more than 6000 unique transcripts from drought-treated root tissue and identified several genes that had been characterized previously, including dehydrins and LEA gene products. A later publication that utilized an improved microarray of 26,496 cDNA sequences demonstrated 2445 DE genes in response to drought stress in loblolly pine roots (Lorenz et al. 2011). Among the DE genes a number of central nodes were identified including 9-cis-epoxycarotenoid dioxygenase (NCED), which is involved in the biosynthesis of ABA, zeatin O-glucosyltransferase, which regulates the cytokinin homeostasis, and ABA-responsive protein (Lorenz et al. 2011). Microarray analysis of the molecular response to drought stress in Pinus pinaster and Pinus pinea revealed 113 inducible genes. Genes that were common in both species were considered as general candidate genes and included glycosyltransferases, galactosidases, sugar transporters, dehydrins and transcription factors (Perdiguero et al. 2013).
The large size of the genomes of members of the Pinaceae (22–30 Gb) presents challenges, with the result that research in Pinaceae tree genomics has lagged behind that of other systems (Neale and Kremer 2011, Plomion et al. 2011, Prunier et al. 2015). However, the development of next-generation sequencing (NGS) technologies, which began in the last decade, has facilitated Pinaceae genomic research (Neale et al. 2013). Moreover, NGS allows us to investigate the transcriptome (all expressed genes in a given tissue at a given time) of any species even in the absence of any other genetic information (Goodwin et al. 2016). This approach provides expression profiles, gene networks and regulators, which are useful for the dissection of temporal and spatial responses to drought at the molecular level. Next-generation sequencing has been used to uncover the transcriptomic profile of several pine species including P. pinaster, Pinus patula, Pinus densiflora and Pinus tabuliformis (Mackay et al. 2012, Niu et al. 2013, Canales et al. 2014, Liu et al. 2015, Visser et al. 2015). These profiles were generated from various tissues including cones, cambium and needles under normal growth conditions or in response to disease in the case of P. patula. However, NGS tools to study the response to drought stress and recovery in pines have not been extensively used.
Marginal forest populations may contain unique genetic properties that allow for plant survival under otherwise unsuitable conditions (Channell and Lomolino 2000, Holliday et al. 2012). These populations provide a unique opportunity to study tree adaptation to climate change (Fady et al. 2016). A planted forest in a semi-arid region of Israel (annual precipitation of less than 300 mm) was extensively established during the last century with P. halepensis, a species known for its eco-physiological capabilities for drought tolerance (Schiller 2000, Rotenberg and Yakir 2010, Klein et al. 2011, Ungar et al. 2013). Although planted forests have grown normally over the years, the impact of climate change is now evident in increased tree mortality in Israeli forests, similar to that which has been reported for other Mediterranean Pinus spp. plantations (Körner et al. 2005, Dorman et al. 2013, Gazol et al. 2017). The ability of P. halepensis to grow under suboptimal climate conditions makes it an ideal candidate for the study of drought responses in conifers (Maseyk et al. 2008, Klein et al. 2011, David-Schwartz et al. 2016). The molecular response of P. halepensis roots to drought stress was studied by Sathyan et al. (2005). They used a cDNA library of 21,500 clones from the root of P. halepensis and revealed 212 DE clones, of which only 14 clones were further characterized. These included LEA gene and genes that code for a chitinase, a cyclophilin (CYC), sucrose synthase (SUS), an inorganic pyrophosphatase and an MYB transcription factor (TF).
In this study, we aimed to comprehensively characterize the dynamics of the physiological and molecular responses to drought stress in P. halepensis needles and to identify key factors in this response. To that end, we utilized a high-throughput experimental platform to simultaneously and continuously evaluate physiological responses. Six physiological stages were selected for transcriptome analysis: pre-stomatal closure, partial stomatal closure, minimum transpiration, post-irrigation, partial recovery and full recovery. Changes in RNA transcript profiles over the course of the experiment were indicative of the dynamic response of P. halepensis to drought stress and recovery.
Materials and methods
Plant material and clonal propagation
In order to avoid genetic variation and to preserve mature-tree characteristics, we selected one tree for clonal propagation. To ensure that the genotype to be studied would possess drought-adaptation mechanisms, we selected a 20-year-old tree with a relatively large diameter at breast height from the Yatir Forest in Israel, which is a semi-arid forest characterized by suboptimal conditions (270 mm of annual precipitation and an aridity index of 0.18). Shoots were harvested in December 2013 and were used for clonal propagation through rooted cuttings, as described by J. Riov (personal communication). In short, 15-cm twigs that each possessed a vegetative shoot apex were collected from the upper part of the tree and stored at 4 °C in a humid environment. After 4 weeks, the twigs were cut and their lowermost part was immersed in rooting solution [5 ppm 2-(2,4-dichlorophenoxy) propionic acid-glycine conjugate, 400 ppm indole-3-butiric acid (IBA), 0.025% Amistar fungicide (Syngenta)] for 4 h. The twigs were set in a closed rooting bed that contained 1:1 vermiculite:styrofoam white beads. They were kept at 25 °C and every 10 min fogging was applied for 10 s. Rooted cuttings were obtained after 6–8 weeks and moved into separate small pots. A total of 10 rooted clones were grown for 18 months before being subjected to the drought experiment. At the beginning of the drought experiment, the average height of the clones was 31.65 ± 3.2 cm and their average weight was 167 ± 7.73 g. The rooting procedure was conducted on additional three genotypes that were included in the experiment for validation purposes (see Figure S1 available as Supplementary Data at Tree Physiology Online).
Drought experiment
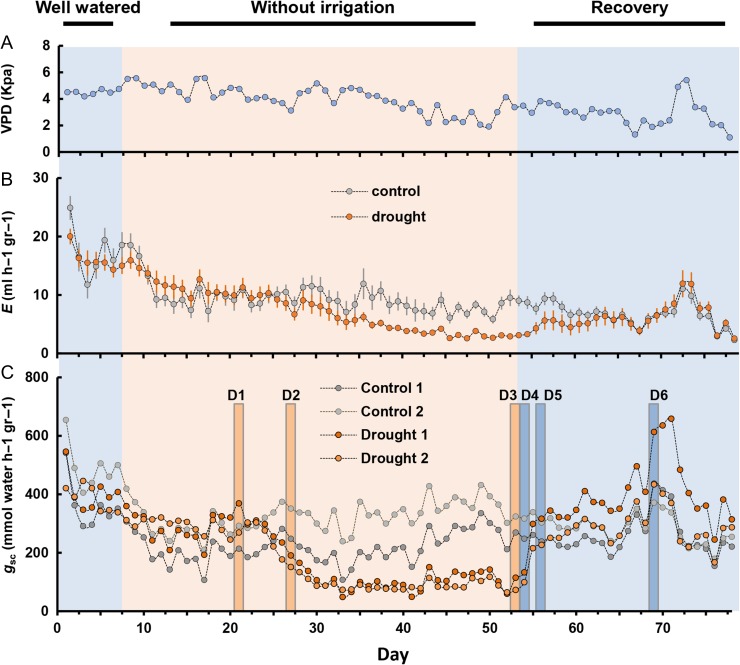
The physiological experiment was carried out for 79 days during which time five clones were subjected to the drought treatment. That treatment consisted of stopping irrigation at Day 7 for 34 days. At the same time, five control clones were regularly watered (Figure 1). Plants were watered every day at 4:00 a.m. to field capacity. Physiological parameters were monitored using a high-throughput system of lysimeters (temperature-compensated load cells) set up in a greenhouse at the Faculty of Agriculture, Rehovot, Israel (Attia et al. 2015, Halperin et al. 2016). The plants were grown in 3.8-l pots in a greenhouse with semi-controlled conditions from late August through early November 2015. The soil surface was covered with white plastic to prevent water loss from soil. The weight of each pot was monitored every 30 s and an average read for each 3-min period was logged with a Campbell Scientific CR1000 Data Logger (Campbell Scientific, Logan, UT, USA). Simultaneously, temperature, ambient radiance and the vapor pressure deficit (VPD) in the greenhouse were continuously recorded. At the end of the experiment, all of the needles were removed from each tree and their weight (at full turgor) was used to normalize the observed transpiration rate. Logged data were used to calculate the whole-plant transpiration rate (E) and the stomatal conductance of the whole canopy (gsc) (Attia et al. 2015, Halperin et al. 2016). Midday E and gsc were calculated as the average values between 1:00 and 3:00 p.m. Well-irrigated trees were used as a control throughout the experiment.
Figure 1.
Whole-plant transpiration rate and canopy stomatal conductance of P. halepensis in response to drought and during recovery. (A) Vapor pressure deficit (VPD, blue circles) over the course of the experiment. (B) Midday transpiration rate (E) was determined for control (gray) and drought-treated trees (orange). Data are means ± SE (n = 5). When not visible, SE is smaller than the symbol. (C) Whole-canopy stomatal conductance (gsc) of the four clones selected for the transcriptome analysis; two control plants (light gray and dark gray) and two drought-treated plants (light and dark orange). D1–D6, selected days over the course of the experiment. D1, pre-stomatal response; D2, partial stomatal closure; D3, minimum transpiration; D4, post-irrigation; D5, partial recovery; D6, full recovery.
Analysis of physiological data
The whole-plant transpiration rate (E) was normalized to the total needle fresh weight (ml h−1 g−1). Whole-canopy stomatal conductance (gsc; mmol s−1 g−1) was calculated using Eq. (1), in which Patm is the atmospheric pressure (101.3 kPa).
| (1) |
The VPD is the difference (in KPa) between the vapor pressure of the saturated air and the vapor pressure of the ambient air. In plants, this refers to the difference between the pressure in the substomatal cavities and the atmospheric pressure.
| (2) |
In Eq. (2), T is the air temperature (°C), RH is relative humidity (0–1), 0.611 is the saturation vapor pressure at 0 °C and 17.502 and 240.97 are constants (Buck 1981). The temperature and relative humidity in the greenhouse were monitored using sensors (HC2-S3-L; Rotronic, Bassersdorf, Switzerland).
RNA extraction, library preparation and sequencing
To minimize the effect of any circadian rhythm, mature needles (50–100 mg) were collected in liquid nitrogen every 3 days between 7:45 and 8:45 a.m. Based on physiological data (see Results), two drought-treated and two control clones were selected for transcriptome analysis at six physiological stages. Samples of the post-irrigation stage were collected from the drought-treated trees only (4 h after re-watering) and were compared with samples of the control trees that were collected at the minimum transpiration stage, which were collected a day before. Altogether, there were 22 samples. Total RNA was extracted using Spectrum™ Plant Total RNA Kit (SIGMA, St Louis, MO, USA) following the manufacturer’s instructions. RNA quantity was determined using a NanoDrop 1000 spectrophotometer and RNA quality was analyzed using the 2200 TapeStation System (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer’s protocol. All RNA samples presented 260/280 and 260/230 purity and an RNA Integrity Number (RIN) >7 in the 2200 TapeStation System measurements. Library preparation and sequencing were done by the Genomics Unit at the Grand Israel National Center for Personalized Medicine (G-INCPM) at the Weizmann Institute of Science (Rehovot, Israel). Libraries were prepared using the INCPM-RNA-seq. Briefly, the polyA fraction (mRNA) was purified from 500 ng of total RNA following by fragmentation and the generation of double-stranded cDNA. Then, end repair, A base addition, adapter ligation and PCR amplification steps were performed. Libraries were evaluated by Qubit and TapeStation. Sequencing libraries were constructed with barcodes to allow the multiplexing of 10 samples in one lane. Between 16.5 and 20 million paired-end 125 reads and between 18.4 and 20.6 million single-end 60-bp reads were sequenced per sample (see Results) using an Illumina HiSeq 2500 V4 instrument. The number of reads was similar for all samples.
Transcriptome analysis: mapping and assembly
The collected raw short-reads were subjected to a filtering and cleaning procedure. The SortMeRNA tool was used to filter out rRNA (Kopylova et al. 2012). Then, the FASTX Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/index.html, version 0.0.13.2) was used to trim read-end nucleotides with quality scores <30, using the fastq_quality_trimmer, and to remove reads with less than 70% base pairs with a quality score ≤30 using the Fastq_quality_filter. Due to low read quality, one control sample at partial stomatal closure (D2) and one drought-treated sample at partial recovery stage (D5) were eliminated from further analysis. The Bowtie2 version 2.1 alignment tool was used to map cleaned reads on the previously published transcriptome of P. halepensis (Pinosio et al. 2014).
Reads not aligned with the reference P. halepensis transcriptome (~140 M reads) were assembled de novo using Trinity software (version: v2.1.1; Grabherr et al. 2011) with the trimmomatic option to remove adaptors (Bolger et al. 2014) and 25 mer k-mer size. The assembled contigs were used as query terms in a BLASTn search against the pita IU genome, to extract only contigs for which a hit was found in that genome (with at least 50% identity over 100 aa overlaps and an E-value threshold of 10–5). CD-HIT (http://www.bioinformatics.org/cd-hit/) with 97% global sequence identity was used to cluster the sequences of both transcriptomes (the de novo assembled transcriptome and the published P. halepensis transcriptome) in order to remove any redundancy.
Sequence similarity and functional annotation
The merged transcriptome was used as a query term for a search of the NCBI non-redundant (nr) protein database that was carried out with the DIAMOND program (Buchfink et al. 2015). Homologous sequences were also identified by searching the Swiss-Prot database with the BLASTx tool (Altschul et al. 1990) and an E-value threshold of 10–5. The search results were imported into Blast2GO version 4.0 (Conesa et al. 2005) for gene ontology (GO) assignments. Enzyme codes and KEGG pathway annotations were based on the mapping of GO terms to their enzyme codes. The KAAS tool (Kegg Automatic Annotation Server; http://www.genome.jp/tools/kaas/) was used for KEGG Orthology and KEGG pathway assignments. Gene ontology enrichment analysis was carried out using Blast2GO (Conesa et al. 2005) program based on Fisher’s Exact Test (Upton 1992) with multiple testing correction of false discovery rate (FDR; Benjamini and Hocberg 1995). Threshold was set as FDR with corrected P-value of less than 0.05. Gene ontology analysis was done by comparing the GO terms in the test sample to the GO terms in a background reference. Differentially expressed transcripts of three physiological stages (D2–D4) were displayed on diagrams of regulation overview map using MapMan (Usadel et al. 2009).
Abundance estimation and differential expression analysis
The transcript quantification (the number of reads per gene) from the RNA-Seq data was performed using the Bowtie2 aligner (Langmead et al. 2009) and the Expectation-Maximization method (RSEM), by estimating maximum likelihood expression levels (Li and Dewey 2011) via the perl script align_and_estimate_abundance.pl with --est_method RSEM from Trinity protocol (Haas et al. 2013). Differential expression analysis was done using the edgeR R package (Robinson et al. 2010). Each time point was tested separately using the perl script run_DE_analysis.pl from Trinity protocol (Haas et al. 2013). Transcripts that varied from the control plants more than twofold, with an adjusted P-value of no more than 0.05 at least at one physiological stage, were considered differentially expressed (Benjamini and Hocberg 1995). Cluster analysis of the DE transcripts based on the fold-change value, was conducted using Expander 7 software (Ulitsky et al. 2010) with the K-means algorithm (Shamir et al. 2005). The number of clusters was set to 30. Venn diagrams were generated using the online tool at bioinformatics.psb.ugent.be/webtools/Venn/.
Results
Physiological response to drought stress and recovery
We evaluated the physiological response of plants that were grown under changing watering regimes. Initially the plants were well-irrigated for 7 days, followed by suspended irrigation for 46 days to impose increasing drought stress after which irrigation was resumed. Two main parameters were monitored to determine drought severity: E at midday and gsc at midday. The drought treatment induced an intense reduction in midday E and gsc at Day 26 in comparison with control plants, 19 days after irrigation was stopped (Figure 1). Although the E in Figure 1B is the average of five replicates, and the gsc in Figure 1C is of individual plants, the pattern of E matched the pattern of gsc throughout the experiment and both of these parameters were correlated with daily changes in VPD (Figure 1). As the drought persisted, the gsc of the drought-treated trees gradually decreased from 319 at Day 21 to 171 at Day 26, and dropped to 94 mmol s−1 g−1 toward the end of the drought period (Figure 1C). After irrigation was resumed, the E and gsc of the drought-treated trees were restored back to the levels recorded prior to the drought treatment, indicating that the drought treatment was not terminal (Negin and Moshelion 2016). Two drought-treated and two irrigated (control) trees were selected for molecular study at six different physiological stages based on the gsc. The stages selected for transcriptome profiling (Figure 1C) were as follows: (D1) pre-stomatal response at Day 21 with 319 gsc; (D2) partial stomatal closure at Day 27 with 171 gsc; (D3) minimum transpiration at Day 53 with 94 gsc; (D4) post-irrigation at Day 54 with 116 gsc; (D5) partial recovery at Day 56 with 271 gsc; and (D6) full recovery at Day 69 with gsc of 523 mmol s−1 g−1.
Transcriptome sequencing and annotation
Twenty-two cDNA libraries yielded approximately 186.5 million 125-bp paired-end reads and 243.8 million 60-bp single-end reads (see Table S1 available as Supplementary Data at Tree Physiology Online). Quality trimming and filtration yielded 410.6 million cleaned reads that were mapped to the reference P. halepensis transcriptome. Unaligned reads (143.5 million) were assembled using Trinity and merged with the P. halepensis transcriptome (see Materials and methods) to generate 98,091 contigs for the transcriptome catalog. The average contig length was 906.47 bp, with an N50 size of 1269 bp corresponding to a total length of 88.8 Mb.
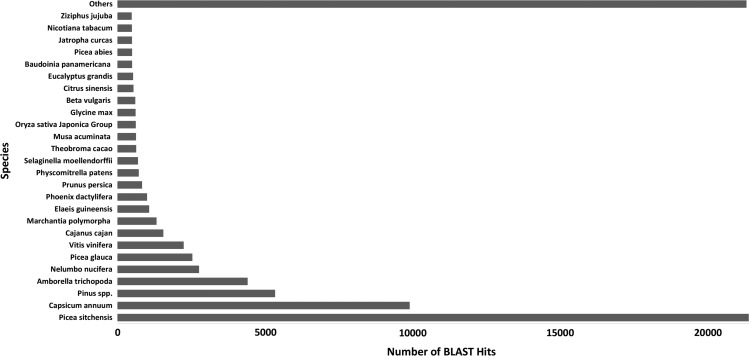
Annotating the transcriptome catalog by aligning the contigs to the NCBI non-redundant (nr) protein database resulted in 76,145 out of 98,091 contigs (77.6%) with at least one DIAMOND hit to a protein. About a quarter of these contigs (26.8%) matched sequences from the genomes of Picea sitchensis, followed by 12.3% matching with Capsicum annuum and 5.5% match with Amborella trichopoda. Pinus taeda was the fourth best hit (1.9%), while the other 97 pine species showed a low number of hits, including 39 hits with P. halepensis. Following aggregation of all hits that matched with pine species, Pinus spp. was the third best hit (6.6%, Figure 2). Blast2GO assigned GO terms to 67,266 contigs (88.3%). The GO terms were mapped to their enzyme code equivalents, which were assigned to 14,371 contigs associated with 144 different KEGG pathways (see Table S2 available as Supplementary Data at Tree Physiology Online).
Figure 2.
Protein BLAST top-hit species distribution for the P. halepensis transcriptome. To identify homologous proteins, the merged transcriptome of P. halepensis proteins was compared with the non-redundant (nr) database using Blast2GO with an E-value threshold of 10–5.
Identification of differentially expressed transcripts
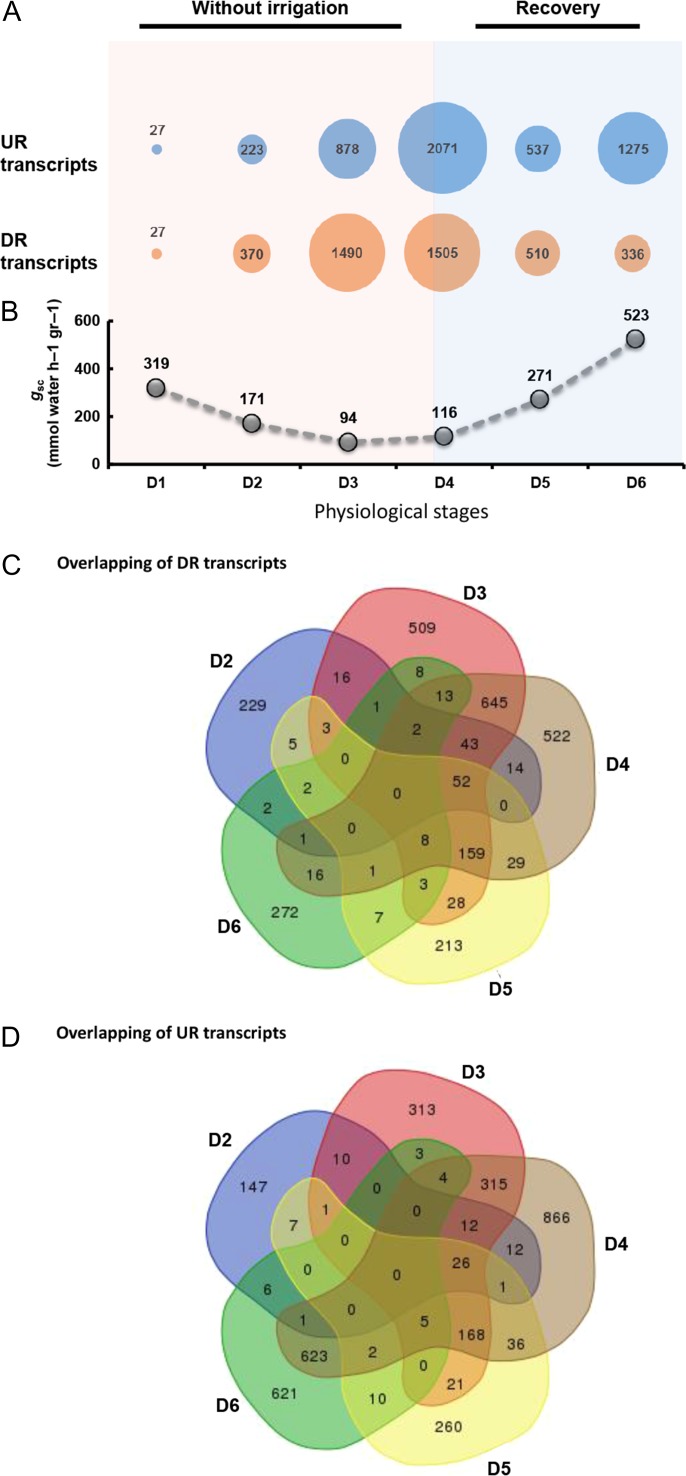
Gene-expression profiling of drought-treated and control trees at the six above-mentioned physiological stages allowed us to analyze transcripts that are differentially expressed between drought-treated and control plants (Figure 3). In general, the number of DE transcripts increased gradually as the drought progressed and decreased gradually during recovery. Transcript down-regulation was dominant in response to drought, while transcript up-regulation was dominant following re-watering and throughout the recovery period (Figure 3). Only 27 up-regulated and 27 down-regulated transcripts were identified among the drought-treated and control clones at Stage D1 (pre-stomatal response, Figure 1C). In agreement with the physiological data collected at Stage D2, at which point a reduction in gsc was noted (Figure 3B), 223 transcripts were up-regulated while 370 transcripts were down-regulated. At Stage D3, at the time of the lowest observed transpiration rate and gsc (Figures 1B and 3B), the up-regulation of 878 transcripts and down-regulation of 1490 transcripts was noted. Several hours after re-watering, at Stage D4, 2071 transcripts were up-regulated while 1505 transcripts were down-regulated. These numbers were reduced to 537 up-regulated transcripts and 510 down-regulated transcripts at Stage D5 (partial recovery). The number of up-regulated transcripts increased dramatically to 1275 at Stage D6 (full recovery), while the number of down-regulated transcripts had declined to 336 by that point (Figure 3A). Altogether, the P. halepensis transcriptome was found to contain 6035 DE transcripts, of which 2466 were previously reported (Pinosio et al. 2014) and 3567 were de novo assembled. The complete transcriptome data set is presented in Table S2 available as Supplementary Data at Tree Physiology Online. The transcriptome includes 1035 (17%) contigs without annotation and 650 (10.1%) contigs related to retrotransposable elements. In order to identify transcripts common among the different physiological stages, two Venn diagrams were generated separately for the up-regulated and for the down-regulated transcripts at Stages D2–D6 (Figure 3C and D; see Table S3 available as Supplementary Data at Tree Physiology Online). The highest overlap of the down-regulated transcripts was between Stages D3 and D4, which showed >50% common transcripts suggesting gradual recovery after re-watering. The highest overlap of the up-regulated transcripts was between D4 and D6, with a majority of them being retrotransposons. The second highest overlap of the up-regulated transcripts was between D3 and D4, suggesting a gradual recovery after re-watering.
Figure 3.
Differentially expressed (DE) transcripts in response to drought and recovery. (A) The total number of transcripts that were up-regulated (blue) and down-regulated (orange) in response to drought over the course of the experiment. Circle size represents the number of DE transcripts, as compared with the control at each time point (D1–D6). UR, up-regulated; DR, down-regulated. (B) Whole-canopy stomatal conductance (gsc, gray circles) at six physiological stages, D1–D6. D1, pre-stomatal response; D2, partial stomatal closure; D3, minimum transpiration; D4, post-irrigation; D5, partial recovery; D6, full recovery. Venn diagram analysis of down-regulated (C) and up-regulated (D) transcripts at D2–D6 sampling time points.
Transcriptome profiling prior to stomatal response
In order to untangle genes that might be upstream of any stomatal response, we first analyzed the transcriptome of Stage D1 and found 27 up-regulated and 27 down-regulated transcripts at that stage (Figures 1C and 3A). The up-regulated transcripts are associated with pathways and processes including oxidative defense (catalase and ferredoxin thioredoxin reductase), transcriptional repression (PLATZ transcription factor (TF)) and the biosynthesis of phenylalanine, tyrosine and tryptophan (arogenate dehydrogenase and shikimate dehydrogenase), and also include a spliceosome-related sequence (SART-1) and membrane-related components (synaptobrevin, ADP-ribosylation factor GTPase-activating protein 1). In addition, heat shock proteins 70 and 90 (HSP70 and HSP90) and gamma-aminobutyrate (GABA) transaminase 1 accumulated significantly.
The transcripts down-regulated at this stage are associated with processes including photosynthesis (photosystem II D1), chlorophyll biosynthesis (magnesium-chelatase subunit chloroplastic), flavonoid biosynthesis (chalcone synthase) and oxidation-reduction (S-norcoclaurine synthase 1-like, peroxidases and sphingolipid desaturase), and also include transcripts related to membranes (glycerol-3-phosphate transporter 4, transcriptional corepressor SEUSS-like, transmembrane 33 homolog and peroxisomal membrane 22 kDa family isoform 1) and RNA-related processing (zinc finger CCCH domain-contain, threonine tRNA mitochondrial and RNase_T,zf-GRF).
Gene ontology enrichment analysis of differentially expressed transcripts
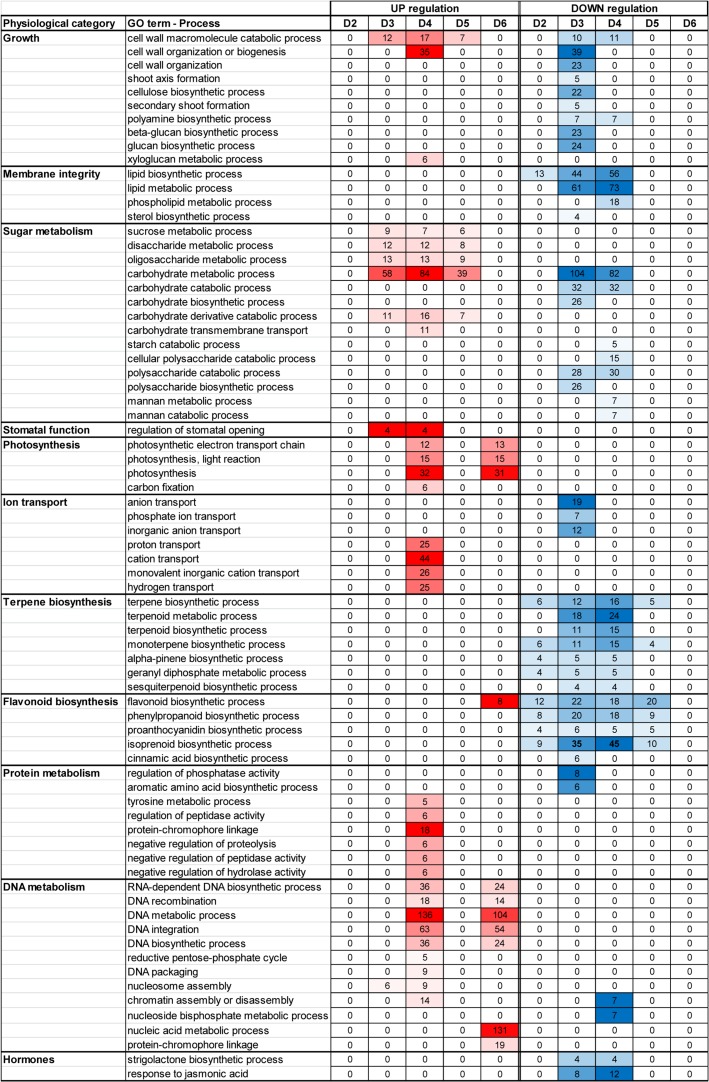
In order to decipher the major biological processes that are affected by the drought stress, we have performed a GO enrichment analysis of up-regulated as well as of down-regulated DE transcripts at each of the five physiological stages, D2–D6 (see Table S4 available as Supplementary Data at Tree Physiology Online). Based on the GO enrichment results, we have generated a Heatmap that reflects the dynamic response of different biological processes at 10 drought-related physiological categories. These categories included growth, membrane integrity, sugar metabolism, stomatal function, photosynthesis, ion transport, terpene biosynthesis, flavonoid biosynthesis, protein metabolism, DNA metabolism and hormones (Figure 4). Processes that promote growth were mainly enriched in the down-regulated transcripts at D3, while the cell wall organization process was enriched at D4 after re-watering. Lipid biosynthetic process was enriched at D2 down-regulated transcripts, and was further enriched at D3 and D4 suggesting reduced membrane formation in response to drought. Disaccharide and oligosaccharide metabolic processes were enriched in the up-regulated transcripts at D3–D5, while carbohydrate metabolic processes were enriched in the up-regulated as well as in the down-regulated transcripts at D3 and D4. Catabolic processes of starch, polysaccharide and mannan were enriched in the down-regulated transcripts at D4, suggesting decline in degradation processes. Transcripts related to stomatal opening were enriched in the up-regulated transcripts at D3 and D4. Photosynthesis related processes were enriched in the up-regulated transcripts only, at D4 after re-watering and at D6 at full recovery. Processes related to anion transport were enriched in the down-regulated transcripts at D3, while processes related to cation transport were enriched in the up-regulated transcripts at D4. The majority of processes related to terpene and flavonoid biosynthesis were commonly enriched in the down-regulated transcripts at D2–D5. Protein metabolism-related processes including regulation of phosphatase activity and aromatic amino acid biosynthetic process were enriched in the down-regulated transcripts at D3, while processes related to negative regulation of protein degradation were enriched at D4, suggesting decline in protein degradation after re-watering. Processes related to DNA metabolism were enriched mainly in the up-regulated transcripts at D4 and D6, while chromatin-related processes were enriched in the down-regulated transcripts at D4. Hormone-related processes showed enrichment for strigolactone biosynthetic process and response to jasmonic acid in the down-regulated transcripts at D3 and D4.
Figure 4.
Heatmap diagram reflecting the dynamics of enriched biological processes in drought-related physiological categories over the course of the experiment as obtained from GO enrichment analysis (see Table S4 available as Supplementary Data at Tree Physiology Online). Enriched processes with FDR less than 0.05 were included in the heatmap. Red and blue represent positive and negative change, respectively, and the intensity of the color reflects the number of transcripts as indicated at each physiological stage (D2–D6) of each processes.
Cluster analysis of differentially expressed transcripts
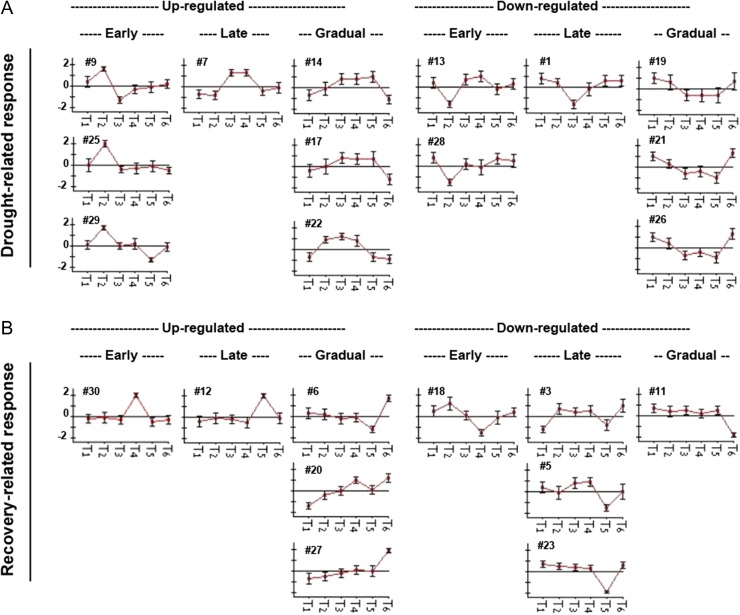
In an effort to further understand the dynamic of molecular response under drought stress and during recovery, the 6035 DE transcripts were grouped into 30 clusters based on their expression trends over the course of the experiment (seeFigure S2 available as Supplementary Data at Tree Physiology Online). Twenty-three clusters were selected based on their expression trends and categorized as ‘drought-related’ or ‘recovery-related’. We further categorized the drought-related or recovery-related as ‘early’, ‘late’ or ‘gradually’ responsive transcripts (Figure 5). Below, we describe the main biological processes and related transcripts that are involved in the drought- and recovery-related responses.
Figure 5.
Cluster analysis of DE transcripts over the course of the experiment. The transcripts were grouped based on their patterns of expression during the drought and recovery periods. (A) Drought-related response clusters were subdivided into early, late and gradual responses. (B) Recovery-related response clusters were subdivided into early, late and gradual responses. T1–T6 on the x-axis of each cluster correspond to D1–D6. Numbers correspond to cluster number as was obtained by cluster analysis. Clusters were identified by K-means clustering.
Early response to drought includes transcripts that were induced or reduced at partial stomatal closure stage, D2. Up-regulated transcripts at this stage were obtained from Clusters 9, 25 and 29, and include transcripts related to processes such as chlorophyll degradation (chlorophyllase), protein degradation (E3 ubiquitin-ligases and proteases), ABA signaling (aspartic protease, phosphatase 2 C (PP2C) and calcium-dependent kinase), DNA repair and a membrane-related transcripts (thaumatin). The analysis of early down-regulated transcripts included data obtained from Clusters 13 and 28. The transcripts of these clusters include transcripts related to cellulose and lignin synthesis, DNA replication (DNA replication licensing factor MCM6), cell division (mitogen-activated kinase homolog MMK1), chloroplast/photosynthesis components, sugar metabolism and stomatal movement (serine threonine-kinase HT1).
Late response to drought included transcripts that were found to be significantly changed at Stage D3. Transcripts that were significantly up-regulated at D3 were grouped together in Cluster 7 and include transcripts related to ABA signaling (PP2C 25 and PP2C 37), protein degradation (E3 ubiquitin-ligase RGLG1), hydrogen peroxide generation (20 kDa chloroplastic-positive regulator of superoxide dismutase), carbohydrate metabolism (sucrose synthase) and a chloroplast proteases (Do-like protease).
Transcripts that are down-regulated at Stage D3 were grouped together in Cluster 1 and include transcripts that are involved in protein degradation (ubiquitin carboxyl-terminal hydrolase and E3 SUMO ligase SIZ1), sugar transport (sugar transporter ERD6), response to auxin (auxin-induced 15 A), DNA replication (DNA replication licensing factor MCM4), DNA repair (CISIN), oxidation-reduction (ferredoxin-NADP+ reductase) and protein degradation (Do-like 9).
The analysis of transcripts involved in gradual responses to drought included transcripts whose expression gradually increased or decreased over the course of the drought treatment, as compared with the control. This pattern is similar to the stomata behavior as measured by the gsc and therefore, some of the processes are expected to correlate with stomatal conductance. Gradually up-regulated transcripts were grouped in Clusters 14, 17 and 22, and are up-regulated mostly at Stages D2–D5. This group includes transcripts related to ROS scavenging (superoxide dismutase), detoxification of ROS via AsA-independent thiol-mediated pathways (glutathione peroxidases, glutaredoxin and thioredoxin), carbon fixation (ribulose-bisphosphate carboxylase units), sucrose degradation (sucrose synthase), chlorophyll degradation (chlorophyllases), ethylene response (ethylene-responsive transcription factors), stomatal movement (RING finger and CHY zinc finger protein1) and ABA signaling (serine threonine-kinase SRK2E and PP2C 37, ABI5). This category also includes up-regulated transcripts related to aromatic amino acid biosynthetic processes (3-phosphoshikimate 1-carboxyvinyltransferase 2 and phenylalanine hydroxylases), lipid hydrolysis (lipase-like PAD4), DNA binding (lysine-specific histone demethylase 1 homolog 3) and auxin responses (auxin aluminum responsive, auxin down-regulated partial and auxin-repressed). In addition, HSPs, inositol transporters, dehydrins and LEA transcripts were also up-regulated. Increased expression of sucrose synthases and thaumatin, as well as phenylalanine ammonia-lyase was negatively correlated with stomatal conductance.
Gradually down-regulated transcripts were grouped in Clusters 19, 21 and 26, which include transcripts that are down-regulated mainly at Stages D3–D5. The down-regulated transcripts are related to the synthesis of terpenoid backbone (geranylgeranyl diphosphate synthase), mono- and diterpenoids, flavonoids (chalcone isomerase (CHI) and dihydroflavonol 4-reductase), phenylalanine, tyrosine and tryptophan. We also noted the down-regulation of transcripts related to the AsA-GSH cycle of ROS removal (ascorbate peroxidase and monodehydroascorbate reductases), cytokinin degradation (cytokinin dehydrogenase), methylation (lysine-specific demethylase JMJ706-like isoform x1), starch degradation (isoamylases, alpha and beta amylases), cellulose metabolism (cellulase and cellulose synthases), cell division (leucine-rich repeat receptor-like serine threonine-kinase BAM1), sugar transport (sugar transporter ERD6), stomatal activity (SLAC), chlorophyll-related proteins and ABA signaling (PYL2 and PYL8). Decreased expression of chalcone synthases and dihydroflavonol 4-reductase was positively correlated with stomatal conductance.
Recovery-related early responses were measured 4 h following re-watering. Early responsive transcripts were grouped in Clusters 30 and 18, which include transcripts that are up- or down-regulated at Stage D4, respectively. Cluster 30 includes transcripts associated with oxidation-reduction processes (peroxidases), ABA catabolism (ABA 8-hydroxylase), auxin flux (auxin transporter 1), fatty-acid biosynthesis (lipoxygenases), membranes and the organization of the microtubule cytoskeleton and cell wall-related transcripts (expansin). Cluster 18 includes down-regulated transcripts associated with RNA processing, cytokinin-activated signaling (histidine-containing phosphotransfer 1) and terpene synthesis (pinene synthase).
Late responsive transcripts were those transcripts for which significant up- or down-regulation was noted at Stage D5, at which point gsc had partially recovered (Figure 1C). This category includes Cluster 12, which consists of transcripts that were up-regulated at Stage D5 and Clusters 3, 5 and 23, which together include transcripts that were down-regulated at Stage D5. The up-regulated transcripts include three different peroxidases and transcripts associated with sucrose metabolism (sucrose-phosphate synthase), photosynthetic acclimation (K+ efflux antiporter chloroplastic protein), the ABA-activated signaling pathway (calcium-dependent kinase 17), stomatal movement (serine threonine-kinase HT1), cell division and DNA repair and sugar transport (SWEET1). The down-regulated transcripts include transcripts that are associated with water transport (aquaporin, aquaporin TIP4-3), cellulose synthesis (cellulose synthases) and polar auxin transport (auxin transport protein BIG).
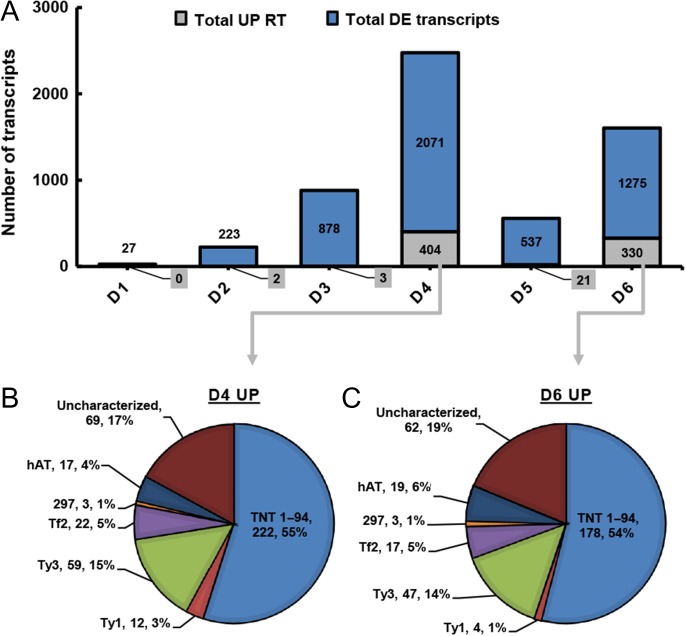
Clusters 6, 20 and 27 include transcripts that were up-regulated at Stage D6, at which point we observed fully recovered gsc (Figures 1C and 5). Cluster 20 is dominated by transcripts that are up-regulated at Stages D4 and D6. This cluster includes 1405 transcripts, of which 646 transcripts are without annotation. This cluster also includes 489 transcripts that are associated with retrotransposons. These induced retrotransposons belong to families HAT, Ty1, Ty3 and Tf2 (Figure 6). However, the most dominant retrotransposon, with 243 related transcripts, was Tnt1-94 (Figure 6B and C). Among the remaining 275 transcripts of this cluster, 8 are TFs, 44 are associated with photosynthesis, 22 peroxidase-64 transcripts, 18 ribonuclease (RNase) H transcripts and 10 endoribonuclease dicer transcripts. A similar pattern of 28 retrotransposable elements was seen in Clusters 6 and 27, which together include 130 transcripts.
Figure 6.
Expression pattern of retrotransposons in P. halepensis during drought and recovery. (A) The number of retrotransposons (RT, gray) that were up-regulated out of the total number of up-regulated DE transcripts (blue). D1–D6 represent six physiological stages: D1, pre-stomatal response; D2, partial stomatal closure; D3, minimum transpiration; D4, post-irrigation; D5, partial recovery; and D6, full recovery. (B and C) Pie charts showing the distribution of DE retrotransposons by family at D4 (B) and D6 (C). Each description includes a name, number of transcripts and percentage.
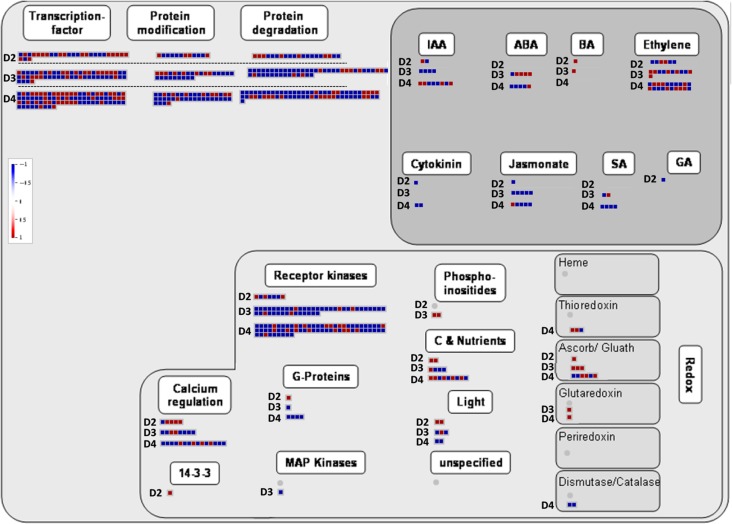
In addition to the cluster analysis, a MapMan regulation overview map analysis was performed on Stages D2–D4 (Figure 7). Transcripts related to the hormones auxin, cytokinin, jasmonic acid and salicylic acid are down-regulated upon drought stress, while ABA and brassinosteroids related transcripts are up-regulated. Auxin seemed to be induced upon re-watering. For ethylene response, the picture seemed more complexed. The regulation overview map also suggests that Redox-related transcripts belong to the Prx/Trx and GPX/GST pathways are activated upon drought stress.
Figure 7.
MapMan regulation overview map showing transcript level changes in partial stomatal closure (D2), minimum transpiration (D3) and post-irrigation (D4) stages. In color scale, blue represents lower transcript level, while red represents higher transcript level in comparison with control plants. IAA, indole-3-acetic acid (auxin); ABA, abscisic acid; BA, brassinosteroids; SA, salicylic acid; GA, gibberellic acid; Ascorb/Gluath, glutathione peroxidase.
Finally, to validate some of the RNA-seq results, a quantitative real-time polymerase chain reaction (RT-PCR) analysis was conducted on four individuals from the major genotype and two more individuals from each of three additional genotypes (seeFigure S1 available as Supplementary Data at Tree Physiology Online). Genes in the RT-PCR analysis included pinene synthase, monofunctional diterpene synthase, chalcone synthase, anthocyanidin reductase, thioredoxin and sugar transporter SWEET3, all of which showed results in agreement with the RNA-seq (see Figure S3 available as Supplementary Data at Tree Physiology Online).
Discussion
This study describes the molecular responses that accompany the physiological response of pine needles to drought stress. The reduction in E was as expected from physiological response to drought stress in this experiment. Selected plants for the molecular analysis also showed reduction in gsc in response to drought. The association between the E and gsc that was observed in this study (Figure 1) is similar to those reported for peach (Prunus persica) and grapevine (Vitis vinifera), in which correlations between soil water content and stem water potential have been documented (Mata et al. 1999, Williams et al. 2012). Whole-canopy stomatal conductance (gsc) has previously been found to correlate with stem water potential in deciduous and evergreen broadleaf tree species (Zhang et al. 2013). Based on that previous finding, in the current study, gsc was used as a stress indicator for the molecular-response analysis.
The transcriptome catalog that was obtained in this study comprehensively represents drought responsive genes in P. halepensis. The protein BLAST top-hit species distribution showed that a quarter of the contigs were matched to P. sitchensis in agreement to previous reports for P. densiflora and Ginkgo biloba transcriptome annotations (Han et al. 2015, Liu et al. 2015). This result was probably obtained due to high proportion of high quality P. sitchensis protein sequences in the NCBI nr protein database relative to pine species (Ralph et al. 2008). Annotating the previous P. halepensis transcriptome by Pinosio et al. (2014) resulted in Glycine max as the major hit. Similarly, in the current study, the second best match was to C. annuum, probably due to improved representation of its proteins at the current database. As most pine species are poorly represented in the database, only Pinus spp. showed a meaningful match of 6.6% (Figure 2).
Drought-related genes
Currently, hundreds of genes have been identified in relation to drought response, while specific drought-related function has been identified for only a portion of them. The challenge is to identify genes that can be used in breeding programs and biotechnological approaches aimed at improving drought resistance, especially among forest tree species, which are completely dependent on environmental conditions. The genotype that was used in this study originated from a semi-arid area with suboptimal growth conditions and thus represents drought-resistance capacity. Although this single genotype does not represent the species genetic variation, it contributes comprehensive information regarding new candidate genes for drought resistance in P. halepensis.
The examination of the transcriptome at Stage D1 enabled us to identify molecular responses before any changes in stomatal conductance could be noted and to elucidate the hierarchy of responses to drought stress in pine. The limited number of DE transcripts at Stage D1 includes the strongly down-regulated photosystem II (PSII) protein D1 (psbA), which is a reaction-center-core protein responsible for electron transport that is an integral part of photosynthesis (Lu 2016). This down-regulation impairs PSII function, leading to reduced CO2 fixation and increased ROS production, which in turn restricts D1 protein synthesis and further damages the routine repair of PSII (Takahashi and Murata 2008, Nishiyama et al. 2011, Noctor et al. 2014). Strong expression of the ROS-scavenging enzymes catalase (CAT) and ferredoxin thioredoxin reductase (FTRC) at this stage is probably induced to mitigate this damage (Nishiyama et al. 2011). Alternatively, this mechanism might also be activated to generate ROS signaling, which promotes the activation of downstream pathways (Noctor et al. 2014, Thomas et al. 2016). Up-regulated transcripts at Stage D1 also included HSP70 and HSP90, which have been shown to modulate transcriptional and physiological responses to ABA and are required for stomatal closure in Arabidopsis (Clément et al. 2011). In agreement with that report, in this work, these genes seem to be upstream of the ABA-induced stomatal closure. PLATZ TF, which was also observed at this stage, has been shown to suppress transcription via non-specific binding to A/T-rich sequences (Nagano et al. 2001). In the current experiment, PLATZ TF might have been partially responsible for the massive down-regulation of transcripts at Stages D2 and D3.
Another up-regulated gene is GABA transaminase, which has been suggested to be involved in the maintenance of mitochondrial GABA pools that are associated with energy production via the tricarboxylic acid (TCA) cycle, to maintain normal cellular metabolism (Fait et al. 2008). GABA accumulation has been reported in Pinus radiata subjected to drought stress and may function as a long-term carbon and nitrogen reserve that facilitates recovery and confers a priming effect against future stress (De Diego et al. 2013).
The major flavonoid biosynthesis-related transcript, chalcone synthase, is down-regulated at Stage D1, preceding massive down-regulation of flavonoid biosynthesis as drought continues (Figure 4). In contrast, up-regulation of transcripts involved in phospholipid biosynthesis and lipid transport, as noted at Stage D1, suggests membrane enrichment toward stabilization and the prevention of membrane leakage. Phospholipids are central components of cell membranes and a decrease in phospholipid levels upon drought stress leads to membrane leakage (Lin et al. 2015). Drought might also lead to membrane shrinkage and turgor loss, which is known to induce the expression of HK1, which may act as an osmosensor (Tran et al. 2007). Two putative homologs of HK1 were identified in our study. However, neither of them was differentially expressed over the course of the experiment (data not shown).
The genes described above are probably some of the upstream regulators that sense the very initial stages of the drought stress and promote further responses to prevent tissue damage and the loss of water from cells. Therefore, each of these genes is a potential candidate for breeding programs and biotechnological approaches towards drought-resistant plants. Stages D2 and D3 of drought stress and Stages D4, D5 and D6 of recovery are associated with many more DE transcripts that reflect various processes that may or may not be connected to one another. Through our GO enrichment and cluster analyses of gene-expression patterns (Figures 4 and 5), we were able to illustrate the dynamic of these processes over the course of drought stress and recovery. In the current study, each physiological stage was shown to be characterized by differential expression of various TFs, which are key regulators of the activation or repression of DE transcripts (Singh et al. 2002, Saibo et al. 2009, Lindemose et al. 2013). Expression of various TFs was similar to what was reported for other species, such the up-regulation of nuclear transcription factor Y subunit B in poplar (Cohen et al. 2010). However, other annotated members belonging to the same gene families have been demonstrated to have opposite expression pattern (see Table S2 available as Supplementary Data at Tree Physiology Online). This observation is due to the NGS technique, which is not biased towards a specific member of a gene family, but rather detects the whole gene range of expressed genes, thus better reflecting the complex regulation of the response to drought. Determining the specific DE transcripts that are affected by each of these TFs is beyond the scope of the current study.
The drought transcriptome that was generated in this study includes a high proportion (17%) of transcripts without annotation, which apparently restricts its potential to reflect the complete toolkit of drought-resistance mechanisms in P. halepensis.
Molecular response at partial stomatal closure includes ROS production, which is induced under drought stress due to the interruption of metabolic processes. Reactive oxygen species play an important role in signaling, but excess ROS are targeted by various mechanisms to prevent cell damage (Kapoor et al. 2015). The conversion of O2– is catalyzed by superoxide dismutase to produce H2O2, while various pathways reduce H2O2 to H2O. These pathways include CAT, the AsA-GSH pathway and two AsA-independent thiol-mediated pathways, the Prx/Trx and the GPX/GST (Noctor et al. 2014). CAT, which is mainly expressed in the peroxisome, is induced early in drought and is down-regulated during recovery (see Supplementary Table S2 available as Supplementary Data at Tree Physiology Online). The AsA-GSH pathway is induced in many species to scavenge ROS under drought stress (Sofo et al. 2005, Cruz de Carvalho 2008, Caverzan et al. 2012, Wang et al. 2016), while other species activate the AsA-independent thiol-mediated pathways in response to drought (Cruz de Carvalho 2008, Kapoor et al. 2015). This study shows that under drought conditions, P. halepensis activates genes related to the Prx/Trx and GPX/GST pathways (Figure 7), while expression levels of genes from the AsA-GSH pathway are reduced (see Table S2 available as Supplementary Data at Tree Physiology Online).
Stomatal closure upon drought stress is known to be induced by the phytohormone ABA (Tardieu and Davies 1992). The major ABA biosynthetic gene NCED (9-cis-epoxycarotenoid dioxygenase), which has been shown to be expressed in response to drought in many species, was not found to be differentially expressed in the current experiment. This might be due to the sampling time, which may have been too late or too early during Stage D2. Alternatively, NCED might be up-regulated and cause ABA to be synthesized in a different organ (such as the root) and then transferred to leaves through the transpiration stream. Another possible explanation for the absence of NCED transcripts is that active ABA is accumulated through the hydrolysis of ABA-GE (glucose-ester), which might be predominant in needles. Few transcripts that were annotated as beta-glucosidases (which releases glucose from ABA-GE) were up-regulated at Stages D2 and D3. However, the actual role of these transcripts in ABA-GE hydrolysis should be examined.
Indications for ABA signaling in the current study come from ABA-responsive transcripts such as the TF ABI5, which is up-regulated at Stage D3, and from the up-regulation of two transcripts of SnRK2 at Stages D3 and D4. There were fewer DE PYL8 homologs at the point at which the minimum transpiration rate was reached (Stage D3), but only one PYL8 homolog was up-regulated at that stage. The PYL8 receptor has been shown to regulate root growth in response to drought in Arabidopsis (Dupeux et al. 2011, Zhao et al. 2014, Xing et al. 2016). A report of poplar response to drought stress demonstrated down-regulation of PYL expression (Cohen et al. 2010); however, not much information is available regarding ABA receptors in trees in response to drought. The fact that PYL8 was the active ABA receptor at Stage D3 suggests that it may play a role in ABA signaling in needles.
In addition, PP2C genes that are known to inhibit ABA signaling by blocking SnRK2 activity were differentially expressed in the current study. While some were down-regulated, others were up-regulated. Although most of the PP2C genes have been shown to act as negative regulators of ABA signaling, some have been shown to be positive regulators (Reyes et al. 2006). Interestingly, E3 ubiquitin-ligase RGLG1, which is up-regulated at Stage D3, has been recently shown, together with RGLG5, to promote ABA signaling by mediating PP2C degradation (Wu et al. 2016). It would be interesting to investigate the effects of RGLG1 on the various PP2C transcripts that were differentially expressed in this study. Abscisic acid degradation following re-watering is probably mediated by ABA 8-hydroxylase, whose transcript levels increased at Stage D4.
The cluster analysis implies that ABA accumulates in P. halepensis needles following drought stress. In P. radiata, foliar ABA was reported to accumulate gradually as drought stress progresses and to remain high as long as the stomata remain closed (Brodribb and McAdam 2013). Upon rehydration, stomatal conductance increased after 48–72 h, that is, only after ABA levels fell to basal levels (Brodribb and McAdam 2013). This type of ABA dynamic is common in pine species that exhibit an R-type (rising) ABA response and an ABA-dependent stomatal closure response (Brodribb et al. 2014). The gradual up-regulation during drought and slow down-regulation during recovery of the CHY ZINC-FINGER AND RING FINGER PROTEIN1 (CHYR1) gene support the slow response of P. halepensis stomata, since this gene has been shown to be essential for ABA-mediated stomatal closure (Ding et al. 2015). In contrast, SLAC1, which is also involved in stomatal closure (Vahisalu et al. 2008), was down-regulated during the drought treatment, demonstrating the complexity of the ABA response over long periods of exposure to drought. Nonetheless, the similarity in stomatal behavior of this species with that observed in P. radiata and with the accompanying ABA-related transcription patterns suggest that the ABA response in P. halepensis is of the R-type.
In general, levels of transcripts related to the biosynthesis of flavonoids were gradually reduced significantly over the drought period and were gradually up-regulated after re-watering, reaching normal levels only at Stage D6 (Figure 4). It has been suggested that flavonoids protect plants against abiotic stress and their accumulation is also induced rapidly upon drought (Liu et al. 2013, Nakabayashi et al. 2014). However, decreased expression of CHI in rice (Oryza sativa) and tea (Camellia sinensis) under drought stress has been documented (Pandey et al. 2010, Rani et al. 2012). A similar pattern of down-regulation was observed for transcripts associated with terpene biosynthesis (Figure 4). Terpenes benefit the plant under conditions of biotic stress. As opposed to the current study, drought stress has been shown to induce an increase in volatile terpenes in Q. ilex and P. halepensis (Blanch et al. 2009). However, a recent paper demonstrated a dramatic decrease in terpene emission in P. halepensis under severe drought conditions in the Yatir forest (Llusia et al. 2016). This discrepancy should be investigated; however, down-regulation of terpenes might explain the high sensitivity of drought-stressed trees to biotic stress. The pattern of down-regulation observed in our study may suggest that the plant limits the energy-intensive biosynthesis of secondary metabolites in favor of drought-related survival mechanisms.
Other genes, such as thaumatin, were differentially expressed in the current experiment in response to drought. Thaumatin-like proteins (TLPs) have been shown to be involved in responses to biotic and abiotic stresses such as salinity, cold, osmotic stress and drought (Piggott et al. 2004, Munis et al. 2010, Ahmed et al. 2013, Wang et al. 2013). Ten TLPs have been identified in western white pine (Pinus monticola) and shown to play a role in responses to pathogens (Liu et al. 2010). Here, we show that TLPs are differentially expressed in P. halepensis in response to drought stress. Thaumatin shares sequence homology with osmotin and is known to play a role in responses to osmotic and drought stress (Singh et al. 1987, Misra et al. 2016). As it is an integral component of membranes, we suggest that aside from its role upon pathogen attack, thaumatin might also be involved in preventing membrane damage.
Recovery-related genes
Processes related to growth, photosynthesis, ion transport and biosynthesis of proteins and DNA were all induced following re-watering (Figure 4). Cation transporters, such as K+ efflux antiporter chloroplastic protein, which is known to increase photosynthetic efficiency in response to light stress (Armbruster et al. 2014), were dominantly induced during recovery, suggesting accelerated photosynthetic activity (Figure 4). The most striking result was the strong transcription of retrotransposons that was observed during the recovery from drought (at Stages D4 and D6, Figure 6). Expressed retrotransposons included Ty1/Copia, Ty3/Gypsy, 297 and members of hAT families. The Copia element Tnt1–94 was by far the most dominant retrotransposon at Stages D4 and D6 and accounted for more than 10% of the total up-regulated genes at those stages (Figure 6A and B). It was recently reported that retrotransposons (mainly Ty1/Copia and Ty3/Gypsy elements) account for 62% of the 23-Gb P. taeda genome (Wegrzyn et al. 2013, Neale et al. 2014). Retrotransposons from Ty1/Copia, Ty3/Gypsy, hAT, 297 and Tf2 families have been characterized previously in many plant species (Rubin et al. 2001). Tnt1 retrotransposons were first identified in tobacco (Nicotiana tabacum) and are widely used to induce mutagenesis in plants (Grandbastien et al. 1989, D’Erfurth et al. 2003). The involvement of transposable elements in stress responses was previously suggested in the Solanaceae (Grandbastien et al. 2005). A recent study demonstrated differential expression of retrotransposons in Pinus sylvestris in response to heat stress, pine woolly aphids and treatment with salicylic acid and ABA (Voronova et al. 2014). A specific response of Tnt1 to biotic and abiotic factors was reported in tobacco (Mhiri et al. 1997, Melayah et al. 2001, Hernández-Pinzón et al. 2009). Sequence variation in the 3′ untranslated region of Tnt1 has been shown to confer a specific response to various stress inducers in tobacco. While methyl jasmonate induces transcription of the Tnt1 A subfamily, salicylic acid and auxin activate the Tnt1 C subfamily (Beguiristain et al. 2001). Tnt1 has been shown to be integrated into gene-rich regions and, therefore, it has been suggested to contribute to genetic variation (Hernández-Pinzón et al. 2009). It has been shown that epigenetic mechanisms suppress retrotransposon activity upon heat stress in Arabidopsis (Ito et al. 2011). Epigenetic activity was noted for several genes in the current study, including those controlling the methylation of the transcriptional repressor H3K9me and the transcriptional activator H3K4me (Xiao et al.2016). The lysine-specific demethylase JMJ706-like isoform x1 (see Table S2 available as Supplementary Data at Tree Physiology Online), which decreases at D3, specifically reverses di- and trimethylation of H3K9 (Sun and Zhou 2008). In a complementary manner, the H3K9-specific methyltransferase SUVH1 increases at D3, suggesting an increase in H3K9me, which would lead to transcriptional suppression of the affected genes under drought. At the stage of post-irrigation, a dramatic decrease of the specific H3K9 methyltransferase SUVR5 (Caro et al. 2012) is observed, suggesting reduction in H3K9me and hence transcriptional activation. Transcriptional suppression during drought stress is also evident in the dramatic increase (~130-fold; see Table S2 available as Supplementary Data at Tree Physiology Online) in the expression of the lysine-specific histone demethylase 1 homolog 3, which specifically demethylates H3K4me, the positive regulator of gene expression. In the current study, we assume that genes regulated by H3K4me were strongly suppressed at Stages D2 and D3. However, those genes are probably reactivated during recovery, as the reduction in the expression of the lysine-specific-histone demethylase 1 homolog 3 at Stage D5 was dramatic (~130-fold; see Table S2 available as Supplementary Data at Tree Physiology Online). It was recently suggested that H3K4me may be related to acclimation or ‘stress memory’ in plants (Kim et al. 2012). In Arabidopsis, increased levels of H3K4me3 during recovery from drought indicated a form of transcriptional memory that persists for several days and results in higher expression levels upon repeated drought stress (Ding et al. 2012). The differential expression of these methylation-related transcripts implies that there is epigenetic regulation of gene expression during drought stress in P. halepensis that might be partially related to transposable elements activation. A growing body of evidence suggests that transposable elements may play a significant role in responses to environmental stimuli (Casacuberta and González 2013). It is not yet clear whether the retrotransposons found in this study are active. However, we speculate that the potential activity of retrotransposons during recovery from drought stress might increase tissue genetic variation, thereby improving the plant’s ability to adapt to future stress that may be imposed by the changing environment.
Conclusions
Climate change, which is leading to increased mean temperatures and is expected to decrease annual precipitation levels in the Eastern Mediterranean region, is also characterized by large inter-annual variations in rainfall, which are becoming increasingly common (Giorgi and Lionello 2008). Adaptation of trees to drought and the ability to efficiently utilize sporadic rain events are crucial for forest tree survival. The available genomic tools allow the elucidation of molecular responses of drought-adaptive species for which we have no prior genomic information. The current study demonstrates this approach and exposes mechanisms that otherwise would not have been exposed. In addition, the large number of transcripts without annotation noted in this study underscores the untapped potential for the identification of novel functional genes in non-model species.
Supplementary Material
Acknowledgments
We thank Prof. Joseph Riov and Dr Einat Sadot for their valuable help with the rooting technique and Robert Sitbun, Guy Halperin and Gil Lerner for their technical help. We thank Shlomit Gilad and Sima Benjamin from the Genomics Unit at the Grand Israel National Center for Personalized Medicine (G-INCPM) for the calibration of the library-preparation method.
Supplementary Data
Supplementary Data for this article are available at Tree Physiology Online.
Conflict of interest
None declared.
Funding
This work was made possible by financial support from the Israeli Forest Service (Jewish National Fund-KKL). H.F. acknowledges financial support from the Rene Karshon Foundation and Appleby Foundation.
References
- Ahmed NU, Park JI, Jung HJ et al. (2013) Molecular characterization of thaumatin family genes related to stresses in Brassica rapa. Sci Hortic 152:26–34. [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. [DOI] [PubMed] [Google Scholar]
- Anderegg WRL, Klein T, Bartlett M et al. (2016) Meta-analysis reveals that hydraulic traits explain cross-species patterns of drought-induced tree mortality across the globe. Proc Natl Acad Sci USA 113:5024–5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armbruster U, Carrillo LR, Venema K et al. (2014) Ion antiport accelerates photosynthetic acclimation in fluctuating light environments. Nat Commun 5:5439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Attia Z, Domec JC, Oren R, Way DA, Moshelion M (2015) Growth and physiological responses of isohydric and anisohydric poplars to drought. J Exp Bot 66:4373–4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert Y, Vile D, Pervent M, Aldon D, Ranty B, Simonneau T, Vavasseur A, Galaud J-P (2010) RD20, a stress-inducible caleosin, participates in stomatal control, transpiration and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol 51:1975–1987. [DOI] [PubMed] [Google Scholar]
- Beguiristain T, Grandbastien MA, Puigdomènech P, Casacuberta JM (2001) Three Tnt1 subfamilies show different stress-associated patterns of expression in tobacco. Consequences for retrotransposon control and evolution in plants. Plant Physiol 127:212–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hocberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc 57:289–300. [Google Scholar]
- Blanch JS, Peñuelas J, Sardans J, Llusià J (2009) Drought, warming and soil fertilization effects on leaf volatile terpene concentrations in Pinus halepensis and Quercus ilex. Acta Physiol Plant 31:207–218. [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina Sequence Data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodribb TJ, McAdam SA (2013) Abscisic acid mediates a divergence in the drought response of two conifers. Plant Physiol 162:1370–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodribb TJ, McAdam SAM, Jordan GJ, Martins SCV (2014) Conifer species adapt to low-rainfall climates by following one of two divergent pathways. Proc Natl Acad Sci USA 111:14489–14493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH (2015) Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. [DOI] [PubMed] [Google Scholar]
- Buck AL. (1981) New equations for computing vapor pressure and enhancement factor. J Appl Meteorol 20:1527–1532. [Google Scholar]
- Butelli E, Licciardello C, Zhang Y et al. (2012) Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell. 3:1242–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cailleret M, Jansen S, Robert EM et al. (2016) A synthesis of radial growth patterns preceding tree mortality. Glob Chang Biol 23:1675–1690. [DOI] [PubMed] [Google Scholar]
- Canales J, Bautista R, Label P et al. (2014) De novo assembly of maritime pine transcriptome: implications for forest breeding and biotechnology. Plant Biotechnol J 12:286–299. [DOI] [PubMed] [Google Scholar]
- Casacuberta E, González J (2013) The impact of transposable elements in environmental adaptation. Mol Ecol 22:1503–1517. [DOI] [PubMed] [Google Scholar]
- Caro E, Stroud H, Greenberg MVC, Bernatavichute YV, Feng S, Groth M, Vashisht AA, Wohlschlegel J, Jacobsen SE (2012) The SET-domain protein SUVR5 mediates H3K9me2 deposition and silencing at stimulus response genes in a DNA methylation-independent manner. PLoS Genet 8:e1002995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caverzan A, Passaia G, Rosa SB, Ribeiro CW, Lazzarotto F, Margis-Pinheiro M (2012) Plant responses to stresses: Role of ascorbate peroxidase in the antioxidant protection. Genet Mol Biol 35:1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SJ, Puryear JD, Dias M, Funkhouser EA, Newton RJ, Cairney J (1996) Gene expression under water deficit in loblolly pine (Pinus taeda): isolation and characterization of cDNA clones. Physiol Plant 97:139–148. [Google Scholar]
- Channell R, Lomolino MV (2000) Dynamic biogeography and conservation of endangered species. Nature 403:84–86. [DOI] [PubMed] [Google Scholar]
- Choat B, Jansen S, Brodribb TJ et al. (2012) Global convergence in the vulnerability of forests to drought. Nature 491:752–756. [DOI] [PubMed] [Google Scholar]
- Clément M, Leonhardt N, Droillard MJ et al. (2011) The cytosolic/nuclear HSC70 and HSP90 molecular chaperones are important for stomatal closure and modulate abscisic acid-dependent physiological responses in Arabidopsis. Plant Physiol 156:1481–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen D, Bogeat-Triboulot MB, Tisserant E, Balzergue S, Martin-Magniette ML, Lelandais G (2010) Comparative transcriptomics of drought responses in Populus: a meta-analysis of genome-wide expression profiling in mature leaves and root apices across two genotypes. BMC Genomics 11:630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. [DOI] [PubMed] [Google Scholar]
- Cruz de Carvalho MH. (2008) Drought stress and reactive oxygen species. Plant Signal Behav 3:156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David-Schwartz R, Paudel I, Mizrachi M et al. (2016) Indirect evidence for genetic differentiation in vulnerability to embolism in Pinus halepensis. Front Plant Sci 7:768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Diego N, Pérez-Alfocea F, Cantero E, Lacuesta M, Moncaleán P (2012) Physiological response to drought in radiata pine: phytohormone implication at leaf level. Tree Physiol 32:435–449. [DOI] [PubMed] [Google Scholar]
- De Diego N, Sampedro MC, Barrio RJ, Saiz-Fernández I, Moncaleán P, Lacuesta M (2013) Solute accumulation and elastic modulus changes in six radiata pine breeds exposed to drought. Tree Physiol 33:69–80. [DOI] [PubMed] [Google Scholar]
- D’Erfurth I, Cosson V, Eschstruth A, Lucas H, Kondorosi A, Ratet P (2003) Efficient transposition of the Tnt1 tobacco retrotransposon in the model legume Medicago truncatula. Plant J 34:95–106. [DOI] [PubMed] [Google Scholar]
- Ding S, Zhang B, Qin F (2015) Arabidopsis RZFP34/CHYR1, a ubiquitin E3 Ligase, regulates stomatal movement and drought tolerance via SnRK2.6-mediated phosphorylation. Plant Cell 27:3228–3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Fromm M, Avramova Z (2012) Multiple exposures to drought ‘train’ transcriptional responses in Arabidopsis. Nat Commun 3:740. [DOI] [PubMed] [Google Scholar]
- Dorman M, Svoray T, Perevolotsky A, Sarris D (2013) Forest performance during two consecutive drought periods: diverging long-term trends and short-term responses along a climatic gradient. For Ecol Manage 310:1–9. [Google Scholar]
- Dupeux F, Santiago J, Betz K et al. (2011) A thermodynamic switch modulates abscisic acid receptor sensitivity. EMBO J 30:4171–4184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fady B, Aravanopoulos FA, Alizoti P et al. (2016) Evolution-based approach needed for the conservation and silviculture of peripheral forest tree populations. For Ecol Manage 375:66–75. [Google Scholar]
- Fait A, Fromm H, Walter D, Galili G, Fernie AR (2008) Highway or byway: the metabolic role of the GABA shunt in plants. Trends Plant Sci 13:14–19. [DOI] [PubMed] [Google Scholar]
- Gazol A, Ribas M, Gutiérrez E, Camarero JJ (2017) Aleppo pine forests from across Spain show drought-induced growth decline and partial recovery. Agric For Meteorol 232:186–194. [Google Scholar]
- Giorgi F, Lionello P (2008) Climate change projections for the Mediterranean region. Glob Planet Change 63:90–104. [Google Scholar]
- Goodwin S, McPherson JD, McCombie WR (2016) Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 17:333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandbastien MA, Spielmann A, Caboche M (1989) Tnt1, a mobile retroviral-like transposable element of tobacco isolated by plant cell genetics. Nature 337:376–380. [DOI] [PubMed] [Google Scholar]
- Grandbastien MA, Audeon C, Bonnivard E et al. (2005) Stress activation and genomic impact of Tnt1 retrotransposons in Solanaceae. Cytogenet Genome Res 110:229–241. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J (2013) De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halperin O, Gebremedhin A, Wallach R, Moshelion M (2016) High-throughput physiological phenotyping and screening system for the characterization of plant–environment interactions. Plant J 89:839–850. [DOI] [PubMed] [Google Scholar]
- Han S, Wu Z, Jin Y, Yang W, Shi H (2015) RNA-Seq analysis for transcriptome assembly, gene identification, and SSR mining in ginkgo (Ginkgo biloba L.). Tree Genet Genomes 11:37. [Google Scholar]
- Harfouche A, Meilan R, Altman A (2014) Molecular and physiological responses to abiotic stress in forest trees and their relevance to tree improvement. Tree Physiol 34:1181–1198. [DOI] [PubMed] [Google Scholar]
- Hernández-Pinzón I, de Jesús E, Santiago N, Casacuberta JM (2009) The frequent transcriptional readthrough of the tobacco Tnt1 retrotransposon and its possible implications for the control of resistance genes. J Mol Evol 68:269–278. [DOI] [PubMed] [Google Scholar]
- Holliday JA, Suren H, Aitken SN (2012) Divergent selection and heterogeneous migration rates across the range of Sitka spruce (Picea sitchensis). Proc Biol Sci 279:1675–1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H, Gaubert H, Bucher E, Mirouze M, Vaillant I, Paszkowski J (2011) An siRNA pathway prevents transgenerational retrotransposition in plants subjected to stress. Nature 472:115–119. [DOI] [PubMed] [Google Scholar]
- Kapoor D, Sharma R, Handa N et al. (2015) Redox homeostasis in plants under abiotic stress: role of electron carriers, energy metabolism mediators and proteinaceous thiols. Front Environ Sci 3:13. [Google Scholar]
- Kim J-M, To TK, Ishida J, Matsui A, Kimura H, Seki M (2012) Transition of chromatin status during the process of recovery from drought stress in Arabidopsis thaliana. Plant Cell Physiol 53:847–856. [DOI] [PubMed] [Google Scholar]
- Kim J-M, Sasaki T, Ueda M, Sako K, Seki M (2015) Chromatin changes in response to drought, salinity, heat, and cold stresses in plants. Front Plant Sci 6:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein T, Cohen S, Yakir D (2011) Hydraulic adjustments underlying drought resistance of Pinus halepensis. Tree Physiol 31:637–648. [DOI] [PubMed] [Google Scholar]
- Kopylova E, Noe L, Touzet H (2012) SortMeRNA: fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28:3211–3217. [DOI] [PubMed] [Google Scholar]
- Körner C, Sarris D, Christodoulakis D (2005) Long-term increase in climatic dryness in the East-Mediterranean as evidenced for the island of Samos. Reg Environ Change 5:27–36. [Google Scholar]
- Krasensky J, Jonak C (2012) Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J Exp Bot 63:1593–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, Zhang Z, Wu H et al. (2014) Single-base-resolution methylomes of Populus trichocarpa reveal the association between DNA methylation and drought stress. BMC Genet 15:S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Yc Liu, Nakamura Y (2015) The choline/ethanolamine kinase family in arabidopsis: essential role of CEK4 in phospholipid biosynthesis and embryo development. Plant Cell 27:1497–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemose S, O’Shea C, Jensen M, Skriver K (2013) Structure, function and networks of transcription factors involved in abiotic stress responses. Int J Mol Sci 14:5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, Gendrel AV, Black M et al. (2004) Role of transposable elements in heterochromatin and epigenetic control. Nature 430:471–476. [DOI] [PubMed] [Google Scholar]
- Liu JJ, Zamani A, Ekramoddoullah AK (2010) Expression profiling of a complex thaumatin-like protein family in western white pine. Planta 231:637–651. [DOI] [PubMed] [Google Scholar]
- Liu L, Zhang S, Lian C (2015) De novo transcriptome sequencing analysis of cDNA library and large-scale unigene assembly in Japanese Red Pine (Pinus densiflora). Int J Mol Sci 16:26139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Li X, Liu Y, Cao B (2013) Regulation of flavanone 3-hydroxylase gene involved in the flavonoid biosynthesis pathway in response to UV-B radiation and drought stress in the desert plant, Reaumuria soongorica. Plant Physiol Biochem 73:161–167. [DOI] [PubMed] [Google Scholar]
- Liu W, Fairbairn DJ, Reid RJ, Schachtman DP (2001) Characterization of two HKT1 homologues from Eucalyptus camaldulensis that display intrinsic osmosensing capability. Plant Physiol 127:283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llusia J, Roahtyn S, Yakir D, Rotenberg E, Seco R, Guenther A, Peñuelas J (2016) Photosynthesis, stomatal conductance and terpene emission response to water availability in dry and mesic Mediterranean forests. Trees 30:749–759. [Google Scholar]
- Lorenz WW, Sun F, Liang C et al. (2006) Water stress-responsive genes in loblolly pine (Pinus taeda) roots identified by analyses of expressed sequence tag libraries. Tree Physiol 26:1–16. [DOI] [PubMed] [Google Scholar]
- Lorenz WW, Alba R, Yu YS, Bordeaux JM, Simões M, Dean JF (2011) Microarray analysis and scale-free gene networks identify candidate regulators in drought-stressed roots of loblolly pine (P. taeda L.). BMC Genomics 12:264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. (2016) Identification and roles of photosystem II assembly, stability, and repair factors in Arabidopsis. Front Plant Sci 7:168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay J, Dean JFD, Plomion C et al. (2012) Towards decoding the conifer giga-genome. Plant Mol Biol 80:555–569. [DOI] [PubMed] [Google Scholar]
- Maseyk KS, Lin T, Rotenberg E, Grünzweig JM, Schwartz A, Yakir D (2008) Physiology–phenology interactions in a productive semi-arid pine forest. New Phytol 178:603–616. [DOI] [PubMed] [Google Scholar]
- Mata M, Girona J, Goldhamer DA, Fereres E, Cohen M, Johnson RS (1999) Water relations of lysimeter-grown peach trees are sensitive to deficit irrigation. Calif Agric 53:17–20. [Google Scholar]
- McKiernan AB, Hovenden MJ, Brodribb TJ, Potts BM, Davies NW, O’Reilly-Wapstra JM (2014) Effect of limited water availability on foliar plant secondary metabolites of two Eucalyptus species. Environ Exp Bot 105:55–64. [Google Scholar]
- McKiernan AB, Potts BM, Brodribb TJ, Hovenden MJ, Davies NW, McAdam SAM, Ross JJ, Rodemann T, O’Reilly-Wapstra JM (2016) Responses to mild water deficit and rewatering differ among secondary metabolites but are similar among provenances within Eucalyptus species. Tree Physiol 36:133–147. [DOI] [PubMed] [Google Scholar]
- Melayah D, Bonnivard E, Chalhoub B, Audeon C, Grandbastien MA (2001) The mobility of the tobacco Tnt1 retrotransposon correlates with its transcriptional activation by fungal factors. Plant J 28:159–168. [DOI] [PubMed] [Google Scholar]
- Mhiri C, Morel JB, Vernhettes S, Casacuberta JM, Lucas H, Grandbastien MA (1997) The promoter of the tobacco Tnt1 retrotransposon is induced by wounding and by abiotic stress. Plant Mol Biol 33:257–266. [DOI] [PubMed] [Google Scholar]
- Misra RC, Sandeep, Kamthan M, Kumar S, Ghosh S (2016) A thaumatin-like protein of Ocimum basilicum confers tolerance to fungal pathogen and abiotic stress in transgenic Arabidopsis. Sci Rep 6:25340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita P, Boeke JD (2016) How retrotransposons shape genome regulation. Curr Opin Genet Dev 37:90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munis MF, Tu L, Deng F et al. (2010) A thaumatin-like protein gene involved in cotton fiber secondary cell wall development enhances resistance against Verticillium dahliae and other stresses in transgenic tobacco. Biochem Biophys Res Commun 393:38–44. [DOI] [PubMed] [Google Scholar]
- Nagano Y, Furuhashi H, Inaba T, Sasaki Y (2001) A novel class of plant-specific zinc-dependent DNA-binding protein that binds to A/T-rich DNA sequences. Nucleic Acids Res 29:4097–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakabayashi R, Mori T, Saito K (2014) Alternation of flavonoid accumulation under drought stress in Arabidopsis thaliana. Plant Signal Behav 9:e29518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale D, Kremer A (2011) Forest tree genomics: growing resources and applications. Nat Rev Genet 12:111–122. [DOI] [PubMed] [Google Scholar]
- Neale D, Langley C, Salzberg S, Wegrzyn J (2013) Open access to tree genomes: the path to a better forest. Genome Biol 14:120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale DB, Wegrzyn JL, Stevens KA et al. (2014) Decoding the massive genome of loblolly pine using haploid DNA and novel assembly strategies. Genome Biol 15:R59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi P, Rai AN, Suprasanna P (2016) Moving through the stressed genome: emerging regulatory roles for transposons in plant stress response. Front Plant Sci 7:1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negin B, Moshelion M (2016) The advantages of functional phenotyping in pre-field screening for drought-tolerant crops. Funct Plant Biol 44:107–118. [DOI] [PubMed] [Google Scholar]
- Niinemets Ü. (2001) Global-scale climatic controls of leaf dry mass per area, density, and thickness in trees and shrubs. Ecology 82:453–469. [Google Scholar]
- Niinemets Ü. (2016) Uncovering the hidden facets of drought stress: secondary metabolites make the difference. Tree Physiol 36:129–132. [DOI] [PubMed] [Google Scholar]
- Nishiyama Y, Allakhverdiev SI, Murata N (2011) Protein synthesis is the primary target of reactive oxygen species in the photoinhibition of photosystem II. Physiol Plant 142:35–46. [DOI] [PubMed] [Google Scholar]
- Niu S-H, Li Z-X, Yuan H-W, Chen X-Y, Li Y, Li W (2013) Transcriptome characterisation of Pinus tabuliformis and evolution of genes in the Pinus phylogeny. BMC Genomics 14:263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Mhamdi A, Foyer CH (2014) The roles of reactive oxygen metabolism in drought: not so cut and dried. Plant Physiol 164:1636–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey A, Rajamani U, Verma J et al. (2010) Identification of extracellular matrix proteins of rice (Oryza sativa L.) involved in dehydration-responsive network: A proteomic approach. J Proteome Res 9:3443–3464. [DOI] [PubMed] [Google Scholar]
- Peleg Z, Blumwald E (2011) Hormone balance and abiotic stress tolerance in crop plants. Curr Opin Plant Biol 14:290–295. [DOI] [PubMed] [Google Scholar]
- Perdiguero P, MdC Barbero, Cervera MT, Collada C, Soto Á (2013) Molecular response to water stress in two contrasting Mediterranean pines (Pinus pinaster and Pinus pinea) Plant Physiol. Biochem 67:199–208. [DOI] [PubMed] [Google Scholar]
- Piggott N, Ekramoddoullah AK, Liu JJ, Yu X (2004) Gene cloning of a thaumatin-like (PR-5) protein of western white pine (Pinus monticola D. Don) and expression studies of members of the PR-5 group. Physiol Mol Plant Pathol 64:1–8. [Google Scholar]
- Pinosio S, Gonzalez-Martinez SC, Bagnoli F et al. (2014) First insights into the transcriptome and development of new genomic tools of a widespread circum-Mediterranean tree species, Pinus halepensis Mill. Mol Ecol Resour 14:846–856. [DOI] [PubMed] [Google Scholar]
- Plomion C, Bousquet J, Kolen CP (2011) Genetics, genomics and breeding of conifers. CRC Press, New York, NY. [Google Scholar]
- Prunier J, Verta JP, MacKay JJ (2015) Conifer genomics and adaptation: at the crossroads of genetic diversity and genome function. New Phytol 209:44–62. [DOI] [PubMed] [Google Scholar]
- Ralph SG, Chun HJ, Kolosova N et al. (2008) A conifer genomics resource of 200,000 spruce (Picea spp.) ESTs and 6,464 high-quality, sequence-finished full-length cDNAs for Sitka spruce (Picea sitchensis). BMC Genomics 9:484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rani A, Singh K, Ahuja PS, Kumar S (2012) Molecular regulation of catechins biosynthesis in tea [Camellia sinensis (L.) O. Kuntze]. Gene 495:205–210. [DOI] [PubMed] [Google Scholar]
- Rey O, Danchin E, Mirouze M, Loot C, Blanchet S (2016) Adaptation to global change: a transposable element–epigenetics perspective. Trends Ecol Evol 31:514–526. [DOI] [PubMed] [Google Scholar]
- Reyes D, Rodríguez D, González-García MP et al. (2006) Overexpression of a protein phosphatase 2C from Beech seeds in Arabidopsis shows phenotypes related to abscisic acid responses and gibberellin biosynthesis. Plant Physiol 141:1414–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotenberg E, Yakir D (2010) Contribution of semi-arid forests to the climate system. Science 327:451–454. [DOI] [PubMed] [Google Scholar]
- Rubin E, Lithwick G, Levy AA (2001) Structure and evolution of the hAT transposon superfamily. Genetics 158:949–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saibo NJM, Lourenco T, Oliveira MM (2009) Transcription factors and regulation of photosynthetic and related metabolism under environmental stresses. Ann Bot 103:609–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathyan P, Newton RJ, Loopstra CA (2005) Genes induced by WDS are differentially expressed in two populations of aleppo pine (Pinus halepensis). Tree Genet Genomes 1:166–173. [Google Scholar]
- Savard L, Li P, Strauss SH, Chase MW, Michaud M, Bousquet J (1994) Chloroplast and nuclear gene sequences indicate late Pennsylvanian time for the last common ancestor of extant seed plants. Proc Natl Acad Sci USA 91:5163–5167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiller G. (2000) Eco-physiology of Pinus halepensis Mill. and Pinus brutia Ten. In: Ne'eman G, Trabaud L (eds) Ecology, biogeography and management of Mediterranean pine forest ecosystems (Pinus halepensis and Pinus brutia). Backhuys Publishers, Amsterdam, pp 51–65. [Google Scholar]
- Shamir R, Maron-Katz A, Tanay A et al. (2005) EXPANDER—an integrative program suite for microarray data analysis. BMC Bioinformatics 6:232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh KB, Foley RC, Onate-Sanchez L (2002) Transcription factors in plant defense and stress responses. Curr Opin Plant Biol 5:430–436. [DOI] [PubMed] [Google Scholar]
- Singh NK, Bracker CA, Hasegawa PM et al. (1987) Characterization of osmotin: a thaumatin-like protein associated with osmotic adaptation in plant cells. Plant Physiol 85:529–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofo A, Dichio B, Xiloyannis C, Masia A (2005) Antioxidant defences in olive trees during drought stress: changes in activity of some antioxidant enzymes. Funct Plant Biol 32:45–53. [DOI] [PubMed] [Google Scholar]
- Sun Q, Zhou D-X (2008) Rice jmjC domain-containing gene JMJ706 encodes H3K9 demethylase required for floral organ development. Proc Natl Acad Sci USA 105:13679–13684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi S, Murata N (2008) How do environmental stresses accelerate photoinhibition? Trends Plant Sci 13:178–182. [DOI] [PubMed] [Google Scholar]
- Tardieu F, Davies WJ (1992) Stomatal response to abscisic acid is a function of current plant water status. Plant Physiol 98:540–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B, Murphy DJ, Murray BG (2016) Encyclopedia of applied plant sciences, 2nd edn Elsevier Science, New York, NY. [Google Scholar]
- Tran LSP, Urao T, Qin F et al. (2007) Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc Natl Acad Sci USA 104:20623–20628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Maron-Katz A, Shavit S et al. (2010) Expander: from expression microarrays to networks and functions. Nat Protoc 5:303–322. [DOI] [PubMed] [Google Scholar]
- Ungar ED, Rotenberg E, Raz-Yaseef N, Cohen S, Yakir D, Schiller G (2013) Transpiration and annual water balance of Aleppo pine in a semiarid region: implications for forest management. For Ecol Manage 298:39–51. [Google Scholar]
- Upton GJG. (1992) Fisher’s exact test. J R Stat Soc Ser A Stat Soc 155:395–402. [Google Scholar]
- Usadel B, Poree F, Nagel A, Lohse M, Czedik-Eysenberg A, Stitt M (2009) A guide to using MapMan to visualize and compare Omics data in plants: a case study in the crop species, Maize. Plant Cell Environ 32:1211–1229. [DOI] [PubMed] [Google Scholar]
- Vahisalu T, Kollist H, Wang YF et al. (2008) SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452:487–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser EA, Wegrzyn JL, Steenkmap ET, Myburg AA, Naidoo S (2015) Combined de novo and genome guided assembly and annotation of the Pinus patula juvenile shoot transcriptome. BMC Genomics 16:1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronova A, Belevich V, Jansons A, Rungis D (2014) Stress-induced transcriptional activation of retrotransposon-like sequences in the Scots pine (Pinus sylvestris L.) genome. Tree Genet Genomes 10:937–951. [Google Scholar]
- Wang L, Yang L, Zhang J et al. (2013) Cloning and characterization of a thaumatin-like protein gene PeTLP in Populus deltoides × P. euramericana cv.‘Nanlin895’. Acta Physiol Plant 35:2985–2998. [Google Scholar]
- Wang X, Cai X, Xu C, Wang Q, Dai S (2016) Drought-responsive mechanisms in plant leaves revealed by proteomics. Int J Mol Sci 17:1706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkinson JI, Sioson AA, Vasquez-Robinet C et al. (2003) Photosynthetic acclimation is reflected in specific patterns of gene expression in drought-stressed Loblolly Pine. Plant Physiol 133:1702–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn JL, Lin BY, Zieve JJ et al. (2013) Insights into the loblolly pine genome: Characterization of BAC and fosmid sequences. PLoS ONE 8:e72439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LE, Baeza P, Vaughn P (2012) Midday measurements of leaf water potential and stomatal conductance are highly correlated with daily water use of Thompson Seedless grapevines. Irrigation Sci 30:201–212. [Google Scholar]
- Wu Q, Zhang X, Peirats-Llobet M et al. (2016) Ubiquitin ligases RGLG1 and RGLG5 regulate abscisic acid signaling by controlling the turnover of phosphatase PP2CA. Plant Cell 28:2178–2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Lee US, Wagner D (2016) Tug of war: adding and removing histone lysine methylation in Arabidopsis. Curr Opin Plant Biol 34:41–53. [DOI] [PubMed] [Google Scholar]
- Xing L, Zhao Y, Gao J, Xiang C, Zhu JK (2016) The ABA receptor PYL9 together with PYL8 plays an important role in regulating lateral root growth. Sci Rep 6:27177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Meinzer FC, Qi JH, Goldstein G, Cao KF (2013) Midday stomatal conductance is more related to stem rather than leaf water status in subtropical deciduous and evergreen broadleaf trees. Plant Cell Environ 36:149–158. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Xing L, Wang X et al. (2014) The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci Signal 7:ra53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.