This was the first birth cohort study with active surveillance of sapovirus infection in a developing country. High incidences of sapovirus infection and associated diarrhea during the first 2 years of life were reported. Sapovirus reinfection is common but rare with the same genotype.
Keywords: sapovirus, birth cohort, diarrhea, gastroenteritis, epidemiology
Abstract
Background
Sapovirus is one of the primary viral causes of acute gastroenteritis (AGE), especially where rotavirus vaccination has been implemented. The characteristics and impact of natural infection at the community level, however, have not been well documented.
Methods
Stool samples were analyzed from 100 children randomly selected from a community-based birth cohort study in Peru. All diarrheal and 1 nondiarrheal stools collected trimonthly from children up to age 2 years (n = 1669) were tested for sapovirus detection. Viral shedding duration was determined by testing additional weekly samples (n = 440) collected before and after a sapovirus-positive sample.
Results
The incidence of sapovirus infection in the first and second years of life was 4.3 and 11.1 per 100 child-months, respectively. By age 2 years, 82% of children had at least 1 sapovirus infection, and 64% had at least 1 sapovirus-associated diarrhea episode. The median shedding period was 18.5 days. In 112 of 175 infections, 14 genotypes from 4 genogroups (GI, GII, GIV, and GV) were determined. Among genogroups, GI were more frequently found in symptomatic infections than in asymptomatic infections (odds ratio, 3.1; 95% confidence interval, 1.3–7.4). Fifty-nine children had serial sapovirus infections, but only 3 had repeated infection of the same genotype.
Conclusions
Sapovirus was frequently detected in children with AGE at the community level during the first 2 years of life. Serial sapovirus infections by multiple genotypes in a child suggest genotype-specific immunity from each infection, which needs to be taken into account for vaccine development.
Diarrhea is the second leading cause of child mortality worldwide, causing an estimated 800,000 deaths annually [1]. Rotavirus was the major cause of severe diarrhea in infants prior to the introduction of vaccination programs [2]. Today, in countries where rotavirus vaccination coverage is high, norovirus, sapovirus, adenovirus, and astrovirus have emerged as the most important causes of viral gastroenteritis [3, 4]. In a community-based study in Nicaragua, after norovirus, sapovirus was identified as the second most frequent cause of acute gastroenteritis (AGE) in children, following norovirus but more frequent than rotavirus [3]. A similar pattern was also observed among children in a Peruvian hospital (unpublished data, S.B. Ballard).
Sapovirus, a member of the Caliciviridae family, is a single-stranded positive sense RNA virus [5] with 4 genogroups, GI, GII, GIV, and GV, that infect humans [6–8]. Sapovirus infections primarily affect children aged <5 years, causing mild to moderate diarrhea [9, 10] and outbreaks in all age groups [11–14]. Though research on sapovirus outbreaks [15–17] and hospital-based case-control studies [18–20] have been carried out, only a few community-based studies with active surveillance have examined the natural history of sapovirus. These include a study in the Netherlands that detected 5%–9% of sapovirus in children with gastroenteritis aged 0.5–11 months [9] and a study in Brazil that detected 3% in children with gastroenteritis aged <5 years by conventional reverse transcription polymerase chain reaction (RT–PCR) [21]. Using RT real-time quantitative PCR (RT-qPCR), the study in Nicaragua reported that 17% of children aged<5 years with AGE had sapovirus infection [3].
We previously reported significant attribution of sapovirus to AGE in a case-control study [22] using selected stool samples collected from a Peruvian birth cohort [23]. Based on the those results, we randomly selected 100 children from the same cohort to determine the following: prevalence of sapovirus in diarrheal and nondiarrheal samples, incidence of sapovirus infection and reinfection during the first 2 years of life, genogroup and genotype distribution, and duration of viral shedding. Elucidating these epidemiological parameters at the community level is key to the development of effective control strategies, including vaccine development.
METHODS
Sample Selection for Sapovirus Infection
In a previous cohort study conducted from June 2007 to April 2011, 291 children were followed from birth until age 2 years for norovirus infection [23]. For this sapovirus study, we randomly selected 100 children from the norovirus cohort study who completed 2 years of follow-up and analyzed archived stool samples. The stool samples tested were diarrheal samples collected during each episode of diarrhea (n = 877) and nondiarrheal samples (n = 792) collected every 3 months. These nondiarrheal stools were chosen systematically from archived weekly collected stools; 8 samples were missing. Samples were selected independently regardless of previous norovirus and sapovirus results from the same cohort [22, 23]. However, 709 samples for the present study overlapped with samples tested in the norovirus cohort study, from which 476 were diarrhea stool samples [23]. Also, 205 stool samples overlapped with the samples tested in the previous case-control study [22].
All samples (n = 1669) were collected prior to addition of the rotavirus vaccine Rotarix to Peru’s national vaccine schedule.
Definitions
A day of diarrhea was defined by the presence of ≥3 liquid or semiliquid stools in 24 hours. For infants aged <2 months, the definition also required that the mother or caregiver considered the child to have diarrhea. The start date for a symptomatic infection was defined by the first day with diarrhea [23]. An episode of diarrhea was considered complete when the infant had 2 consecutive days without diarrhea [23].
A sample was considered positive for sapovirus and an infection was defined if the sample was positive by RT-qPCR. If a positive sample was detected later in the same child, it was considered a new infection if 1 of the following conditions applied: identification of a new sapovirus genotype in a participant’s sample at any time; sapovirus with at least 2 intervening negative samples unless the same genotype was detected within 30 days; or, if only 1 negative sample existed, a 30-day interval was required to define a new sapovirus infection.
A “sapovirus symptomatic infection” was defined as a positive RT-qPCR result in a stool taken during or within 7 days of the beginning or end of the diarrheal episode. An “asymptomatic” infection occurred when no symptoms were reported within 7 days of the positive specimen. “Undefined” infection occurred when sapovirus was detected in a stool specimen more than 7 days before or after diarrhea onset but less than 14 days.
Duration of Viral Shedding
Shedding was evaluated for a subset of the infections detected in the cohort testing described above. Eligible infections were those for which genotyping was successful and at least 2 weekly specimens were available before and after sapovirus was detected in a stool sample. The weekly samples tested in addition to the cohort samples described above are referred to as “shedding samples” (n = 440). The beginning and end of the shedding period were defined based on 2 consecutive negative results, as described elsewhere [23]. Positive shedding samples were sequenced to confirm that they belonged to the same infection by the same genotype if the samples are taken more than 7 days apart.
Reverse Transcription and qPCR for Sapovirus Detection
Archived stool samples, stored at −80°C, were allowed to thaw at 4°C. A 10% stool suspension was prepared and centrifuged at 5000 × g for 10 minutes. Viral RNA was extracted using 140 μL of the supernatant using the QIAamp viral RNA kit and the QIAcube (Qiagen, Hilden, Germany). The complementary DNA (cDNA) was synthesized using the SuperScript III First-Strand Synthesis System with random primers, according to the manufacturer’s protocol (Invitrogen, Carlsbad, California).
The RT-qPCR targeting the sapovirus polymerase-viral protein 1 (VP1) junction, using previously described primers and probes [24], was carried out using the LightCycler 96 thermocycler (Roche, Germany). The RT-qPCR reaction was performed in a final volume of 10 uL, containing 1 uL of cDNA, 1X of TaqMan Fast Advanced Master Mix (Life Technologies, Foster City, California), 10 pmol of each primer (Sapovirus124F, Sapovirus1F, Sapovirus5F, and Sapovirus1245R), and 10 pmol of each probe (Sapovirus124TP and Sapovirus5TP). Amplification conditions were 50°C for 2 minutes, 95°C for 15 minutes, 45 cycles each consisting of 95°C for 30 seconds and 60°C for 30 seconds, and finally 60°C for 2 minutes.
The detection limit was determined using a standard curve based on a 10-fold serial dilution (107–101) of GII.4 plasmid provided by the Department of Virology, Tohoku University (Sendai, Japan). The detection limit was 10 copies corresponding to a cycle of quantification (Cq) of 38.0. A sample with a Cq value ≤38.0 and a sigmoidal amplification curve in the RT-qPCR was considered positive. Samples with Cq values >38.0 and <40.0 were considered positive if conventional PCR for genotyping was positive and its genotype was identified.
Norovirus and Rotavirus Detection by qPCR
Sapovirus diarrheal samples were tested to determine the proportion of coinfection with norovirus and rotavirus using methods described previously [23, 25].
Genotyping and Sequence Analysis
In this study, the partial capsid region was used for genotyping. All sapovirus-positive samples by RT-qPCR were amplified by a nested PCR targeting a partial sequence of the VP1 gene (400 nucleotides). The first PCR reaction was performed using primers F13/F14 and R13/R14, while the second PCR reaction was performed using primers F22/R2 [26]. Cycling conditions for the first and second rounds of nested PCR were 94°C for 5 minutes, 35 cycles each consisting of 94°C for 30 seconds, 52°C for 40 seconds, and 72°C for 1 minute and then 1 cycle of 72°C for 10 minutes. Final amplification products were sent for sequencing to Macrogen USA (Rockville, Maryland). Sequencing was performed using primers F22/R2. Sequencing data were aligned and trimmed using the bioinformatics software Geneious R9.1 [27]. Genogroups and genotypes were assigned using BLAST and constructing phylogenetic trees as previously described [22], including the sequences cited by Oka et al [28], and using maximum likelihood and a 2000 bootstrap performed using Geneious R9.1.
Statistical Analyses
Associations were tested using logistic regression models with generalized estimating equations to account for repeated measures in the same individuals. The attributable fraction of sapovirus infection was estimated based on the comparison of sapovirus prevalence in diarrheal and nondiarrheal stool samples [29]. Median Cq values of sapovirus in diarrheal and nondiarrheal stool samples were compared using the Mann-Whitney U test. Cumulative incidence rates of sapovirus infection and symptomatic sapovirus infections were estimated by Kaplan-Meier survival analyses. To assess the associations of specific genogroups of sapovirus infection and diarrhea, the odds ratio (OR) was calculated for each genogroup and genotype. The shedding period was estimated as described above and compared by genogroup using the Mann-Whitney U test or the Kruskal-Wallis test. A P value <.05 was considered statistically significant.
RESULTS
Sapovirus Infection
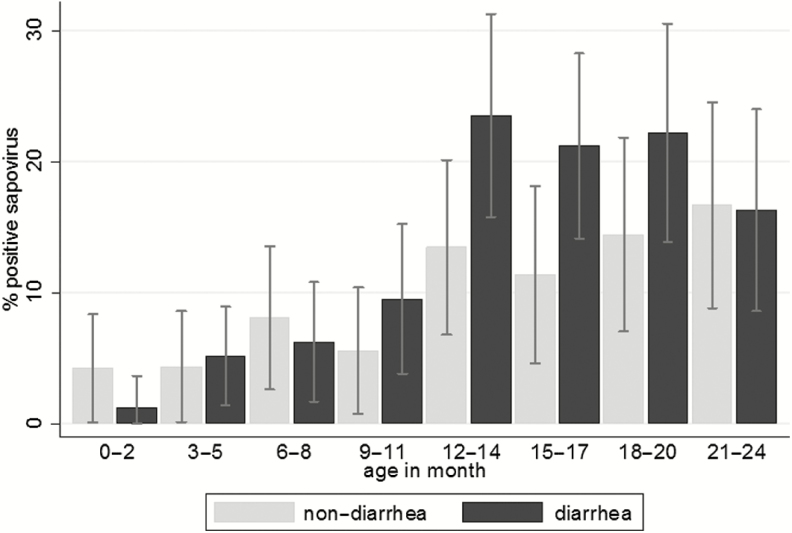
Sapovirus was detected in 192 of 1669 samples, 13.5% (118/877) of diarrheal stool samples and 9.7% (73/748) of nondiarrheal stool samples (P = .008). If symptom data were incomplete 7 days before or after stool sampling, the data were not included in the analysis of the association between sapovirus infection and diarrhea (n = 44). The prevalence of sapovirus in diarrheal stool samples was significantly higher during the second year of life compared to the first: 21.0% and 5.8%, respectively (P < .001; Figure 1). Overall, the attributable fraction was 0.2% and 7.9% in the first and second years of life, respectively. Median Cq values of RT-qPCR in diarrheal samples were lower than in nondiarrheal samples (34.1 vs 35.6, P = .017, respectively).
Figure 1.
Prevalence of sapovirus in diarrheal and nondiarrheal stool samples by age group. Prevalences were based on cross-sectional analysis of sapovirus detected by reverse transcription real-time polymerase chain reaction in 877 diarrheal and 748 nondiarrheal stool samples. Forty-four nondiarrheal samples, which were missing symptom data within 7 days from the sampling date, were excluded.
Based on 192 positive cohort samples, 167 sapovirus infections were identified in 82 children. By testing the 440 samples to determine the duration of shedding, 8 additional sapovirus infections were identified. Therefore, 175 sapovirus infections were identified in 82 children, comprising 97 symptomatic, 68 asymptomatic, and 10 infections classified as undefined.
Of 118 diarrheal samples positive for sapovirus, 111 were tested for norovirus and 106 were tested for rotavirus. The proportion of sapovirus diarrheal samples with coinfection with norovirus GI, GII, or rotavirus were 12.6% (14/111), 1.8% (2/111), and 3.6% (4/106), respectively, corresponding to 15% (15/97) of sapovirus symptomatic infections (coinfection with norovirus GI [n = 2], norovirus GII [n = 10], and rotavirus [n = 3]).
The incidence of sapovirus infection and diarrhea was 7.8 (95% confidence interval [CI], 6.7–9.0) and 4.3 (CI, 3.5–5.3) per 100 child-months, respectively. The incidence of sapovirus infection was higher among children aged 12–24 months (11.1 [CI, 9.1–13.1] per 100 child-months) compared to children aged <12 months (4.3 [CI, 3.2–5.7] per 100 child-months; P < .001).
Overall, 82 of the 100 children in our cohort had at least 1 sapovirus infection during the 2-year follow-up, while 59 experienced 2 or more sapovirus infections (Figure 2A). Furthermore, 64 children had at least 1 and 22 had 2 or more sapovirus symptomatic infections (Figure 2B).
Figure 2.
Cumulative incidence of first and subsequent sapovirus infections (A) and sapovirus-associated diarrhea (B) in a birth cohort of 100 children. A, Survival curves show cumulative incidence of first through fifth sapovirus infection during the first 2 years of life. B, Curves show the cumulative incidence of the first through fifth infection of sapovirus-associated diarrhea. Of the 100 children, 23 had 1 sapovirus infection and 59 had more than 1 infection (34 had 2, 20 had 3, 3 had 4, 1 had 5, and 1 had 6).
Genogroup and Genotype Relation to Diarrhea
Of 175 infections, we successfully genotyped 112 (64%) infections in 66 children. For 1 other sample, its genogroup (GII) was defined but not its genotype. All 4 sapovirus genogroups that infect humans were detected. Genotyping was more successful in samples with higher viral loads, that is, 91% in samples with Cq <30 vs 50% in samples with Cq ≥30 (50%; P < .001).
The most frequently detected genogroup from 113 infections was GI (52/113, 46.0%), followed by GII (44/113, 38.9%), GIV (9/113, 8.0%), and GV (8/113, 7.1%). Among 113 infections with genogroup information, 69 were symptomatic, 36 were asymptomatic, and 8 were undefined. Significantly, GI viruses were found more frequently in symptomatic infections (38/69, 55.1%) than in asymptomatic infections (10/36, 27.8%; OR, 3.1 [CI, 1.3–7.4]; P = .011; Figure 3). GIV viruses were found in 6 (8.7%) of 69 symptomatic infections and in 2 (5.4%) of 36 asymptomatic infections (OR, 1.9 [CI, 0.4–8.9]; P = .396). GV viruses were found only in symptomatic infections. In contrast, GII viruses were less common in symptomatic (18/69, 26.1%) than in asymptomatic infections (24/36, 64.7%; OR, 0.2 [CI, 0.1–0.5]; P < .001). A significant difference in median Cq values between symptomatic and asymptomatic samples was detected among those with GI viruses (P = .037), but not in samples with GII viruses (P = .383).
Figure 3.
Proportion (%) of sapovirus genotypes detected by symptomatic and asymptomatic infection. Proportions (%) were based on the number of sapovirus infections whose genotypes were identified. Abbreviations: GI, genogroup I; GII, genogroup II; GIV, genogroup IV; GV, genogroup V.
Fourteen of 19 known genotypes [28] were identified in 112 samples (Supplementary Table S1). There was no dominant genotype over the 2 years of follow-up (Supplementary Figure S1). GI.1 (28/112, 25.0%) was the most frequently reported genotype, followed by GI.2 (13/112, 11.6%) and GII.1 (13/112, 11.6%). Viruses with genotype GI.1, GI.2, GI.6, and GII.1 were more frequent in symptomatic than asymptomatic infections, albeit not significantly so (Figure 3).
Duration of Sapovirus Shedding
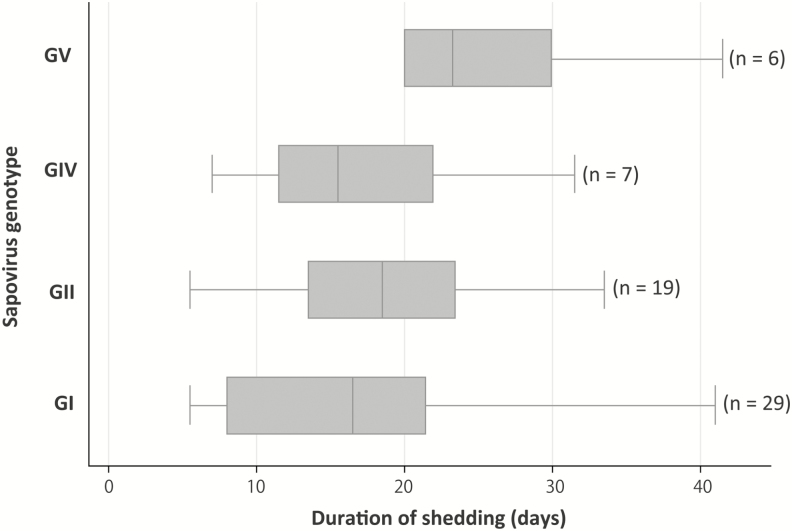
A total of 440 weekly collected stool samples from 61 sapovirus infections in 47 children were tested to determine the duration of viral shedding (Supplementary Figure S2). The median duration of sapovirus shedding was 18.5 days (interquartile range [IQR], 12.5–23.5) with a range of 5.5–41.5 days. The index samples corresponded to 38 symptomatic, 17 asymptomatic, and 6 undefined infections. There was no difference in the duration of sapovirus shedding between symptomatic and asymptomatic infections (n = 38, median = 16 days [IQR, 11.5–21.5] vs n = 17, median = 20.5 days [IQR, 12.0–23.5]; P = .357). The median shedding periods were 16.5 and 18.5 days for GI and GII viruses, respectively (Figure 4). There was no significant difference in the median shedding time among the 4 genogroups (P = .158) even after adjusting for symptomatic status.
Figure 4.
Duration of sapovirus shedding by reverse transcription real-time polymerase chain reaction in 61 infections. Sixty-one sapovirus infections in 47 children were tested to determine the duration of viral shedding. The boxes represent the 25th percentile, median, and 75th percentile, and the whiskers show the minimum and maximum value of the duration of shedding in days. For 2 infections for which the child’s last available specimen was positive, we estimated the duration of shedding based on the date of the last positive sample (28.5 days and 41.5 days). There were 8 infections with a shedding period of 30 days or longer, 2 of which were symptomatic. Abbreviations: GI, genogroup I; GII, genogroup II; GIV, genogroup IV; GV, genogroup V.
Reinfection
Of the 100 children, 23 had 1 infection and 59 had more than 1 infection (Figure 2A). Of those 59, 152 infections were detected. Genogroup and genotypes were determined in 99 (65%) and 98 samples (64%), respectively (Supplementary Table S2). We were able to compare 81 genotypes in 35 children with multiple infections. Reinfection with the same genogroup was found in 51% (18/35) of those children (GI = 10 and GII = 8). Only 3 children had repeated infections with the same genotype of GI.1, which had >99% of identity of the nucleotide sequence, in the partial capsid region.
DISCUSSION
To our knowledge, this is the first birth cohort study done prior to rotavirus vaccine implementation to evaluate the epidemiology of sapovirus infection in a low- to middle-income country. In this cohort, sapovirus, followed by norovirus as the next most common, was associated with both asymptomatic and symptomatic enteric viral infection during the first 2 years of life. Using RT-qPCR, the proportion of children who experienced at least 1 sapovirus symptomatic infection was similar (64%) to that for a norovirus infection (71%) [23]. More than half of the children experienced sapovirus reinfection. Reinfection with the same genogroup was common but was rare with the same genotype.
In contrast to norovirus infection, which occurs frequently after an infant reaches age 6 months [23], sapovirus infections were generally delayed until the second year of life. Cumulative incidence of the first sapovirus infection was approximately half that of norovirus during the first year of life. This finding is concordant with previous studies conducted in Nicaragua and Japan [3, 30, 31].
Among previous community-based studies with active surveillance, in Nicaragua GI, GII, and GIV viruses were reported using the same primers for VP1 region; in Brazil GI and GII were reported using polymerase region [3, 21]. In our study, sapovirus was genetically highly diverse. All 4 human genogroups and 14 of the 19 known genotypes that affect humans were identified. This diversity was higher than that reported in hospital settings in Vietnam and Bangladesh using primers for a similar region of VP1 [32, 33] and in the United States and India using the same primers [19, 20].
In our study, slightly more GI viruses than GII viruses were detected, which is similar to findings in a recent community-based study in Nicaragua [3]. GI viruses were more frequently detected than other genogroups in symptomatic infections, suggesting that GI viruses may be more pathogenic.
Reinfection of sapovirus has been reported in only a small number of cases from 1 clinic-based study in Japan [34]. A study that used antisera raised in animals showed limited cross-reactivity between GI.1 and GI.5 viruses, even within the same genogroup [35]. Our study also demonstrated that the repeated infection of different genotypes in the same genogroup was frequent and that repeated infection by the same genotype was rare. Although greater sample size will be necessary to demonstrate genotype-specific and/or cross immunity in each combination of genotypes, this area of research will be particularly important for sapovirus vaccine development.
Our finding of a median shedding duration of 18.5 days for sapovirus was similar to the 15 days identified in older children and adults in Japan [17]. The median duration for norovirus GII was nearly twice as long (35 days) in the same population [23]. This difference suggests that the immune response to sapovirus may be more effective in limiting shedding compared to norovirus or that norovirus is more pathogenic than sapovirus. The difference in sensitivity of diagnostic methods must be taken into account. Our detection limit was similar for both sapovirus and norovirus, yet cDNA was used for detection of sapovirus while RNA was used for norovirus.
In this study, we were able to identify genotypes in more than 60% of sapovirus-positive samples. High viral loads in a specimen were important for obtaining sequences for genotyping.
Study limitations include the relatively small number of children sampled (Supplementary Table S3) and the long 3-month interval for nondiarrheal stool sampling due to logistic limitations. Therefore, the cumulative incidence of sapovirus may be underestimated. In addition, genotyping was based on a partial region of the VP1 gene, and not all genotypes were determined among sapovirus-positive samples. Therefore, recombinant strains or variants within the same genotypes were not evaluated during analysis of repeated infections.
In summary, sapovirus frequently caused infection and diarrhea in young children, even prior to the implementation of a rotavirus vaccine, which may increase the relative importance of sapovirus as a diarrheal pathogen. Findings from this study as well as the study on norovirus from the same population strongly suggest the need for a triple vaccine that targets rotavirus, norovirus, and sapovirus to prevent diarrhea among children.
Supplementary Data
Supplementary materials are available at Clinical Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
Notes
Acknowledgments We thank the community of San Juan de Miraflores and the field staff for their collaboration. We also acknowledge J. Rothstein, K. Roman, H. Torres, C. Puemape, and M. Varela for their technical support.
The Sapovirus Working Group includes all present authors and Mayra Ochoa, Macarena Vittet, and Alejandra Pando from Department of Cellular and Molecular Sciences, Universidad Peruana Cayetano Heredia.
Financial support This work was supported by the European Union, Project CONTENT, Sixth Framework Programme (INCO-CT-2006–032136), the National Institute of Allergy and Infectious Diseases at the National Institutes of Health (1R21AI099737-01, 3R01AI108695), the Centers for Disease Control and Prevention, China Scholarship Council, Consejo Nacional de Ciencia, Tecnología e Innovación Tecnológica (CONCYTEC, 110-2015-FONDECyT), and Japan Society for the Promotion of Science (16H02772).
Potential conflicts of interest All authors report no potential conflict of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Contributor Information
Sapovirus Working Group:
References
- 1. Liu L, Oza S, Hogan D et al. . Global, regional, and national causes of child mortality in 2000-13, with projections to inform post-2015 priorities: an updated systematic analysis. Lancet 2015; 385:430–40. [DOI] [PubMed] [Google Scholar]
- 2. Tate JE, Burton AH, Boschi-Pinto C, Parashar UD, World Health Organization–Coordinated Global Rotavirus Surveillance Network. Global, regional, and national estimates of rotavirus mortality in children <5 years of age, 2000–2013. Clin Infect Dis 2016; 62:S96–S105. [DOI] [PubMed] [Google Scholar]
- 3. Bucardo F, Reyes Y, Svensson L, Nordgren J. Predominance of norovirus and sapovirus in Nicaragua after implementation of universal rotavirus vaccination. PLoS One 2014; 9:e98201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Heusinkveld M, Mughini-Gras L, Pijnacker R et al. . Potential causative agents of acute gastroenteritis in households with preschool children: prevalence, risk factors, clinical relevance and household transmission. Eur J Clin Microbiol Infect Dis 2016; 35:1691–700. [DOI] [PubMed] [Google Scholar]
- 5. Alan Radford AWS, Hansman G, Meyers G et al. . Caliciviridae. In King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. Virus Taxonomy. Oxford: Elsevier, 2011:977–86. [Google Scholar]
- 6. Hong-ying Z, Li-min S, Wei L et al. . Molecular epidemiology of genogroup II noroviruses infection in outpatients with acute gastroenteritis in Nanjing, China (2010–2013). Biomed Res Int 2014; 2014:620740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Romani S, Azimzadeh P, Mohebbi SR, Bozorgi SM, Zali N, Jadali F. Prevalence of sapovirus infection among infant and adult patients with acute gastroenteritis in Tehran, Iran. Gastroenterol Hepatol Bed Bench 2012; 5:43–8. [PMC free article] [PubMed] [Google Scholar]
- 8. Liu X, Yamamoto D, Saito M et al. . Molecular detection and characterization of sapovirus in hospitalized children with acute gastroenteritis in the Philippines. J Clin Virol 2015; 68:83–8. [DOI] [PubMed] [Google Scholar]
- 9. Rockx B, De Wit M, Vennema H et al. . Natural history of human calicivirus infection: a prospective cohort study. Clin Infect Dis 2002; 35:246–53. [DOI] [PubMed] [Google Scholar]
- 10. Leblanc D, Inglis GD, Boras VF, Brassard J, Houde A. The prevalence of enteric RNA viruses in stools from diarrheic and non-diarrheic people in southwestern Alberta, Canada. Arch Virol 2017; 162:117–28. [DOI] [PubMed] [Google Scholar]
- 11. Pang XL, Lee BE, Tyrrell GJ, Preiksaitis JK. Epidemiology and genotype analysis of sapovirus associated with gastroenteritis outbreaks in Alberta, Canada: 2004–2007. J Infect Dis 2009; 199:547–51. [DOI] [PubMed] [Google Scholar]
- 12. Lee LE, Cebelinski EA, Fuller C et al. . Sapovirus outbreaks in long-term care facilities, Oregon and Minnesota, USA, 2002–2009. Emerg Infect Dis 2012; 18:873–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bon F, Ambert-Balay K, Giraudon H et al. . Molecular epidemiology of caliciviruses detected in sporadic and outbreak cases of gastroenteritis in France from December 1998 to February 2004. J Clin Microbiol 2005; 43:4659–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Iritani N, Kaida A, Abe N et al. . Detection and genetic characterization of human enteric viruses in oyster-associated gastroenteritis outbreaks between 2001 and 2012 in Osaka City, Japan. J Med Virol 2014; 86:2019–25. [DOI] [PubMed] [Google Scholar]
- 15. Svraka S, Vennema H, van der Veer B et al. . Epidemiology and genotype analysis of emerging sapovirus-associated infections across Europe. J Clin Microbiol 2010; 48:2191–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Torner N, Martinez A, Broner S et al. . Epidemiology of acute gastroenteritis outbreaks caused by human calicivirus (norovirus and sapovirus) in Catalonia: a two year prospective study, 2010–2011. PLoS ONE 2016; 11:e0152503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iwakiri A, Ganmyo H, Yamamoto S et al. . Quantitative analysis of fecal sapovirus shedding: identification of nucleotide substitutions in the capsid protein during prolonged excretion. Arch Virol 2009; 154:689–93. [DOI] [PubMed] [Google Scholar]
- 18. Amar CF, East CL, Gray J, Iturriza-Gomara M, Maclure EA, McLauchlin J. Detection by PCR of eight groups of enteric pathogens in 4,627 faecal samples: re-examination of the English case-control Infectious Intestinal Disease Study (1993–1996). Eur J Clin Microbiol Infect Dis 2007; 26:311–23. [DOI] [PubMed] [Google Scholar]
- 19. Chhabra P, Payne DC, Szilagyi PG et al. . Etiology of viral gastroenteritis in children <5 years of age in the United States, 2008–2009. J Infect Dis 2013; 208:790–800. [DOI] [PubMed] [Google Scholar]
- 20. Lasure N, Gopalkrishna V. Epidemiological profile and genetic diversity of sapoviruses (SaVs) identified in children suffering from acute gastroenteritis in Pune, Maharashtra, Western India, 2007–2011. Epidemiol Infect 2017; 145:106–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aragão GC, Mascarenhas JD, Kaiano JH et al. . Norovirus diversity in diarrheic children from an African-descendant settlement in Belém, Northern Brazil. PLoS One 2013; 8:e56608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Liu X, Jahuira H, Gilman RH et al. . Etiological role and repeated infections of sapovirus among children aged less than 2 years in a cohort study in a peri-urban community of Peru. J Clin Microbiol 2016; 54:1598–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Saito M, Goel-Apaza S, Espetia S et al. . Multiple norovirus infections in a birth cohort in a Peruvian periurban community. Clin Infect Dis 2014; 58:483–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oka T, Katayama K, Hansman GS et al. . Detection of human sapovirus by real-time reverse transcription-polymerase chain reaction. J Med Virol 2006; 78:1347–53. [DOI] [PubMed] [Google Scholar]
- 25. Freeman MM, Kerin T, Hull J, McCaustland K, Gentsch J. Enhancement of detection and quantification of rotavirus in stool using a modified real-time RT-PCR assay. J Med Virol 2008; 80:1489–96. [DOI] [PubMed] [Google Scholar]
- 26. Okada M, Yamashita Y, Oseto M, Shinozaki K. The detection of human sapoviruses with universal and genogroup-specific primers. Arch Virol 2006; 151:2503–9. [DOI] [PubMed] [Google Scholar]
- 27. Kearse M, Moir R, Wilson A et al. . Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 2012; 28:1647–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Oka T, Wang Q, Katayama K, Saif LJ. Comprehensive review of human sapoviruses. Clin Microbiol Rev 2015; 28:32–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Blackwelder WC, Biswas K, Wu Y et al. . Statistical methods in the Global Enteric Multicenter Study (GEMS). Clin Infect Dis 2012; 55:S246–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Becker-Dreps S, Bucardo F, Vilchez S et al. . Etiology of childhood diarrhea after rotavirus vaccine introduction: a prospective, population-based study in Nicaragua. Pediatr Infect Dis J 2014; 33:1156–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Thongprachum A, Khamrin P, Maneekarn N, Hayakawa S, Ushijima H. Epidemiology of gastroenteritis viruses in Japan: prevalence, seasonality, and outbreak. J Med Virol 2016; 88:551–70. [DOI] [PubMed] [Google Scholar]
- 32. Dey SK, Phan TG, Nguyen TA et al. . Prevalence of sapovirus infection among infants and children with acute gastroenteritis in Dhaka City, Bangladesh during 2004–2005. J Med Virol 2007; 79:633–8. [DOI] [PubMed] [Google Scholar]
- 33. Hansman GS, Doan LT, Kguyen TA et al. . Detection of norovirus and sapovirus infection among children with gastroenteritis in Ho Chi Minh City, Vietnam. Arch Virol 2004; 149:1673–88. [DOI] [PubMed] [Google Scholar]
- 34. Harada S, Oka T, Tokuoka E et al. . A confirmation of sapovirus re-infection gastroenteritis cases with different genogroups and genetic shifts in the evolving sapovirus genotypes, 2002–2011. Arch Virol 2012; 157:1999–2003. [DOI] [PubMed] [Google Scholar]
- 35. Hansman GS, Saito H, Shibata C et al. . Outbreak of gastroenteritis due to sapovirus. J Clin Microbiol 2007; 45:1347–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.