Abstract
Cordyceps guangdongensis is an edible fungus which was approved as a novel food by the Chinese Ministry of Public Health in 2013. It also has a broad prospect of application in pharmaceutical industries, with many medicinal activities. In this study, the whole genome of C. guangdongensis GD15, a single spore isolate from a wild strain, was sequenced and assembled with Illumina and PacBio sequencing technology. The generated genome is 29.05 Mb in size, comprising nine scaffolds with an average GC content of 57.01%. It is predicted to contain a total of 9150 protein-coding genes. Sequence identification and comparative analysis indicated that the assembled scaffolds contained two complete chromosomes and four single-end chromosomes, showing a high level assembly. Gene annotation revealed a diversity of transposons that could contribute to the genome size and evolution. Besides, approximately 15.57% and 12.01% genes involved in metabolic processes were annotated by KEGG and COG respectively. Genes belonging to CAZymes accounted for 3.15% of the total genes. In addition, 435 transcription factors, involved in various biological processes, were identified. Among the identified transcription factors, the fungal transcription regulatory proteins (18.39%) and fungal-specific transcription factors (19.77%) represented the two largest classes of transcription factors. This genomic resource provided a new insight into better understanding the relevance of phenotypic characters and genetic mechanisms in C. guangdongensis.
Keywords: Cordyceps, chromosome, transporters, transcription factors, Genome Report
Cordyceps guangdongensis T. H. Li, Q. Y. Lin & B. Song was discovered in Southern China (Lin et al. 2008), and has been successfully cultivated (Lin et al. 2010). Its fruiting body is nontoxic, and was approved as the second novel food of Cordyceps species by the Ministry of Public Health of China in 2013. This fungus is rich in nutrients and bioactive compounds, such as cordycepic acid, adenosine and polysaccharides (Lin et al. 2009; 2010); these contents are similar to those of the traditional Chinese invigorant, Ophiocordyceps sinensis (=Cordyceps sinensis) (Lin et al. 2009). Previous research by the authors’ group indicated that the fruiting bodies of C. guangdongensis possessed various therapeutic properties, including antioxidant activity (Zeng et al. 2009), longevity-increasing activity (Yan et al. 2011), anti-fatigue effect (Yan et al. 2013), curative effect on chronic renal failure (Yan et al. 2012), and anti-inflammatory effect (Yan et al. 2014). These active effects provided great potential for its application in food and medicinal industries. Therefore, it is a matter of cardinal significance to further understand the fruiting body development and metabolic mechanisms of C. guangdongensis, as well as its evolutionary relationship with other related species.
In recent years, whole genome sequencing (WGS) has been widely used to analyze the relevance of phenotypic characters and genetic mechanisms. The rapid development of advanced sequencing techniques and bioinformatic methods makes it more convenient to further explore the mechanisms of fungal development, metabolism, systematic taxonomy, and evolution at the molecular level. To date, numerous genomes of fungi within the order Hypocreales have been published in the Ensembl fungus database (http://fungi.ensembl.org/index.html). Based on the available genome sequences, researchers not only ascertained evolutionary relationship of many fungi (Zheng et al. 2011; Bushley et al. 2013; Xia et al. 2017), but also identified almost 40 medically active product producing gene clusters in different Cordyceps species, such as cyclosporine, oosporein, beauvericin, efrapeptins, 2-pyridone alkaloids, equisetin, emericellamide, tolypin (Zheng et al. 2011; Bushley et al. 2013; Quandt et al. 2015; Kramer and Nodwell 2017; Lu et al. 2017). Meanwhile, various transcription factors (TFs), including bZIP TFs, zinc finger TFs and fungal-specific TFs, were proven to be involved in fruiting body development by transcriptome analysis on the basis of genome sequences (Zheng et al. 2011; Yin et al. 2012; Yang et al. 2016). However, the whole genome sequence for C. guangdongensis is still lacking.
In order to acquire abundant molecular information to effectively explore the genetic characteristics of C. guangdongensis, the whole genome of C. guangdongensis was sequenced for the first time in this study. At the assembly level, the types of transposable elements and transcriptional factors (TFs) were further analyzed. This genomic resource provids a new insight to better understand the relevance of phenotypic characters and genetic mechanisms in C. guangdongensis.
Material and Methods
Fungal strains and DNA extraction
The sample used for the whole genome sequencing and assembly was isolated from the strain GDGM30035 (wild fruiting bodies of C. guangdongensis). The strain was cultured on PDA medium at 23 ± 1° for four weeks. Aqueous suspensions of fungal spores were prepared by pouring sterile distilled water onto the sporulated cultures and gently scrubbing the agar surface. The spore suspension was collected by passing the aqueous fungal suspensions through four layers of sterile cheesecloth to remove mycelial fragments. The spore suspension was diluted to 1×103 conidia ml/L using a hemocytometer, and was coated on the PDA medium covered with cellophane. A single colony (C. guangdongensis GD15) was transferred onto a new PDA medium and was cultured three times. For DNA isolation, the strain was cultured on a PDA medium, which was covered with cellophane in advance. Genomic DNA from a 7-day-old fungal colony was extracted using CTAB-based extraction buffer (Watanabe et al. 2010). The DNA concentration was determined using UV-Vis spectrophotometer (BioSpec-nano), the integrity of the DNA was detected using 0.8% agarose gel, and the purity of the DNA was analyzed with PCR using 16S rDNA primers.
Genome sequencing and assembly
The genome of C. guangdongensis GD15 was sequenced at the Beijing Genomics Institute at Shenzhen with the hybrid of Illumina HiSequation 2000 and the PacBio sequencing platform. The PacBio sequencing approach could provide previously unprecedented sequencing read lengths (>2kb), and get better sequencing depth. The next generation sequencing approach has also been widely used in various species. It has lower mismatch rate, with shorter sequencing read lengths. To cater for mismatch rate and get better sequencing read lengths, we combined the next generation sequencing approach (Illumina HiSeq) and the PacBio sequencing approach for the genome sequencing. DNA libraries with 500 bp inserts were constructed and sequenced with the Illumina HiSeq2000 Genome Analyzer. Long insert SMRTbell template libraries were prepared according to PacBio protocols. The unqualified raw reads obtained by PacBio were filtered out, the subreads (≥1000bp) were corrected by Proovread 2.12 (https://github.com/BioInf-Wuerzburg/proovread) (Hackl et al. 2014), and were initially assembled by SMRT Analysis v.2.3.0 (Chin et al. 2013). The preliminary assembly results were further corrected using small Illumina reads by GATK v1.6-13 (http://www.broadinstitute.org/gatk/) (De Summa et al. 2017), and the scaffolds were assembled and optimized using long Illumina reads by SSPACE Basic v2.0 (http://www.baseclear.com/genomics/bioinformatics/basetools/SSPACE) and PBJelly2 v15.8.24 (https://sourceforge.net/projects/pb-jelly/) (English et al. 2012). The completeness of this assembly was assessed using the BUSCO analysis described by Simão et al. (2015).
Genome components analysis
The characteristic telomeric repeats (TTAGGG/CCCTAA) were searched for on both ends of each scaffold within 100bp length. Repetitive elements included tandem repeats and transposable elements (TEs). Tandem repeats were searched for in all scaffolds with Tandem Repeats Finder (TRF 4.04), as described by Benson (1999). TEs annotation was performed with RepeatMasker 4.06 (Smit et al. 2014) based on the Repbase database (http://www.girinst.org/repbase/). The tRNAs were predicted using tRNAscan-SE 1.3.1 (Lowe and Eddy 1997), rRNAs were identified using RNAmmer 1.2 (Lagesen et al. 2007), and sRNA were predicted with Infernal based on the Rfam database (Gardner et al. 2009). Genes were annotated based on sequence homology and de novo gene predictions. The homology approach was based on the reference genomes downloaded from EnsemblFungi (http://fungi.ensembl.org/index.html) including the protein sequences of C. militaris, O. sinensis and Cordyceps ophioglossoides (=Tolypocladium ophioglossoides). The de novo gene predictions were performed with Genemark-ES 4.21 (Ter-Hovhannisyan et al. 2008).
Functional annotation
Structural and functional annotations of genes were performed according to various databases of ARDB (Antibiotic Resistance Genes Database) (Liu and Pop 2009), CAZymes (Carbohydrate-Active enZYmes Database) (Cantarel et al. 2009), COG (Cluster of Orthologous Groups) (Tatusov et al. 2003), GO (Gene Ontology) (Ashburner et al. 2000), KEGG (Kyoto Encyclopedia of Genes and Genomes) (Kanehisa et al. 2006), NR (Non-Redundant Protein Database) (Yu and Zhang 2013), P450 (Magrane et al. 2011), PHI (Pathogen Host Interactions) (Torto-Alalibo et al. 2009), SwissProt (Magrane et al. 2011), T3SS (Type III Secretion System Effector protein) (Vargas et al. 2012), TrEMBL (O’Donovan et al. 2002), VFDB (Virulence Factors of Pathogenic Bacteria) (Chen et al. 2016), IPR (InterPro Protein Families Database) (Mitchell et al. 2015), KOG (Eukaryotic Orthologous Groups) (Tatusov et al. 2003), and NOG (Non-supervised Orthologous Groups) (Huerta-Cepas et al. 2016). Transcription factors were annotated according to their InterPro IDs in the Fungal Transcription Factor Database (Wilson et al. 2008).
Data availability
The genome sequencing project has been deposited at GenBank under the accession number NRQP00000000. The BioProject designation for this project is PRJNA399600. Figure S1 shows the length and quality distributions of PacBio reads. Figure S2 shows the BUSCO analysis of the completeness of the assembly results. Figure S3 shows the distribution of gene length predicted in the C. guangdongensis genome. Table S1 shows the genome sequences used to analyze the chromosome. Table S2 shows the genome annotation of proteins in C. guangdongensis genome. Table S3 shows the transposable element classification in C. guangdongensis. Supplemental material available at Figshare: https://figshare.com/s/6ad20d1f75328ca704ca.
Results and Discussion
Whole-genome assembly
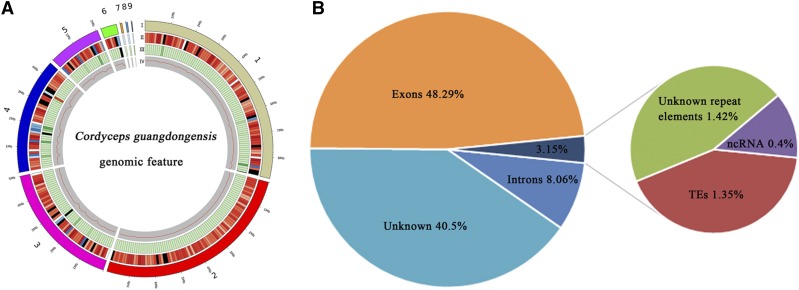
A total of 3,926,378,523 reads representing a cumulative size of 3.926 Gb were generated, including 13,392,532 and 3,912,985,991 reads from Illumina and PacBio sequencing platforms, respectively. The PacBio sequencing results showed high quality polymerase reads and subreads (Figure S1). After filtering out the low quality reads, a total of 3,484,503,143 reads were assembled into nine scaffolds with N50 of 7.88 Mb from ∼183 average coverage. In addition, a total length of 29.05 Mb with a 57.01% GC content was obtained (Figure 1A). Based on the Illumina sequencing data, the predicted genome size by K-mer analysis was 31.58Mb; the total size of the combined assembly closely matched this estimated size (91.98%). Compared to the previously reported draft genomes listed in Table 1. The completeness of the assembly results was evaluated by comparing with the BUSCO set of 1315 fungal orthologs. According to the results, a total of 1295 set appeared complete in the C. guangdongensis gene sets; this indicated an estimated completeness of 98.5%, with only 0.99% missing (Figure S2). These results indicated that our assembly is relatively contiguous.
Figure 1.
General genomic features of Cordyceps guangdongensis. A, I, scaffolds, the different colors represented different scaffolds; II, gene density, represented as the number of genes per 100 kb, increased in color intensity from light blue, to dark blue, dark, dark red, and light red. The density of non-coding RNA increased in color intensity from dark blue, to light blue, white, light red, and dark red; III, percentage of coverage of repetitive sequences, increased in color intensity from light green, to dark green, dark, dark red, and light red; IV, GC content estimated by the percentage of G + C in 100 kb. B, Genomic element density including genic and nongenic features of the overall genome assembly length including 40.5% non-annotated sequences.
Table 1. Assembly summary statistics of Cordyceps guangdongensis GD15 compared to other Cordyceps genomes.
| Species | C. guangdongensis | C. militaris CM01 | O. sinensis CO18 | C. cicadae CCAD02 |
|---|---|---|---|---|
| NCBI Bio Project | NRQP00000000 | AEVU00000000 | ANOV00000000 | MWMN00000000 |
| Assembly size (Mb) | 29.0 | 32.2 | 78.5 | 33.9 |
| Coverage fold | 183x | 147x | 241x | 80x |
| No. of Scaffold | 9 | 32 | 10603 | 599(>1 kb) |
| N50 | 7.88 Mb | 0.11 Mb | 5.39 kb | 0.21 Mb |
| GC% | 57.0 | 51.4 | 46.1 | 53.0 |
| Repeat content (%) | 2.77 | 3.04 | 37.98 | 3.19 |
| Gene density (genes per Mb) | 315 | 301 | 87 | 286 |
Chromosome analysis
Sequence analysis of telomeric repeats was used to estimate the number of chromosomes in the C. guangdongensis genome according to the method described by Zheng et al. (2011). The characteristic telomeric repeats (TTAGGG/CCCTAA)n were found at either 5 ʹ or 3 ʹ terminal of six scaffolds, of which the telomeric repeats were found at both ends of scaffolds one and three, suggesting that the two scaffolds are complete chromosomes. The lengths of the two complete chromosomes were about 8.81 Mba and 5.00 Mba, respectively. Single-ended telomeric repeats were found at four scaffolds, including the start of scaffolds four and six, the ends of scaffolds two and five, suggesting that these four scaffolds extended to the telomeres. The lengths of the four candidate scaffolds were about 7.88, 4.50, 2.05, and 0.61 Mba, respectively. The remaining three scaffolds contained no telomeric repeats, possibly due to incompleteness of the scaffold sequence data (Table 2, Table S1). Furthermore, on scaffold seven, 14 genes were identified which belonged to the core genes of mitochondrial genome, indicating that this scaffold represents mitochondrial genome sequence. Previous researches showed that the haploid genome of C. militaris contains seven chromosomes (Kramer and Nodwell 2017), and Cordyceps subsessilis (=Tolypocladium inflatum) also contains seven chromosomes (Stimberg et al. 1992). Taking into consideration the chromosome number of these related species and the present telomeric repeats analysis, it was inferred that C. guangdongensis may also possibly contain seven chromosomes. This hypothesis should be further proved with karyotype analysis.
Table 2. Chromosome analysis of Cordyceps guangdongensis GD15 genomic sequence.
| Scaffold | Size (bp) | Start Telomere | End Telomere | Judge | chromosome |
|---|---|---|---|---|---|
| Scaffold1 | 8,817,043 | CCCTAA | TTAGGG | double-end | Complete chromosome |
| Scaffold2 | 7,881,840 | No | TTAGGG | single-ended | Chromosome fragment |
| Scaffold3 | 5,000,199 | CCCTAA | TTAGGG | double-end | Complete chromosome |
| Scaffold4 | 4,508,454 | CCCTAA | No | single-ended | Chromosome fragment |
| Scaffold5 | 2,058,248 | No | TTAGGG | single-ended | Chromosome fragment |
| Scaffold6 | 614,660 | CCCTAA | No | single-ended | Chromosome fragment |
| Scaffold7 | 75,887 | No | No | No | Mitochondrial genome |
| Scaffold8 | 68,140 | No | No | No | Fragment |
| Scaffold9 | 31,250 | No | No | No | Fragment |
Genome features and annotation
As shown in Table 3, a total of 9150 protein-coding genes were predicted in the genome, including 31 rRNA, 111 tRNA, 121 sRNA, 25 snRNA and 26 miRNA. The cumulative length of the total number of genes accounted for 56.35% of the whole genome sequence length, and the lengths of most genes were in the range of 200-5000bp (Figure S3). There was a large proportion of exons (48.29%), with a maximum number of 29,548, and the number of introns was 20,398, with a total length of 2.34 Mba (8.06%). The total number of ncRNA was 314, representing 0.4% of the genome assembly; this suggested that ncRNA formed only a small proportion of the overall genome size (Figure 1B).
Table 3. Genome annotation features of Cordyceps guangdongensis GD15.
| Feature | Total number | Total length (bp) | Average length (bp) | Length/ genome length (%) |
|---|---|---|---|---|
| gene | 9,150 | 16,372,278 | 1,789.32 | 56.35 |
| Exons | 29,548 | 14,031,735 | 474.88 | 48.29 |
| CDS | 9,150 | 14,031,735 | 1,533.52 | 48.29 |
| Introns | 20,398 | 2,340,543 | 114.74 | 8.06 |
| tRNA | 111 | 9,519 | 85.75 | 0.03 |
| rRNA | 31 | 95,875 | 3,092.74 | 0.33 |
| sRNA | 121 | 7,369 | 60.9 | 0.025 |
| snRNA | 25 | 2,908 | 116.32 | 0.01 |
| miRNA | 26 | 1,766 | 67.92 | 0.006 |
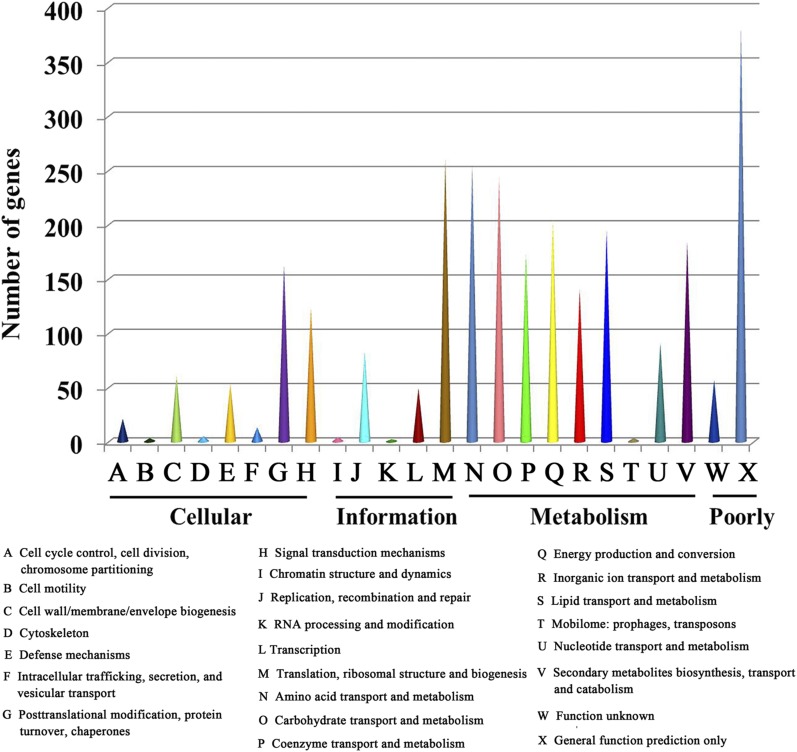
Of the 9150 identified genes, 8486 genes (92.74%) were annotated using the databases described in the methods section (Table S2). This present paper mainly focused on the genes involved in metabolic processes. Among all the genes predicted, approximately 48.90% (4475) were annotated by KEGG pathway, and in these genes, 15.57% (1425) of the total predicted genes were involved in metabolism accounted for the major proportion. Genes classified into functional categories based on the COG analysis accounted for 24.43% (2236), and in these genes, 12.01% (1099) of the total predicted genes were involved in metabolic processes, and approximately 2.01% (184) of the total predicted genes were related to the biosynthesis, transportation, and catabolism of secondary metabolites (Figure 2). The percentage of genes encoding CAZymes was 3.15% (289); these genes were contributed to substrate degradation processes in nutrition for fungal development and reproduction. Among the genes related to CAZymes, 103 genes encoding glycoside hydrolases (GHs) accounted for the largest proportion (1.12%) of the total predicted genes, followed by 78 genes encoding carbohydrate-binding modules (CBMs) (0.85%), and then 66 genes encoding glycosyl transferases (GTs) (0.72%). Genes acted as auxiliary activities (AAs) accounted for 0.32%. In addition, genes belonging to the carbohydrate esterases (CEs) and polysaccharide lyases (PLs) had much lower percentages of about 0.13% and 0.01% of the total predicted genes, respectively. Since the genes relevant to CAZymes in Pleurotus eryngii were not only involved in decomposition of organic materials, but also primordium differentiation and fruiting body development (Xie et al. 2018), the CAZymes genes identified in C. guangdongensis could likely be also involved in primordium differentiation and fruiting body development. These results are beneficial and provide the basis to further study the genetic and molecular mechanisms underlying fruiting body development.
Figure 2.
COG functional classification of proteins in the Cordyceps guangdongensis genome.
Repetitive elements
The cumulative sequences of repetitive elements identified in C. guangdongensis genome occupied 2.77% of the assembly sequences. The tandem repeats represented 1.42% of the genome assembly, with a total length of 412,989 bp; and the TEs represented 1.35% of the genome assembly with a total length of 393,608 bp. The total number of TE families analyzed with RepeatMasker in the genome assembly was 1534, of which 1527 (99.5%) belonged to the known TEs, including 1033 retrotransposons (Class I) and 494 DNA transposons (Class II); the remaining TEs could not be classified at the time of this study (Table 4 and Table S3).
Table 4. Transposable element repeat class analysis in Cordyceps guangdongensis.
| Repeat element family | Number of unique elements in family | cumulative length (bp) | % of genome assembly | ||
|---|---|---|---|---|---|
| Class I - Retrotransposons | 1,033 | 319,324 | 1.09898 | ||
| LINE | 343 | 57,004 | 0.19618 | ||
| L1 | 14 | 660 | |||
| others | 329 | 56,344 | |||
| LTR | 661 | 259,862 | 0.89435 | ||
| Copia | 269 | 174,266 | |||
| DIRS | 4 | 324 | |||
| ERV1 | 17 | 1,170 | |||
| Gypsy | 314 | 79,093 | |||
| Pao | 17 | 1,741 | |||
| others | 40 | 3,268 | |||
| SINE | 15 | 1,304 | 0.00448 | ||
| Alu | 1 | 51 | |||
| Others | 14 | 1,253 | |||
| others | 14 | 1,154 | 0.00397 | ||
| Class II - DNA Transposons | 494 | 73,739 | 0.25378 | ||
| CMC-EnSpm | 44 | 3,001 | |||
| Dada | 8 | 485 | |||
| Ginger | 2 | 141 | |||
| hAT | 4 | 440 | |||
| Merlin | 3 | 108 | |||
| MULE-MuDR | 29 | 4,628 | |||
| P | 2 | 134 | |||
| PIF-Harbinger | 11 | 729 | |||
| PiggyBac | 63 | 19,337 | |||
| Sola | 29 | 1,840 | |||
| TcMar-Tc1 | 14 | 3,780 | |||
| Zisupton | 1 | 88 | |||
| others | 284 | 39,028 | |||
| Unclassified | 7 | 545 | 0.00187 | ||
| Total | 1,534 | 393,608 | 1.35466 | ||
Class I retrotransposons can be mainly divided into three groups of TEs, including LINE, LTR, and SINE; each group contains several subgroups. Retrotransposons, particularly L1, Copia, DIRS, ERV1, Gypsy, Pao, Alu, etc., are the easiest to be annotated; they are also the most abundant transposons in fungi (Suarez et al. 2018). Class II DNA transposons contain a lot of known groups and some unclassified members. Among the transposons, hAT, MULE, PIF-Harb and Tc1-Mariner were reported to be extraordinarily abundant in fungi, whereas transposons CMC and piggyBac have limited taxonomic distribution and seem to exist in only a few fungal taxa (Muszewska et al. 2017). Other transposons, including P, Sola, Dada, Ginger, Zisupton, and Merlin, had been identified only in a handful of species (Kojima and Jurka 2013; Iyer et al. 2014; Majorek et al. 2014).
Previous studies indicated that TEs contributed to genome size expansion and evolution (Cordaux and Batzer 2009; Sun et al. 2012) and played crucial roles in a wide range of biological events, including organism development (Kano et al. 2009; Garcia-Perez et al. 2016), regulation (Elbarbary et al. 2016) and differentiation (Morales-Hernández et al. 2016). In addition, they sometimes act as novel promoters to activate the transcription process (Faulkner et al. 2009; Mita and Boeke 2016). Therefore, the abundance in C. guangdongensis would be more significant in this regard; they should be noted and further studied for their application in fungal taxonomy and regulatory roles in fruiting body development.
Transcription factors
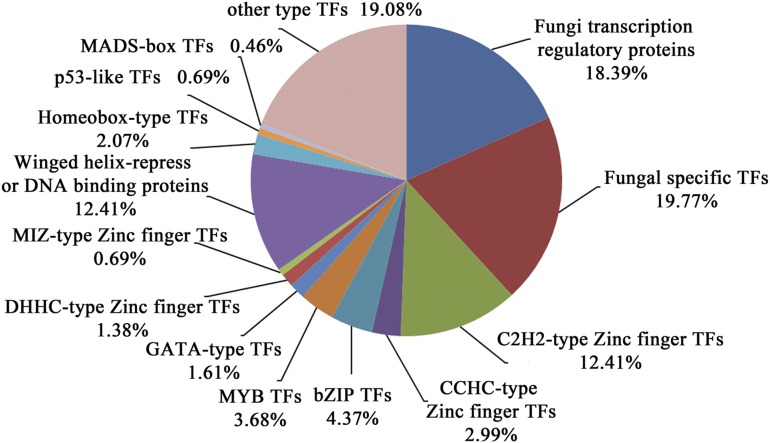
Transcription factors (TFs) are essential for modulating diverse biological processes by regulating gene expression and playing central roles in organism development and evolution. In this study, functional annotation identified 435 genes of TFs in C. guangdongensis, accounting for 4.75% of the total predicted genes. Like other fungi, genes encoding fungal-specific TFs (86 members) and fungal transcription regulatory proteins (80 members) represented the two largest classes of TFs in C. guangdongensis, accounting for approximately 19.77% and 18.39% of the total predicted TFs, respectively; and followed by C2H2-type zinc finger TFs (54 members) and winged helix-repressor DNA binding proteins (54 members), accounting for approximately 12.41% and 12.41%, respectively (Figure 3).
Figure 3.
Transcription factors analysis in the Cordyceps guangdongensis genome.
Moreover, other different types of zinc finger TFs were identified, including 13 CCHC-type zinc finger TFs (2.98%), 6 DHHC-type TFs (0.06%) and 3 MIZ-type TFs (0.03%). Apart from these, there were 19 bZIP TFs (0.21%), 16 MYB TFs (0.17%), 7 GATA TFs (0.08%) and 9 homeobox-type TFs (0.10%). In C. militaris, majority of TFs, such as Zn2Cys6-type TFs, GATA-type TFs, bZIP TFs, and CHCC-type TFs, were differentially expressed during fruiting body developmental stages (Zheng et al. 2011). Hence, this information could help explore the regulatory mechanisms of TFs in fruiting body development in C. guangdongensis.
Conclusion
High quality genome sequencing of C. guangdongensis was presented in this study. Two complete chromosomes and four single-end chromosomes were assembled through genome sequence analysis. In the genomic sequences, diverse transposable elements were identified, which may contribute to genome size and evolution. Moreover, transcription factors in the genome of C. guangdongensis were identified and classified; these transcription factors may facilitate further studies of fruiting body development. And above all, knowledge about the genome sequence of C. guangdongensis will reveal more detailed molecular information and facilitate further studies of fruiting body development and identification of secondary metabolites in C. guangdongensis.
Acknowledgments
This work was supported by the Science and Technology Planning Project of Guangzhou, China (Nos. 201804020018, 201504291620324), Natural Science Foundation of Guangdong Province, China (2017A030310533), the Science and Technology Planning Project of Guangdong Province, China (No. 2015A030302052), the National Natural Science Foundation of China (No. 31470155), and GDAS’ Special Project of Science and Technology Development (No. 2017GDASCX-0822). The authors sincerely thank the professor Chengshu Wang in Shanghai Institute for Biological Sciences for providing the guidance of genome sequencing.
Footnotes
Supplemental material available at Figshare: https://figshare.com/s/6ad20d1f75328ca704ca.
Communicating editor: A. Rokas
Literature Cited
- Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., et al. , 2000. Gene Ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25: 25–29. 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G., 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27: 573–580. 10.1093/nar/27.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushley K. E., Raja R., Jaiswal P., Cumbie J. S., Nonogaki M., et al. , 2013. The Genome of Tolypocladium inflatum: Evolution, Organization, and Expression of the Cyclosporin Biosynthetic Gene Cluster. PLoS Genet. 9: e1003496 10.1371/journal.pgen.1003496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., et al. , 2009. The Carbohydrate-Active EnZymes database (CAZy): an expert resource for Glycogenomics. Nucleic Acids Res. 37: D233–D238. 10.1093/nar/gkn663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. H., Zheng D. D., Liu B., Yang J., Jin Q., 2016. VFDB 2016: hierarchical and refined dataset for big data analysis--10 years on. Nucleic Acids Res. 44: D694–D697. 10.1093/nar/gkv1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin C. S., Alexander D. H., Marks P., Klammer A. A., Drake J., et al. , 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods 10: 563–569. 10.1038/nmeth.2474 [DOI] [PubMed] [Google Scholar]
- Cordaux R., Batzer M. A., 2009. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 10: 691–703. 10.1038/nrg2640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Summa S., Malerba G., Pinto R., Mori A., Mijatovic V., et al. , 2017. GATK hard filtering: tunable parameters to improve variant calling for next generation sequencing targeted gene panel data. BMC Bioinformatics 18: 119–127. 10.1186/s12859-017-1537-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbarbary R. A., Lucas B. A., Maquat L. E., 2016. Retrotransposons as regulators of gene expression. Science 351: aac7247 10.1126/science.aac7247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- English A. C., Richards S., Han Y., Wang M., Vee V., et al. , 2012. Mind the Gap: Upgrading Genomes with Pacific Biosciences RS Long-Read Sequencing Technology. PLoS One 7: e47768 10.1371/journal.pone.0047768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner G. J., Kimura Y., Daub C. O., Wani S., Plessy C., et al. , 2009. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 41: 563–571. 10.1038/ng.368 [DOI] [PubMed] [Google Scholar]
- Garcia-Perez J. L., Widmann T. J., Adams I. R., 2016. The impact of transposable elements on mammalian development. Development 143: 4101–4114. 10.1242/dev.132639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner P. P., Daub J., Tate J. G., Nawrocki E. P., Kolbe D. L., et al. , 2009. Rfam: updates to the RNA families database. Nucleic Acids Res. 37: D136–D140. 10.1093/nar/gkn766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackl T., Hedrich R., Schultz J., Förster F., 2014. proovread: large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics 30: 3004–3011. 10.1093/bioinformatics/btu392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J., Szklarczyk D., Forslund K., Cook H., Heller D., et al. , 2016. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44: D286–D293. 10.1093/nar/gkv1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer L. M., Zhang D., de Souza R. F., Pukkila P. J., Rao A., et al. , 2014. Lineage-specific expansions of TET/JBP genes and a new class of DNA transposons shape fungal genomic and epigenetic landscapes. Proc. Natl. Acad. Sci. USA 111: 1676–1683. 10.1073/pnas.1321818111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S., Hattori M., Aoki-Kinoshita K. F., Itoh M., et al. , 2006. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 34: D354–D357. 10.1093/nar/gkj102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kano H., Godoy I., Courtney C., Vetter M. R., Gerton G. L., et al. , 2009. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 23: 1303–1312. 10.1101/gad.1803909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K. K., Jurka J., 2013. A superfamily of DNA transposons targeting multicopy small RNA genes. PLoS One 8: e68260 10.1371/journal.pone.0068260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer G. J., Nodwell J. R., 2017. Chromosome level assembly and secondary metabolite potential of the parasitic fungus Cordyceps militaris. BMC Genomics 18: 912–921. 10.1186/s12864-017-4307-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagesen K., Hallin P. F., Rødland E. A., Staerfeldt H. H., Rognes T., et al. , 2007. RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 35: 3100–3108. 10.1093/nar/gkm160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q. Y., Li T. H., Song B., 2008. Cordyceps guangdongensis sp. nov. from China. Mycotaxon 103: 371–376. [Google Scholar]
- Lin Q. Y., Li T. H., Song B., Huang H., 2009. Comparison of selected chemical component levels in Cordyceps guangdongensis, C. sinensis and C. militaris. Acta Edulis Fungi. 16: 54–57. [Google Scholar]
- Lin Q. Y., Song B., Huang H., Li T. H., 2010. Optimization of selected cultivation parameters for Cordyceps guangdongensis. Lett. Appl. Microbiol. 51: 219–225. [DOI] [PubMed] [Google Scholar]
- Liu B., Pop M., 2009. ARDB--Antibiotic Resistance Genes Database. Nucleic Acids Res. 37: D443–D447. 10.1093/nar/gkn656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe T. M., Eddy S. R., 1997. tRNAscan-SE: A Program for Improved Detection of Transfer RNA Genes in Genomic Sequence. Nucl Acids Res. 25: 0955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Y. Z., Luo F. F., Cen K., Xiao G. H., Yin Y., et al. , 2017. Omics data reveal the unusual asexual fruiting nature and secondary metabolic potentials of the medicinal fungus Cordyceps cicadae. BMC Genomics 18: 668–682. 10.1186/s12864-017-4060-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrane M., UniProt Consortium , 2011. UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011: bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majorek K. A., Dunin-Horkawicz S., Steczkiewicz K., Muszewska A., Nowotny M., et al. , 2014. The RNase H-like superfamily: new members, comparative structural analysis and evolutionary classification. Nucleic Acids Res. 42: 4160–4179. 10.1093/nar/gkt1414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mita P., Boeke J. D., 2016. How retrotransposons shape genome regulation. Curr. Opin. Genet. Dev. 37: 90–100. 10.1016/j.gde.2016.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell A., Chang H. Y., Daugherty L., Fraser M., Hunter S., et al. , 2015. The InterPro protein families database: the classification resource after 15 years. Nucleic Acids Res. 43: D213–D221. 10.1093/nar/gku1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Hernández A., González-Rico F. J., Román A. C., Rico-Leo E., Alvarez-Barrientos A., et al. , 2016. Alu retrotransposons promote differentiation of human carcinoma cells through the aryl hydrocarbon receptor. Nucleic Acids Res. 44: 4665–4683. 10.1093/nar/gkw095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muszewska A., Steczkiewicz K., Stepniewska-Dziubinska M., Ginalski K., 2017. Cut-and-Paste transposons in fungi with diverse lifestyles. Genome Biol. Evol. 9: 3463–3477. 10.1093/gbe/evx261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donovan C., Martin M. J., Gattiker A., Gasteiger E., Bairoch A., et al. , 2002. High-quality protein knowledge resource: SWISS-PROT and TrEMBL. Brief. Bioinform. 3: 275–284. 10.1093/bib/3.3.275 [DOI] [PubMed] [Google Scholar]
- Quandt C. A., Bushley K. E., Spatafora J. W., 2015. The genome of the truffle-parasite Tolypocladium ophioglossoides and the evolution of antifungal peptaibiotics. BMC Genomics 16: 553–556. 10.1186/s12864-015-1777-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31: 3210–3212. 10.1093/bioinformatics/btv351 [DOI] [PubMed] [Google Scholar]
- Smit A. F. A., Hubley R., Green P. 2014. RepeatMasker Open-4.0. 2013–2015. URL http://www. repeatmasker. org.
- Stimberg N., Walz M., Schörgendorfer K., Kiick U., 1992. Electrophoretic karyotyping from Tolypocladium inflatum and six related strains allows differentiation of morphologically similar species. Appl. Microbiol. Biotechnol. 37: 485–489. [Google Scholar]
- Suarez N. A., Macia A., Muotri A. R., 2018. LINE-1 Retrotransposons in healthy and diseased human brain. Dev. Neurobiol. 78: 434–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C., Shepard D. B., Chong R. A., Lopez Arriaza J., Hall K., et al. , 2012. LTR retrotransposons contribute to genomic gigantism in plethodontid salamanders. Genome Biol. Evol. 4: 168–183. 10.1093/gbe/evr139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatusov R. L., Fedorova N. D., Jackson J. D., Jacobs A. R., Kiryutin B., et al. , 2003. The COG database: an updated version includes eukaryotes. BMC Bioinformatics 4: 41–54. 10.1186/1471-2105-4-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter-Hovhannisyan V., Lomsadze A., Chernoff Y. O., Borodovsky M., 2008. Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 18: 1979–1990. 10.1101/gr.081612.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torto-Alalibo T., Collmer C. W., Gwinn-Giglio M., 2009. The Plant-Associated Microbe Gene Ontology (PAMGO) Consortium: community development of new Gene Ontology terms describing biological processes involved in microbe-host interactions. BMC Microbiol. 9: S1 10.1186/1471-2180-9-S1-S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M., Lee K., Goto K., Kumagai S., Sugita-Konishi Y., et al. , 2010. Rapid and effective DNA extraction method with bead grinding for a large amount of fungal DNA. J. Food Prot. 73: 1077–1084. 10.4315/0362-028X-73.6.1077 [DOI] [PubMed] [Google Scholar]
- Wilson D., Charoensawan V., Kummerfeld S. K., Teichmann S. A., 2008. DBD--taxonomically broad transcription factor predictions: new content and functionality. Nucleic Acids Res. 36: D88–D92. 10.1093/nar/gkm964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas W. A., Martín J. M., Rech G. E., Rivera L. P., Benito E. P., et al. , 2012. Plant defense mechanisms are activated during biotrophic and necrotrophic development of Colletotricum graminicola in maize. Plant Physiol. 158: 1342–1358. 10.1104/pp.111.190397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia E. H., Yang D. R., Jiang J. J., Zhang Q. J., Liu Y., et al. , 2017. The caterpillar fungus, Ophiocordyceps sinensis, genome provides insights into highland adaptation of fungal pathogenicity. Sci. Rep. 7: 1806–1816. 10.1038/s41598-017-01869-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C., Gong W., Zhu Z., Yan L., Hu Z., et al. , 2018. Comparative transcriptomics of Pleurotus eryngii reveals blue-light regulation of carbohydrate-active enzymes (CAZymes) expression at primordium differentiated into fruiting body stage. Genomics. 110: 201–209. 10.1016/j.ygeno.2017.09.012 [DOI] [PubMed] [Google Scholar]
- Yan W. J., Li T. H., Jiang Z. D., 2011. Anti-fatigue and life-prolonging effects of Cordyceps guangdongensis. Food R & D. 32: 164–167. [Google Scholar]
- Yan W. J., Li T. H., Jiang Z. D., 2012. Therapeutic effects of Cordyceps guangdongensis on chronic renal failure rats induced by adenine. Junwu Xuebao 31: 432–442. [Google Scholar]
- Yan W. J., Li T. H., Lao J. H., Song B., Shen Y. H., 2013. Anti-fatigue property of Cordyceps guangdongensis and the underlying mechanisms. Pharm. Biol. 51: 614–620. 10.3109/13880209.2012.760103 [DOI] [PubMed] [Google Scholar]
- Yan W. J., Li T. H., Zhong Z. Y., 2014. Anti-inflammatory effect of a novel food Cordyceps guangdongensis on experimental rats with chronic bronchitis induced by tobacco smoking. Food Funct. 5: 2552–2557. 10.1039/C4FO00294F [DOI] [PubMed] [Google Scholar]
- Yang T., Guo M. M., Yang H. J., Guo S. P., Dong C. H., 2016. The blue-light receptor CmWC-1 mediates fruit body development and secondary metabolism in Cordyceps militaris. Appl. Microbiol. Biotechnol. 100: 743–755. 10.1007/s00253-015-7047-6 [DOI] [PubMed] [Google Scholar]
- Yin Y., Yu G., Chen Y., Jiang S., Wang M., et al. , 2012. Genome-wide transcriptome and proteome analysis on different developmental stages of Cordyceps militaris. PLoS One 7: e51853 10.1371/journal.pone.0051853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Zhang T., 2013. Construction of Customized Sub-Databases from NCBI-nr Database for Rapid Annotation of Huge Metagenomic Datasets Using a Combined BLAST and MEGAN Approach. PLoS One 8: e59831 10.1371/journal.pone.0059831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng H. B., Li T. H., Song B., Lin Q. Y., Huang H., 2009. Study on antioxidant activity of Cordyceps guangdongensis. Nat Prod Res Dev. 21: 201–204. [Google Scholar]
- Zheng P., Xia Y., Xiao G., Xiong C. H., Hu X., et al. , 2011. Genome sequence of the insect pathogenic fungus Cordyceps militaris, a valued traditional Chinese medicine. Genome Biol. 12: R116 10.1186/gb-2011-12-11-r116 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The genome sequencing project has been deposited at GenBank under the accession number NRQP00000000. The BioProject designation for this project is PRJNA399600. Figure S1 shows the length and quality distributions of PacBio reads. Figure S2 shows the BUSCO analysis of the completeness of the assembly results. Figure S3 shows the distribution of gene length predicted in the C. guangdongensis genome. Table S1 shows the genome sequences used to analyze the chromosome. Table S2 shows the genome annotation of proteins in C. guangdongensis genome. Table S3 shows the transposable element classification in C. guangdongensis. Supplemental material available at Figshare: https://figshare.com/s/6ad20d1f75328ca704ca.