Abstract
Background
Lung adenocarcinomas (LUADs) lead to the majority of deaths attributable to lung cancer. We performed whole-exome sequencing (WES) and immune profiling analyses of a unique set of clinically annotated early-stage LUADs to better understand the pathogenesis of this disease and identify clinically relevant molecular markers.
Methods
We performed WES of 108 paired stage I-III LUADs and normal lung tissues using the Illumina HiSeq 2000 platform. Ten immune markers (PD-L1, PD-1, CD3, CD4, CD8, CD45ro, CD57, CD68, FOXP3 and Granzyme B) were profiled by imaging-based immunohistochemistry (IHC) in a subset of LUADs (n = 92). Associations among mutations, immune markers and clinicopathological variables were analyzed using ANOVA and Fisher’s exact test. Cox proportional hazards regression models were used for multivariate analysis of clinical outcome.
Results
LUADs in this cohort exhibited an average of 243 coding mutations. We identified 28 genes with significant enrichment for mutation. SETD2-mutated LUADs exhibited relatively poor recurrence- free survival (RFS) and mutations in STK11 and ATM were associated with poor RFS among KRAS-mutant tumors. EGFR, KEAP1 and PIK3CA mutations were predictive of poor response to adjuvant therapy. Immune marker analysis revealed that LUADs in smokers and with relatively high mutation burdens exhibited increased levels of immune markers. Analysis of immunophenotypes revealed that LUADs with STK11 mutations exhibited relatively low levels of infiltrating CD4+/CD8+ T-cells indicative of a muted immune response. Tumoral PD-L1 was significantly elevated in TP53 mutant LUADs whereas PIK3CA mutant LUADs exhibited markedly down-regulated PD-L1 expression. LUADs with TP53 or KEAP1 mutations displayed relatively increased CD57 and Granzyme B levels indicative of augmented natural killer (NK) cell infiltration.
Conclusion(s)
Our study highlights molecular and immune phenotypes that warrant further analysis for their roles in clinical outcomes and personalized immune-based therapy of LUAD.
Keywords: lung adenocarcinoma, whole-exome sequencing, immune profiles
Introduction
Lung adenocarcinoma (LUAD) is the most prevalent subtype of non-small cell lung cancer (NSCLC) [1]. LUAD exhibits relatively poor prognosis warranting the need for better predictors of clinical outcome and treatment strategies [1]. These advances are restrained by a limited understanding of LUAD pathobiology [2]. Smoker LUADs commonly (∼25%) exhibit KRAS mutations [2, 3]. In contrast, mutations in EGFR are more prevalent in nonsmoker LUADs and predict response to epidermal growth factor receptor (EGFR) tyrosine kinase inhibitors (TKIs) [2, 3]. Using next-generation sequencing and profiling methods, various point mutations and copy number variations (CNVs) have been described in LUADs [3–5]. While these alterations are implicated in canonical cancer signaling pathways, their relevance to the clinical outcome of early-stage LUAD is poorly understood.
The dynamic relationship between tumor cells and immune microenvironment plays key roles in cancer pathogenesis [6, 7]. This is largely due to failure of deployment of cytotoxic T-cell lymphocytes (CTLs) or to suppression by immune inhibitory checkpoints (e.g. CTLA4 or PD-1/PD-L1) [6, 7]. Markers of immune response are prognostic and predictive in lung cancer patients, for example, increased levels of tumor-infiltrating CTLs are a favorable prognostic indicator [8]. The role of the immune system in lung oncogenesis is accentuated by studies demonstrating clinical responses to checkpoint blockade immunotherapies [9, 10]. Recently, studies have pointed to association of expression levels of immune markers with response to immune-based therapy [11, 12]. It is not clear which molecular alterations in LUAD cells could influence immune marker expression and, potentially, response to immune-based therapies [11].
We sought to characterize molecular phenotypes of early-stage LUAD that are significant to the pathogenesis of the disease by querying those that impact several important clinicopathological features, namely prognosis, response to therapy and immunophenotypes. To do so, we surveyed the mutation spectrum of 108 richly annotated early-stage surgically resected LUADs by whole-exome sequencing (WES) and immune profiles of 92 of these LUADs using imaging-based immunohistochemistry (IHC). Here we report alterations that are associated with clinical outcome in early-stage LUAD patients and point mutations that are significantly associated with expression levels of effector and inhibitory checkpoint immune markers. These mutations may play a key role in personalized strategies for targeted immunotherapy.
Methods
Details on methods and codes pertaining to statistical analyses are found in supplementary Methods, available at Annals of Oncology online and Sweave Reports (supplement Data, available at Annals of Oncology online).
Early-stage LUAD cohort
LUADs and matched normal lung tissues (n = 108 pairs) (supplementary Table S1, available at Annals of Oncology online) were obtained from the MD Anderson Cancer Center (MD Anderson) Lung Cancer SPORE tissue bank and from the PROSPECT cohort [13]. Tumors were classified using the 2004 WHO classification system as described previously [14]. All specimens were obtained from patients who were evaluated at MD Anderson and following informed consent under protocols approved by Institutional Review Board. Paired LUAD and normal tissues were obtained snap-frozen. The percentage of malignant tissue was calculated by histological examination following Hematoxylin-Eosin staining. All LUADs analyzed comprised at least 30% malignant lung cells.
Whole-exome sequencing
Genomic DNA was captured on the NimbleGen 2.1M human exome array according to the manufacturer’s instructions and subjected to 75-bp paired-end sequencing using the Illumina HiSeq2000 platform as described previously [15]. Sequence reads were mapped to the reference genome (hg19) and analyzed as described previously [16].
Identification of somatic point mutations
Somatic mutations were identified as described previously [16, 17]. Briefly, somatic variants were identified by statistically comparing reference and nonreference reads in LUADs relative to reads in corresponding normal lung tissues. Somatic small insertions and deletions (indels) were identified as reported previously [16]. Significantly mutated genes were identified based on displaying an overall increased somatic mutation burden than that expected by chance.
IHC and image analysis
Four-micron-thick sequential tumor sections were obtained from formalin-fixed paraffin-embedded tumor blocks of 92 LUADs studied by WES. IHC was performed using an automated staining system (BOND-MAX; Leica Microsystems) with antibodies against PD-L1, PD-1, CD3, CD4, CD8, CD45RO, CD57, Granzyme B, FOXP3 and CD68. Details on the antibodies used and on IHC and image-based analysis are included in supplementary Methods, available at Annals of Oncology online.
Results
WES of early-stage LUADs
Early-stage (stages I-III) LUADs and paired normal lung tissues (108 pairs) were studied by WES (supplementary Table S1, available at Annals of Oncology online). Eighty-six percent of patients were ever-smokers. Median follow-up times to survival and recurrence were 50.6 (range: 1–172) and 35 (range: 1–162) months, respectively. By design, LUADs were sequenced at a greater depth compared to normal lung tissues. Targeted regions in LUADs and normal samples were read by an average of 221 X and 119 X, respectively (supplementary Table S2, available at Annals of Oncology online). We then mapped loss-of-heterozygosity (LOH) segments by computing differences in B-allele frequencies of common germline variants in tumor-normal pairs. The LUADs exhibited a median of four events per tumor and an overall LOH rate of 10.6% across the genome (supplementary Figure S1, available at Annals of Oncology online).
The LUADs harbored a mean of 243 somatic mutations. In concordance with previous reports [4, 5], smoker LUADs exhibited significantly more somatic coding mutations compared to nonsmoker tumors (P < 0.01; supplementary Figure S2, available at Annals of Oncology online). Also consistent with previous reports [4, 5], the most common base substitutions were C > T transitions and C > A transversions (supplementary Figure S3, available at Annals of Oncology online) with smoker LUADs exhibiting significantly more C > A transversions than nonsmoker LUADs (supplementary Figure S3, available at Annals of Oncology online; P < 0.001). Early-stage LUADs with relative lower (less than the median) somatic mutation number exhibited a trend for poorer recurrence-free survival (RFS) compared to patients with higher mutation rates (supplementary Figure S4, available at Annals of Oncology online).
Identification of single-nucleotide variants and CNVs in early-stage LUAD
To identify genes with overall significantly increased somatic mutation burden, we first combined MD Anderson cohort LUADs (n = 108) with LUADs from the cancer genome atlas (TCGA; n = 387) (total of 495 LUADs). Probabilities of mutations being present in each gene were adjusted in the combined cohort in order to prioritize genes for analysis in the MD Anderson cohort (supplementary Methods, available at Annals of Oncology online). We found 28 genes with significantly increased mutation burden based on the genome-wide threshold of P < 2.4 × 10 − 6 (supplementary Table S3, available at Annals of Oncology online). Consistent with previous reports [4, 5], these included TP53, KEAP1, STK11, NF1, ATM, KRAS, EGFR, PIK3CA, BRAF, SMARCA4, SETD2, RBM10 and U2AF1 (supplementary Table S3, available at Annals of Oncology online). Mutations in TP53 were most frequent (supplementary Table S3, available at Annals of Oncology online). LUADs with mutant EGFR exhibited significantly lower somatic mutational burdens compared to EGFR wild type (WT) tumors (P < 0.001; supplementary Figure S5, available at Annals of Oncology online). Additional significantly mutated genes included VCAN, ROBO2, BAZ2B, FOLH1, COL12A1, HEPACAM2, TRHDE, UBA6, INHBA, SPATA18, ZNF479, EPRS, NFATC2, LRRIQ3 and ALS2CR11 (supplementary Table S3, available at Annals of Oncology online). The gene mutation frequencies were significantly correlated between the MD Anderson and TCGA cohorts (R = 0.94) (supplementary Table S3, available at Annals of Oncology online).
CNVs affecting broad regions in chromosome (chr) 1 and the short arm of chr 8 were the most frequently observed gains and losses, respectively (47% and 35%, respectively; supplementary Figure S6, Tables S4 and S5, available at Annals of Oncology online). We observed significant gains in the oncogenes MCL1, TERT, EGFR, CDK6, MYC, MUC5AC, AKT1, ERBB2 and BCL2L1 and losses in the suppressors SETD2, APC, PRDM1, TSC1, CDKN2A, TP53, STK11 and SMARCA4 (supplementary Figure S6, available at Annals of Oncology online). We also found that the identified CNVs patterns in the MD Anderson cohort were significantly and positively correlated (R = 0.72 and P < 2.2 × 10 − 16) with profiles in the TCGA set (supplementary Figure S7, available at Annals of Oncology online).
Analysis of mutations in the context of clinical outcome in early-stage LUAD
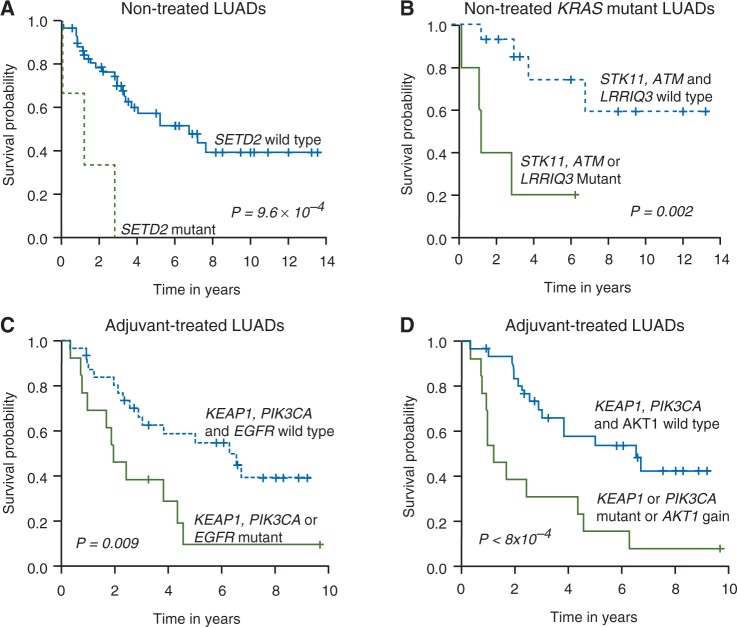
Mutant genes and CNVs that were associated with clinical outcome at the univariate level are depicted in the supplementary Sweave Reports, available at Annals of Oncology online. We sought to survey alterations or groups of alterations that are significantly associated with prognosis (in patients who did not receive therapy, n = 64) and response to therapy (in patients who received adjuvant therapy, n = 44). Akaike information criterion was applied stepwise to filter out insignificant terms during building of models incorporating mutations alone or both mutations and CNVs. Multivariate analysis revealed that among LUAD patients who did not receive adjuvant therapy (nontreated), mutations in SETD2 were significantly associated with poorer RFS (P = 9.6 × 10 − 4; Figure 1A). KRAS mutant LUAD patients with co-occurring mutations in STK11, ATM or LRRIQ3 exhibited poorer RFS compared to patients who were WT for these events (P = 0.002; Figure 1B). Among LUAD patients who received adjuvant therapy, mutations in KEAP1, PIK3CA or EGFR were significantly predictive of poorer response to adjuvant therapy (P = 0.009; Figure 1C). When CNVs were incorporated into the model, KEAP1 or PIK3CA mutations or focal gains in chr14 (AKT1) were predictive of poor response to therapy (P < 0.001; Figure 1D).
Figure 1.
Prognostic and predictive significantly mutated genes and CNVs in early-stage LUAD. (A) RFS of early-stage LUADs who did not receive adjuvant therapy (nontreated) based on mutational status of SETD2. (B) Multivariate analysis demonstrating association of mutations in either STK11, ATM or LRRIQ3 with relatively poor RFS in nontreated KRAS mutant early-stage LUADs. (C) Identification by multivariate analysis of mutated genes (C) or a combination of mutated genes and CNVs (D) that were associated with relatively poor RFS in early-stage LUADs who received adjuvant therapy. P-values were obtained using the log-rank test and the Kaplan–Meier method for estimation of survival probability. CNV, copy number variation; LUAD, lung adenocarcinoma; RFS, recurrence-free survival.
Analysis of immune marker expression in early-stage LUAD
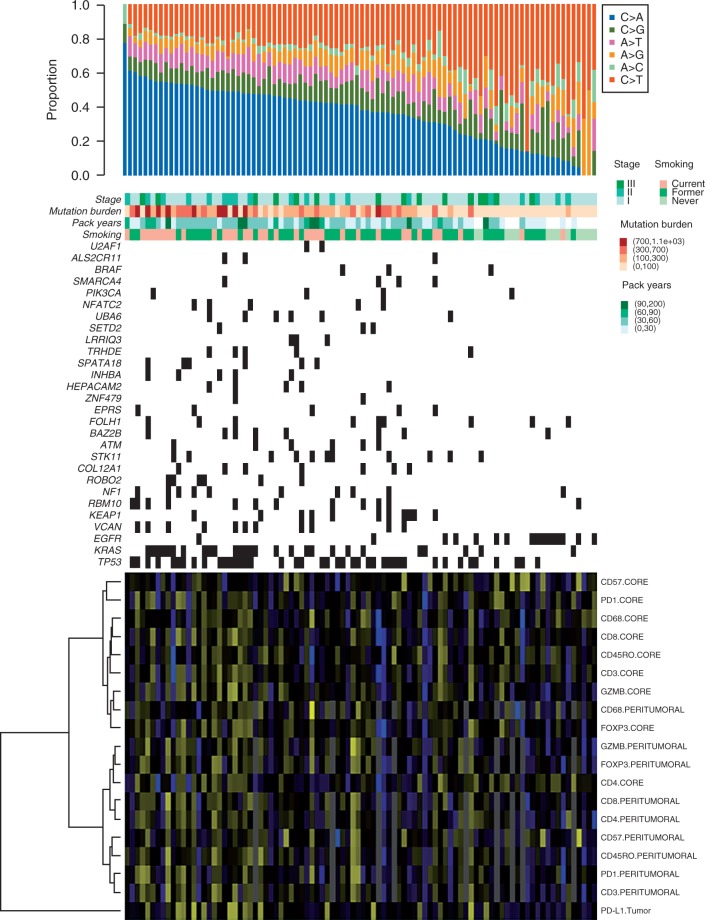
We then sought to understand immunophenotypes that are particularly associated with specific molecular subsets of LUAD. We assessed the intratumoral (tumor core) and peritumoral (surrounding the tumor) expression of PD-1, CD3, CD4, CD8, CD45ro, CD57, CD68, FOXP3 and Granzyme B as well as the tumoral expression of PD-L1 (supplementary Table S6, available at Annals of Oncology online) in 92 out of the 108 early-stage LUADs. We first integrated and correlated different mutational features with the expression of the immune markers (Figure 2). LUADs that developed in smokers, displayed relatively high mutation burdens, were enriched with C > A transversions and/or tumors that harbored KRAS and TP53 mutations (all molecular features of smoker LUADs) displayed generally elevated immune marker levels (Figure 2 and Table 1). Tumoral PD-L1 was the most elevated immune marker in smoker LUADs (P = 0.0006; Table 1 and supplementary Figure S8, available at Annals of Oncology online). Expression levels of PD-L1, CD3, CD4, CD8, CD45ro, FOXP3, Granzyme B, PD-1 were significantly (P < 0.05) positively correlated with somatic mutation burden (Table 1).
Figure 2.
Mutation signatures and immune marker expression in 108 early-stage LUADs. Exonic somatic mutations were identified as described in supplementary Methods, available at Annals of Oncology online. Columns represent LUADs, arranged from left to right by decreasing proportion of C > A transversions (top panel). Middle rows indicate pathological stage, mutation burden categorized into four groups, pack years and smoking status. Rows represent significantly mutated genes that are ordered vertically in decreasing order of NS mutation frequency. Corresponding (bottom heatmap) expression levels of immune markers were log-transformed and analyzed by hierarchical clustering using Gower’s distance and ward’s linkage method (yellow, relatively up-regulated; blue, relatively down-regulated). LUADs, lung adenocarcinomas; NS, non-silent.
Table 1.
Differential immune marker expression in early-stage lung adenocarcinomas based on smoking status and somatic mutation burden
| Smokers vs nonsmokers |
With somatic mutation burden |
|||
|---|---|---|---|---|
| Immune marker | Fold change | P-value | Spearman coefficient | P-value |
| PD-L1 tumoral | 3.63 | 0.00064 | 0.256 | 0.01368 |
| Granzyme B (tumor core) | 2.05 | 0.00231 | 0.321 | 0.00183 |
| Granzyme B (peritumoral) | 1.75 | 0.0046 | 0.312 | 0.00412 |
| FOXP3 (peritumoral) | 1.35 | 0.17067 | 0.299 | 0.00610 |
| CD57 (peritumoral) | 1.3 | 0.30872 | 0.114 | 0.30390 |
| CD45ro (peritumoral) | 1.27 | 0.12464 | 0.160 | 0.14883 |
| CD68 (peritumoral) | 1.25 | 0.35142 | 0.053 | 0.63537 |
| CD3 (peritumoral) | 1.24 | 0.2348 | 0.339 | 0.00173 |
| CD8 (peritumoral) | 1.18 | 0.16296 | 0.289 | 0.00811 |
| FOXP3 (tumor core) | 1.17 | 0.41822 | 0.103 | 0.32981 |
| CD8 (tumor core) | 1.12 | 0.51534 | 0.164 | 0.11738 |
| CD57 (tumor core) | 1.09 | 0.82812 | −0.023 | 0.82542 |
| CD4 (peritumoral) | 1.06 | 0.74455 | 0.230 | 0.03657 |
| CD3 (tumor core) | 1.06 | 0.75984 | 0.089 | 0.39985 |
| CD45ro (tumor core) | 1.02 | 0.93189 | −0.090 | 0.39334 |
| PD-1 (tumor core) | 0.98 | 0.88961 | 0.082 | 0.43869 |
| PD-1 (peritumoral) | 0.91 | 0.22802 | 0.091 | 0.41090 |
| CD68 (tumor core) | 0.88 | 0.5043 | 0.008 | 0.93602 |
| CD4 (tumor core) | 0.8 | 0.11511 | −0.183 | 0.08027 |
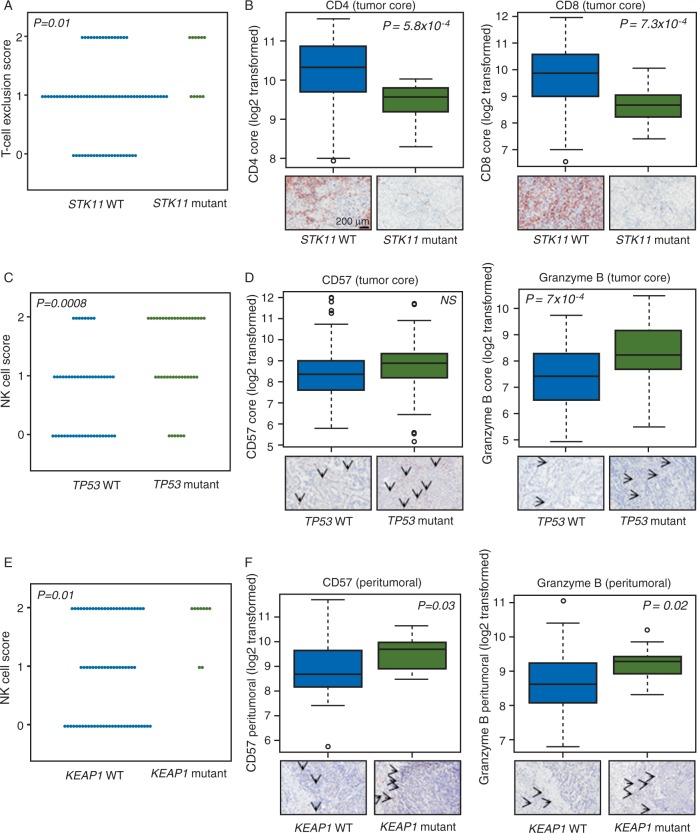
We then probed immunophenotypes based on the identified significantly mutated genes. We derived immune scores (see supplementary Methods, available at Annals of Oncology online) indicative of T-cell exclusion (comprised of CD4/CD8 and PD-L1 expression) and natural killer (NK) cell (CD57 and Granzyme B) infiltration. LUADs with mutations in STK11 displayed relatively high levels of the T-cell exclusion score, i.e. low expression of CD4 and CD8 indicative of a muted immune response (P < 0.05; Figure 3A and B and supplementary Table S7, available at Annals of Oncology online) and relatively high tumoral PD-L1. Tumoral PD-L1 expression was significantly elevated in TP53 mutant LUADs (P = 0.002; supplementary Figure S9A, available at Annals of Oncology online). In contrast, PIK3CA mutant LUADs exhibited marked suppression of the immune checkpoint (P < 0.001; supplementary Figure S9B, available at Annals of Oncology online). LUADs with mutated TP53 (Figure 3C and D) and KEAP1 (Figure 3E and F) exhibited elevated tumor core and peritumoral NK scores, respectively, indicative of increased infiltration of NK cells (P < 0.05). These findings point to immunophenotypes that are differentially expressed among different molecular subsets of LUAD.
Figure 3.
Differential expression of immunophenotypes and immune markers based on gene mutation status. (A) Score indicative of T-cell exclusion was computed as described in the supplementary Methods, available at Annals of Oncology online and statistically analyzed by STK11 mutation status using Fisher’s exact test (A). Significantly increased levels of expression of tumor core CD4 (B, left) and CD8 (B, right) in STK11 mutant LUADs compared with STK11-WT LUADs. A score indicative NK cell infiltration (C and E) was computed as described in supplementary Methods, available at Annals of Oncology online and statistically compared and contrasted between the indicated subgroups using Fisher’s exact test. Significantly elevated tumor core and peritumoral Granzyme B levels in TP53 mutant and KEAP1 mutant LUADs, respectively (D and F, right panels). Significantly elevated peritumoral CD57 in KEAP1 mutant relative to KEAP1 WT LUADs (F, left panel). Tumor core CD57 followed the same trend in TP53 mutant relative to TP53 WT LUADs albeit not reaching statistically significance (D, left panel). Upper panels in (B, D and F) represent graphical summaries (box plots) of the log2-transformed expression of the indicated immune markers. Lower panels comprise representative photomicrographs of the immunohistochemical expression of the immune markers. P-values in panels B, D and F were obtained by t-tests; values < 0.05 were considered statistically significant. LUADs, lung adenocarcinomas; WT, wild type; NK, natural killer.
Discussion
Our WES analysis pinpointed significant mutated genes and recurrent CNVs in early-stage LUAD. We identified prognostic alterations and those that were predictive of response to adjuvant therapy. IHC- and image-based profiling pointed to markers of immune response that were modulated based on smoking patterns and somatic mutation status. Of note, we found immunophenotypes that were differentially modulated among different molecular subsets of LUAD. STK11 mutant LUADs exhibited significantly reduced CD4/CD8 expression and, thus, infiltration of T-cells whereas LUADs with mutated TP53 or KEAP1 displayed increased levels of markers associated with NK cell infiltration. We also found that PIK3CA mutant LUADs, relative to WT tumors, exhibited substantially reduced expression of the immune inhibitory checkpoint PD-L1. Our findings point to alterations that may influence clinical outcome of early-stage LUAD and to mutations that impact the interaction between LUADs and the host immune system.
Our WES analysis recapitulated reported findings on the mutational landscape of LUAD [4, 5]. For instance, we found that smoker LUADs exhibited significantly higher numbers of somatic coding mutations and differential mutation spectra (increased C > A transversions) compared with nonsmoker tumors. Our WES analysis also pinpointed somatic significant mutant genes and CNVs that corroborated recent efforts and reports by the TCGA [4]. Additional candidate genes also demonstrated statistical enrichment for mutation in our early-stage LUAD cohort (Figure 2). These additional putative candidate genes have been previously implicated in cancer pathogenesis [18–20]. For example, significant somatic alterations in the tumor suppressor ROBO2 were described in pancreatic cancer [20] and in gastric and colorectal tumors [18]. INHBA, a member of the transforming growth factor-β superfamily, was demonstrated to be a significantly mutated driver candidate gene in endometrial cancer [19]. Our current findings, and the aforementioned reported observations, point to an expanded repertoire of candidate mutant genes involved in LUAD pathogenesis.
Our clinicopathological analysis revealed alterations that were indicative of poor RFS (e.g. SETD2 mutations) in early-stage LUAD. Of note, inactivating mutations in SETD2 were found to be significantly associated with disease progression and poor disease-free survival in 609 patients with clear cell renal cell carcinoma (ccRCC) [21]. Our analysis also identified aberrations (PIK3CA, KEAP1, EGFR mutations and AKT1 gain) that were predictive of poor response to adjuvant therapy. Our data on KEAP1 mutations are consistent with earlier reports on the associations of mutations in the NRF2-KEAP1 axis with poor response to adjuvant chemotherapy [22]. Also, previous reports have revealed a significant association between PIK3CA mutations and resistance to therapies targeting EGFR and ERBB2 [23, 24]. Certainly, how early-stage LUAD patients with these alterations would respond to inhibitors of the PIK3CA pathway is a question that will need to be answered in the future. It is worthwhile to note that our analysis of clinical outcomes based on the identified mutation profiles and molecular phenotypes is limited by the cohort size and by the number of patients exhibiting specific mutated genes. In this context, our findings generate hypotheses on the role of mutations and CNVs in the clinical outcome of early-stage LUAD that warrant further analysis and validation in different cohorts.
It is still not clear what molecular factors may influence patient response to immune-based therapies and whether different molecular subsets of LUADs exhibit differential immune responses. In this context, we sought to analyze the association of immune marker expression with the mutational phenotype in early-stage LUAD. TP53 mutant LUADs exhibited both an overall increased immune response including significantly elevated expression of PD-L1 as well as relatively high-somatic mutation burdens. Of note, LUADs with a higher nonsynonymous mutation burden in NSCLCs treated with the anti-PD-1 antibody pembrolizumab exhibited improved objective response [25]. It is plausible that the elevated immune marker expression in TP53 mutant LUADs is linked to the increase somatic mutation burden in these tumors and not to the tumor suppressor itself. Indeed, we also found in TP53 mutant LUADs elevated expression levels of Granzyme B and CD57 which were also significantly and positively correlated with somatic mutation burdens.
We also found that STK11 mutant LUADs exhibited significantly suppressed levels of tumor-infiltrating CD4+ and CD8+ T-cells. PIK3CA or KRAS mutant LUADs with co-occurring STK11 mutations also displayed down-regulated expression of PD-L1. Our data on the association of STK11 mutations with reduced immune response are consistent with a recent study probing co-occurring mutations and immune alterations in KRAS-mutant LUADs [26]. It is plausible that mutations in STK11 or PIK3CA affect the interaction of LUADs with the immune microenvironment through specific contextual cues. Our group has demonstrated that the PD-L1-targeting antibody MPDL3280A (atezolizumab) was most effective when tumors exhibited high levels of PD-L1 [12], suggesting that the status of preexisting immunity is an important predictor of response to immunotherapies. Thus, somatic mutations such as PIK3CA and STK11 that may alter preexisting antitumor immunity, e.g. PD-L1 signaling, might serve as viable biomarkers for response to immunotherapies such as PD-L1-targeting antibodies. Efforts are underway to probe and validate the clinical utility of these mutations in predicting early-stage LUAD patients who will benefit from anti-PD-L1 therapy.
In conclusion, our WES and immune profiling analysis identified prognostic genomic alterations as well as mutations and CNVs that were predictive of response to adjuvant chemotherapy in early-stage LUAD. Our study also reveals dynamic relationships between the tumor and host immune response that define unique subsets of early-stage LUAD with potentially varying vulnerability to immune-based therapies. Lastly, our findings highlight mutations that could serve as candidate biomarkers for targeted immunotherapies, particularly anti-PD-L1 therapy.
Funding
This work was supported by Yale Gilead Program [R.S.H, R.P.L. and J.S.], Department of Defense (DoD) PROSPECT grant W81XWH-07-1-0306 [I.I.W.]; The Cancer Prevention and Research Institute of Texas (CPRIT) multi-investigator research award RP120713 [I.I.W.]; CPRIT individual investigator research award RP150079 [H.K.]; The University of Texas Specialized Programs of Research Excellence (SPORE) in lung cancer grant P50CA70907 [I.I.W]; Yale University SPORE in lung cancer grant P50CA196530 [R.S.H. and E.K.] and R01CA155196 [R.S.H].
Disclosure
IIW is an advisor to Genentech/Roche, Genecentric, GlaxoSmithKline, GE Healthcare, Lilly, Clovis Oncology, Bristol-Myers Squibb and Celgene and has received funding from Genetech, Merck, OncoPlex Diagnostics, Myriad Genetics, Bayer and Jounce Therapeutics outside the submitted work. RSH is an advisor to Koltan pharmaceuticals, DaiTech Oncology, and Biothera, received funding from Genentech and Gilead Sciences and has received Honoraria from Lily, Merck, Boehringer Ingelheim, Pfizer, AstraZeneca/MedImmune, Genetech and Bristol-Myers Squibb outside the submitted work. DR reports stock in Metamark Genetics, received honoraria from Amgen, ACD Inc, Astrazeneca and Bristol-Myers Squibb, is an advisor to Biocept, Cernostics, Genoptix and Perkin Elmer and received funding from Gilead Sciences, Cepheid, Koltan Pharmaceuticals, OncoPlex Diagnostics and Perkin Elmer outside the submitted work. SS is an advisor to GlaxoSmithKline outside the submitted work. JPT reports stock or ownership in Penrose Care, is an advisor to Merck, Sanofi Pasteur and Vitality outside the submitted work and has received funding from Gilead Sciences. ZM and SGG received funding from Gilead Sciences. JF received funding from Astellas Pharma outside the submitted work. TJL reports stock or ownership in and received honoraria from Bristol-Myers Squibb outside the submitted work. All remaining authors have declared no conflicts of interest.
Supplementary Material
References
- 1. Herbst RS, Heymach JV, Lippman SM. Lung cancer. N Engl J Med 2008; 359: 1367–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kadara H, Kabbout M, Wistuba II. Pulmonary adenocarcinoma: a renewed entity in 2011. Respirology 2012; 17: 50–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Devarakonda S, Morgensztern D, Govindan R. Genomic alterations in lung adenocarcinoma. Lancet Oncol 2015; 16: e342–351. [DOI] [PubMed] [Google Scholar]
- 4. Cancer Genome Atlas Research N. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014; 511: 543–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Imielinski M, Berger AH, Hammerman PS. et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell 2012; 150: 1107–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dranoff G. Immunotherapy at large: balancing tumor immunity and inflammatory pathology. Nat Med 2013; 19: 1100–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Quezada SA, Peggs KS, Simpson TR, Allison JP. Shifting the equilibrium in cancer immunoediting: from tumor tolerance to eradication. Immunol Rev 2011; 241: 104–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schalper KA, Brown J, Carvajal-Hausdorf D. et al. Objective measurement and clinical significance of TILs in non-small cell lung cancer. J Natl Cancer Inst 2015; 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brahmer JR. Immune checkpoint blockade: the hope for immunotherapy as a treatment of lung cancer? Semin Oncol 2014; 41: 126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Topalian SL, Hodi FS, Brahmer JR. et al. Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. N Engl J Med 2012; 366: 2443–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Henick BS, Herbst RS, Goldberg SB. The PD-1 pathway as a therapeutic target to overcome immune escape mechanisms in cancer. Expert Opin Ther Targets 2014; 18: 1407–1420. [DOI] [PubMed] [Google Scholar]
- 12. Herbst RS, Soria JC, Kowanetz M. et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014; 515: 563–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cardnell RJ, Behrens C, Diao L. et al. An integrated molecular analysis of lung adenocarcinomas identifies potential therapeutic targets among TTF1-negative tumors, including DNA repair proteins and Nrf2. Clin Cancer Res 2015; 21: 3480–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tang X, Kadara H, Behrens C. et al. Abnormalities of the TITF-1 lineage-specific oncogene in NSCLC: implications in lung cancer pathogenesis and prognosis. Clin Cancer Res 2011; 17: 2434–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Choi M, Scholl UI, Ji W. et al. Genetic diagnosis by whole exome capture and massively parallel DNA sequencing. Proc Natl Acad Sci USA 2009; 106: 19096–19101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhao S, Choi M, Overton JD. et al. Landscape of somatic single-nucleotide and copy-number mutations in uterine serous carcinoma. Proc Natl Acad Sci USA 2013; 110: 2916–2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Choi M, Scholl UI, Yue P. et al. K+ channel mutations in adrenal aldosterone-producing adenomas and hereditary hypertension. Science 2011; 331: 768–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Je EM, Gwak M, Oh H. et al. Frameshift mutations of axon guidance genes ROBO1 and ROBO2 in gastric and colorectal cancers with microsatellite instability. Pathology 2013; 45: 645–650. [DOI] [PubMed] [Google Scholar]
- 19. Liang H, Cheung LW, Li J. et al. Whole-exome sequencing combined with functional genomics reveals novel candidate driver cancer genes in endometrial cancer. Genome Res 2012; 22: 2120–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Waddell N, Pajic M, Patch AM. et al. Whole genomes redefine the mutational landscape of pancreatic cancer. Nature 2015; 518: 495–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hakimi AA, Ostrovnaya I, Reva B. et al. Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: a report by MSKCC and the KIRC TCGA research network. Clin Cancer Res 2013; 19: 3259–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Solis LM, Behrens C, Dong W. et al. Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res 2010; 16: 3743–3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hanker AB, Pfefferle AD, Balko JM. et al. Mutant PIK3CA accelerates HER2-driven transgenic mammary tumors and induces resistance to combinations of anti-HER2 therapies. Proc Natl Acad Sci USA 2013; 110: 14372–14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Majewski IJ, Nuciforo P, Mittempergher L. et al. PIK3CA mutations are associated with decreased benefit to neoadjuvant human epidermal growth factor receptor 2-targeted therapies in breast cancer. J Clin Oncol 2015; 33: 1334–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rizvi NA, Hellmann MD, Snyder A. et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015; 348: 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Skoulidis F, Byers LA, Diao L. et al. Co-occurring genomic alterations define major subsets of KRAS-mutant lung adenocarcinoma with distinct biology, immune profiles, and therapeutic vulnerabilities. Cancer Discov 2015; 5: 860–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.