Abstract
Temporal variation in microbiome measurements can reduce statistical power in research studies. Quantification of this variation is essential for designing studies of chronic disease. We analyzed 16S ribosomal RNA profiles in paired biological specimens separated by 6 months from 3 studies conducted during 1985–2013 (a National Cancer Institute colorectal cancer study, a Costa Rica study, and the Human Microbiome Project). We evaluated temporal stability by calculating intraclass correlation coefficients (ICCs). Sample sizes needed in order to detect microbiome differences between equal numbers of cases and controls for a nested case-control design were calculated on the basis of estimated ICCs. Across body sites, 12 phylum-level ICCs were greater than 0.5. Similarly, 11 alpha-diversity ICCs were greater than 0.5. Fecal beta-diversity estimates had ICCs over 0.5. For a single collection with most microbiome metrics, detecting an odds ratio of 2.0 would require 300–500 cases when matching 1 case to 1 control at P = 0.05. Use of 2 or 3 sequential specimens reduces the number of required subjects by 40%–50% for low-ICC metrics. Relative abundances of major phyla and alpha-diversity metrics have low temporal stability. Thus, detecting associations of moderate effect size with these metrics will require large sample sizes. Because beta diversity for feces is reasonably stable over time, smaller sample sizes can detect associations with community composition. Sequential prediagnostic specimens from thousands of prospectively ascertained cases are required to detect modest disease associations with particular microbiome metrics.
Keywords: epidemiologic methods, microbiome, microbiota, statistical power, temporal stability
Editor’s note: An invited commentary on this article appears on page 1291.
The human microbiota comprise the collection of microbes inhabiting the human body, including bacteria, archaea, fungi, and other eukaryotic microbes. Advances in low-cost high-throughput sequencing (1) and bioinformatic analyses (2–4) now allow the characterization of human microbial communities. Importantly, the human microbiome has been recently shown to be associated with selected health conditions: the fecal microbiota with obesity (5–7), colorectal cancer (CRC) (8), estrogen levels (9), postmenopausal breast cancer (10), and inflammatory bowel disease (11); the oral microbiota with pancreatic (12), oral (13), and gastrointestinal (13) cancers; the vaginal microbiota with bacterial vaginosis (14); and the skin microbiota with atopic dermatitis and other skin diseases (15–17). Interventions with medications or diet produce alterations in microbial communities (17–19). Together, these results demonstrate the potential of microbiome research for elucidating the etiology, prevention, and possibly treatment of complex human diseases. While promising, some important questions need to be answered in order for large-scale epidemiologic studies to clarify the role of microbiota in human health (20).
The aim of a prospective epidemiologic study is to identify whether selected factors predict a disease outcome. Due to cost, compliance, and degree of invasiveness, many exposures are typically measured only at baseline, which works well if the factor is temporally stable. At the community level, the microbiome has been reported to be reasonably stable over time (21–23). In contrast, other studies found substantial temporal variability in composition for particular bacteria at different body sites (15, 23–26), but between-person variability was still much larger than differences over time. If a variable exposure is measured at a single time point, its temporal instability dramatically reduces the study’s statistical power to detect associations and also can bias the estimate of its effect (27). It is therefore crucial to systematically evaluate the temporal stability of microbiome measures, including the relative abundances of taxa, alpha diversity, and beta diversity, in order to accurately determine sample sizes required for future studies.
We evaluated the temporal stability of specific microbiome metrics, based on 16S ribsomal RNA (rRNA) gene profiles, in 3 populations of persons who provided 2 biological specimens separated by approximately 6 months: 1) hospital-based controls from a National Cancer Institute CRC study (28, 29); 2) randomly sampled adults in the Guanacaste region of Costa Rica (Dr. Paula González, Guanacaste Epidemiology Project–INCIENSA Foundation (San José, Costa Rica), unpublished data, 2015); and 3) participants in the National Institutes of Health’s Human Microbiome Project (HMP) (30–32). Our objective was to estimate the degree of temporal microbiome variability within individuals and the impact that will have on sample size requirements for future studies. We focused on case-control studies nested within prospective studies, since sequencing the microbiome for an entire cohort would be prohibitively expensive. We estimated intraclass correlation coefficients (ICCs) in these 3 studies; higher ICCs imply larger statistical power and smaller bias in estimating an exposure’s effect. This research is critical for designing large-scale epidemiologic studies of complex human diseases, including cancer (33, 34).
METHODS
CRC case-control study—feces
We used fecal specimens from hospital-based controls of a National Cancer Institute CRC case-control study (1985–1987) (8, 28, 29). Control subjects were recruited from patients awaiting elective surgery for nononcological, nongastrointestinal conditions at 3 Washington, DC-area hospitals. Before hospitalization and treatment, participants provided 2-day fecal specimens that were freeze-dried. Forty-four controls made 2 study visits separated by approximately 6 months (Table 1). DNA extraction, sequencing, and bioinformatic processing were conducted at New York University School of Medicine (New York, New York) and were described in an earlier publication (8). Briefly, DNA was extracted from fecal specimens using the MO BIO PowerSoil DNA Isolation Kit (MO BIO Laboratories, Inc., Carlsbad, California). We generated 16S rRNA gene amplicons covering variable regions V3–V4 using the 454 Roche GS-FLX Titanium pyrosequencing system (Roche Diagnostics Corporation, Indianapolis, Indiana).
Table 1.
Sample Sizes Used in Replicate Measurements of Microbiota Collected in 3 Studies, 1985–2013
| Study | Sample Size, no. |
|---|---|
| NCI CRC case-control study (1985–1987) (28, 29) | |
| Stool | 44 |
| Costa Rica study (2010–2012)a | |
| Stool | 116 |
| Saliva | 42 |
| HMP (2008–2013) (30) | |
| Stool | 107 |
| Saliva | 94 |
| Mouth (oral average) | 103 |
| Airways (anterior nares) | 78 |
| Skin (average) | 63 |
| Vagina (average) | 37 |
Abbreviations: CRC, colorectal cancer; HMP, Human Microbiome Project; NCI, National Cancer Institute.
a Dr. Paula González, INCIENSA Foundation, unpublished data, 2015.
Costa Rica study—feces and saliva
In the Costa Rica study, fecal (n = 116) and saliva (n = 42) specimens were collected at 2 study visits separated by 6 months (2010–2012) in a randomly selected population in Costa Rica (Table 1). For the fecal collection, participants were provided with a self-collection kit with detailed instructions on collecting specimens from the first stool of the day. Fecal specimens were collected in 20-mL screw-top Sarstedt tubes (Sarstedt AG & Company KG, Nümbrecht, Germany) that had been prefilled with 5 mL of RNAlater (QIAGEN, Valencia, California) (9). After collection, the participant stored the fecal specimen in a thermal container with dry ice. The study staff collected specimens from the participants’ homes or the participants brought them to the clinic, where they were transferred for storage in liquid nitrogen within 24 hours.
Saliva specimens were collected at the time of the clinic visits. Participants were instructed to let saliva collect in the mouth for at least 30 seconds and then spit the saliva into a sterile collection tube. The process was repeated until the target collection volume of 2–3 mL was reached. In the laboratory, the saliva samples were mixed with 5 mL of RNAlater and frozen in liquid nitrogen.
DNA was extracted from both the fecal and saliva samples and sequenced at the Institute of Genome Sciences, University of Maryland School of Medicine (Baltimore, Maryland), as described previously (9, 35). Briefly, the samples were extracted with a modification of the QIAamp DNA Stool Mini Kit (QIAGEN) (9). An approximately 469-base-pair segment of the 16S rRNA gene V3–V4 hypervariable region of the DNA was amplified with primers that included a linker sequence (suitable for the MiSeq 300PE Illumina sequencer; Illumina, Inc., San Diego, California), a 12-base-pair index sequence, a heterogeneity spacer (to minimize bias with low-diversity amplicons), and 16S rRNA gene universal primers 319F/806R. The amplicons were sequenced in a single pool in 1 run on the MiSeq instrument using the 300PE protocol, generating approximately 2.22 GB of data.
Human Microbiome Project—multiple body sites
The HMP was established in 2008 and completed in 2013 (30–32). We downloaded the HMP 16S rRNA gene database with its associated mapping files from the HMP Data Analysis and Coordination Center (36). The HMP subjects were sampled for microbiota at 15 body sites (for males) or 18 body sites (for females), including feces, oropharynx (buccal mucosa, hard palate, keratinized gingiva, palatine tonsils, saliva, subgingival plaque, supragingival plaque, throat, and tongue dorsum), anterior nares, skin (left and right antecubital fossa, left and right retroauricular creases), and vagina (midvagina, posterior fornix, and vaginal introitus). A subset of HMP subjects (stool: n = 107; saliva: n = 94; mouth: n = 103; airways: n = 78; skin: n = 63; vagina: n = 37) were sampled at a second time point that was separated from the first by an average of 219 (standard deviation, 69) days (30), which we included for this analysis. Our analysis focused on the V3–V5 sequence data.
16S sequence data processing and microbiota measurements
The Quantitative Insights Into Microbial Ecology analysis pipeline (2) was used to assemble and filter the 16S rRNA sequence reads, removing reads with low-quality scores and reads judged to be chimeras or to have sequencing artifacts. The retained, high-quality sequence reads that had at least 97% sequence identity were clustered into operational taxonomic units. Taxonomy was assigned using closed reference operational taxonomic unit picking in comparison with the Greengenes reference set, version 12_10 (37, 38). Reads that did not match a reference sequence at ≥97% sequence identity were discarded.
We calculated the relative abundance of the top 5 bacterial phyla (Actinobacteria, Bacteroidetes, Firmicutes, Fusobacteria, and Proteobacteria), 4 alpha-diversity metrics (number of observed species (Sobs) or operational taxonomic unit, Chao1, Shannon index, and phylogenetic distance—whole tree (PD_tree)), and 2 beta-diversity metrics (unweighted and weighted UniFrac distance). Alpha-diversity metrics were calculated as the average of 20 rarefaction subsampling repeats to 1,000 sequencing reads to retain individuals analyzed using 454 pyrosequencing. We also performed rarefaction to 5,000 sequencing reads as a sensitivity analysis (data not reported). Similar to alpha diversity, beta-diversity metrics were also calculated after rarefying to 1,000 sequencing reads. No existing statistical methods are available with which to appropriately quantify temporal stability for beta-diversity distance matrices. Thus, we performed a principal coordinates analysis (PCoA) to calculate the top factors that captured a large proportion of the information of the distance matrix to be used for analysis.
ICCs of microbiome measurements
The value of an ICC ranges from 0 (no reproducibility) to 1 (perfect reproducibility). For each microbiome metric, we calculated ICCs to quantify the biological variability for each of the microbiome metrics, defined as
Here represents between-individual variability and represents the variance component capturing both technical variability and temporal instability. Note that the 2 variance components related to technical reproducibility and temporal instability can be separated if technical replicates are available. Although we did not separate the 2 components in the current article due to a lack of technical replicates, the ICC reflects the effective sample size after accounting for both technical reproducibility and temporal stability. The parameters and were estimated by a linear mixed-effects model using the R package “lme” (R Foundation for Statistical Computing, Vienna, Austria). For the CRC and Costa Rica data, we adjusted for sex and age in the model. For the HMP data, we adjusted for sex, age, and sequencing center to account for the variability introduced by the sequencing centers. For a given microbiome metric, the ICC is directly related to the statistical power of testing an association in an epidemiologic study. The standard error for the estimated ICC was approximated by bootstrapping subjects at the cluster level—for example, keeping the relationship of sample pairs for each subject unchanged.
For HMP data, we report ICCs for feces, saliva, and anterior nares. We also present averaged ICCs for 3 sites—the oropharynx (buccal mucosa, hard palate, keratinized gingiva, palatine tonsils, subgingival plaque, supragingival plaque, throat, and tongue dorsum), skin (left and right antecubital fossa, left and right retroauricular creases), and vagina (midvagina, posterior fornix, and vaginal introitus)—since population studies would most likely evaluate these microbiomes from a combined-organ perspective rather than by subsite within the organ (e.g., vagina rather than midvagina, posterior fornix, and vaginal introitus) in relation to the outcome. The ICCs for the subsites are presented separately in Web Tables 1–4 (available at https://academic.oup.com/aje). The standard error for the average ICC was approximated by bootstrapping based on 1,000 random samples with replacement. To keep the correlation between body sites, bootstrapping was performed by sampling subjects, and ICCs across body sites were calculated on the basis of the same set of bootstrapped subjects.
Estimating required sample sizes for future studies
Our second objective was to evaluate the effect of variability on statistical power to inform future epidemiologic study design. We wanted to estimate how much power would be gained by using longitudinal repeated specimens as compared with a single-specimen design. We used the estimated ICCs from the fecal specimens to determine the sample sizes that would be necessary to detect associations between various microbiome metrics and disease, given specific effect sizes. We estimated the numbers of individuals that would be needed for both a 1:1 matched case-control study and a 1:3 matched case-control study (nested within a cohort or as an independent study) in order to have 80% power to detect an association between microbiome metrics and a disease at P values equal to 0.05, 0.001, 0.0001, and 0.00001.
For estimation of the required sample size based on the calculated ICCs, let y denote the case-control status and x denote a “long-term” (multiple values averaged over an extended time period) microbiome metric (e.g., relative abundance of a taxon, alpha diversity, or PCoA scores based on a beta-diversity matrix) (see Web Appendix). We assume a logistic regression model . Let K denote the number of repeat specimens for each subject. We assume that we can use the average of the K repeats to estimate the “long-term” metric. In addition, suppose that the study has N subjects with cases and controls. In the Web Appendix, we derive the noncentrality parameter for a score statistic testing :
where and are the expectation of the microbiome metric in the case group and the control group, respectively.
Let be the quantile for the standard normal distribution. The power of detecting an association with P value threshold is given by
Thus, to achieve 80% power,
Combing the above 2 equations, we derive the sample size required to achieve 80% power:
In this analysis, we considered sample size requirements for the relative abundances of 5 taxa, 4 alpha-diversity metrics, and the top PCoA scores for the beta-diversity metrics (unweighted and weighted UniFrac distances) to detect differences between cases and an equal number of controls. Calculations were based on critical values of 0.05, 0.001, 0.0001, and 0.00001. All calculations were performed in R. The R code for power calculation is provided in the Web Appendix.
Let be the 25% quantile and be the 75% quantile of the microbiome metric x. In the Web Appendix, we show that the power (and required sample size) is a function of the odds ratio for the top 25% microbiome metric versus the bottom 25% microbiome metric.
RESULTS
Temporal stability of fecal samples
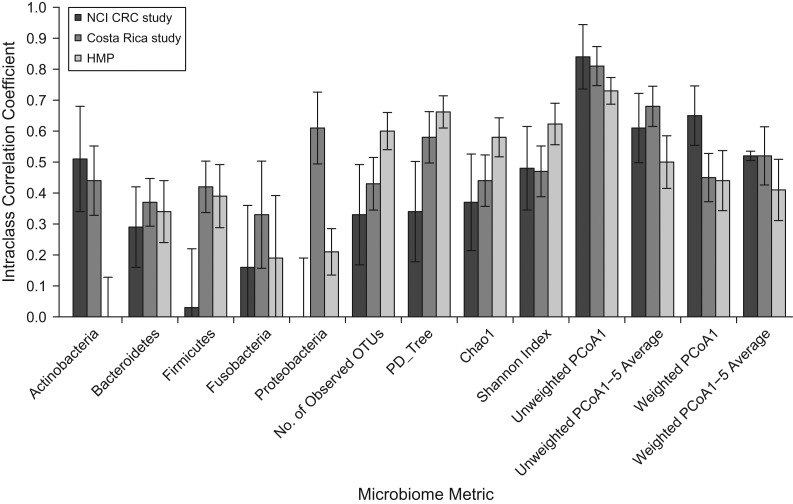
The ICCs for the relative abundance of 5 common phyla, 4 estimates of alpha diversity, and the top PCoA scores based on unweighted and weighted UniFrac distance matrices are presented in Figures 1–3 and Web Tables 1–4. The proportions of explained variance of the top 5 PCoA scores are reported in Web Tables 5 and 6. For the stool microbiome (Figure 1), the ICCs had a wide range, from 0.00 for Proteobacteria to 0.84 for unweighted UniFrac first principal coordinates analysis (PCoA1). Higher ICCs (>0.50) were noted for the average of the top 5 PCoA scores based on unweighted UniFrac in all 3 studies. For relative abundance at the phylum level across all 3 studies, 12 ICCs were 0.50 or greater (Web Table 1). However, as Figures 1–3 show for each study, ICCs were very low for the relative abundance of Actinobacteria and Fusobacteria (ICC = 0.00 and ICC = 0.19, respectively) in the HMP, and in the CRC study, the ICCs for the relative abundance of Firmicutes, Fusobacteria, and Proteobacteria were also low (ICC = 0.03, ICC = 0.16, and ICC = 0.00, respectively). The ICCs for PD_ tree were 0.66 in the HMP, 0.58 in the Costa Rica study, and 0.34 in the CRC study.
Figure 1.
Within-subject stability intraclass correlation coefficients for fecal samples in 3 studies (a National Cancer Institute (NCI) colorectal cancer (CRC) study (28, 29), a Costa Rica study (Dr. Paula González, INCIENSA Foundation, unpublished data, 2015), and the Human Microbiome Project (HMP) (30)) for 5 phyla, 4 alpha-diversity metrics, and the first principal coordinate (PCoA1) and average of 1–5 principal coordinates (PCoA1–5) of 2 beta-diversity metrics. OTUs, operational taxonomic units; PCoA, principal coordinates analysis; PD_Tree, phylogenetic distance—whole tree. Bars, 95% confidence intervals.
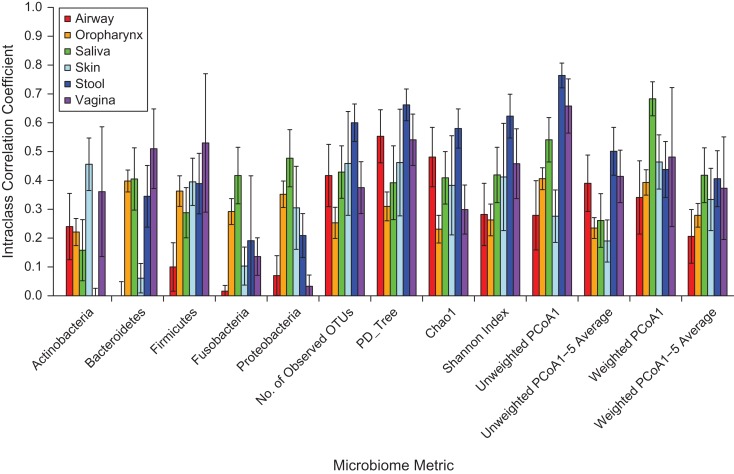
Figure 3.
Within-subject stability intraclass correlation coefficients for samples of fecal, saliva, average oropharynx, nares, average skin, and average vagina microbiota in the Human Microbiome Project (HMP) (30) for 5 phyla, 4 alpha-diversity metrics, and the first principal coordinate (PCoA1) and average of 1–5 principal coordinates (PCoA1–5) of 2 beta-diversity metrics. OTUs, operational taxonomic units; PCoA, principal coordinates analysis; PD_Tree, phylogenetic distance—whole tree. Bars, 95% confidence intervals.
Temporal stability of saliva samples
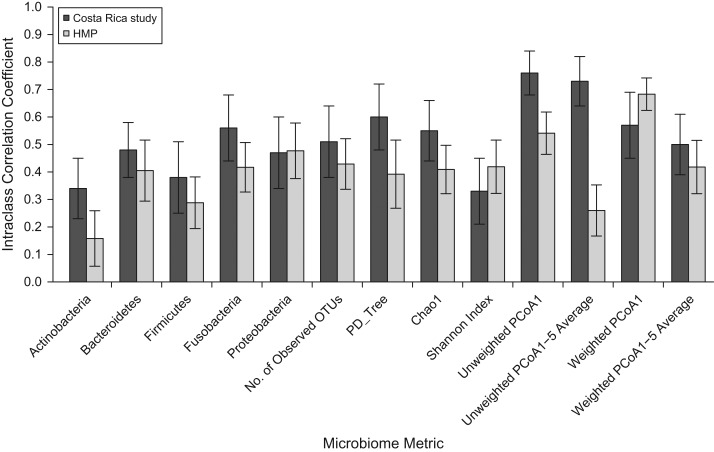
Repeat saliva samples were obtained in the Costa Rica study and the HMP. For all saliva microbiome metrics, ICCs were higher in Costa Rica than in the HMP, often by 10%–50% (Figure 2). The lowest saliva ICC was for the relative abundance of Actinobacteria (ICC = 0.16) in the HMP, and the highest ICC was for the average of the top 5 PCoA scores for unweighted UniFrac (ICC = 0.73) in the Costa Rica study.
Figure 2.
Within-subject stability intraclass correlation coefficients for saliva samples in 2 studies (a Costa Rica study (Dr. Paula González, INCIENSA Foundation, unpublished data, 2015) and the Human Microbiome Project (HMP) (30)) for 5 phyla, 4 alpha-diversity metrics, and the first principal coordinate (PCoA1) and average of 1–5 principal coordinates (PCoA1–5) of 2 beta-diversity metrics. OTUs, operational taxonomic units; PCoA, principal coordinates analysis; PD_Tree, phylogenetic distance—whole tree. Bars, 95% confidence intervals.
Temporal stability of samples from other body sites
Data for additional body sites were available only from the HMP (Figure 3). We also included stool and saliva samples in Figure 3 for comparison with other body sites. ICCs for various body sites in the HMP varied enormously (Figure 3). For the relative abundance of the different bacterial phyla, the ICCs were relatively low, except for the relative abundance of Bacteroidetes and Firmicutes in the vagina, which had ICCs of 0.57 and 0.59, respectively. Alpha-diversity ICCs were highest for stool samples compared with other sites. Unweighted UniFrac PCoA1 ICCs were high for stool (ICC = 0.76), saliva (ICC = 0.54), and averaged vagina (ICC = 0.68). Unweighted UniFrac PCoA1 ICCs were 0.25 for skin and 0.28 for nares. Averaged unweighted UniFrac top-5 PCoA ICCs were generally lower than only PCoA1 ICCs for all sites. Weighted UniFrac ICCs were also lower than unweighted UniFrac ICCs overall, except for the saliva sample for the PCoA1 ICC.
Estimates of sample size requirements for a fecal microbiome study
For a large association (i.e., odds ratio = 3.5) with 1 fecal specimen at P = 0.05, approximately 100–400 cases would be sufficient for all microbiome metrics when matching 1 case to 1 control. In contrast, smaller associations (i.e., odds ratio = 1.5) could only be detected in studies that were 6-fold larger (approximately 1,000–3,000 cases; Table 2). As expected, fewer cases are needed for metrics with higher ICCs (i.e., lower within-subject variability), particularly for unweighted UniFrac. The required sample size is also lower when multiple specimens per subject over time would be collected, and the benefit is substantial for low-ICC metrics. Detecting an odds ratio of 1.5 for the relative abundance of Fusobacteria (ICC = 0.18) would require 3,659 cases with 1 specimen, 2,158 cases with a second specimen, and 1,658 cases with a third specimen. Detection of an odds ratio of 1.5 in unweighted UniFrac (ICC = 0.81) would require only 813 cases, which decreases to 735 cases if a second specimen is available and 710 cases if a third specimen is available. In the Web tables, we have provided detailed sample-size calculations for 1:1 case-control matching (Web Tables 7–10) and 1:3 case-control matching (Web Tables 11–14) at different levels of significance (P’s = 0.05–0.00001).
Table 2.
Numbers of Cases Required to Detect a Significant Associationa in 1, 2, or 3 Fecal Specimens Using an Intraclass Correlation Coefficient From 3 Fecal Sample Populations
| Measure and No. of Specimens | Estimated ICCb | Odds Ratioc | ||||
|---|---|---|---|---|---|---|
| 1.5 | 2.0 | 2.5 | 3.0 | 3.5 | ||
| Relative Abundance of Phylum-Level Taxa | ||||||
| Actinobacteria | 0.44 | |||||
| 1 | 1,496 | 498 | 273 | 190 | 151 | |
| 2 | 1,077 | 359 | 196 | 136 | 109 | |
| 3 | 938 | 312 | 171 | 119 | 94 | |
| Bacteroidetes | 0.37 | |||||
| 1 | 1,780 | 593 | 325 | 226 | 180 | |
| 2 | 1,219 | 406 | 222 | 154 | 123 | |
| 3 | 1,032 | 344 | 188 | 131 | 104 | |
| Firmicutes | 0.42 | |||||
| 1 | 1,568 | 522 | 286 | 199 | 158 | |
| 2 | 1,113 | 371 | 203 | 141 | 112 | |
| 3 | 961 | 320 | 175 | 122 | 97 | |
| Fusobacteria | 0.18 | |||||
| 1 | 3,659 | 1,219 | 668 | 464 | 370 | |
| 2 | 2,158 | 719 | 394 | 274 | 218 | |
| 3 | 1,658 | 552 | 302 | 210 | 167 | |
| Proteobacteria | 0.29 | |||||
| 1 | 2,271 | 757 | 414 | 288 | 229 | |
| 2 | 1,464 | 488 | 267 | 186 | 148 | |
| 3 | 1,196 | 398 | 218 | 151 | 121 | |
| Alpha Diversity | ||||||
| PD_tree | 0.58 | |||||
| 1 | 1,135 | 378 | 207 | 144 | 114 | |
| 2 | 897 | 299 | 163 | 113 | 90 | |
| 3 | 817 | 272 | 149 | 103 | 82 | |
| Chao1 | 0.44 | |||||
| 1 | 1,496 | 498 | 273 | 190 | 151 | |
| 2 | 1,077 | 359 | 196 | 136 | 109 | |
| 3 | 938 | 312 | 171 | 119 | 94 | |
| No. of species | 0.43 | |||||
| 1 | 1,531 | 510 | 279 | 194 | 155 | |
| 2 | 1,095 | 365 | 199 | 139 | 110 | |
| 3 | 949 | 316 | 173 | 120 | 96 | |
| Shannon index | 0.47 | |||||
| 1 | 1,401 | 467 | 255 | 177 | 141 | |
| 2 | 1,030 | 343 | 188 | 130 | 104 | |
| 3 | 906 | 302 | 165 | 115 | 91 | |
| Beta Diversity | ||||||
| Unweighted UniFrac PCoA1 | 0.81 | |||||
| 1 | 813 | 271 | 148 | 103 | 82 | |
| 2 | 735 | 245 | 134 | 93 | 74 | |
| 3 | 710 | 236 | 129 | 90 | 71 | |
| Weighted UniFrac PCoA1 | 0.48 | |||||
| 1 | 1,372 | 457 | 250 | 174 | 138 | |
| 2 | 1,015 | 338 | 185 | 128 | 102 | |
| 3 | 896 | 298 | 163 | 113 | 90 | |
Abbreviations: ICC, intraclass correlation coefficient; PCoA1, first principal coordinates analysis; PD_tree, phylogenetic distance—whole tree.
a Number of cases (assuming an equal number of controls) required to detect an association at a significance level of 0.05 with 80% power, based on 1, 2, or 3 fecal specimens per subject and the ICC estimated from the fecal samples from the 3 populations. Disease prevalence = 1%.
b ICCs were the median values of 3 estimates from 3 studies: a National Cancer Institute colorectal cancer study (28, 29), the Human Microbiome Project (30), and a Costa Rica study (Dr. Paula González, INCIENSA Foundation, unpublished data, 2015).
c Odds ratio for the top 25% microbiome metric versus the bottom 25% microbiome metric.
DISCUSSION
In this analysis of the temporal stability of relative abundance, alpha diversity, and beta diversity, we found that ICCs for temporal stability over a period of 6 months were generally 0.5 or below for the majority of the phylum-level relative abundances and alpha-diversity metrics across different types of specimens. This finding implies that even nominally significant associations with these unstable metrics should be interpreted with caution and that sample sizes need to be quite large for these types of analyses. In contrast, unweighted UniFrac, one measure of beta diversity, was relatively stable not only for stool specimens but also for oral, oropharynx, and vagina specimens. Unweighted UniFrac is the pairwise comparison of the sizes and shapes of each specimen’s phylogenetic tree without consideration of relative abundances. In contrast, weighted UniFrac, which considers the relative abundances of taxa and also utilizes the phylogenetic tree, had lower ICCs. This observation favors unweighted UniFrac as a useful metric for identifying disease associations or predictions based on overall differences in detection of the many different microbes, as compared with the relative abundance of particular phyla. For a single collection with most microbiome metrics, detecting an odds ratio of 2.0 would require 300–500 cases when matching 1 case to 1 control at P = 0.05. Smaller case numbers would be required to detect associations between particular pathogenic bacteria that directly cause disease, as is known for Mycobacterium tuberculosis or Clostridium difficile, since the hypothesized association would be large.
To be successful in conducting an epidemiologic study of the microbiome, investigators must have knowledge of both technical reproducibility and the temporal stability of human microbiota; this can be quantified by the ICC, which is directly related to statistical power for testing associations. Technical reproducibility has been reported in some studies for fecal samples (39, 40). In our recent work, we have found that the technical reproducibility in fecal samples is very high (taxa: ICCs >80%; alpha/beta diversity: ICCs >90%) (41, 42). However, temporal stability ICCs have not been systematically investigated across body sites using a large number of subjects. The many low ICCs that we found contribute to the substantial challenges in replicating novel associations with the microbiome. These results have important implications for designing and scaling epidemiologic studies.
Previous studies have evaluated the temporal stability of the microbiome. In 2 individuals examined daily for 1 year, overall community composition was stable (43). However, next-day correlations with fiber intake and marked alterations associated with international travel or enteric infection were also noted. Zhou et al. (26) and Ding and Schloss (44) investigated temporal variation of microbiota in the HMP subjects. In the Zhou et al. study (26), the Spearman correlation between 2 vectors of taxon relative abundances corresponding to 2 study visits was calculated for each subject. By averaging across subjects, Zhou et al. concluded that oral and fecal samples were the most stable temporally, whereas skin and vaginal samples were the most unstable. However, their calculations did not reflect the instability of individual microbiome metrics across participants and thus did not provide quantified information necessary for prospective epidemiologic studies. With such limitations, there is concern that these conclusions might be used erroneously to guide the design of epidemiologic studies. For example, it was stated that vaginal samples were very unstable, with Spearman correlations close to zero, suggesting no power for association studies. However, in our analysis, the ICC for unweighted UniFrac was reasonably high (averaged across 3 vaginal sites, ICC = 0.68 for PCoA1), implying only moderate power loss due to temporal variation. In addition, temporal stability evaluated with their approach provides little or no information relevant to the temporal stability of within-subject alpha diversity or community composition (i.e., beta diversity). Ding and Schloss (44) developed a simple Markov model to characterize the temporal stability; however, the inferred model parameters cannot be used directly to calculate the effective sample size for an epidemiologic study.
There are 3 methods with which to increase statistical power for microbiome analyses in epidemiologic studies, particularly for case-control studies nested within prospective cohort studies. One method for improving power is to have more endpoints in the study, which could be accomplished by recruiting a larger cohort or following the cohort for a longer time. The second approach is to collect and test specimens at multiple time points for each subject and to average across the specimens. Our analyses of stability ICCs revealed that a second or third specimen per subject is only moderately beneficial for a stable (high-ICC) metric such as unweighted UniFrac, whereas additional specimens greatly reduce the required sample size for unstable (low-ICC) metrics, such as the relative abundance of Fusobacteria. The multiple-specimen approach also affords the opportunity to detect temporal changes in the metric that may predict disease onset. The optimal design depends on the hypothesis (microbiome metric) to be tested (average vs. change) and the relative costs of microbiome sequencing and subject recruitment and retention. When the cost of recruiting subjects is much higher than the laboratory costs, sequencing of multiple specimens over time is expected to have better power. The third method for improving statistical power is to identify factors that contribute to the instability of a microbiome metric and adjust for these factors in association studies. Except for antibiotic use, which has a potent but unquantified effect, such factors are currently under debate.
Investigators in prospective cohort studies need to perform cost-benefit analyses and decide whether it is more expensive to recruit more participants into the study or collect multiple specimens from the same individual. Related methods for increasing the numbers could include pooling or meta-analysis of participants from different studies. It may also be possible to collect multiple samples from a subset of a cohort and use these data to correct the association, as has been done in nutritional data (45, 46). However, if specimens have been collected or analyzed by different methods in various studies, the microbiome metric may not be conducive to being pooled or meta-analyzed. We have found that different methods of collecting samples show bias in the microbiome data (41, 42), and study-specific differences in DNA extraction, polymerase chain reaction amplification, and sequencing also contribute to heterogeneity, making it difficult to pool studies (47). We have evaluated herein a real-world situation in which the ICCs were not standardized for study-specific differences in collection, extraction, amplification, and sequencing.
Our study had several limitations. First, subjects made only 2 study visits which were separated by approximately 6 months. A longitudinal study with multiple sampling over a given time period (months and years) and fixed sampling methods would provide more accurate estimates, especially for unstable (low-ICC) metrics. Second, the number of subjects was low for some body sites (e.g., antecubital fossa and vagina), and subjects originated from only 1 study, the HMP, for many of the body sites. Third, lack of technical replicates prevented the separation of temporal variation from technical variability. However, technical reproducibility is usually high in experienced laboratories, and typical epidemiologic studies are based on single-point sampling with no technical replicates; thus, the ICCs estimated here reflect both technical variability and temporal instability and are directly applicable to statistical power for such epidemiologic studies. However, we have found that the technical reproducibility is very high; thus, the inclusion of multiple aliquots of the same samples will not substantially improve the statistical power for detecting associations (41, 42). Fourth, because we focused on microbial communities, as estimated by 16S rRNA gene amplicons, our work has limited application for research on specific bacterial species or strains or functional pathways that employ other laboratory methods, such as whole-genome shotgun metagenomics. However, because we found that stability ICCs were lowest for the low-abundance phyla, it is highly likely that ICCs for uncommon or rare genera, species, or strains would be as low or even lower.
Identifying which alterations of microbial populations or functions contribute to disease, treatment response, or remission will hinge on comparisons of specimens that are collected prospectively. Researchers conducting prospective studies need to consider sampling needs for adequate statistical power for the various microbiome metrics. In this article, we have quantified temporal variation in microbiome measurements and provided the sample size requirements to help cohort study investigators plan for microbiome analyses.
Supplementary Material
ACKNOWLEDGMENTS
Author affiliations: Division of Cancer Epidemiology and Genetics, National Cancer Institute, Bethesda, Maryland (Rashmi Sinha, James J. Goedert, Emily Vogtmann, Xing Hua, Guoqin Yu, Joshua Sampson, Jianxin Shi); Costa Rican Agency for Biomedical Research-INCIENSA Foundation, San José, Costa Rica (Carolina Porras); Division of Epidemiology, Department of Population Health, School of Medicine, New York University, New York, New York (Richard Hayes, Jiyoung Ahn); and Department of Medical and Scientific Affairs, Roche Molecular Systems, Inc., Pleasanton, California (Mahboobeh Safaeian).
This work was funded by the Intramural Research Program of the National Cancer Institute and by National Cancer Institute grant R03CA159414.
This study utilized the high-performance computational capabilities of the Biowulf Linux cluster (https://hpc.nih.gov/systems/) at the National Institutes of Health (Bethesda, Maryland).
Conflict of interest: none declared.
Abbreviations
- CRC
colorectal cancer
- HMP
Human Microbiome Project
- ICC
intraclass correlation coefficient
- PCoA
principal coordinates analysis
- PCoA1
first principal coordinates analysis
- PD_tree
phylogenetic distance—whole tree
- rRNA
ribosomal RNA
REFERENCES
- 1. Metzker ML. Sequencing technologies—the next generation. Nat Rev Genet. 2010;11:31–46. [DOI] [PubMed] [Google Scholar]
- 2. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Edgar RC, Haas BJ, Clemente JC, et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bokulich NA, Subramanian S, Faith JJ, et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods. 2013;10:57–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Turnbaugh PJ, Hamady M, Yatsunenko T, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ley RE. Obesity and the human microbiome. Curr Opin Gastroenterol. 2010;26(1):5–11. [DOI] [PubMed] [Google Scholar]
- 7. Flier JS, Mekalanos JJ. Gut check: testing a role for the intestinal microbiome in human obesity. Sci Transl Med. 2009;1(6):6ps7. [DOI] [PubMed] [Google Scholar]
- 8. Ahn J, Sinha R, Pei Z, et al. Human gut microbiome and risk for colorectal cancer. J Natl Cancer Inst. 2013;105(24):1907–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Flores R, Shi J, Fuhrman B, et al. Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: a cross-sectional study. J Transl Med. 2012;10:253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goedert JJ, Jones G, Hua X, et al. Investigation of the association between the fecal microbiota and breast cancer in postmenopausal women: a population-based case-control pilot study. J Natl Cancer Inst. 2015;107(8):djv147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sokol H, Seksik P, Furet JP, et al. Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis. 2009;15(8):1183–1189. [DOI] [PubMed] [Google Scholar]
- 12. Farrell JJ, Zhang L, Zhou H, et al. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut. 2012;61(4):582–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ahn J, Chen CY, Hayes RB. Oral microbiome and oral and gastrointestinal cancer risk. Cancer Causes Control. 2012;23(3):399–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Spear GT, Sikaroodi M, Zariffard MR, et al. Comparison of the diversity of the vaginal microbiota in HIV-infected and HIV-uninfected women with or without bacterial vaginosis. J Infect Dis. 2008;198(8):1131–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Grice EA, Kong HH, Conlan S, et al. Topographical and temporal diversity of the human skin microbiome. Science. 2009;324(5931):1190–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Grice EA, Segre JA. The skin microbiome. Nat Rev Microbiol. 2011;9(4):244–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kong HH, Oh J, Deming C, et al. Temporal shifts in the skin microbiome associated with disease flares and treatment in children with atopic dermatitis. Genome Res. 2012;22(5):850–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu GD, Chen J, Hoffmann C, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Smith MI, Yatsunenko T, Manary MJ, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013;339(6119):548–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goedert JJ. The microbiota and human health: beyond exploration. Eur J Clin Invest. 2013;43(7):657–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oh J, Byrd AL, Park M, et al. Temporal stability of the human skin microbiome. Cell. 2016;165(4):854–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. DiGiulio DB, Callahan BJ, McMurdie PJ, et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc Natl Acad Sci USA. 2015;112(35):11060–11065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Caporaso JG, Lauber CL, Costello EK, et al. Moving pictures of the human microbiome. Genome Biol. 2011;12(5):R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Costello EK, Lauber CL, Hamady M, et al. Bacterial community variation in human body habitats across space and time. Science. 2009;326(5960):1694–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gajer P, Brotman RM, Bai G, et al. Temporal dynamics of the human vaginal microbiota. Sci Transl Med. 2012;4(132):132ra52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhou Y, Gao H, Mihindukulasuriya KA, et al. Biogeography of the ecosystems of the healthy human body. Genome Biol. 2013;14:R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Laird NM, Ware JH. Random-effects models for longitudinal data. Biometrics. 1982;38(4):963–974. [PubMed] [Google Scholar]
- 28. Schiffman MH, Van Tassell RL, Robinson A, et al. Case-control study of colorectal cancer and fecapentaene excretion. Cancer Res. 1989;49(5):1322–1326. [PubMed] [Google Scholar]
- 29. Schiffman MH, Andrews AW, Van Tassell RL, et al. Case-control study of colorectal cancer and fecal mutagenicity. Cancer Res. 1989;49(12):3420–3424. [PubMed] [Google Scholar]
- 30. Human Microbiome Project Consortium A framework for human microbiome research. Nature. 2012;486(7402):215–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486(7402):207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Turnbaugh PJ, Ley RE, Hamady M, et al. The Human Microbiome Project. Nature. 2007;449(7164):804–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bultman SJ. Emerging roles of the microbiome in cancer. Carcinogenesis. 2013;35(2):249–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schwabe RF, Jobin C. The microbiome and cancer. Nat Rev Cancer. 2013;13(11):800–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fadrosh DW, Ma B, Gajer P, et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014;2:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. National Institutes of Health NIH Human Microbiome Project. https://www.hmpdacc.org/HMQCP/. Published 2010. Accessed May 1, 2016.
- 37. DeSantis TZ, Dubosarskiy I, Murray SR, et al. Comprehensive aligned sequence construction for automated design of effective probes (CASCADE-P) using 16S rDNA. Bioinformatics. 2003;19(12):1461–1468. [DOI] [PubMed] [Google Scholar]
- 38. DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wu GD, Lewis JD, Hoffmann C, et al. Sampling and pyrosequencing methods for characterizing bacterial communities in the human gut using 16S sequence tags. BMC Microbiol. 2010;10:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Flores R, Shi J, Gail MH, et al. Assessment of the human faecal microbiota: II. Reproducibility and associations of 16S rRNA pyrosequences. Eur J Clin Invest. 2012;42(8):855–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sinha R, Chen J, Amir A, et al. Collecting fecal samples for microbiome analyses in epidemiology studies. Cancer Epidemiol Biomarkers Prev. 2016;25(2):407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Vogtmann E, Chen J, Amir A, et al. Comparison of collection methods for fecal samples in microbiome studies. Am J Epidemiol. 2017;185(2):115–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. David LA, Materna AC, Friedman J, et al. Host lifestyle affects human microbiota on daily timescales. Genome Biol. 2014;15(7):R89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ding T, Schloss PD. Dynamics and associations of microbial community types across the human body. Nature. 2014;509(7500):357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Norat T, Bingham S, Ferrari P, et al. Meat, fish, and colorectal cancer risk: the European Prospective Investigation into Cancer and Nutrition. J Natl Cancer Inst. 2005;97(12):906–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Slimani N, Bingham S, Runswick S, et al. Group level validation of protein intakes estimated by 24-hour diet recall and dietary questionnaires against 24-hour urinary nitrogen in the European Prospective Investigation into Cancer and Nutrition (EPIC) calibration study. Cancer Epidemiol Biomarkers Prev. 2003;12(8):784–795. [PubMed] [Google Scholar]
- 47. Sinha R, Abnet CC, White O, et al. The Microbiome Quality Control Project: baseline study design and future directions. Genome Biol. 2015;16:276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.